Abstract
The purpose of this manuscript is to provide a summary of the evaluation done by the Throughput and Multiplexing subteam on five emerging technologies: Single molecule array (Simoa™), Optimiser™, CyTOF® (Mass cytometry), SQIDLite™, and iLite™. Most of the information is presented with a minimum amount of published data and much is based on discussions with users and the vendor, to help provide the reader with an unbiased assessment of where the subteam sees each technology fitting best in the bioanalysis of large molecules. The evaluation focuses on technologies with advantages in throughput and multiplexing, but is wide enough to capture their strengths in other areas. While all platforms may be suited to support bioanalysis in the discovery space, because of their emergent nature, only Optimiser and SQIDLite are currently ready to be used in the regulated space. With the exception of Optimiser, each instrument/technology requires an up-front investment from the bioanalytical lab that will need justification during capital budget discussions. Ultimately, the platform choice should be driven by the quality of data, project needs, and the intended use of the data generated. In a time- and resource-constrained environment, it is not possible to evaluate all emergent technologies available in the market; we hope that this review gives the reader some of the information needed to decide which technology he/she may want to consider evaluating to support their drug development program in comparison to the options they already have in their hands.
KEY WORDS: emerging, high throughput, ligand binding assay, multiplexing, technology
INTRODUCTION
The Throughput and Multiplexing subteam assesses newly emerging technologies for their potential practical applications in analyte multiplexing and/or maximizing throughput for ligand binding assays (LBA). The subteam is formed by members from different pharmaceutical and instrument companies with experience in bioanalysis of biotherapeutics (biomarkers for pharmacodynamic (PD), pharmacokinetic (PK), and immunogenicity (IMG) determinations). This collaborative effort helped establish a series of attributes to identify strengths and weakness of the technologies presented here and to compare across technology platforms. The definitions for each of these attributes are laid out in the introductory article to this theme issue.
In an effort to increase awareness and share knowledge within the LBA bioanalytical community, the team identified five emerging technologies that have the potential to positively impact the way we do bioanalysis in terms of throughput and multiplexing. They include Single molecule array (Simoa™), Optimiser™, CyTOF® (Mass cytometry), SQIDLite™, and iLite™. The information presented here has been drawn from vendors, user experiences, and scientific literature (where available) and not based on a large body of data. Therefore, as the technologies are applied to various biological matrices, various therapeutic molecules and used to answer a spectrum of scientific questions, they may or may not perform as expected. Furthermore, although each of these technologies provides advantages in some respects, they may not be needed in other areas and therefore not applied across the board until they have a strong track record.
SINGLE MOLECULE ARRAY (Simoa)
Simoa is a microparticle-based technology developed by Quanterix© (1). Founded in 2007, Quanterix is a private company based in Lexington, MA. They are the exclusive licensee of a broad intellectual property portfolio initially developed at Tufts University by Dr. David Walt, scientific founder of Quanterix and Illumina® (NASDAQ: ILMN). Their mission is to develop ground-breaking tools in high-definition diagnostics.
Quanterix has used Simoa to develop a highly sensitive, single molecule immunoassay, also known as digital enzyme-linked immunosorbent assay (ELISA). The assay is based upon the capture of low abundance proteins via specific antibodies coated onto paramagnetic dye-coded beads. The immunocomplexes are labeled with an enzyme capable of generating fluorescent signal. The bead and the complex immobilized on them are isolated in very small (50 fl) chambers that are designed to hold only a single-complexed bead (see Fig. 1). This approach has been shown to detect enzyme-labeled complexes at concentrations ranging from 0.1 aM to 10 pM (2). This increase in sensitivity compared to standard ligand binding platforms has great potential for analyte quantitation (3).
Fig. 1.
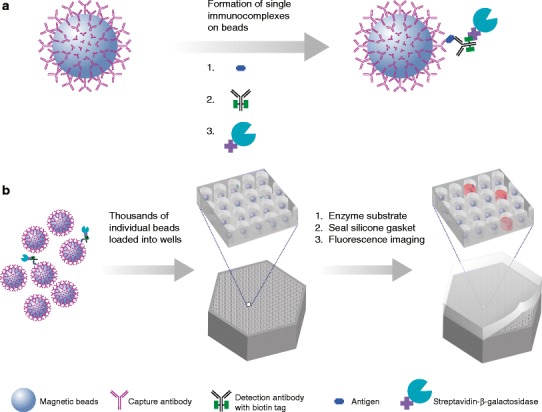
Representation of how analyte detection is achieved on the Simoa platform
Quanterix claims several advantages for their platform:
Sensitivity (up to 1,000 greater than ELISA)
Precision (coefficient of variation (CV) typically under 10% for replicates)
Dynamic range (>4 logs)
Low sample volume requirements
Robust multiplexing capabilities (up to 10-plex)
Automated (all mixing and washing, analyst just loads reagents and selects program)
Throughput (30 min to first sample result, others every 45 s after)
Although the technology has similarities to both the Singulex® Erenna® and Luminex® platforms, Simoa has differentiated itself by combining the single molecule counting of the Erenna with the multiplex capability of Luminex in a walk-away platform, where only the sample preparation is done off-line. Simoa multiplexing is achieved by encoding beads with dye and mixing different bead types together in an assay where each encoded bead type is associated with a unique capture antibody. The bead mixture is then deposited onto a single array, and each bead that is loaded is decoded to determine its identity. Each of the decoded groups is analyzed in parallel (4).
Quanterix offers the flexibility for customers to develop their own assays, offers custom assay development, and is currently building a library of assay kits. The kits currently available are predominantly for quantification of interleukins and various Alzheimer’s biomarkers.
Based on the available literature (1–4) and on conversations with the vendor, the technology was evaluated in a wide range of attributes summarized in Table I. Given the low-end sensitivity and potential for a high level of multiplexing, this technology seems to fit well for biomarker measurements. The potential to evaluate several key analytes from a single common sample is particularly appealing when dealing with precious or rare matrices. After sample loading, the assay process is completely automated as the instrument contains its own washer, reaction cuvettes, single molecule reader, and waste containers. This automation not only increases assay precision by removing the element of human error, but also enables laboratory personnel to pursue other work concurrently. Simoa technology shares some common consumables with standard ELISA such as sample plates and buffers, but has some that are unique such as microwell discs and sealing oil.
Table I.
Summary of the Technology Attributes Evaluated by This Emerging Technology Discussion Group on the Technologies Presented Here
| Technology attributes | Simoa | CyTOF | Optimiser | SQI |
|---|---|---|---|---|
| Format | Single molecule array | Mass cytometer | Microfluidic ELISA | Protein microarray |
| Assay sensitivity | Double digit fg/mL | Up to 500 Ab/cella | Low pg/mL to fg/mL reported | Double digit ng/mL (IMG) Low pg/mL (Biomarker) |
| Automation | Automated sample processing | Automated sample processing | Can be coupled to automated liquid handling/sample processing | Automated sample preparation and processing |
| Detection | Integrated | Integrated | Not integrated, plate reader of original assay may be used | Integrated |
| LIMS connectivity | Yes (LIMS brand TBD) | Possible with some manipulation | Not applicable | Available feature |
| Multiplexing capacity | Up to 10-fold | Up to 121-fold (dependent on availability of labels) | Not capable of multiplexing | Yes, for both capture and detection. Up to 90-fold |
| Throughput | 575 results per hour (10 plex) | Dependent upon acquisition time (AT); 640 results/hour with 3 min AT | 96 results in 1–2 h | 8,640 results in 3 h |
| • Batch sizing | 96 samples per plate | Dependent upon acquisition time | 96 samples per plate | 96 samples per plate |
| • Sample volume requirement (neat) | ∼5 μL | ∼5 μL (dependent on cell count, introduced through a 500-μL loop) | ∼5 μL | 100 μL |
| Performance | Precision equal to or better than ELISA | Precision equal to or better than flow cytometry | Precision equal to or better than ELISA | Precision equal to or better than ELISA; drug tolerance better than ELISA |
| Application—biomarkers, immunogenicity, PK | Biomarkers and other low abundance analytes | Biomarkers | PK, biomarkers when improvements in sensitivity are needed without the need to make major changes to platform | Biomarkers and multiple drug or anti-drug antibody targets need to be interrogated |
| Regulatory fit | Currently RUO only | RUO only (no plans for IVD) | Currently RUO only | FDA approved for IVD, 21 CFR part 11 compliant software |
| Development and automation tools | Assay development tools available | Assay development tools available | Assay development tools available | Assay development tools available |
| Secondary detector label technology | Single fluorescence label | Proprietary metal label | Single fluorescence label | Fluorescence label, 3 different |
| Sample type (e.g., serum, CSF, tissue, etc.) | All biological fluids | All biological fluids | All biological fluids | All biological fluids |
| Matrix toleranceb | Good | Good | Good | Good |
| Technology cost | Instrument US$150,000 | US$630,000 instrument; US$45,000 autosampler | US$5.40/plate for reagents. No additional instrument necessary | US$50,000–120,000 instrument cost depending on system choice |
| Technology vendor | ||||
| • Single vendor technology? | Yes | Yes | Yes | Yes |
| • Availability at CROs | No | No | No | Yes, 1CRO |
| • Technology flexibility—open/closed format | Open flexible format; assay development flexibility | Open flexible format; assay development flexibility | Open flexible format; assay development flexibility | Open flexible format. Limited user assay development flexibility |
| • Potential business sustainability | Venture capital backed, biotech/technology provider | Biotech innovators | Venture capital backed, biotech/technology provider | Equity capital backed, publicly traded biotechnology/technology provider |
| • Vendor reputation | So far very good | So far very good | So far very good | So far very good |
| Areas of positive impact | Sensitivity, throughput, multiplexing and automated | Good for discovery; multiplexing | Sensitivity, throughput and automated | Sensitivity, specificity, drug tolerance, throughput, multiplexing and automated |
iLite is not described as most of the parameters listed below are not applicable to the technology
aDue to the nature of the technology, it cannot be compared in similar type units
bNone of the evaluations focused on comparing the new technology to ELISA or MSD in terms of matrix tolerance. It was just determined to be good as the MRD was not high
The current instruments are for research use only (RUO) and lack compliance with 21 Code of Federal Regulations (CFR) Part 11; therefore, it is not currently recommended for the support of regulated studies. But, Quanterix has a goal of providing in vitro diagnostic (IVD) instruments within a few years of this publication.
OPTIMISER
The Optimiser technology was developed by Siloam Biosciences, Inc. (www.siloambio.com). Founded in 2004, Siloam Biosciences is primarily focused on the development of diagnostic platforms based on microfluidic immunoassay systems. Siloam is a privately held firm with funding sources from private investors and government agencies (National Institutes of Health (NIH) and the US army) as well as contracts from international multinationals.
Optimiser has a 4.5 μL spiral reaction microchannel etched in each plate well in order to increase the surface area in the familiar 96-well plate layout. Figure 2 illustrates the technology principle. The assay workflow is similar to an ELISA except the reagent volume needs are significantly lower, each step requiring 5–10 μL of reagent/sample. Flow of reagents/sample via the microchannel is driven by capillary forces. Excess reagent is siphoned onto an absorbent pad that sits beneath the microplate. The same capture and detection antibody pairs used in ELISA can be easily adapted for this technology platform. The detection antibody is fluorescently labeled and a plate reader capable of measuring fluorescence intensity is all the equipment needed.
Fig. 2.
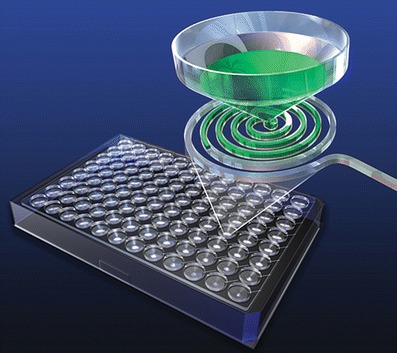
Representation of how sample or reagent is added to the Optimiser plate wells and flow through the channels into an absorbent pad (printed with permission from Siloam Biosciences)
Assay sensitivity can be enhanced by repeat addition of the sample to the microchannel, with reports on improvements in assay sensitivity ranging between 10 and 1,000-fold (Kai J, Santiago N, PuntambekarA, Lee SH, et. al. Tunable Sensitivity to Attogram/mL levels for Cytokine Assays using a Novel Microfluidics Microplate Combined with Conventional ELISA Reagents and Instrumentation, Abstract at SLAS Conference. 2012. San Diego, CA) (5). The large differences in reported sensitivity were attributed to the repeat addition of sample to the same well (Kai J, Puntambekar A, Sehy D, Brescia P and Banks P, Amplifying Immunoassay Sensitivity, Genetic Engineering and Biotechnology News. 2011; 31(18)).
Given that the Optimiser is a modified 96-well ELISA plate, it precludes the need to purchase specialized equipment and the associated expenses of computer system validation, software upgrades, and preventive maintenance. This is a significant advantage to the resource-constrained laboratory in an era when we are always challenged to do more with less. The caveat is the need to utilize a liquid handler to dispense the small volumes with the precision needed to give confidence in the data generated and meet regulatory expectations (Kai J, Puntambekar A, Sehy D, Brescia P and Banks P. Amplifying Immunoassay Sensitivity, Genetic Engineering and Biotechnology News. 2011; 31(18) AND Comley J. ELISA Assays—Recent innovations take analyte detection to new levels. Drug Discovery World. 2012). If the laboratory does not already have these systems, it would be an added cost. Although the vendor publicizes compatibility with a lower cost semi-automated instrumentation, this subteam is not aware of any bioanalytical laboratories with experience in these alternative systems.
The assay workflow, while similar to a typical ELISA, does not need significant incubation times due to the increased surface area and the lack of wash steps (Comley J. ELISA Assays—Recent innovations take analyte detection to new levels. Drug Discovery World.2012). This leads to a complete assay in about 1.5 h (when sample containing analyte is loaded only once) which is a significant time savings and can lead to higher throughput. In addition, the need for small sample volumes (∼5 μL) makes this technology attractive for precious samples or rare matrices and reduces reagent consumption per sample. The technology allows for sensitivity “tuning” by repeat additions of the samples. Literature reports for the measurement of interleukin-4 (IL-4) from cell culture supernatants demonstrate an approximate 10-fold increase in sensitivity in direct proportion to the number of analyte loadings (Kai J, Puntambekar A, Sehy D, Brescia P and Banks P. Amplifying Immunoassay Sensitivity, Genetic Engineering and Biotechnology News. 2011; 31(18)). The trade-off is an increase in assay time. To gain the ∼100-fold sensitivity, the assay time was increased from ∼1.5 to 10 h (very comparable to a typical ELISA). There are also reports from users seeing nearly a 10-fold improvement in sensitivity when switching an assay from an electrochemiluminesce (ECL) format (∼4 h) to the Optimiser (∼2 h) and adding a secondary antibody for detection (for the Optimiser assay direct labeling did not work). (6) User feedback for this technology platform while limited has been positive overall. In a specific user case study on the development of a PK assay to measure a monoclonal antibody (6), the user reported that making the switch from the existing assay to the Optimiser platform was relatively straightforward. The key to improving reproducibility was to use a robotic liquid handling system (Comley J. ELISA Assays—Recent innovations take analyte detection to new levels. Drug Discovery World. 2012) (6). One of the main advantages for the implementation of a liquid handler is that Siloam provides their own Tecan scripts (at this time limited to Tecan and not other brands) including liquid class and repetitive additions. The scripts only needed small modifications to allow implementation and was all performed by a medium-experienced Tecan user. Implementing the Optimiser technology in conjunction with the liquid handler resulted in increased sensitivity and acceptable precision (%CV < 20%), although the assay range was lower (2.6–625 ng/mL) than that of the ECL technology platform (10–10,000 ng/mL) (6). Based on our evaluation (Table I), the technology platform currently fits well in the discovery realm as there is insufficient user published information pertaining to parameters (e.g., precision and accuracy) more extensively evaluated prior to the support of regulated bioanalysis. As no new instrumentation is required, there is no need to check for LIMS interfacing and 21 CFR part 11 compliance, which may help smooth the applicability in the regulated space.
CyTOF® MASS CYTOMETER
DVS Sciences Inc. (recently acquired by Fluidigm) produces and markets the CyTOF 2 Mass Cytometer and the MaxPar® system of metal conjugated reagents to enable mass cytometry single cell proteomics applications. The founders of the company conceived the technology while working at MDS Sciex in conjunction with PerkinElmer Health Sciences, Inc. and the CyTOF was further developed in collaboration with academic researchers at the University of Toronto in 2005. Currently, CyTOF has not been adopted by any CROs; however, the company reports that the technology has been introduced into leading pharmaceutical and academic laboratories in the USA, Canada, Europe, and Asia.
The CyTOF 2 mass cytometer analyzes single cells labeled with metal isotope tags using inductively coupled plasma and time-of-flight (TOF) mass spectrometry. The CyTOF system contains over 120 detection channels which allow simultaneous resolution of multiple elemental probes per cell at high acquisition rates without the need for compensation, thereby maximizing the per-cell information obtained from a single sample. Issues with spectral overlap can limit the capabilities of traditional flow cytometry, whereas CyTOF can simultaneously measure over 40 different antigens in a single panel. Analysis takes place by incubating metal-labeled probes with the sample, (normally a cell suspension), followed by introduction into an inductively coupled plasma, where individual cells are atomized and ionized, then introduced into the ion optics and the TOF regions. The masses are counted in time-separated detector channels based on the metal-tagged probes. This results in the generation of a file that records the type and amount of each probe on the cells. Data are analyzed using traditional flow methods, such as 2D dot plots, as well as heat maps and tree plots using Spanning-tree Progression Analysis of Density-normalized Events (SPADE) analysis. SPADE clusters phenotypically similar cells into a hierarchy that allows high throughput, multidimensional analysis of heterogeneous samples.
As with many emerging technologies, customer experience or the ability to provide customer information is limited. Early adopters at Pfizer, Inc. utilized CyTOF for high dimensional analysis of cells derived from human blood samples and multiplexing of samples via surface markers and intracellular markers. Pfizer’s users also investigated activation of T cells and changes in intracellular phosphorylation states in stimulated and unstimulated whole blood. They reported that when analyzing heterogenous mixtures of peripheral blood mononuclear cells (PBMC) in quadruplicate, the CyTOF provides robust data. When the population frequency of a specific cell type was ≥5%, the CV between results was less than 2%. However, if the population frequency is <5%, for example B cells and dendritic cell subsets, the CV increased close to 10%. One Stanford University lab has used the technology to examine human bone marrow for several parameters, including binding of several antibodies, viability, and relative cell size (7). When comparing levels of cytokine-mediated signaling responses, specifically phosphorylated signal transducer and activator of transcription 3 and 5 (pSTAT3 and pSTAT5) in unstimulated and stimulated cells as measured by CyTOF and flow cytometry, the correlation between signaling induction was r = 0.92; P < 0.000001 for pSTAT3 and r = 0.89; P < 0.000001 for pSTAT5. This data illustrates that the CyTOF is capable of generating equivalent data as flow cytometry, while providing increased parameter analysis and lower background due to the absence of autofluorescence. CyTOF has also been combined with other techniques in order to investigate cell-cell interactions in tumor tissue. An imaging mass cytometry approach, which combines CyTOF with immunocytochemistry (ICC) and immunohistochemistry (IHC) as well as laser ablation, was developed to gain spatial understanding of tissue microenvironments (8). The investigators determined the validity of this approach by comparing to immunofluorescence microscopy (IFM) results. They found that tumor cell expression of specific biomarkers on serial tissue sections was similar with both techniques. Specifically, the percent expression as measured by IFM vs. imaging mass cytometry was, respectively, 100 and 100% for H3, 75 and 79% for HER2, and 63 and 66% for cytokeratin. Using the CyTOF coupled with additional imaging techniques allows for accurate measurement of cell markers in a multiplexed format vs. single-plex (IHC) and duplex (IFM) techniques.
Having only a single detector and utilizing metals that have similar signal efficiency allow for limited experimental setup and fewer adjustments for variability in signal intensity on the CyTOF as compared to traditional flow cytometry. The absence of multiple detectors translates into fewer controls per experiment. However, control beads, which are labeled with four distinct metals, are necessary for signal normalization. Users report that due to buildup of cellular debris in glass components between the nebulizer and inductive plasma of the CyTOF as well as metal deposits at the interface between the plasma and TOF regions, signal is lost over time. Aside from routine maintenance of the instrument, this loss in signal can be corrected through the use of the control beads. Fluidigm provides specifications on each lot of beads that can be entered directly into the software.
It is also important to note that while spectral overlap is substantially lower with the metal tags compared to fluorophores, some amount of overlap is still possible (Fig. 3). Many commercially available reagents, although enriched, will have purities less than 100%. Dependent upon the sensitivity threshold, there may be some overlap if the label has a purity of only 95%, for example. To a lesser extent, an overabundance of metal label in the sample as well as oxidation of the metals in the instrument can also affect the resolution of the results. However, multiplexing capability is still vastly improved over other methods.
Fig. 3.
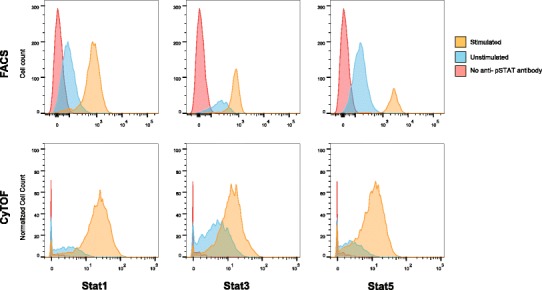
Reduction in spectral overlap between CyTOF and flow cytometry. Stimulated CD4+ cells from human whole blood were compared to unstimulated and an antibody negative control sample using flow cytometry (FACS) and CyTOF. Stimulated samples were incubated with interferon alpha and IL-10 for 15 min at 37°C and then analyzed for levels of phosphorylated Stat1, Stat3, and Stat5. Increased space between signal peaks indicates a reduction in spectral overlap
For potential users who are accustomed to flow cytometry, one major disadvantage is the inability to recover sample for sorting. This drawback is not as problematic for scientists analyzing samples using other formats, such as ELISA. CyTOF technology is also slower than flow cytometry—samples are run at approximately 500 cells/s vs. 5,000–10,000 cells/s, respectively. However, this decrease in throughput can be abrogated by the use of an autosampler and overnight running of samples.
Based on our assessment (Table I), CyTOF appears to be most relevant for a discovery setting. Its ability to screen for multiple analytes makes it a good candidate for high throughput binning, identification of key analytes of interest, target selection, and drug discovery structure activity relationship (SAR). Although the consumables’ costs are similar to those of traditional flow cytometry, the cost of the instrument (US$630,000) might put it out of reach for some. The lack of any immediate plans to develop an IVD instrument can also be an issue for any subsequent intended use in a regulated environment.
SQI’s Dlite SYSTEM
SQI Diagnostics is a Canadian company based in Toronto. The company was founded in 2003 and for the first 8 years built their platform technology to support assays to be Food and Drug Administration (FDA) cleared for in vitro diagnostic (IVD) testing; two immunoassay systems, Rheumatoid Arthritis 3-plex and Celiac 3-plex (immunoassay + instrument), are currently cleared (9).
In 2012, SQI broadened their market focus to include pharmaceutical anti-drug antibody (ADA) and biomarker multiplex testing using the same platform technology that was built for FDA-cleared tests. The company has several collaborations and projects in place with large pharmaceutical companies, two of which have been publicly shared: Isis Pharmaceuticals and Bristol-Myers Squibb (http://tinyurl.com/sqidiagnostics-bms). The technology is currently adopted by one CRO, Algorithme Pharma in Montreal, and other pharmaceutical customers are starting to drive the technology into their CRO partners.
The technology, represented in Fig. 4, is an automated microarray immunoassay platform that enables dual layer multiplexing of ADAs at the therapeutic protein and the anti-drug antibody level, i.e., at the capture reagent and detection level. The process starts with neat serum (a minimum of 10 μL is required) loaded on a 96-well microtiter plate, and an onboard liquid-handler/automation completes the dilutions, incubations, washing, and scanning. SQI’s technology prints microarray spots on an activated glass surface. These spots consist of multiple capture therapeutic proteins (drugs) in the case of detecting ADAs or one of the antibodies in the antibody pair in a cytokine (biomarker assay). Diluted patient serum is incubated on the microarray, washed/rinsed, followed by the addition of a secondary reporter antibody with a fluorescent label, followed by a wash/rinse/dry with a vapor phase removal system. The consumable plate is scanned with multiple wavelengths of fluorescence and filtered by precise emission/detection filters, and the captured image is processed using a unique algorithm to complete the analysis. Results are reported in either titers or units of concentration interpolated from a standard curve on board. The key to the multiplexing rests in the covalently bonded capture protein spots that do not interfere with each other as their location is fixed, combined with the multiple fluorescent signals that detect the differentially labeled secondary antibodies. The degree of simultaneous multiplexing equals up to 30 different capture proteins multiplied with 3 different detector reagents labeled with 3 different non-interfering fluorophores.
Fig. 4.
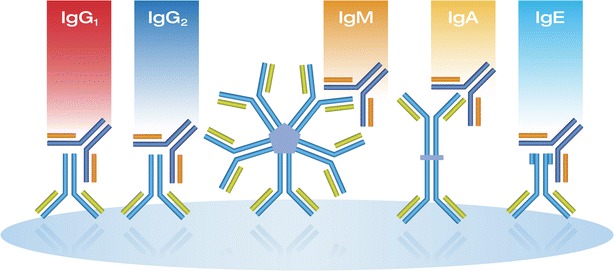
Representation of how the SQI Dlite platform isotypes ADAs capture on a well printed with the biotherapeutic
The configurable flexibility of the onboard assay development toolkit software provided by the company allows the user to optimize and control the different steps of the assay, including dilutions, incubations, washing, drying, scanning, reporting, LIMS reporting, and complete workflow integration. The software is 21 CFR Part 11 compliant. Assay incubation times are comparable to ELISA and one plate can be completed in less than 3 h.
The company reports several advantages with their technology:
Multiplexing at two levels, therapeutic protein(s) and/or metabolites subunits (used for epitope mapping) and secondly at the isotype and/or subclass level in the same well.
The platform has been reported to have equivalent to or higher drug tolerance as compared to competing ADA testing platforms (Nesman S, Gokemeijer J, et. al., Poster Presentation, Bioassays Conference 2013, Berkeley, CA).
Early data regarding improved drug tolerance may suggest that acid dissociation may not be required as much as is required in a traditional ELISA approach.
The company sells a range of platforms from a semi-automatic bench top system through to a high throughput fully automated system—kits can be used interchangeably between these systems
ADA testing and biomarker testing are executed in a similar manner.
One limitation is that customer’s assay development (including any reagent labeling and plate printing) is initially completed by SQI, which means the consumable must be purchased from the company, versus allowing the user to build the assay internally. Reagents need to be built to a specification provided by SQI using the customers’ assay-specific reagents; however, most users have all the reagents needed for buffer and diluent preparation. The company reports that in two recent projects involving multiple drug capture reagents and multiple detectors that the assay was built and functionally available to their customer in 6–8 weeks.
As SQI is new in the ADA/biomarker/epitope mapping space, their customer experience has been limited to six industry customers to date. Here, the company shared a recent presentation given by Bristol-Myers Squibb (Gokemeijer J. Podium Presentation, Bioassays Conference, 2013, Berkeley, CA) describing the multiplexed isotyping of a therapeutic protein in a single well. In this case study, the drug molecule was coated on the microarray and anti-IgG/IgM/IgA antibodies were used as a secondary reagent. In this study, equivalent or improved sensitivity and drug tolerance were shown compared to existing methods (Table II). Precision across multiple plates, days, and operators were acceptable (Table III). Agreement for sample testing with the existing methods was also good. All while providing quantitative results for multiple anti-drug antibody isotypes in a single test, which has the opportunity to improve workflow and reduces labor costs.
Table II.
Sensitivity and Drug Tolerance: Comparison to Existing Methods for ADA Assay on SQI Microassay Platform
| Anti-adnectin-Fc Fusion | ||
|---|---|---|
| Sensitivity (monoclonal) | Drug tolerance (polyclonal serum) | |
| ELISA | 125 ng/ml | 200 ng/ml @ 1/64K |
| ECL bridging assay | a500 ng/ml | 0.1 ng/ml @ 1/160 |
| SQI | b62.5 ng/ml | 250 ng/ml @ 1/128K |
aThe ECL Bridging assay employs a different format requiring bivalent binding and is typically less sensitive than ELISA
bThe sensitivity of the printed microarray format, wherein densely packed epitopes concentrate the detection signal, typically provides comparable or improved sensitivity relative to a standard ELISA, even in the absence of enzymatic amplification
Table III.
Reproducibility for an Immunogenicity Assay on the SQIDlite
| Reproducibility (n = 72 replicates performed inter-plate, inter-day, inter-operator) | ||||||
|---|---|---|---|---|---|---|
| Sample | Paba 2 1/200 | Pab2 1/800 | Pab2 1/256K | Pab2 1/512K | Pab6 1/400 | |
| % CV | IgA | 14.2 | 11.9 | 14.7 | 17.8 | 21.7 |
| IgG | 2.7 | 6.8 | 6.8 | 12.5 | 2.9 | |
| IgM | 14.3 | 14.7 | 14.7 | 11.8 | 11.3 | |
aPolyclonal antibody (Pab)
The ADA results from Bristol-Myers Squibb’s in-house ELISA were compared to the SQI results for several pre-clinical samples. The BMS ELISA does not isotype the response and combines the non-differentiated detection of immunoglobulin (Ig) isotype, IgG and IgM (no IgA). For 17 blinded monkey sera (ultimately revealed to be 8 positive and 9 negative), the isotyping assay demonstrated 100% agreement for IgG/IgM, and all true negatives also tested negative for IgA.
Algorithme Pharma, a CRO with experience using the technology, reported (Masse R, Sawyer J, Podium Presentation, Immunogenicity Conference, 2012, Boston, MA) that SQI’s collaborative development model enabled them to provide customers information with quick development time and multiplexing at both the protein and the isotype and subclass levels simultaneously. Algorithme Pharma has developed a multiplexed assay for detection of IgG, IgM, and IgA immune response to heparin, with assay sensitivity equivalent to a previously developed single-plex ELISA.
The technology appears to be suited for immunogenicity testing, in particular when there is a need for multiplexing assays: multiplexed isotyping, multiplexed screening assays, or assays to compare multiple therapeutic compounds in one test. Assay results are available in a semi-quantitative fashion with a built-in standard curve, or quantitative result if World Health Organization (WHO) standards are available. In addition, the platform can be used for biomarker testing (http://tinyurl.com/sqidiagnostics-cytokine).
There is a cost associated with the up-front investment of the platform (Table I), as well as vendor-specific consumables and the assay development which is initially performed by the vendor for the first assay. Subsequent assays can be developed by the customer with plate printing performed by SQI at a cost. However, the use of a fully automated system in combination with multiplexing can reduce the full time employee (FTE) cost by reducing the hands-on work needed.
iLITE
Biomonitor A/S is a Danish biotechnology company that combines the development of proprietary bioassays with the capabilities of a clinical reference laboratory. Biomonitor’s iLite technology represents reporter gene assays that improve the throughput as well as precision of traditional cell-based assays. Typically, cell-based assays are used to detect potency-related neutralization effects (10) and the increasing complexity of biology for newly emerging therapeutics leads to the need for more complex assay requirements. It is recognized that cell-based assays have low throughput due to the need for hands-on scientist involvement in cell culture and time required for biological effects to manifest themselves. This coupled with the impact of environmental and unique method-specific factors too often results in assays that are challenging to control. As a result, assays have evolved over time in the search for improved precision and greater throughput (11).
Biomonitor’s iLite reporter gene technology is based upon conventional gene-reporter assay format that has been modified and adapted for commercial applications in the diagnostic and clinical research market segments. In this approach, genetic engineering techniques are used to put firefly luciferase under a specific promoter and modifications are made to maximize reporter gene expression (12). Figure 5 illustrates an example of the iLite construct as applied to tumor necrosis factor alpha (TNF-α) (13). In this example, in the presence of TNF-α in the sample, luciferase enzyme is produced. After the required incubation, the cells are lysed, substrate is added, and the luminescence is measured. The amount of luciferase enzyme produced over time is directly proportional to the amount of TNF-α in the sample.
Fig. 5.
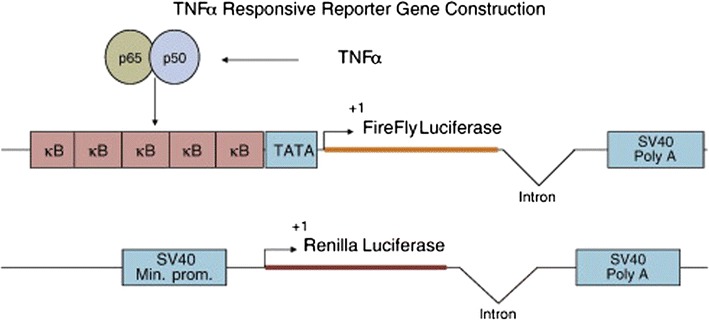
TNFα responsive reporter gene construction. κB, NFκB recognition sequence; TATA, TATA box; Firefly Luciferase, coding region of the Firefly luciferase gene; Intron, Intron from the human β-globulin gene; SV40 Poly A, SV40 polyadenylation site; SV40 Min. Prom, SV40 minimal promoter; Renilla Luciferase, coding region of the Renilla luciferase gene. Reprinted from Journal of Immunological Methods (13), with permission from Elsevier
Reporter gene assays are becoming a more common and acceptable approach for neutralization assays. Biomonitor A/S reports that the enhanced precision and throughput experienced by the end user is due not only to genetic engineering, but also in part to the clonal population of cells. When available, the standards are calibrated to international cytokine calibrators. As additional reference, the company’s website (http://www.biomonitor.dk/) provides a list of scientific publications employing this technology for methods to detect neutralization of interferon alpha and beta and TNF-α.
In order to evaluate the technology, a comparison was made between a traditional cell-based assay format in which a potency-based assay signal is neutralized by antibody and the comparable iLite cell line approach. In this application, the iLite assay required a total of 5 hours’ time for execution as compared to a 3-day method for the traditional assay, allowing for improvement in throughput, faster turnaround time for data reporting, as well as allows the operator to focus on other activities. In addition to the time saving, this method enhanced the assay’s precision both within and between plates; this allowed to reduce the number of required replicates from six (in the proliferation assay) to two (iLite approach). Although not a part of this evaluation, the Biomonitor menu of reporter gene assays includes both TNF-α and interferon beta (IFN-ß) assays that use dual-luciferase signal generation in which Renilla luciferase activity is under the promoter of a constitutive gene. Measuring each luciferase signal independently allows for normalization to a constitutive gene (Fig. 5).
iLITE kits available from Biomonitor include kits designed for RUO as well as kits that are 510(k) cleared. In addition, Biomonitor provides customized reporter gene cell line and assay development services.
As with any technology, the end user must have a full understanding of the potential drawbacks as well. Some users have reported viability problems with the growth-arrested cell lines, and Biomonitor is the sole vendor of iLite technology and consequently also serves as the sole CRO. Currently, the technology does not provide multiplexing capabilities, and only a small menu of cytokines (TNFα, TNFß, IFNß) is available. However, the vendor is able to perform custom design.
CONCLUSION
The drive for increased efficiencies within the bioanalytical laboratory is fostering collaborations between the vendors developing these innovative technologies and the end users, as well as collaborations transcending company boundaries, such as the one fostered by the LBAFG, Emerging Technology Multiplexing and High Throughput discussion group. Both efforts have culminated in this communication, where we presented an in-depth and balanced evaluation of the five promising emerging technologies. Each of the five technologies offers some unique advantages to the bioanalytical scientist. However, it is important that the end user have a thorough understanding of not only the advantages, but also the costs and risks associated with implementation of newly emerging and single source technologies. The platform choice should be driven by project need and intended use of the data generated, but in today’s reality, it must also include some information on return on investment. It is our hope that the reader finds the information gathered in Table I helpful in determining which emergent technology is best suited on their next evaluation to address their bioanalytical needs.
Acknowledgments
The authors acknowledge Frank Zambito from BMS and Melissa Wojick from Biogen Idec for their user perspective on the Optimiser technology, Andreas Jeromin for his perspective on the Simoa technology, Kondala Atkuri and Robert Smith from Pfizer for their user perspective on the CyTOF technology, and Steven Piccoli for his review and comments on the manuscript.
Footnotes
The opinions presented in this manuscript reflect those of the authors and do not reflect the opinions of the companies represented here.
References
- 1.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28(6):595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rissin DM, Fournier DR, Piech T, Kan CW, Campbell TG, Song L, et al. Simultaneous detection of single molecules and singulated ensembles of molecules enables immunoassays with broad dynamic range. Anal Chem. 2011;83(6):2279–2285. doi: 10.1021/ac103161b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang L, Rissin DM, Fournier DR, Piech T, Patel PP, Wilson DH, et al. Single molecule enzyme-linked immunosorbent assays: theoretical considerations. J Immunol Methods. 2012;378(1–2):102–115. doi: 10.1016/j.jim.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rissin DM, Kan CW, Song L, Rivnak AJ, Fishburn MW, Shao Q, et al. Multiplexed single molecule immunoassays. Lab Chip. 2013;13(15):2902–2911. doi: 10.1039/c3lc50416f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kai J, Puntambekar A, Santiago N, Lee SH, Sehy DW, Moore V, et al. A novel microfluidic microplate as the next generation assay platform for enzyme linked immunoassays (ELISA) Lab Chip. 2012;12(21):4257–4262. doi: 10.1039/c2lc40585g. [DOI] [PubMed] [Google Scholar]
- 6.Zambito F KA, Zhang Y, DeSilva B. Development of a PK immunoassay for a therapeutic monoclonal antibody using OptimiserTM technology. AAPS National Biotechnology Conference; San Diego, CA, USA, 2013.
- 7.Bendall SC, Simonds EF, Qiu P, el Amir AD, Krutzik PO, Finck R, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 9.Lea P, Keystone E, Mudumba S, Kahama A, Ding SF, Hansen J, et al. Advantages of multiplex proteomics in clinical immunology: the case of rheumatoid arthritis: novel IgXPLEX: planar microarray diagnosis. Clin Rev Allergy Immunol. 2011;41(1):20–35. doi: 10.1007/s12016-009-8189-z. [DOI] [PubMed] [Google Scholar]
- 10.Gupta S, Indelicato SR, Jethwa V, Kawabata T, Kelley M, Mire-Sluis AR, et al. Recommendations for the design, optimization, and qualification of cell-based assays used for the detection of neutralizing antibody responses elicited to biological therapeutics. J Immunol Methods. 2007;321(1–2):1–18. doi: 10.1016/j.jim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Tovey MG, Lallemand C. Improved analytical methods for the detection and quantification of neutralizing antibodies to biopharmaceuticals. Bioanalysis. 2012;4(17):2179–2190. doi: 10.4155/bio.12.186. [DOI] [PubMed] [Google Scholar]
- 12.Lallemand C, Meritet JF, Erickson R, Grossberg SE, Roullet E, Lyon-Caen O, et al. Quantification of neutralizing antibodies to human type I interferons using division-arrested frozen cells carrying an interferon-regulated reporter-gene. J Interferon Cytokine Res. 2008;28(6):393–404. doi: 10.1089/jir.2007.0142. [DOI] [PubMed] [Google Scholar]
- 13.Lallemand C, Kavrochorianou N, Steenholdt C, Bendtzen K, Ainsworth MA, Meritet JF, et al. Reporter gene assay for the quantification of the activity and neutralizing antibody response to TNFalpha antagonists. J Immunol Methods. 2011;373(1–2):229–239. doi: 10.1016/j.jim.2011.08.022. [DOI] [PubMed] [Google Scholar]


