Abstract
This report demonstrates the use of major histocompatibility complex (MHC) class II dextramers for detection of autoreactive CD4 T cells in situ in myelin proteolipid protein (PLP) 139-151-induced experimental autoimmune encephalomyelitis (EAE) in SJL mice and cardiac myosin heavy chain-α (Myhc) 334-352-induced experimental autoimmune myocarditis (EAM) in A/J mice. Two sets of cocktails of dextramer reagents were used, where dextramers+ cells were analyzed by laser scanning confocal microscope (LSCM): EAE, IAs/PLP 139-151 dextramers (specific)/anti-CD4 and IAs/Theiler’s murine encephalomyelitis virus (TMEV) 70-86 dextramers (control)/anti-CD4; and EAM, IAk/Myhc 334-352 dextramers/anti-CD4 and IAk/bovine ribonuclease (RNase) 43-56 dextramers (control)/anti-CD4. LSCM analysis of brain sections obtained from EAE mice showed the presence of cells positive for CD4 and PLP 139-151 dextramers, but not TMEV 70-86 dextramers suggesting that the staining obtained with PLP 139-151 dextramers was specific. Likewise, heart sections prepared from EAM mice also revealed the presence of Myhc 334-352, but not RNase 43-56-dextramer+ cells as expected. Further, a comprehensive method has also been devised to quantitatively analyze the frequencies of antigen-specific CD4 T cells in the ‘Z’ serial images.
Keywords: Immunology, Issue 90, dextramers; MHC class II; in situ; EAE; brain; EAM; heart; confocal microscopy.
Introduction
The use of major histocompatibility complex (MHC) class II tetramers for detection of antigen-specific CD4 T cells has been a challenge 1-7. To enhance the detection sensitivity of MHC class II tetramers, the research team has created next-generation tetramers, designated “dextramers” 8. Dextramers contain dextran backbones in which multiple streptavidin-fluorophore moieties are conjugated, such that several peptide-tethered, biotinylated MHC monomers can be attached to one dextran molecule9. Thus, large aggregates of MHC dextramers that contain several MHC-peptide complexes can interact with multiple T cell receptors (TCRs).
This group recently generated dextramers for various self-antigens and demonstrated that the detection sensitivity of dextramers was approximately five-fold more than tetramers 8. The experimental systems include experimental autoimmune encephalomyelitis (EAE) induced with proteolipid protein (PLP) 139-151 in SJL mice; EAE induced with myelin oligodendrocyte glycoprotein (MOG) 35-55 in C57Bl/6 mice; and experimental autoimmune myocarditis (EAM) induced with cardiac myosin heavy chain-α (Myhc) 334-352 in A/J mice. This study demonstrates the use of PLP 139-151- and Myhc 334-352 dextramers to detect antigen-specific CD4 T cells in situ with specificity by developing a direct staining protocol, thereby eliminating the need to amplify the signals from using fluorophore antibodies, an approach commonly employed with the use of tetramers. In addition, a comprehensive method of evaluating tissues has also been devised to reliably enumerate the frequencies of antigen-specific CD4 T cells in situ 10. Detection of antigen-specific T cells in situ permits delineation of events caused by specific antigenic stimuli from those caused by bystander activation. Likewise, the in situ technique also provides opportunities to track the kinetics of antigen-specific T cell infiltrations in the target organs.
Protocol
Ethics Statement:
All animal procedures were conducted in accordance with the guidelines for the Care and Use of Laboratory Animals and approved by the University of Nebraska-Lincoln, Lincoln, NE.
1. Induction of EAE
To induce EAE, prepare the peptide/complete Freund’s adjuvant (CFA) emulsion as described in 11 with the following modifications: Step 1.1: prepare PLP 139-151 at a concentration of 1 mg/ml in 1x phosphate buffered saline (PBS). Step 1.4: use 200 µl of CFA emulsion containing 100 µg of PLP 139-151 per animal. Step 1.4.1: to immunize ten animals, prepare 2 ml of emulsion by mixing 1 ml of PLP 139-151 (1 mg/ml in 1x PBS) and an equal volume of CFA. Step 1.5: inject 200 μl of emulsion in split doses subcutaneously in the two inguinal regions (~75 μl each) and sternal region (~50 μl) of five-to-six-week-old female SJL/J mice. Steps 1.7 and 1.8 are not applicable for induction of EAE.
2. Clinical Scoring of EAE Mice and Harvesting of Cerebrums
Monitor the animals for appearance of clinical signs, and score the disease-severity daily from day 9 postimmunization as follows: 0 - healthy; 1 - limp tail or hind limb weakness but not both; 2 - limp tail and hind limb weakness; 3 - partial paralysis of hind limbs; 4 - complete paralysis of hind limbs; 5 - moribund or dead 12.
Euthanize the mice when they reach a neurological score of 3 or 4 using CO2. To euthanize the mice, pre-fill the euthanasia chamber first with CO2 by turning the regulator of the CO2 tank not exceeding 6 psi and allow animals to stay within the chamber for approximately 5 min to achieve complete asphyxia. Soak the euthanized animals in 70% alcohol. Immediately, place the animals on an absorbent pad and using a pair of sterile forceps and scissors expose the hearts by cutting through the thoracic cavity. Puncture the right auricles and perfuse by injecting 10 ml of cold 1x PBS into the left ventricles of the hearts.
Excavate the skull by snipping the cranium in a circular fashion using a pair of curved scissors, and carefully, flip the skull bones with forceps. Harvest the brain by blunt dissection and using a clean scalpel, separate cerebrum from the cerebellum. Place the cerebrum in a tube containing cold 1x PBS and leave the tube on ice until processing.
3. In Situ Staining of Cerebral Sections with MHC Class II (IAs)/PLP 139-151 Dextramers
Prepare 4% PBS-buffered low-melting agarose, melt by heating at 50 °C and leave it in a 40 °C water bath until use.
Place the cerebrum in a disposable deep base mold histology cassette kept over ice, and pour molten (40 °C) agarose. Immediately, press the cerebrum gently with forceps to submerge the tissue. Leave the cassette on ice for 15-20 min until the solidification is complete.
Fill the vibratome bath with chilled 1x PBS. Fill the space around vibratome bath with ice cubes to maintain the temperature of PBS between 0 and 2 °C. Fix a clean razor blade into the vibratome and set the blade angle to 15°.
- Placement of tissue in vibratome. Trim excess agarose from all the sides of the embedded tissue and slightly expose the ventral surface of the cerebrum.
- Smear vibratome tissue block with super glue and transfer the agarose-embedded tissue over the glued surface by placing the exposed ventral surface of the cerebrum over the glue for transverse sectioning. Do not move the tissue, once the tissue is set over the block.
- Place the vibratome block containing the agarose-embedded tissue on ice, and allow the glue to dry for 15 min.
- In the meantime, place the tissue chambers one per well of a 24-well plate. Fill the wells containing the tissue chambers with 1 ml of cold 1x PBS, and leave the plate on ice.
- Prepare the tissue chambers by attaching a thin nylon mesh to the cut-end of a 25 ml polypropylene pipette using super glue; after trimming the excess mesh around the pipette, cut the pipette to a length of 1 cm from the mesh.
Lower the vibratome blade to the level of the top surface of tissue; set the vibratome speed to 0.22 mm/sec and start sectioning with a forward movement to make 200 µm thick sections. Transfer one cerebral section per well using a watercolor soft brush into the tissue chambers placed within the wells of a 24-well plate. Keep the plate on ice until sectioning is complete to prevent tissue degradation. Note: Follow these steps to obtain in total, three paired sections from three different regions of the brain dorsal to the ventral surface (Figure 2). One section from each pair (in total three sections) placed in three individual tissue chambers are designated for staining with specific dextramers (IAs/PLP 139-151) and anti-CD4. Likewise, another set of three sections that are placed in three individual tissue chambers are designated for staining with control dextramers (IAs/TMEV 70-86) and anti-CD4.
Transfer the three tissue chambers containing the sections designated for staining with specific dextramers into three wells in a 24-well plate with one chamber per well in which, 300 µl of a cocktail containing allophycocyanin (APC)-conjugated PLP 139-151 dextramers (2.5 μg/ml) and anti-CD4-phycoerythrin (PE; 5 µg/ml) in blocking solution (1x PBS with 2% normal goat serum) was previously added. Cover the wells with a lid.
Similarly, transfer the three tissue chambers containing the sections designated for staining with control dextramers into three wells in a 24-well plate with one chamber per well in which, 300 µl of a cocktail containing APC-conjugated TMEV 70-86 dextramers (2.5 μg/ml) and anti-CD4- PE (5 µg/ml) in blocking solution was previously added. Cover the wells with a lid. Note: Refer to Massilamany et al., 8 for preparation of dextramers, and the dextran molecules to prepare dextramers were kindly provided by Immudex, Copenhagen, Denmark.
Label the wells, and wrap the plate with aluminum foil and place the plate on a rocking platform set to gentle rotations for 1.5 hr in the dark at RT.
After the incubation period, wash tissue sections on the rocking platform, by transferring the tissue chambers to a fresh well containing 1 ml of cold 1x PBS, and avoid dribbling the contents of one well into the other. Repeat washing thrice, allowing at least 10 min per wash. Ensure that the tissue sections do not get dried as they can stick to the sides of the chamber.
Fix the sections by transferring the tissue chambers into a fresh well in 24-well plate containing 1ml of filtered 4% PBS-buffered paraformaldehyde (pH 7.4) on a rocking platform for 2 hr with gentle rotations.
Repeat washing thrice as described in step 3.10.
Place the stained and fixed sections on a clean microscopic slide using a watercolor soft brush. Wipe excess water without touching the section, and mount the section using mounting medium. Cover with a microscopic coverslip and allow the slides to dry at RT O/N in the dark room.
Store the slides in a light-proof slide storage box at 4 °C.
4. Induction of EAM and Collection of Hearts
Induce EAM as described in reference 11.
Euthanize the mice on day 21 postimmunization using clean chamber fitted with CO2 gas cylinder, and soak the animals in 70% alcohol. Immediately, place the animals on an absorbent pad and using a pair of forceps and scissors expose the hearts by cutting through the thoracic cavity. Puncture the right auricles and perfuse by injecting 10 ml of cold 1x PBS into the left ventricles of the hearts. Collect the hearts and place in tubes containing cold 1x PBS.
5. In Situ Staining of Heart Sections with MHC Class II (IAk)/Myhc 334-352 Dextramers
Prepare 4% PBS-buffered low-melting agarose, melt by heating at 50 °C and leave it in a 40 °C water bath until use.
Place the heart in a disposable deep base mold histology cassette kept on ice, and pour molten 40 °C agarose until the tissues are completely submerged as described in step 3.2.
Follow step 3.3.
- Trim excess agarose from all the sides of the embedded tissue and slightly expose the base of the heart. Smear vibratome tissue block with super glue and transfer the agarose-embedded tissue over the glued surface by placing the exposed surface of the heart over the glue. Do not move the tissue, once the tissue is set over the block.
- Place the vibratome block containing the agarose-embedded tissue on ice, and allow the glue to dry for 15 min.
Follow steps 3.5 and 3.6.
Transfer the heart sections using a watercolor soft brush into the tissue chambers placed within the wells of a 24-well plate. Keep the plate on ice until sectioning is complete to prevent degradation of tissue. Note: Follow these steps to obtain in total, eight paired sections from the apex to base of the heart (Figure 2). Place all sections in tissue chambers, allocating up to 3 sections per chamber. One section from each pair (in total eight sections) is designated for staining with specific dextramers (IAk/Myhc 334-352) and anti-CD4. Likewise, another set of eight sections is designated for staining with control dextramers (IAk/RNase 43-56) and anti-CD4.
- Transfer the three tissue chambers containing the sections designated for staining with specific dextramers into three wells in a 24-well plate with one chamber per well in which, 300 µl of a cocktail containing APC-conjugated Myhc 334-352 dextramers (2.5 μg/ml) and anti-CD4-PE (5 µg/ml) in blocking solution was previously added. Cover the wells with a lid.
- Similarly, transfer the three tissue chambers containing the sections designated for staining with control dextramers into three wells in a 24-well plate with one chamber per well in which, 300 µl of a cocktail containing APC-conjugated RNase 43-56 dextramers (2.5 μg/ml) and anti-CD4- PE (5 µg/ml) in blocking solution was previously added. Cover the wells with a lid.
Label the wells, and wrap the plate with aluminum foil and place the plate on a rocking platform set to gentle rotations for 1.5 hr in the dark at RT.
Follow steps, 3.10 to 3.11
Repeat washing thrice as described in step 3.10.
Place the stained and fixed sections (up to three sections/slide) on a clean microscopic slide using a watercolor soft brush. Wipe excess water without touching the sections, and mount the sections using mounting medium. Cover with a microscopic coverslip and allow the slides to dry at RT O/N in the dark room.
Store the slides in a light-proof slide storage box at 4 °C.
6. Analysis of Dextramer+ (dext+) CD4+ T Cells by Laser Scanning Confocal Microscope (LSCM) (Common to Both Brain and Heart Sections)
- Use Nikon A1-Eclipse 90i confocal microscope system for routine scanning and acquisition of images. To start Click open, the NIS Elements AR software (version 4.13).
- Choose “TRITC” and “Cy5” channels from the panel. The excitation/emission wavelengths respectively for TRITC and Cy5 channels, 561.5 nm / 553-618 nm and 640.7 nm / 663-738 nm appear by default.
- Assign the pseudocolors “green” and “red” for the TRITC (CD4-PE) and Cy5 (dextramer-APC) channels, respectively.
- Place the slide to be examined on the microscopic stage and focus manually under 100X magnification. Set the “HV” to 110 and offset to “0”. Click “Focus” and adjust the “voltage” to optimize the lasers.
- Click “Focus”; scan the section from top to bottom, and set the ‘Z’ level for image acquisition. Click “CH series” and acquire the image sequentially. Identify dext+ CD4+ T cells appearing as yellow punctate cells based on co-localization of signals generated from both the red (dextramers) and green (CD4) channels.
- Use Olympus FV500-BX60 confocal microscope for easy quantitative enumeration of dext+ CD4+ T cells. To begin click open, the FLUOVIEW software (version 4.3).
- Select the dyes “Cy3” and “Cy5” from the drop-down menu, and click “apply” to load the respective lasers. Note: the excitation/emission wavelengths for Cy3 (543 nm/pseudocolored “green”; CD4-PE) and Cy5 (633 nm/pseudocolored “red”; dextramer-APC) channels.
- Place the slide to be examined on the microscopic slide, and focus it manually under low magnification (4X or 10X). Click “Focus” while the Cy3 laser is on, and focus the inflammatory focus.
- Change the objective to 100X magnification. Set the file size to “512/512” from the drop-down menu. Click “Focus” and adjust the values for “PMT”, “Gain” and “Offset” parameters to optimize the lasers separately for Cy3 and Cy5.
- Click “Z stage” and then click “Focus” while both the lasers are on. Set the thickness of the “Z stage” by clicking the “up” and “down” arrows. Set the “Z” interval to 2 µm.
- Click “Stop”, and choose “Seq123”. Then, click “XYZ” and start acquiring “Z” series images for the selected area within the inflammatory focus. Save the “Z serial images” as AVI files. To save image files, select the image from the “Z serial image”, and then save as “tiff” format.
- Examine three paired sections from the cerebrum where one set of three sections was stained with IAs/PLP 139-151 dextramers/anti-CD4 and the other set of three sections with IAs/TMEV 70-86 dextramers/anti-CD4 to perform quantitative analysis.
- Similarly, analyze eight paired sections for heart, in which, one set of eight sections was stained with IAk/Myhc 334-352 dextramers/anti-CD4 and another set of eight sections with RNase 43-56 dextramers/anti-CD4.
- Examine 10 to 15 inflammatory foci per section in cerebrum, and in each focus, acquire a set of 15 to 25 serial images sequentially.
- Examine a total of 20 inflammatory foci from all the eight sections of heart and from each focus acquire 10 to 15 serial images sequentially. Take the ‘Z’ serial images by scanning the slides in a ‘horizontal-vertical up-horizontal-vertical down’ movement so that repetition of ‘Z’ series within the same focus is avoided.
- Once ‘Z’ serial images are captured, name the files by identifying the slide numbers, name of the sections, and the number of the ‘Z’ serial images taken.
- Use ‘ImageJ’ software to count dext+ CD4+ T cells in relation to the total number of CD4+ T cells. Select the ‘type of counter’ in ‘ImageJ, check ‘show all’ and start counting by clicking the center of each cell and continue counting in different ‘Z’ serial images by using side arrows. After counting all the CD4 T cells, choose another type of counter and count the CD4+ dext+ T cells. Click the results tab to get the total number of cells counted with the two different counters.
- To obtain the total number, add the number of cells in all the ‘Z’ serial images representing all the three cerebral sections or all the eight heart sections.
Representative Results
In this report, in situ detection of antigen-specific, autoreactive CD4 T cells by direct staining with MHC class II dextramers is described. Freshly cut cerebral sections were derived from EAE mice and stained with cocktails containing PLP 139-151 (specific) or TMEV 70-86 (control) dextramers and anti-CD4. The top panels in Figure 1 shows LSCM analysis of cerebral sections costained with PLP 139-151 dextramers (red) and CD4 antibody (green), where the double-positive cells appeared yellow. Such a feature was absent in sections stained with TMEV 70-86 (control) dextramers (bottom panels). The costained (dext+ CD4+) cells showed punctate dots around the periphery. Quantitative analysis of PLP-specific T cells in the cerebral sections involved the following steps (Figure 2): 1) Sections of three pairs were made, and one section from each pair was stained individually with anti-CD4 and PLP 139-151 dextramers. Likewise, another set of three sections were stained with anti-CD4 and control (TMEV 70-86) dextramers. 2) From a given section, inflammatory foci (10 to 15) were identified based on anti-CD4 staining. 3) A set of ‘Z’ serial images (15 to 25) were taken from each focus sequentially from top to bottom with an interval of 2 µm between each. In essence, 10 to 15 similar sets of ‘Z’ serial images representing equal number of inflammatory foci were analyzed in every section. 4) In individual images, CD4+ cells (green), the cells positive for both dextramers (red) and CD4 (green) which appeared as yellow punctate cells were enumerated by marking them individually using the open source software, ImageJ, and finally, a grand total was obtained for each section by adding the number of cells counted in all the ‘Z’ serial images. Because that the cells were marked, the possibility of recounting the cells in multiple images that are overlapping was eliminated. Thus, reliable enumeration of dext+ cells relative to the total number of cells expressing CD4 can be achieved.
This study was further extended to demonstrate the use of dextramer reagents for detecting antigen-sensitized T cells in situ in EAM induced with Myhc 334-352 in A/J mice, which is also a T cell-mediated disease 13,14. Eight paired heart sections were obtained, each 200 µm thick, from EAM mice (Figure 2). From each pair, one section (with a total of 8 sections) was stained with Myhc 334-352 dextramers and anti-CD4, whereas another set of sections (8 in total) was stained with RNase 43-56 dextramers (control) and anti-CD4. Twenty sets of ‘Z’ serial images corresponding to that many inflammatory foci representing all eight sections were acquired sequentially as above (Figure 2). Essentially, a given set of ‘Z’ serial images consisted of 10 to 15 individual represented one single inflammatory focus. Analysis of these images by LSCM showed the presence of cells positive for Myhc 334-352 dextramers (red), and CD4 (green) and expectedly, the cells bound with both dextramers and CD4 appeared as yellow punctate cells (Figure 3, top panels). The background staining for control dextramers was negligible (Figure 3, bottom panels). The total numbers of dext+ CD4+ T cells and the total number of CD4+ T cells were determined for each section and counts from all the sections were added together to derive the total number for each subset (dext+ CD4+ T cells; CD4+ T cells).
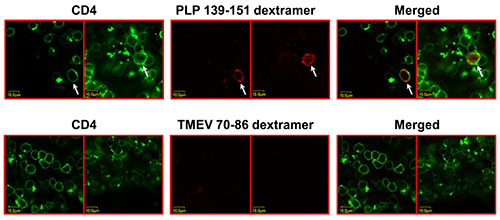 Figure 1. Detection of PLP-specific CD4 T cells by in situ staining with PLP 139-151 dextramers. EAE was induced in SJL mice by immunizing the animals with PLP 139-151 in CFA. At termination, cerebrums collected from EAE mice were embedded in 4% agarose, and the sections were made using vibratome. After staining with cocktails containing either PLP 139-151 dextramers/anti-CD4 or TMEV 70-86 dextramers (control)/anti-CD4 and fixing with 4% PBS-buffered paraformaldehyde, sections were washed and mounted for examination by LSCM. Top panels: two separate sections stained with PLP 139-151 dextramers/anti-CD4. Bottom panels: two separate sections stained with TMEV 70-86 dextramers/anti-CD4. Left panel: CD4, green; Middle panel: dextramers, red; Right panel: merged (arrows, dext+ CD4+ T cells). Original magnification 1,000X (adopted: Massilamany et al., 2014. Direct Staining with Major Histocompatibility Complex Class II Dextramers Permits Detection of Antigen-Specific, Autoreactive CD4 T Cells In Situ. PLOS ONE
9 (1): e87519). Please click here to view a larger version of this figure.
Figure 1. Detection of PLP-specific CD4 T cells by in situ staining with PLP 139-151 dextramers. EAE was induced in SJL mice by immunizing the animals with PLP 139-151 in CFA. At termination, cerebrums collected from EAE mice were embedded in 4% agarose, and the sections were made using vibratome. After staining with cocktails containing either PLP 139-151 dextramers/anti-CD4 or TMEV 70-86 dextramers (control)/anti-CD4 and fixing with 4% PBS-buffered paraformaldehyde, sections were washed and mounted for examination by LSCM. Top panels: two separate sections stained with PLP 139-151 dextramers/anti-CD4. Bottom panels: two separate sections stained with TMEV 70-86 dextramers/anti-CD4. Left panel: CD4, green; Middle panel: dextramers, red; Right panel: merged (arrows, dext+ CD4+ T cells). Original magnification 1,000X (adopted: Massilamany et al., 2014. Direct Staining with Major Histocompatibility Complex Class II Dextramers Permits Detection of Antigen-Specific, Autoreactive CD4 T Cells In Situ. PLOS ONE
9 (1): e87519). Please click here to view a larger version of this figure.
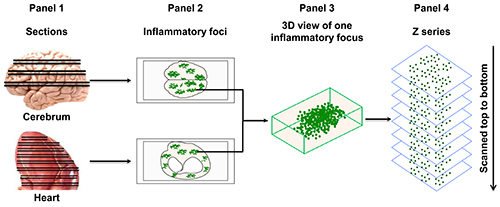 Figure 2. An approach to quantitate dext+ cells in brains and hearts. Paired sections are made from cerebrum or auricle-excised heart as shown (Panel 1). One section from each pair corresponding to the indicated organs was designated for staining with specific dextramers (cerebrum, PLP 139-151 dextramers; heart, Myhc 334-352 dextramers), and the other set of sections for control dextramers (cerebrum, TMEV 70-86; heart, RNase 43-56). After fixing and mounting, the sections were examined to locate inflammatory foci based on staining with CD4 by LSCM (original magnification, 40X) (Panel 2). Each inflammatory focus as visualized in three-dimensional (3D) view (Panel 3) was subjected to a set of ‘Z’ serial images sequentially (Panel 4). In all the images, cells positive for both specific or control dextramers and CD4, or CD4 alone were counted using ‘ImageJ’ software (adopted: Massilamany et al., 2014. Direct Staining with Major Histocompatibility Complex Class II Dextramers Permits Detection of Antigen-Specific, Autoreactive CD4 T Cells In Situ. PLOS ONE
9 (1): e87519). Please click here to view a larger version of this figure.
Figure 2. An approach to quantitate dext+ cells in brains and hearts. Paired sections are made from cerebrum or auricle-excised heart as shown (Panel 1). One section from each pair corresponding to the indicated organs was designated for staining with specific dextramers (cerebrum, PLP 139-151 dextramers; heart, Myhc 334-352 dextramers), and the other set of sections for control dextramers (cerebrum, TMEV 70-86; heart, RNase 43-56). After fixing and mounting, the sections were examined to locate inflammatory foci based on staining with CD4 by LSCM (original magnification, 40X) (Panel 2). Each inflammatory focus as visualized in three-dimensional (3D) view (Panel 3) was subjected to a set of ‘Z’ serial images sequentially (Panel 4). In all the images, cells positive for both specific or control dextramers and CD4, or CD4 alone were counted using ‘ImageJ’ software (adopted: Massilamany et al., 2014. Direct Staining with Major Histocompatibility Complex Class II Dextramers Permits Detection of Antigen-Specific, Autoreactive CD4 T Cells In Situ. PLOS ONE
9 (1): e87519). Please click here to view a larger version of this figure.
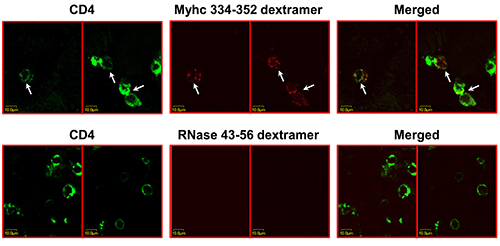 Figure 3. Detection of cardiac myosin-specific CD4 T cells by in situ staining with Myhc 334-352 dextramers. EAM was induced in A/J mice by immunizing the animals with Myhc 334-352 in CFA; at termination on day 21, hearts were collected after perfusion, and the tissue blocks were prepared using 4% PBS-buffered agarose. Eight paired sections of 200 µm thickness each were made using vibratome. One section from each pair (a total of eight sections) was stained with mixtures containing either Myhc 334-352 dextramers/anti-CD4 or RNase 43-56 (control) dextramers/anti-CD4. After fixing with 4% PBS-buffered paraformaldehyde, the sections were washed and mounted for examination by LSCM. Images showing the cells positive for both dextramers and CD4 or CD4 alone are shown. Top panels: two separate sections stained with Myhc 334-352 dextramers/anti-CD4. Bottom panels: two separate sections stained with RNase 43-56 dextramers /anti-CD4. Left panel: CD4, green; middle panel: dextramers, red; right panel: merged (arrows, dext+ CD4+ T cells). Original magnification 1,000 X (adopted: Massilamany et al., 2014. Direct Staining with Major Histocompatibility Complex Class II Dextramers Permits Detection of Antigen-Specific, Autoreactive CD4 T Cells In Situ. PLOS ONE
9 (1): e87519). Please click here to view a larger version of this figure.
Figure 3. Detection of cardiac myosin-specific CD4 T cells by in situ staining with Myhc 334-352 dextramers. EAM was induced in A/J mice by immunizing the animals with Myhc 334-352 in CFA; at termination on day 21, hearts were collected after perfusion, and the tissue blocks were prepared using 4% PBS-buffered agarose. Eight paired sections of 200 µm thickness each were made using vibratome. One section from each pair (a total of eight sections) was stained with mixtures containing either Myhc 334-352 dextramers/anti-CD4 or RNase 43-56 (control) dextramers/anti-CD4. After fixing with 4% PBS-buffered paraformaldehyde, the sections were washed and mounted for examination by LSCM. Images showing the cells positive for both dextramers and CD4 or CD4 alone are shown. Top panels: two separate sections stained with Myhc 334-352 dextramers/anti-CD4. Bottom panels: two separate sections stained with RNase 43-56 dextramers /anti-CD4. Left panel: CD4, green; middle panel: dextramers, red; right panel: merged (arrows, dext+ CD4+ T cells). Original magnification 1,000 X (adopted: Massilamany et al., 2014. Direct Staining with Major Histocompatibility Complex Class II Dextramers Permits Detection of Antigen-Specific, Autoreactive CD4 T Cells In Situ. PLOS ONE
9 (1): e87519). Please click here to view a larger version of this figure.
Discussion
Unlike MHC class I tetramers, the use of MHC class II tetramers for routine applications continues to be challenging, especially to enumerate the precursor frequencies of low-affinity autoreactive CD4 T cells, due in part to their activation dependency 6,15-17. Recently, this problem was circumvented by creating the next generation of tetramers, called “dextramers,” permitting us to detect antigen-specific CD4 T cells for a number of autoantigens, such as PLP 139-151, MOG 35-55 and Myhc 334-352 8. In this report, for the first time, the utility of MHC class II dextramers to detect and quantify self-reactive CD4 T cells in situ has been demonstrated using brain sections from EAE mice, and heart sections from EAM mice. On a reaction basis, dextramers offer the advantage of requiring a small amount of MHC/peptide complexes. In this protocol, each reaction requires only 0.75 µg of MHC/peptide monomers. In addition, staining with dextramers is a one-step reaction, since dextramers are coincubated with anti-CD4, and the whole procedure, from tissue sectioning to staining, can be finished in less than a day. In contrast, the published protocols for in situ staining with conventional tetramers commonly involve amplification procedures in which the fluorescent signals are amplified using secondary antibodies for fluorophores. As a result, staining procedures may take up to three days to complete 18-20.
Furthermore, the use of dextramers in cerebral sections permits detection of antigen-specific T cells deep in the tissue sections, even up to 50 µm. It was noted that PLP 139-151 dext+ CD4+ cells were found to be located both perivascularly and also within the parenchyma. Furthermore, the intensity of signals generated from Myhc 334-352 dextramers was low in cardiac sections when compared with the PLP 139-151 dextramers in brain sections (Figure 1), and such a variation may reflect the characteristics unique to each organ. One such variable is the lipid content which is present in high amounts in the brain tissue as opposed to cardiac tissue that contains mainly striated muscle fibers that can potentially restrict easy diffusion of dextramers into the inflammatory focus. In support of this notion, Myhc 334-352 dext+ cells were detected only to a depth up to 20 µm in majority of cardiac sections. Additionally, Myhc 334-352 dext+ cells were present scattered all through the myocardium suggesting the diffused nature of inflammatory infiltrates in myocarditic lesions.
Generally, two major factors can negatively influence the detection of antigen-specific T cells in situ by LSCM. First, insufficient signals emitted by fluorophores can lead to the need to amplify the signals using fluorophore antibodies. Second, such an indirect staining can undermine the specificity, as the background staining for control reagents also can increase proportionately 18,19. These limitations were circumvented by direct staining with the dextramers. Although, detection of CD4 T cells has been successfully shown with fresh sections, the use of dextramer reagents in frozen tissues requires additional studies, which can be a challenge as the retention of tissue-integrity may be difficult because of the tendency for tissues to get disintegrated during various staining steps 18,19 .
Disclosures
No conflicts of interests were declared.
Acknowledgments
This work was supported by the American Heart Association and the Children’s Cardiomyopathy Foundation (SDG2462390204001), and the National Institutes of Health (HL114669). CM is a recipient of a postdoctoral research fellowship grant awarded by the Myocarditis Foundation, NJ.
References
- Cecconi V, Moro M, Del Mare, Dellabona S, P, Casorati G. Use of MHC class II tetramers to investigate CD4+ T cell responses: problems and solutions. Cytometry A. 2008;73(11):1010–1018. doi: 10.1002/cyto.a.20603. [DOI] [PubMed] [Google Scholar]
- Ferlin W, Glaichenhaus N, Mougneau E. Present difficulties and future promise of MHC multimers in autoimmune exploration. Curr Opin Immunol. 2000;12:670–675. doi: 10.1016/s0952-7915(00)00161-8. [DOI] [PubMed] [Google Scholar]
- Kwok WW, Ptacek NA, Liu AW, Buckner JH. Use of class II tetramers for identification of CD4+ T cells. J Immunol Methods. 2002;268:71–81. doi: 10.1016/s0022-1759(02)00201-6. [DOI] [PubMed] [Google Scholar]
- Landais E, et al. New design of MHC class II tetramers to accommodate fundamental principles of antigen presentation. J Immunol. 2009;183:7949–7957. doi: 10.4049/jimmunol.0902493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepom GT. MHC class II tetramers. J Immunol. 2012;188:2477–2482. doi: 10.4049/jimmunol.1102398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollers SS, Stern LJ. Class II major histocompatibility complex tetramer staining: progress, problems, and prospects. Immunology. 2008;123:305–313. doi: 10.1111/j.1365-2567.2007.02801.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge L, et al. Tricks with tetramers: how to get the most from multimeric peptide-MHC. Immunology. 2009;126:147–164. doi: 10.1111/j.1365-2567.2008.02848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massilamany C, Upadhyaya B, Gangaplara A, Kuszynski C, Reddy J. Detection of autoreactive CD4 T cells using major histocompatibility complex class II dextramers. BMC Immunol. 2011;12:1471–2172. doi: 10.1186/1471-2172-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batard P, et al. Dextramers: new generation of fluorescent MHC class I/peptide multimers for visualization of antigen-specific CD8+ T cells. J Immunol Methods. 2006;310(1-2):136–148. doi: 10.1016/j.jim.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Massilamany C, Gangaplara A, Jia T, et al. Direct staining with major histocompatibility complex class II dextramers permits detection of antigen-specific, autoreactive CD4 T cells in situ. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0087519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massilamany C, Khalilzad-Sharghi V, Gangaplara A, Steffen D, Othman SF, Reddy J. Noninvasive Assessment of Cardiac Abnormalities in Experimental Autoimmune Myocarditis by Magnetic Resonance Microscopy Imaging in the Mouse. J Vis Exp. 2014. p. e51654. [DOI] [PMC free article] [PubMed]
- Massilamany C, Steffen D, Reddy J. An epitope from Acanthamoeba castellanii that cross-react with proteolipid protein 139-151-reactive T cells induces autoimmune encephalomyelitis in SJL mice. J Neuroimmunol. 2010;219(1-2):17–24. doi: 10.1016/j.jneuroim.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Donermeyer DL, Beisel KW, Allen PM, Smith SC. Myocarditis-inducing epitope of myosin binds constitutively and stably to I-Ak on antigen-presenting cells in the heart. J Exp Med. 1995;182(5):1291–1300. doi: 10.1084/jem.182.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao L, Sindhwani R, Leinwand L, Diamond B, Factor S. Cardiac alpha-myosin heavy chains differ in their induction of myocarditis. Identification of pathogenic epitopes. J Clin Invest. 1993;92(6):2877–2882. doi: 10.1172/JCI116909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron TO, Cochran JR, Yassine-Diab B, Sekaly RP, Stern LJ. Cutting edge: detection of antigen-specific CD4+ T cells by HLA-DR1 oligomers is dependent on the T cell activation state. J Immunol. 2001;166(2):741–745. doi: 10.4049/jimmunol.166.2.741. [DOI] [PubMed] [Google Scholar]
- Novak EJ, et al. Activated human epitope-specific T cells identified by class II tetramers reside within a CD4high, proliferating subset. Int Immunol. 2001;13(6):799–806. doi: 10.1093/intimm/13.6.799. [DOI] [PubMed] [Google Scholar]
- Reddy J, et al. Detection of autoreactive myelin proteolipid protein 139-151-specific T cells by using MHC II (IAs) tetramers. J Immunol. 2003;170(2):870–877. doi: 10.4049/jimmunol.170.2.870. [DOI] [PubMed] [Google Scholar]
- Skinner PJ, Daniels MA, Schmidt CS, Jameson SC, Haase AT. Cutting edge: In situ tetramer staining of antigen-specific T cells in tissues. J Immunol. 2000;165(2):613–617. doi: 10.4049/jimmunol.165.2.613. [DOI] [PubMed] [Google Scholar]
- Skinner PJ, Haase AT. In situ tetramer staining. J Immunol Methods. 2002;268(1):29–34. doi: 10.1016/s0022-1759(02)00197-7. [DOI] [PubMed] [Google Scholar]
- Skinner PJ, Haase AT. In situ staining using MHC class I tetramers. Curr Protoc Immunol. 2004;Chapter 17:Unit 17.14.11. doi: 10.1002/0471142735.im1704s64. [DOI] [PubMed] [Google Scholar]


