Abstract
Increasing numbers of natural products have been found to possess anticancer effects. Nuclear factor erythroid-2-related factor-2 (Nrf2) is a master regulator of the antioxidative stress response, and our previous studies found that epigenetic modification of the Nrf2 gene appears to be a critical mechanism. Salvia miltiorrhiza, a Chinese herbal medicine widely used in Asian countries, has been shown to possess anticancer and antioxidant effects. Tanshinone IIA (TIIA), an active component in S. miltiorrhiza, has been reported to activate Nrf2 pathway. The objective of this study was to investigate the epigenetic regulation of Nrf2 by TIIA in mouse skin epidermal JB6 cells and the functional consequences for cell transformation. TIIA was found to induce antioxidant response element-luciferase and upregulate the mRNA and protein levels of Nrf2 and Nrf2 downstream target genes HO-1 and NQO-1. TIIA decreased the colony formation of JB6 cells by approximately 80%. TIIA decreased the protein levels of DNMT1, DNMT3a, DNMT3b, and HDAC3 and inhibited the enzymatic activity of HDACs. Bisulfite genomic sequencing indicated that TIIA demethylated the first five CpGs in the promoter region of the Nrf2 gene. Chromatin immunoprecipitation assays showed that TIIA treatment increased the recruitment of RNA polymerase II at Nrf2 transcription start site but had limited effects on enrichment of Ac-H3 in Nrf2 promoter. Taken together, our results show that TIIA activates the Nrf2 signaling pathway and induces epigenetic demethylation of the CpGs of Nrf2. The epigenetic reactivation of the Nrf2 signaling pathway by TIIA could potentially contribute to the attenuation of JB6 cellular transformation and anticancer effects.
KEY WORDS: epigenetics, Nrf2, skin cancer, tanshinone IIA
INTRODUCTION
The incidence of skin cancer has risen steadily in recent years, and an estimated 82,770 new cases will occur in the USA during 2013 (1). It is evident that oxidative stress plays an important role in carcinogenesis and cancer progression (2). Oxidative stress is caused by an imbalance between systemic reactive oxygen species (ROS) and the body’s capability to neutralize ROS, which may result in genomic instability, genetic mutation, and neoplastic transformation, leading to a higher incidence of carcinogenesis. Endogenous ROS can be generated during normal cellular metabolism, immune reactions, or under pathological conditions, whereas exogenous ROS may originate from the exposure to air pollution, UV irradiation, microorganisms, viruses, and xenobiotics. Skin is the first defensive barrier for the body and is thus susceptible to both endogenous and exogenous ROS (3).
It is estimated that more than two million cases of skin cancers around the world could be prevented by protecting the skin from excessive ROS exposure, such as extensive sun exposure and indoor tanning. To protect the body from ROS-mediated cellular injury, elimination or neutralization of ROS is required to maintain the redox balance in the cell (4).
Nuclear factor erythroid-2-related factor-2 (Nrf2 or NFE2L2) is a critical molecule involved in maintaining a balanced redox potential in the human body and plays an important role in regulating the expression of phase II detoxifying and antioxidative enzymes (29). Nrf2 or NFE2L2 is a basic helix-loop-helix leucine zipper transcription factor that can activate antioxidant genes by binding the antioxidant response element (ARE) in their corresponding promoter areas (5). These enzymes that protect against oxidation include heme oxygenase-1 (HO-1), NAD(P)H:quinone oxidoreductase-1 (NQO-1), UDP-glucuronosyltransferase (UGT), and glutathione-S-transferase (GST) among others. It has been reported that Nrf2-deficient mice display increased susceptibility to tumorigenesis induced by a carcinogen, and cancer chemoprevention is partially correlated with the induction of phase II enzymes. The Nrf2 −/− mice were found to have a higher risk of developing skin cancer when treated with DMBA-TPA (6).
Our recent study demonstrated that the expression of Nrf2 can also be regulated by epigenetic alterations in both the prostate tissue of the transgenic adenocarcinoma mouse (TRAMP) and tumorigenic TRAMP C1 cells (7,8). Epigenetic modifications, defined as the regulation of gene expression without an alteration in the DNA sequence, include DNA methylation, histone modification, nucleosome remodeling, and microRNA silencing and have been reported to contribute to many diseases (9). Accumulating evidence suggests that carcinogenesis can be modulated by epigenetic alterations such as DNA methylation and histone modifications in tumor suppressor genes and oncogenes (10). DNA methylation typically occurs at CpG sites (11,12). Physiologically, 70 to 80% of CpG sites within promoters are methylated, resulting in gene silencing. In contrast, the hypomethylation of CpG sites has been associated with the overexpression of target genes. To maintain normal methylation status, some co-suppressors, including DNA (cytosine-5)-methyltransferase (DNMT), histone deacetylases (HDACs), methyl CpG binding protein 2 (MECP2), the transcriptional repressor sin3, and hBrm are also involved (13). Among the repressors, DNMTs and HDACs are considered as the two major epigenetic effectors for the transcriptional control of gene promoters. Drugs targeting the enzymes responsible for epigenetic silencing, such as 5-azadeoxycytidine (5-aza, a DNMT inhibitor) (14) and trichostatin A (TSA, an HDAC inhibitor), have been widely investigated as cancer therapeutic agents. In addition to Western drugs, phytochemicals have also been found to possess chemoprevention effects via epigenetic alterations (15). For example, mahanine, a carbazole alkaloid found abundantly in some Asian vegetables, has been reported to be a DNA hypomethylation agent that restored the expression of an epigenetically silenced tumor suppressor gene due to the inhibitory effect of DNMTs (16).
Danshen, the rhizome of Salvia miltiorrhiza Bunge, is one of the most widely used Chinese herbs. Danshen has been used for centuries primarily in traditional Chinese medicine (TCM) without obvious side effects for the treatment of coronary heart disease (17). The pharmacological activities of Danshen include anticancer, antioxidant, anti-inflammatory, and antibacterial activity among others. Two major groups of compounds, phenolic acids such as salvianolic acid and lithospermic acid B, and tanshinones, such as tanshinone I, tanshinone IIA, and cryptotanshinone, have been identified in S. miltiorrhiza. Among these components, tanshinone IIA (TIIA) is the major constituent that is officially used as a quality control marker as per Chinese Pharmacopoeia (Fig. 1) and was found to be the major antioxidant component in S. miltiorrhiza. A recent study showed that TIIA was able to modulate the intracellular redox status in human aortic smooth muscle cells by inducing Nrf2 (18).
Fig. 1.
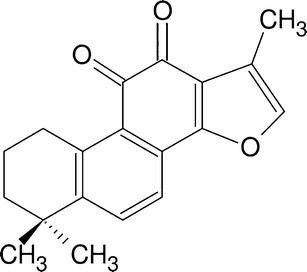
Chemical structure of tanshinone IIA
As mentioned above, overexposure to ROS may cause cellular injury, leading to a higher incidence of cancer. TIIA may be able to protect the cell from ROS-induced carcinogenesis by activating the Nrf2 pathway through epigenetic modulation. Thus, here, an in vitro study was performed to investigate the potential inhibitory effect of TIIA on the neoplastic transformation of JB6 P+ cells (a mouse keratinocyte cell line) when exposed to a carcinogen and to determine the underlying epigenetic mechanisms.
MATERIALS AND METHODS
Materials
TIIA, 5-aza, bacteriological agar, Eagle’s basal medium (BME), puromycin, 12-O-tetradecanoylphorbol-13-acetate (TPA), and TSA were obtained from Sigma-Aldrich (St. Louis, MO). Fetal bovine serum (FBS), minimum essential medium (MEM), and trypsin-EDTA solution were purchased from Gibco Laboratories (Grand Island, NY). The primary antibodies anti-Nrf2, anti-HO-1, anti-NQO-1, anti-UGT1A1, and anti-β-actin were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-DNMT primary antibodies (DNMT1, DNMT3a, and DNMT3b) were obtained from IMGENEX (San Diego, CA), and the anti-HDAC primary antibodies (HDAC1, HDAC2, HDAC3, HDAC4, and HDAC8) were obtained from Cell Signaling (Danvers, MA).
Anchorage-Independent Cell Neoplastic Transformation Assay
Stable mock (vector control) or Nrf2-knockdown JB6 P+ cells were established using lenti virus-mediated short hairpin RNAs (shRNAs) as previously described (19). Both control and Nrf2-depleted JB6 P+ cells were used in the following TPA-induced neoplastic transformation assay. The JB6 P+ cells were transferred to 1 mL of BME containing 0.33% agar over 3 mL of BME containing 0.5% agar with 10% FBS in six-well plates. The cells were maintained with TPA (20 ng/mL) alone or in a combination with TIIA. The cell colonies that formed in soft agar were photographed using a computerized microscope system with the Nikon ACT-1 program (Version 2.20, LEAD Technologies, Charlotte, NC) and counted using the ImageJ program (Version 1.40g, National Institutes of Health).
Cell Culture and Treatment
The human hepatocellular HepG2-C8 cell line was previously established by stable transfection with an ARE-luciferase construct. The cells were cultured and maintained in DMEM supplemented with 10% FBS, 100 units/mL penicillin, and 100 μg/mL streptomycin. JB6 P+ cells from American Type Culture Collection (Manassas, VA) were maintained in MEM containing 5% FBS with the same concentration of antibiotics. JB6 P+ cells stably transfected with shNrf2 were maintained in MEM supplemented with 5% FBS and 2 μg/mL puromycin. The cells were incubated at 37°C in a humidified 5% CO2 atmosphere. DMSO was used as a vehicle in all experiments at a concentration of 0.1%.
Cell Viability Tests
JB6 P+ cells, JB6-shNrf2, and HepG2 were seeded in 96-well plates at density of 6 × 103 cells/well. After incubation for 24 h, the cells were treated with either vehicle or various concentrations of TIIA for another 5 days. The medium was changed every 2 days. Cell viability was determined using a CellTiter 96 Aqueous One Solution Cell Proliferation (MTS) assay kit (Promega, Madison, WI) according to the manufacturer’s instructions.
Luciferase Reporter Activity Assay
The stably transfected HepG2-C8 cells expressing the ARE-luciferase vector were used to study the effects of TIIA on Nrf2-ARE pathways. The ARE-luciferase activity in the HepG2-C8 cells was determined using a luciferase assay kit in accordance with the manufacturer’s instructions (Promega, Madison, WI). Briefly, HepG2-ARE-C8 cells (1.0 × 105 cells/well) were seeded in 12-well plates in 1 mL of medium containing 10% FBS, incubated for 24 h, and then treated with various concentrations of samples. Afterwards, the cells were lysed using the reporter lysis buffer, and 10 μL of the cell lysate supernatant was analyzed for luciferase activity using a Sirius luminometer (Berthold Detection System GmbH, Pforzheim, Germany). Normalization of the luciferase activity was performed based on protein concentrations, which were determined using a BCA protein assay (Pierce Biotech, Rockford, IL, USA). The data were obtained from three independent experiments and are expressed as the inducible fold change compared with the vehicle control.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction
JB6 P+ cells were seeded in 6-cm dishes at a density of 1 × 104 cells/dish. After incubation for 24 h, the cells were treated with TIIA at different concentrations for 5 days. Total RNA was extracted using the RNeasy Mini Kit (QIAGEN, Valencia, CA). The SuperScript III First-Strand cDNA Synthesis System (Invitrogen, Grand Island, NY) was used to synthesize first-strand cDNA. The mRNA expression of Nrf2, HO-1, NQO1, HDACs (HDAC1, HDAC3, HDAC4, and HDAC8), and DNMTs (DNMT1, DNMT3a, and DNMT3b) was determined using quantitative real-time polymerase chain reaction (qPCR). The primer pairs used were described previously.
Western Blotting
After incubation for 24 h, JB6 P+ cells (5 × 103 cells per 6 cm dish) were treated with various concentrations of TIIA. Protein was extracted using RIPA buffer (Cell Signaling Technology®, Danvers, MA), and the protein concentration was determined using the bicinchoninic acid (BCA) kit (Pierce, Rockford, IL). The proteins were separated by 4 to 15% SDS-polyacrylamide gel electrophoresis (Bio-Rad, Hercules, CA) and transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA). After blocking with 5% BSA in Tris-buffered saline-0.1% Tween 20 buffer, the membrane was sequentially incubated with specific primary antibodies and HRP-conjugated secondary antibodies. The SuperSignal enhanced chemiluminescence (ECL) detection system was used to detect the antibody-bound proteins on the membrane. The intensity of the bands was analyzed using densitometry and the ImageJ program (Version 1.40g, National Institutes of Health).
HDAC and DNMT Activity Assay
HDAC and DNMT activity assays were performed using the EpiQuik™ HDAC Activity/Inhibition Assay Kit and DNMT Activity/Inhibition Assay Kit (Epigentek, Farmingdale, NY) following the manufacturer’s protocol, respectively. The nuclear protein fraction was prepared using the NEPER Nuclear and Cytoplasmic Protein Extraction Kit (Thermo scientific, Pittsburgh, PA), and the relative HDAC or DNMT activity was calculated based on the ratio of the HDAC or DNMT activity of the TIIA treatment group compared with that of the control group.
DNA Isolation and Bisulfite Genomic Sequencing
After incubation for 24 h, the cells were treated with TIIA at various concentrations or 5-aza (500 nM) in MEM containing 1% FBS for 5 days, and the medium was refreshed every 48 h. For combination 5-aza and TSA treatment, TSA (100 nM) was added to the medium on day 4. The cells were harvested on day 5. Genomic DNA was isolated from the treated cells using the QIAamp® DNA Mini Kit (Qiagen, Valencia, CA). Bisulfite conversion of genomic DNA was performed using the EZ DNA Methylation Gold Kit (Zymo Research Corp., Orange, CA). The DNA fragment containing the first five CpGs, which are located between −1085 and −1226 in the murine Nrf2 gene (with the translation start site defined as position +1), was amplified from the converted DNA by PCR using Platinum Taq DNA polymerase (Invitrogen, Grand Island, NY). The primer sequences were as follows: sense, 5′-AGT TAT GAA GTA GTA GTA AAA A-3′; anti-sense, 5′-AAT ATA ATC TCA TAA AAC CCC AC-3′. The PCR products were further cloned into a pCR4 TOPO vector using the TOPO™ TA Cloning Kit (Invitrogen, Carlsbad, CA). At least 10 colonies from each treatment group were randomly selected, and the plasmids were extracted using a QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). The target region was analyzed by sequencing (GeneWiz, South Plainfield, NJ).
Chromatin Immunoprecipitation Assay
After treatment of 1 × 107 cells in 150 mm dish with various concentrations of TIIA or TSA (100 nM), the cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Then 1.25 M glycine was added to quench the excess formaldehyde. Next, the cells were washed twice with ice-cold PBS and pelleted by spinning down at 1,000×g for 5 min. Chromatin samples were prepared using EpiSeeker Chromatin Extraction kit (Abcam, Cambridge, MA), then sheared to an average length of 600–1,000 bp using a Bioruptor sonicator (Diagenode, Sparta, NJ). Cross-linked chromatin fragments were subjected to immunoprecipitation with specific antibodies for Ac-H3 and Pol II (Cell Signaling, Danvers, MA) or nonspecific IgG using EpiSeeker chromatin immunoprecipitation (ChIP) one-step kit (Abcam, Cambridge, MA) following manufacturer’s protocol. After precipitation, 2 μL of each purified DNA was used as template for 35 cycles of PCR amplification using primers that cover the first five CpG: sense: 5′-GAG GTC ACC ACA ACA CGA AC-3′; anti-sense, 5′-ATC TCA TAA GGC CCC ACC TC-3′. Another set of primers covering the transcription start site was used to evaluate the recruitment of RNA polymerase complex II to the Nrf2 promoter: sense, 5′-CCT CAC CTC TGC TGC AAG TA-3′; anti-sense, 5′-GGC AAC TCC AAG TCC ATC AT-3′. The PCR products were analyzed by 2.0% agarose gel electrophoresis and visualized using EB staining. Primers covering GAPDH promoter region was used as a control to verify the efficacy of ChIP assays.
Statistical Analysis
The data are presented as the means ± SDs. Statistical analyses were performed using one-way analysis of variance (ANOVA). P < 0.05 or P < 0.01 was considered to be statistically significant.
RESULTS
TIIA Inhibits TPA-Induced JB6 P+ Cell Transformation
The effects of TIIA treatment on the TPA-induced anchorage-independent growth of JB6 P+ and JB6-shNrf2 cells were examined in soft agar. TIIA treatment with concentrations ranging from 0.5 to 5 μM significantly decreased the number of JB6 P+ colonies compared with the TPA-treated control group (positive control) (P < 0.01, one-way ANOVA) (Fig. 2). Treatment with TIIA (1.0–5.0 μM) significantly inhibited the TPA-induced anchorage-independent growth of JB6 P+ cells by approximately 28–56%, indicating that TIIA may have a chemopreventive potential against TPA-induced carcinogenesis in JB6 P+ cells.
Fig. 2.
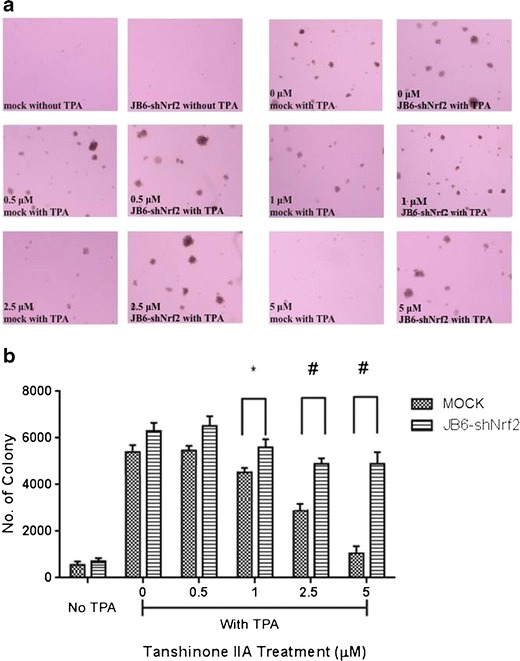
Inhibitory effect of TIIA pretreatment on the TPA-induced transformation of shMock and shNrf2-transfected JB6 P+ cells. Cells were treated with TIIA (0–5.0 μM) for 5 days. The TIIA pretreated cells (at a density of 8,000 cells/well) were then transferred to soft agar containing TPA in six-well plates for an additional 14 days. The colonies exhibiting anchorage-independent growth were counted under a microscope using ImageJ software. a Representative images of each group under microscope; b graphical data are presented as the average of triplicate results from two independent experiments. *P < 0.05; #P < 0.01, indicating a significant increase in colony formation compared with the shMock JB6 P+ cells in each treatment group
The colony formation of JB6-shNrf2 cells in soft agar was significantly increased when compared with the vector control cell line (Fig. 2) (P < 0.01, Student’s t test). In addition, no significant difference was observed between control and TIIA treatment groups (P > 0.05, one-way ANOVA). These results indicate that the protective effect of TIIA in JB6-shNrf2 cells was decreased, suggesting that Nrf2 plays an important role in the inhibitory effects of TIIA on TPA-induced JB6 P+ cell transformation.
Cytotoxicity of TIIA in JB6 P+ and HepG2-C8 Cells
The viability of JB6 P+, JB6-shNrf2, and HepG2-C8 cells after TIIA treatment for 24 h is shown in Fig. 3; IC50 values of 28.9, 20.1, and 26.3 μM for TIIA in JB6 P+, JB6-shNrf2, and HepG2-C8 cells, respectively, were observed. Similar cytotoxicity levels were observed among these three cell lines. To avoid using concentrations that were substantially toxic, no concentration greater than the IC50 was utilized in subsequent cell studies.
Fig. 3.
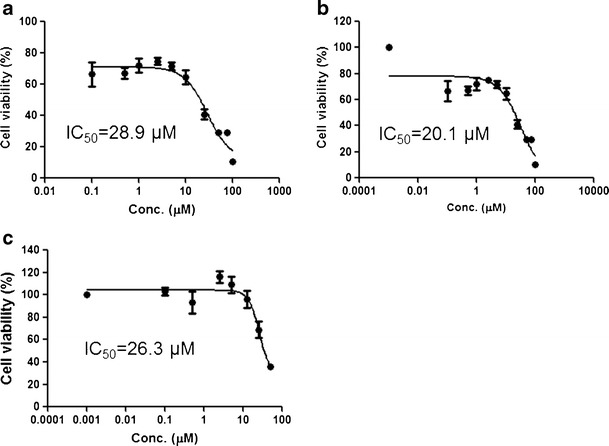
Cell viability of JB6 P+, JB6-shNrf2, and HepG2-C8 cells after treatment by TIIA for 24 h. a JB6 P+; b JB6-shNrf2; c HepG2-C8. The IC50 values were calculated using Emax sigmoid method
TIIA Induces ARE-Luciferase Reporter Activity
The relative fold changes of luciferase activity in cells transfected with the ARE-luciferase reporter vector are shown in Fig. 4. TIIA induced luciferase activity in a dose-dependent manner at concentrations ranging from 5 to 25 μM, although no inductive effect was observed at concentrations lower than 5 μM. This result further verified the effect of TIIA on Nrf2 as reported previously.
Fig. 4.
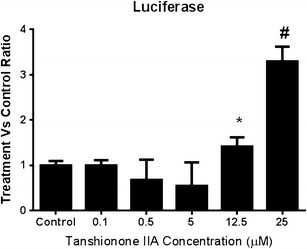
Induction of ARE-luciferase activity by the treatment of TIIA with concentrations from 0 to 25 μM in HepG2-C8 cells expressed with ARE-luciferase vector. The normalization of the luciferase activity was performed based on protein concentrations, which were determined using a BCA protein assay. The data were obtained from three independent experiments and expressed as the inducible fold change compared with the vehicle control. *P < 0.05 and #P < 0.01, respectively, indicate significant differences between the treatment and control group
TIIA Upregulates the mRNA and Protein Levels of Nrf2 Target Enzymes in JB6 P+ Cells
TIIA treatment significantly increased the expression of Nrf2, NQO1, and HO-1 mRNA (Fig. 5a). The upregulation of HO-1 and Nrf2 occurred in a dose-dependent manner at concentrations ranging from 2 to 10 μM. Conversely, the upregulation of NQO1 mRNA occurred in a dose-independent manner, with the highest expression level found at the lowest concentration (2 μM). This inconsistency may be caused by experimental variability or the incubation time. Another possibility is that TIIA may inhibit the expression of NQO1 at higher doses. In contrast, no statistically significant effect of TIIA was observed on UGT1A1, which was slightly decreased by the treatment with TIIA compared with the control group.
Fig. 5.
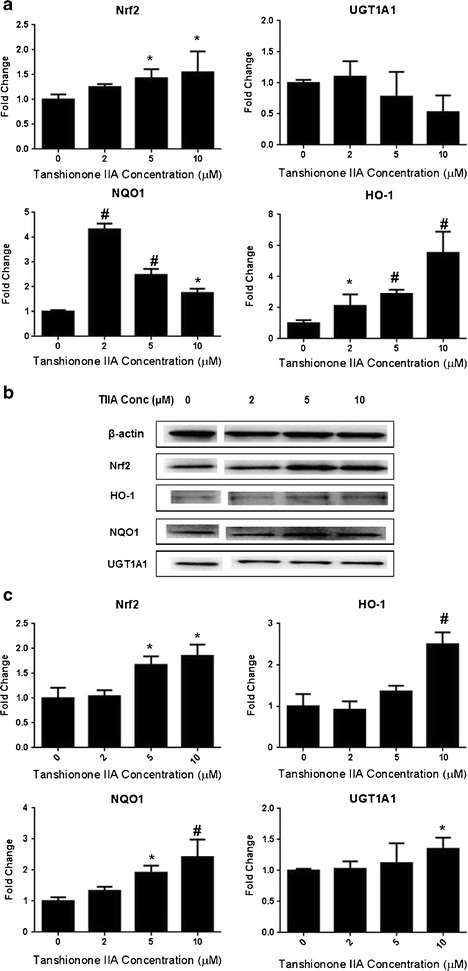
Effect of TIIA on Nrf2 mRNA and protein expression of Nrf2 target genes (HO-1, NQO1, and UGT1A1) in JB6 P+ cells. Cells were incubated with various concentrations of TIIA (2–10 μM) for 5 days. a TIIA increased the mRNA levels of Nrf2 and its downstream representative enzymes including HO-1, NQO1, and UGT1A1; b Western blot images of Nrf2 and its downstream genes including HO-1, NQO1, and UGT1A1; c TIIA (2–10 μM) increased the protein expression of Nrf2 and Nrf2 downstream enzymes. The graphical data are presented as the mean ± SD from three independent experiments. *P < 0.05 and #P < 0.01, respectively, indicate significant differences in each target protein or mRNA compared with its level in non-TIIA-treated cells
The protein levels of Nrf2, HO-1, NQO1, and UGT1A1 were further evaluated in the JB6 P+ cells treated with TIIA using Western blotting. In accordance with the qRT-PCR results, TIIA (2–10 μM) also increased the protein levels of Nrf2, HO-1, and NQO1 in a concentration-dependent manner (Fig. 5b). Unlike the mRNA result, the expression of UGT1A1 at the protein level was also increased. These results indicate that TIIA has the potential to induce the Nrf2 pathway, including antioxidant and detoxifying enzymes, which might be correlated with the increased cellular expression of Nrf2 in JB6 P+ cells.
TIIA Inhibits the mRNA and Protein Expression of Epigenetic Modification Enzymes in JB6 P+ Cells
The effect of TIIA on DNMTs (subtypes of DNMT1, DNMT3a, and DNMT3b) and HDACs (subtypes of HDAC1, HDAC3, HDAC4, and HDAC8) was further examined to investigate the epigenetic mechanism by which TIIA affects promoter demethylation and induces Nrf2 gene transcription. TIIA at concentrations of 2.0–10.0 μM decreased the mRNA of HDAC1, HDAC3, and HDAC8 as well as DNMT1, DNMT3a, and DNMT3b in a concentration-dependent manner in JB6 P+ cells after 5 days of treatment (Fig. 6a). Consistent with the mRNA expression, TIIA also decreased protein levels (Fig. 6b) in a concentration-dependent manner at concentrations ranging from 2 to 10 μM. In addition, TIIA repressed the expression of the HDAC4 protein, although no effect was observed at the mRNA level.
Fig. 6.
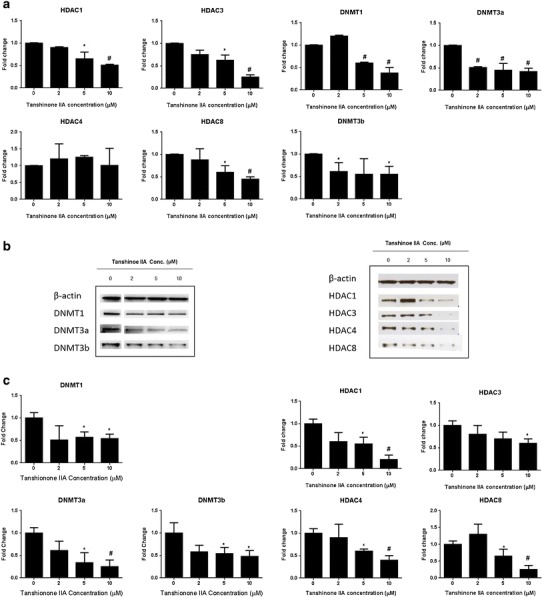
Effect of TIIA (2–10 μM) on DNMT and HDAC mRNA and protein expression in JB6 P+ cells. Cells were incubated with various concentrations of TIIA (2–10 μM) for 5 days. The expression of DNMT1, DNMT3a, and DNMT3b as well as HDAC1, HDAC3, HDAC4, and HDAC8 mRNA and proteins was detected by real-time PCR and Western blotting, respectively. a TIIA decreased the mRNA level of DNMT1, DNMT3a, and DNMT3b, and HDAC1, HDAC3, and HDAC8. b Western blot images of DNMTs including DNMT1, DNMT3a, and DNMT3b, as well as HDAC1, HDAC3, HDAC4, and HDAC8. c TIIA significant inhibit the protein levels of DNMT1, DNMT3a, and DNMT3b, as well as HDAC1, HDAC3, HDAC4, and HDAC8; The graphical data are represented as the mean ± SD from three independent experiments. *P < 0.05 and #P < 0.01, respectively, indicate significant differences in each group compared with its level in non-TIIA-treated cells
These results indicate that TIIA has the potential to epigenetically modify DNA methylation of the Nrf2 promoter; this may be an important mechanism for the induction of Nrf2 as mentioned above.
HDAC and DNMT Activity Assay
Treatment with either 5.0 or 10.0 μM TIIA significantly inhibited relative HDAC activity by 50% (P < 0.01) (Fig. 7). However, no significant inhibition of DNMT activity was observed despite the inhibition of DNMT1, DNMT3a, and DNMT3b expression by TIIA at the concentration mentioned above.
Fig. 7.
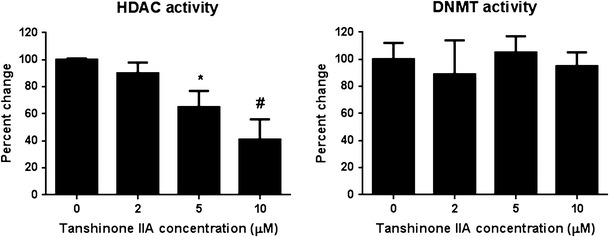
HDAC and DNMT activity assay was performed using EpiQuik Nuclear Extraction Kit. The relative HDAC activity was calculated based on the ratio of the HDAC activity of the TIIA treatment group to that of the control group. TIIA significantly inhibited the relative HDAC activity by 50%. The graphical data are represented as the mean ± SD from three independent experiments. *P < 0.05 and #P < 0.01, respectively, indicate significant differences in each group compared with its level in non-TIIA-treated cells
TIIA Decreases the Proportion of Methylated CpG in the Nrf2 Gene Promoter Region
The Nrf2 promoter region containing the first five CpGs was converted and amplified. The methylation status of the CpGs was then determined using bisulfite sequencing. Hypermethylation of these five CpGs (84%) was observed in the control JB6 P+ cells (Fig. 8), which was consistent with previous reports. In contrast to the control group, the methylation level decreased to 46.0% when cells were treated with TIIA (10 μM), which was similar to the positive control group (46.0%, 5-aza (500 nM) + TSA (100 nM)). These results suggest that TIIA reverses the CpG methylation status of the Nrf2 gene promoter, which may drive the transcriptional re-expression of Nrf2 in JB6 P+ cells.
Fig. 8.
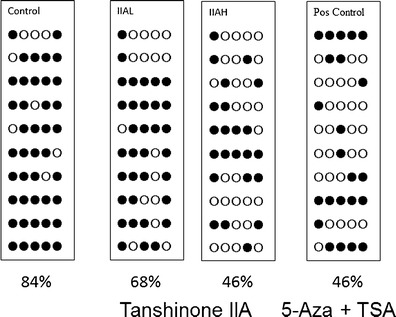
Effect of TIIA on methylation alteration of the Nrf2 promoter regions in JB6 P+ cells. a Genomic DNA from non-treatment cells; b genomic DNA was extracted from TIIA-treated cells after 5 days of treatment; c the 5-aza (500 nM)/TSA (100 n ) combination treatment. The filled and open dots indicate methylated and unmethylated CpGs, respectively. Methylated CpG ratio, the percentage of methylated CpGs was based on the total CpGs in each treatment group. IIAH, TIIA high concentration (10 μM); IIAL, TIIA low concentration (5 μM); Pos Control, positive control (5-aza (500 nM) + TSA (100 nM))
TIIA Increases the Recruitment of RNA Polymerase Complex II at Nrf2 Transcription Start Site
ChIP assays were employed to further examine the proteins that could potentially interact with the Nrf2 promoter in JB6 cells. Based on the methylation pattern of Nrf2 promoter, primers were designed to cover the first five CpGs and the transcription start site (TSS) to detect the enrichment of Ac-H3 and RNA polymerase II associated with the Nrf2 promoter. Although TIIA treatment slightly increased total Ac-H3 level (Fig. 9b), an enriched acetylation of H3 was not observed at the loci where the first five CpGs are located. On the other hand, the recruitment of RNA polymerase II (Pol II) to Nrf2 TSS was significantly increased after exposure of TIIA (Fig. 9d). The specificity of ChIP assays was verified by nonspecific IgG pull down. As a positive control, GAPDH promoter is equally amplified among all the samples associated with Pol II.
Fig. 9.
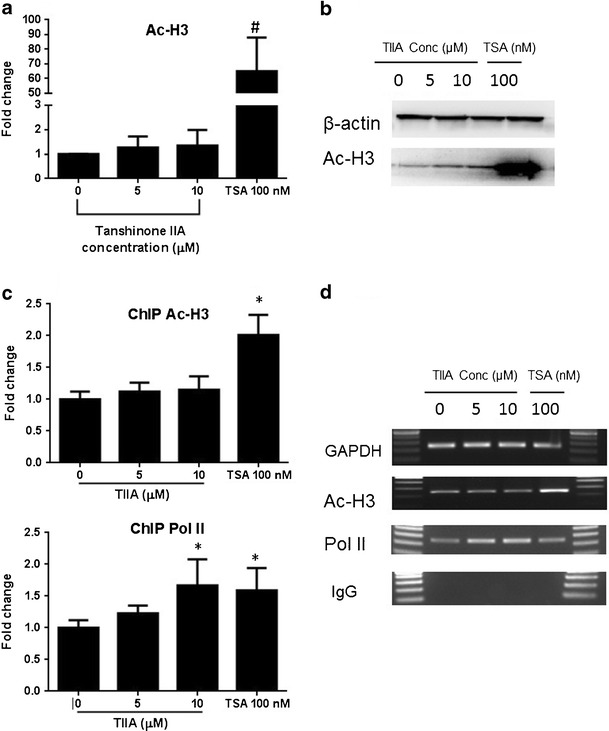
Effect of TIIA on histone modification associated to Nrf2 gene promoter. Cells were incubated with TIIA (5 and 10 μM) for 5 days. The total level of Ac-H3 was detected by Western blotting. a TIIA increased the protein level of Ac-H3. b Western blot images; ChIP assays were performed to analyze the enrichment of DNA binding to Ac-H3 and Pol II using primers cover the first five CpGs or TSS of Nrf2 gene promoter, respectively. c The enrichment of the Ac-H3 and Pol II binding DNA was determined by PCR. Relative ratio was calculated using ImageJ software and present by the fold change compared to control group. Data are expressed as mean ± SD from three independent experiments. d Regular PCR was performed to compare the immunoprecipitated DNA versus control group, a representative result is shown from three independent experiments. Trichostatin A (TSA) was used as positive control. *P < 0.05 and #P < 0.01, respectively
DISCUSSION
In this study, the chemopreventive potential of TIIA in skin cancer and its underlying mechanisms were investigated in an in vitro study using JB6 P+ cells. JB6 P+ is a normal skin keratinocyte cell line that will transform under carcinogenic or environmental challenges including TPA, EGF, and UVB (20). Therefore, this model has been widely used for skin cancer risk assessment. In the present work, we found that TIIA inhibited TPA-induced JB6 P+ cell transformation, which indicated a potential chemopreventive effect of TIIA on skin cancer.
Nrf2 is an important transcription factor regulating phase II detoxifying (or cytoprotective) enzymes and the expression of antioxidant response genes. Numerous studies have provided strong evidence for a relationship between Nrf2 deficiency and increased susceptibility to carcinogenesis, and elimination of carcinogens through induction of cytoprotective enzymes is one important approach to prevent cancer (20). In our previous study, we have shown that the epigenetics reactivation of Nrf2 pathway appears to be an important mechanism resulting in the inhibition of transformation of JB6 P+ cells (19). Therefore, Nrf2 is a key molecular target for the development of chemopreventive agents. A large amount of chemicals, especially those from natural products such as dithiolethiones and sulforaphane, have been extensively studied for their effects on the induction of the Nrf2 pathway (12,21). Several mechanisms have been identified, including an interaction between KEAP1 and NRF2, phosphorylation of Nrf2 by various protein kinases, interaction with other protein partners (p21, caveolin-1), and epigenetic regulation. Among these mechanisms, epigenetic changes such as DNA promoter methylation and chromatin modifications have been found to play an important role in tumorigenesis (5). Nrf2 loss of function, which is associated with promoter hypermethylation, was reported in skin and prostate tumorigenesis (22). The methylation of the first five CpG sites in the Nrf2 promoter has been shown to substantially decrease Nrf2 expression. Radix Angelicae Sinensis extract and Z-ligustlide (one active component) decreased the level of methylation of the first five CpGs, leading to an increase in Nrf2 expression (23). In this study, hypermethylation of the first five CpGs of the Nrf2 promoter was identified in JB6 P+ cells, leading to reduced Nrf2 activity. The epigenetic silencing could be attributed to the epigenetic alterations, e.g., overexpression of HDACs or DNMTs, because these epigenetic modification enzymes were found to be highly expressed in the control cells.
A previous study showed that Nrf2 is involved in the cytoprotective effects of tanshinone IIA by reducing intracellular redox status in human aortic smooth muscle cells and protecting against oxidative stress via the ERK and PKB signaling pathways (24). However, the exact mechanisms by which TIIA regulates Nrf2 have not been identified. Our present study shows for the first time that TIIA epigenetically regulates Nrf2 activation by decreasing promoter DNA methylation. This result correlates with TIIA’s inhibition of the expression of DNMTs (Fig. 6). Although DNMT activity was not affected by TIIA at the cellular level (Fig. 7) [this enzymatic assay of DNMTs included all of the subtypes of DNMTs], the results suggested a specific DNMT rather than the total DNMTs might be crucial for Nrf2 promoter methylation. Furthermore, it has been reported that cellular DNMT level can be affected at both transcription and post-translation levels (25–27), which could lead to changes of gene methylation profile. Thus, it is possible that TIIA alters Nrf2 promoter methylation pattern when total DNMTs activity remains unchanged. On the other hand, DNA methylation has been shown to have crosstalk with histone modifications at multiple levels. In this study, our results show that the expression of HDAC1, HDAC3, HDAC4, and HDAC8, as well as HDAC enzyme activity, were significantly decreased in a dose-dependent manner by TIIA. To our surprise, in spite of the slight induction of the histone marker acetyl-H3 (open chromatin structure) at the global level, TIIA treatment did not increase the enrichment of acetyl-H3 to the promoter region of the Nrf2 gene where the first five CpGs are located. Nevertheless, TIIA treatment enriched the recruitment of RNA polymerase II complex on transcription start site of Nrf2 gene. These findings suggest that TIIA does not directly affect acety-H3 induced chromatin structure modification associated with the enhanced Nrf2 transcription activity. Considering some HDACs could involve in DNMT complex that mediate DNA methylation (28), inhibition of HDACs by TIIA may also indirectly contribute to the demethylation effects of TIIA on Nrf2 promoter. Thus, alteration of the expression of some specific DNMTs and HDACs by TIIA would contribute to its capability of epigenetic modulation.
Danshen has been used in China in patients with cardiovascular diseases for many years, and it is approved by the State Food and Drug Administration of China. Our studies show that TIIA inhibited the transformation of JB6 P+ cells at doses of 5–10 μM (Fig. 2). Previous studies indicated that the blood levels of tanshinones were less than 1 μM after oral administration. This may raise concerns about the in vitro and in vivo relevance because the blood level is far below the in vitro effective concentration level. However, because Danshen has been widely used with a variety of dosage forms and the safety and toxicity of Danshen in human subjects are well known, it is feasible to optimize the formulation to achieve a high circulating concentration in the human body. In addition, because other components in Danshen have not been found to be effective, the development of TIIA as a pure component is highly feasible. Tanshinone I, another active component in Danshen, has similar structure to TIIA. It has been reported that tanshinone I is a potent Keap1-C151-dependent Nrf2 activator which can stabilize Nrf2 by hindering its ubiquitination (29). Although both compounds are able to upregulate Nrf2 pathway, the underlying mechanisms may be different. Keap1 was found to be involved in Nrf2 regulation by tanshinone I(29), and epigenetic modulation could also play a major role and potentially be the main mechanism contributing to the upregulation of Nrf2 by TIIA.
In conclusion, the present work discovered that tanshinone IIA significantly inhibits HDACs (HDAC1, HDAC3, HDAC4, and HDAC8) and DNMTs (DNMT1, DNMT3a, and DNMT3b) at both the mRNA and protein levels. The inhibitory effect of these epigenetic modification enzymes induced Nrf2 activity via demethylation of the Nrf2 gene promoter in JB6 P+ cells, indicating a potential role in chemoprevention of skin cancer.
ACKNOWLEDGMENTS
This study was supported by Grant 81102856 from the National Science Foundation of China to Ling Wang and institutional funding to Ah-Ng Tony Kong. The authors thank all the members in Dr. Kong’s lab for their helpful discussion and preparation of this manuscript.
Footnotes
Ling Wang and Chengyue Zhang contributed equally to this work.
Contributor Information
Ling Wang, Email: rebeccawang312@gmail.com.
Ah-Ng Tony Kong, Phone: (848) 445-6369/8, Email: kongt@pharmacy.rutgers.edu.
REFERENCES
- 1.Cancer facts & figures, American Cancer Society. 2013:1.
- 2.Slattery ML, John EM, Torres-Mejia G, Lundgreen A, Lewinger JP, Stern MC, et al. Angiogenesis genes, dietary oxidative balance and breast cancer risk and progression: the Breast Cancer Health Disparities Study. Int J Cancer. 2014;134(3):629–44. doi: 10.1002/ijc.28377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birch-Machin MA, Russell EV, Latimer JA. Mitochondrial DNA damage as a biomarker for ultraviolet radiation exposure and oxidative stress. Br J Dermatol. 2013;169:9–14. doi: 10.1111/bjd.12207. [DOI] [PubMed] [Google Scholar]
- 4.Finley JW, Kong AN, Hintze KJ, Jeffery EH, Ji LL, Lei XG. Antioxidants in foods: state of the science important to the food industry. J Agric Food Chem. 2011;59(13):6837–46. doi: 10.1021/jf2013875. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Inoyama D, Kong A-NT, Beamer LJ, Hu L. Kinetic analyses of Keap1–Nrf2 interaction and determination of the minimal Nrf2 peptide sequence required for Keap1 binding using surface plasmon resonance. Chem Biol Drug Des. 2011;78(6):1014–21. doi: 10.1111/j.1747-0285.2011.01240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, et al. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66(16):8293–6. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, Khor TO, Shu L, Saw CL, Wu TY, Suh N, et al. A γ-tocopherol-rich mixture of tocopherols maintains Nrf2 expression in prostate tumors of TRAMP mice via epigenetic inhibition of CpG methylation. J Nutr. 2012;142(5):818–23. doi: 10.3945/jn.111.153114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, et al. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS One. 2010;5(1):e8579. doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shu L, Khor TO, Lee JH, Boyanapalli SS, Huang Y, Wu TY, et al. Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J. 2011;13(4):606–14. doi: 10.1208/s12248-011-9300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stefansson OA, Esteller M. Epigenetic modifications in breast cancer and their role in personalized medicine. Am J Pathol. 2013;183(4):1052–63. doi: 10.1016/j.ajpath.2013.04.033. [DOI] [PubMed] [Google Scholar]
- 11.Khor TO, Huang Y, Wu TY, Shu L, Lee J, Kong AN. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem Pharmacol. 2011;82(9):1073–8. doi: 10.1016/j.bcp.2011.07.065. [DOI] [PubMed] [Google Scholar]
- 12.Su ZY, Zhang C, Lee JH, Shu L, Wu TY, Khor TO, et al. Requirement and epigenetics re-programming of Nrf2 in suppression of tumor promoter TPA-induced mouse skin cell transformation by sulforaphane. Cancer Prev Res. 2014;7(3):319–29. doi: 10.1158/1940-6207.CAPR-13-0313-T. [DOI] [PubMed] [Google Scholar]
- 13.Nakao M. Epigenetics: interaction of DNA methylation and chromatin. Gene. 2001;278(1–2):25–31. doi: 10.1016/S0378-1119(01)00721-1. [DOI] [PubMed] [Google Scholar]
- 14.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–41. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Wang Z, Fan J, Liu M, Yeung S, Chang A, Chow MS, et al. Nutraceuticals for prostate cancer chemoprevention: from molecular mechanisms to clinical application. Expert Opin Investig Drugs. 2013;22(12):1613–26. doi: 10.1517/13543784.2013.833183. [DOI] [PubMed] [Google Scholar]
- 16.Agarwal S, Amin K, Jagadeesh S, Baishay G, Rao P, Barua N, et al. Mahanine restores RASSF1A expression by down-regulating DNMT1 and DNMT3B in prostate cancer cells. Mol Cancer. 2013;12(1):99. doi: 10.1186/1476-4598-12-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou L, Zuo Z, Chow MS. Danshen: an overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J Clin Pharmacol. 2005;45(12):1345–59. doi: 10.1177/0091270005282630. [DOI] [PubMed] [Google Scholar]
- 18.Zhang HS, Wang SQ. Nrf2 is involved in the effect of tanshinone IIA on intracellular redox status in human aortic smooth muscle cells. Biochem Pharmacol. 2007;73(9):1358–66. doi: 10.1016/j.bcp.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Su ZY, Zhang C, Lee JH, Shu L, Wu TY, Khor TO, et al. Requirement and epigenetics reprogramming of Nrf2 in suppression of tumor promoter TPA-induced mouse skin cell transformation by sulforaphane. Cancer Prev Res (Phila) 2014;7(3):319–29. doi: 10.1158/1940-6207.CAPR-13-0313-T. [DOI] [PubMed] [Google Scholar]
- 20.Shin JW, Ohnishi K, Murakami A, Lee JS, Kundu JK, Na HK, et al. Zerumbone induces heme oxygenase-1 expression in mouse skin and cultured murine epidermal cells through activation of Nrf2. Cancer Prev Res. 2011;4(6):860–70. doi: 10.1158/1940-6207.CAPR-10-0354. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C, Su ZY, Khor TO, Shu L, Kong AN. Sulforaphane enhances Nrf2 expression in prostate cancer TRAMP C1 cells through epigenetic regulation. Biochem Pharmacol. 2013;85(9):1398–404. doi: 10.1016/j.bcp.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng X, Cui XX, Khor TO, Huang Y, Dipaola RS, Goodin S, et al. Inhibitory effect of a γ-tocopherol-rich mixture of tocopherols on the formation and growth of LNCaP prostate tumors in immunodeficient mice. Cancers. 2011;3(4):3762–72. doi: 10.3390/cancers3043762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su Z-Y, Khor TO, Shu L, Lee JH, Saw CL-L, Wu T-Y, et al. Epigenetic reactivation of Nrf2 in murine prostate cancer TRAMP C1 cells by natural phytochemicals Z-ligustilide and radix Angelica sinensis via promoter CpG demethylation. Chem Res Toxicol. 2013;26(3):477–85. doi: 10.1021/tx300524p. [DOI] [PubMed] [Google Scholar]
- 24.Gong Y, Li Y, Abdolmaleky HM, Li L, Zhou JR. Tanshinones inhibit the growth of breast cancer cells through epigenetic modification of Aurora A expression and function. PLoS One. 2012;7(4):e33656. doi: 10.1371/journal.pone.0033656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan HH, Porter AG. p21(WAF1) negatively regulates DNMT1 expression in mammalian cells. Biochem Biophys Res Commun. 2009;382(1):171–6. doi: 10.1016/j.bbrc.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Peng L, Yuan Z, Ling H, Fukasawa K, Robertson K, Olashaw N, et al. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol Cell Biol. 2011;31(23):4720–34. doi: 10.1128/MCB.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esteve PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, et al. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol. 2011;18(1):42–8. doi: 10.1038/nsmb.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25(3):338–42. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 29.Tao S, Zheng Y, Lau A, Jaramillo MC, Chau BT, Lantz RC, et al. Tanshinone I activates the Nrf2-dependent antioxidant response and protects against As(III)-induced lung inflammation in vitro and in vivo. Antioxid Redox Signal. 2013;19(14):1647–61. doi: 10.1089/ars.2012.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]


