Abstract
Screening of expression libraries for bioactive clones that modulate the growth of mammalian cells has been limited largely to positive selections incapable of revealing growth suppressive or lethal genetic elements. We have developed a technique, selection–subtraction approach (SSA), that allows growth-modulating clones to be isolated based on alterations in their relative abundance in growing cell populations that have been transduced with an expression library. SSA utilizes tagged retroviral libraries in bacteriophage λ vectors (retrophages). Nylon prints from retrophage libraries are used to determine the relative abundance of tags in library-transduced cells to identify biological activity of individual clones. Applications of SSA for gene discovery, target discovery, and generation of mutant proteins have been demonstrated, by using p53 and ataxia telangiectasia mutated (ATM) as models to isolate growth inhibitory proteins, peptides and antisense RNAs, and temperature-sensitive mutant proteins.
Functional gene discovery approaches have long been used as powerful tools to identify genes involved in a diverse number of cellular processes, including malignant transformation or apoptosis (1, 2). However, the use of these tools has been limited to positive selection, allowing for the identification of biologically active clones from expression libraries that confer selective advantages to cells under the conditions of selection. In other words, this situation allows for the detection only of genes conferring selectable phenotypes, i.e., oncogenes, drug resistance genes, and anti-apoptotic genes (2, 3). The isolation of negative regulators (i.e., tumor suppressor, proapoptotic, or drug-sensitizing genes) through functional selection has proved far more challenging because the selectable phenotype results in their elimination from the transduced population. More sophisticated methods have been developed to resolve this problem, including antisense libraries (4, 5), genetic suppressor elements (GSEs) (6–8), and techniques based on selective killing of proliferating cells in combination with inducible expression of growth-arresting elements: selectable expression of transient growth-arrest phenotype (SETGAP) (9). Recently, GSE and SETGAP techniques have been successfully combined to enhance their ability in the identification of targets for cancer treatment (10). However, all of these techniques are based on positive selections and are therefore indirect by definition. The identification of tumor suppressors, proapoptotic genes, and other negative growth regulators, as well as the search for new molecular targets for selective killing of tumor cells, would be greatly facilitated if it were possible to allow for effective identification of clones that suppress growth or survival within transduced cells. Here, we describe a technique that provides a solution for this problem.
Methods
Cell Lines. The human tumor lines H1299 (lung adenocarcinoma), U2-OS, and Saos-2 (osteosarcomas) were obtained from the American Type Culture Collection (ATCC). AmphoPack-293, an amphotropic viral packaging cell line, was from BD Clontech. Cell lines were maintained in DMEM and 10% FBS. Polybrene (8 μg/ml, Sigma) was supplemented during viral infection.
λ Phage and Plasmids. λgt11 was purchased from Promega. pLXSN was kindly provided by A. D. Miller (Imperial College, London). pBabe E6-18 Hygro and pUSIPH were obtained from A. Ivanov and P. M. Chumakov (Cleveland Clinic Foundation). pUSIPH is a variant of pSIR (BD Clontech) where the encephalomyocarditis internal ribosomal entry site driving expression of the tetracycline-controlled transactivator (tTA) was inserted into the multicloning site. In the absence of doxycycline, pUSIPH drives expression of the cloned insert. pConA-luc contains the consensus p53 responsive element and the minimal hsp70 promoter driving luciferase expression.
Construction and Propagation of Retrophage Vector. Two unique SfiI sites (SfiI A, 5′-GGCCATTAAGGCC-3′; SfiI B, 5′-GGCCGCCTCGGCC-3′) were introduced into the multicloning site of pLXSN, resulting in pLXSN-SfiI. Next, a 34-bp LoxP sequence (5′-ATA ACT TCGTATAGCATACAT TATACGAAGTTATAGATCCAATATTAT-3′) was cloned within the SacII site of pLXSN-SfiI. The same LoxP sequence was entered into a modified version of λgt11. In vitro recombination between LoxP pLXSN-SfiI and LoxP λgt11 was done in the presence of Cre recombinase according to the manufacturer's instructions. The reaction was packaged by using Gigapack III XL (Stratagene) and was amplified in the bacterial strain ER1647. A single recombinant plaque containing LXSN (λ-RP-2) was isolated and amplified for subsequent library construction. Conversion of λ phage to circular plasmid was performed by incubating infectious phage with the Cre-expressing bacterial strain BM25.8 and selecting for ampicillin-resistant colonies.
Tag Design. Random-stops [5′-ATGGATCCAGTTCCTCGGTCGATATC-(N)80-GATATCCAGTCACGTCTCATTCATTCA-3′; stop codons are underlined] and Raul-224 [5′-TGAGACGCA ACTATGGTGACGA AGGCCGAGGCGGCCCTAATACGACTCACTATAGGGGAATTCGT(N)80GTAAGCTTGTTGTACTGAGTTCGCTGCACCTGCCCGGGCGGCCGCTCGA(T)30-3′] oligonucleotides were synthesized, PAGE purified, and fractionated through a cesium sulfate gradient to control for GC content.
Construction of Tagged Prostate cDNA Library. The human prostate, full-length cDNA library cloned into pLib was obtained from BD Clontech. PCR was performed by using 5′pLib (5′-AGCCCTCACTCCTTCTCTAG-3′) and Raul-224 for 5 cycles. PCR products between 1 and 5 kb were purified, digested with SfiI, and cloned into the retrophage vector. The resulting ligation was packaged and divided into pools of 50,000 plaque-forming units.
Construction of Tagged ATM GSE Library. The human ATM cDNA was digested by DNaseI, and 100- to 1,000-bp fragments were blunt-end ligated to a mixture of two DNA adaptors obtained by annealing two complementary synthetic oligonucleotides. One adaptor, which contains three stop codons, as well as the 80-bp tag sequence, was created by annealing TAGserv (5′-TGAATGAATGAGACGTGACTGGATATC-3′; stop codons are underlined) and Random-stops. The second adaptor, which contains a Kozak and ATG sequence, as well as the SfiI A restriction site, was created by annealing AdapLibS (5′-TATTGGCCATTAAGGCCACCATG-3′) and AdapLibA (5′-CATGGTGGCCTTAATGGCC-3′). The resulting ligation products were PCR amplified by using AdapLibS and TAGSfiA primers (5′-GAGAAGGCCGAGGCGGCCAGTTCCTCGGTCGATATC-3′), thereby creating the SfiI B restriction site at the tag terminus. After SfiI digestion, the tagged GSE library was cloned into λ-RP-2, packaged, and divided into pools of 50,000 plaque-forming units.
Construction of Tagged p53 Mutant Library. The human p53 cDNA was blunt-end ligated to an adaptor obtained by annealing TAGserv and Random-stops. The ligation product was PCR amplified under standard conditions with the exception of 160 μM MnSO4, 0.2 mM dATP, 0.2 mM dGTP, 1.0 mM dCTP, and 1.0 mM TTP (11, 12). PCR was performed by using the primers AdapLibSp53 (5′-TAT TGGCCAT TA AGGCCACCATGGAGGAGCCGCAGTCA-3′) and TAGSfiA. After SfiI digestion, the tagged mutant library was ligated to λ-RP-2 arms, packaged, and divided into pools of 50,000 plaque-forming units.
Production of Amphotropic Virus. λ DNA from individual library pools was isolated by using a modified ZnCl2 technique in combination with Qiagen (Valencia, CA) DNA Isolation Kit. AmphoPack-293 cells were seeded at 6 × 106 per 10-cm plate the day before transfection. Thirteen micrograms of λ DNA and 2 μg of vesicular stomatitis virus glycoprotein (VSV-G) were transfected in the presence of Lipofectamine 2000 (Invitrogen) according to the manufacturer's directions. Forty-eight hours posttransfection, viral supernatants were collected, filtered, and supplemented with 8 μg/ml polybrene before adding to the target cell line.
Rescue and Labeling of Tag Sequences. Total RNA was isolated by using TRIzol (Sigma) according to the manufacturer's directions. Genomic DNA contamination was removed after digestion with DNaseI (Amersham Pharmacia). The vector-specific primer 3′LXSfi(2s) (5′-TCT TGATCAGATCCGCCTCGACCT-3′) was annealed to the RNA template, and reverse transcription was performed by using SuperScript II (Invitrogen). For cDNA libraries containing Raul-224, PCR was performed by using Raul-224P1 (5′-CTAATACGACTCACTATAGGG-3′) and Raul-224P2 (5′-CACAGTACA ACAAGCTTAC-3′). For GSE and mutant libraries containing Random-stops, PCR using TAGsnerv2 (5′-TGATGAGTGACGTGACTGCATATC-3′) and TAGSfinerv (5′-GAGAAGGCCGAGGCGGCCAGTTCCTC-3′) was done. PCR was carried out for 22–25 cycles and subsequently gel purified. Radiolabeling was performed by completing one round of PCR with the addition of 50 μCi (1 Ci = 37 GBq) of [α-32P]dCTP instead of cold dCTP. The probe was purified by using Probe-Quant G-50 (Amersham Pharmacia) and added to the hybridization solution.
Replica Blot Preparation and Hybridization to Tag Sequences. Ten thousand plaque-forming units from each retrophage library pool were plated onto a 15-cm Petri dish and incubated at 37°C until plaques were 1 mm in diameter. The top agar was allowed to solidify at 4°C for at least 2 hr before using for plaque lifts. Plaque lifts were performed under standard conditions (13) with the exception of prewetting the nylon membranes in 5× SSC to increase phage diffusion. After DNA immobilization by means of both UV crosslinking and baking at 80°C, the membranes were prehybridized in 5× SSC, 5× Denhardt's solution, and 1% SDS at 68°C. Prehybridization was performed for 6 hr before replacing with fresh solution, including the denatured probe and sheared salmon sperm DNA (Eppendorf). After 48 hr, the hybridization was washed under stringent conditions at 68°C and exposed to x-ray film (Fuji Super RX).
Results
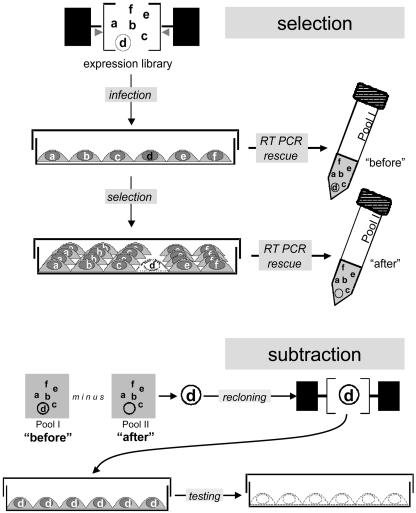
We have created a technique that is equally selective for both growth-promoting and growth-inhibitory clones, by developing the idea presented in Fig. 1, based on monitoring the relative representation of individual clones from an expression retroviral library after its transduction into target cells, followed by selection. The proportion of biologically inactive clones does not change during propagation in cell culture whereas those that affect cell growth in either direction change their relative abundance. Clones are either enriched or diluted in a library-transduced cell population because the encoded proteins promote or inhibit growth, respectively. The library-transduced cell population can also be exposed to selective conditions, such that clones conferring sensitization or resistance to the selective pressure can be isolated. The desired clones can be identified by molecular subtraction (14–16) of library-derived inserts rescued from transduced cells before and after selection. Therefore, the proposed technique is named selection–subtraction approach (SSA).
Fig. 1.
General design of SSA.
Initial experiments indicated three main obstacles. First, PCR rescue is incompatible with maintaining library complexity, resulting in underrepresentation or loss of long or GC-rich inserts. Second, expression libraries commonly contain a mixture of functional and nonfunctional (i.e., truncated or mutated) clones representing the same gene; nonfunctional variants would not be lost during selection and thus would impair the subtraction step. Lastly, to date there is no reliable genetic subtraction procedure free from losses of potentially important differentially expressed sequences. These problems were resolved by the introduction of two technical solutions illustrated in Fig. 1.
The first two problems were overcome by constructing a library where every clone carries a unique untranslated barcode or tag, an idea successfully applied to yeast but never to mammalian cells (17). Tags, consisting of an 80-bp random sequence flanked by universal PCR primers, are introduced during library construction downstream of the stop codon of the insert. All tags are of the same length and have similar (40–60%) GC content. We chose the sequences flanking the tags carefully for effective and specific PCR rescue to circumvent loss of library complexity. These sequences are also long enough to function as highly specific hybridization probes. Thus, instead of inserts, the tags are rescued from the library-transduced cells by RT-PCR. Only those tags marking functional inserts will change in relative abundance whereas those attached to nonfunctional clones will remain unchanged. Therefore, by tagging the expression library, the first two obstacles were resolved.
The problem of effective isolation of differentially represented clones was solved by substituting molecular subtraction by direct quantitative comparison of representational changes for each clone during selection. The simultaneous monitoring of relative abundance of all library sequences can be done by using array hybridization. If each tag within the library is printed on a microarray, one can then compare the relative intensity of hybridization signals for each before and after the selection, as was done for antisense RNA libraries (18). A reduction or increase in the proportion of any given tag relative to the tags in the rest of the library would indicate the relative loss or gain of cells carrying the tagged inserts, thus identifying a clone detrimental or beneficial to cell growth or survival under the conditions applied.
However, it is not realistic to print an array of tags for each individual library. Besides the high cost of such a procedure, a tag microarray would have to be very complex to cover the diversity of clones represented in a well created expression library (up to millions), making their simultaneous detection impossible within achievable specific activities of the probes. To resolve this problem, we used λ phage to create a simpler method of differential hybridization. The use of λ allows one to create several practically identical nylon replicas from a single high-density array of phage plaques growing on a bacterial lawn. The arrays can be hybridized by using standard techniques and are technically equivalent to microarrays. Comparison of hybridization signals of plaques allows each clone to be monitored separately, thereby resulting in no loss of information during the selection process and making it possible to pick clones that are only partially reduced or that increase their abundance during selection. By placing retroviral sequences within the phage λ genome, we created a set of vectors, termed retrophages, differing in their cloning capacities (Fig. 2).
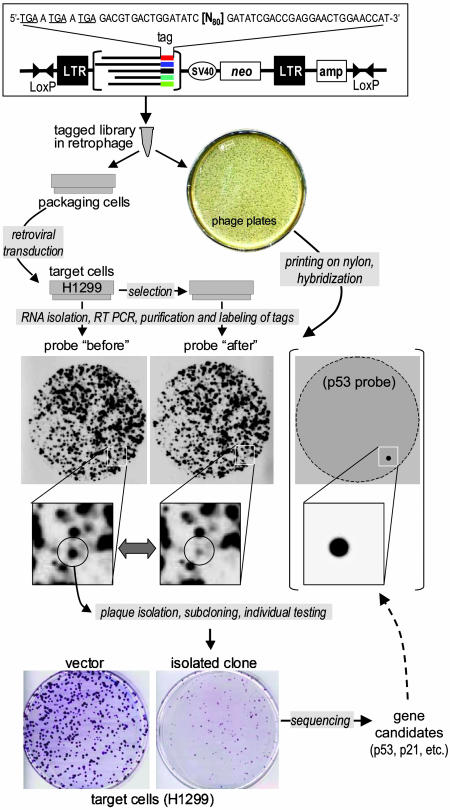
Fig. 2.
Screening of a tagged full-length cDNA library. A tagged prostate cDNA library was created and cloned into the retrophage vector. The retrophage vector consists of retroviral as well as plasmid sequences within a λ phage backbone. Flanking LoxP sites allow for simple plasmid excision in Cre-expressing bacteria. Replication-incompetent virus was produced by transfecting retrophage DNA into AmphoPack-293 and entered into H1299. Total RNA was isolated 48 hr after infection and again after passaging for 7 days. Tags were rescued by RT-PCR, radiolabeled by 32P, and hybridized to replica nylon blots. Clones corresponding to those tags that disappeared on day 7 were isolated, recloned in a retroviral vector with no tags, and individually tested in colony assay. Inserts with confirmed biological activity were sequenced. One of them contained full-length p53 cDNA; this insert was radio-labeled and hybridized with a parallel nylon replica.
A generic scheme of SSA-based library selection is shown in Fig. 2. The intervening retroviral elements within the phage sequence allow the efficient production of replication-incompetent virus after transfection of retrophage DNA into packaging cells, followed by infection of a target cell line. Simultaneously, the same population of phage is represented on multiple identical nylon prints of phage plaques. Before and after selection, RNA is isolated, and the tags are subsequently rescued by RT-PCR. Next, the tags are radiolabeled and hybridized to their corresponding nylon replicas. Clones lost or enriched during the selection process are picked, reconfirmed by hybridization, and validated functionally in biological assays. We were able to successfully screen libraries with complexities of up to 50,000 clones, limited by the efficiency of labeling. Therefore, screening a complex library on the order of 106 clones requires subdividing the library into 20 batches of 50,000 clones each.
Here, we demonstrate three applications of SSA for negative selection: (i) identification of growth-suppressive genes by screening a full-length cDNA library, (ii) isolation of dominant negative mutant proteins from a randomly fragmented gene library, and (iii) isolation of dominant negative and temperature-sensitive mutant proteins from a library of a randomly mutated cDNA. A full-length tagged cDNA library from normal human prostate mRNA was transduced into the human lung adenocarcinoma line H1299 by viral infection under conditions that allowed each clone to be delivered to at least 20 cells. RNA was isolated from a portion of the infected cells 48 hr after transduction, before any change in the representation of biologically active clones had time to occur. After successive growth of the remaining cells for 7 additional days, when the proportion of cytotoxic or cytostatic clones had had time to decrease due to their suppressive effect on growth or survival, RNA was isolated again. The tags were rescued by RT-PCR, radiolabeled, and hybridized to two identical nylon prints of the retrophage library. After comparison of hybridization images, a number of tags were found to be lost during propagation of the library-transduced cell population. The corresponding clones were isolated from the original Petri dishes used for generation of nylon prints, and their activities were confirmed individually by using a colony growth inhibition assay in the same target cells. Among 20 sequenced inserts from confirmed clones, two contained p53 and p21 cDNAs encoding proteins known to be growth-suppressive for H1299 cells, thereby confirming the ability of SSA to identify negative growth regulators effectively. Hybridization of a parallel nylon print with the p53 cDNA probe revealed a single hybridizing plaque that coincided with one of the picked clones (Fig. 2). This result illustrates the discovery power of SSA and its ability to identify natural negative growth inhibitors by functional selection. Other clones isolated along with p53 and p21 have been analyzed (unpublished results).
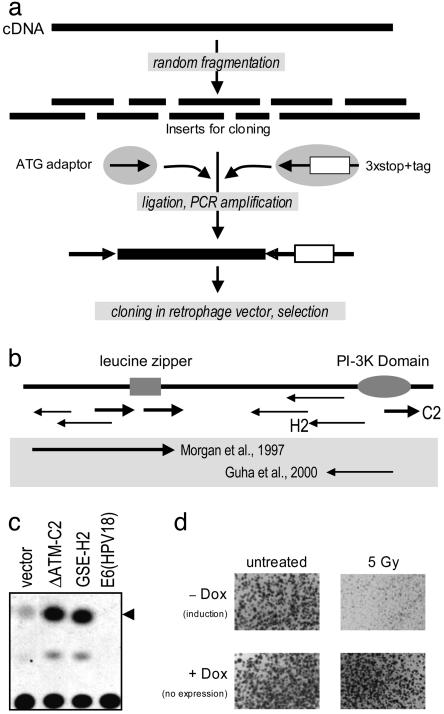
Dominant negative peptides and mutants have been important tools in studying protein structure and function. Random mutagenesis or fragmentation, followed by functional selection, can be used to isolate dominant negative mutants of proteins with poorly defined domain structures. This approach, however, has been limited to genes whose repression results in a selectable phenotype. SSA now extends our ability to isolate dominant negative mutants for genes encoding vitally important proteins. To illustrate this principle, we chose to study the ATM gene (19), a key mediator of the DNA damage response, whose loss has been shown to result in increased radiosensitivity of cells. ATM cDNA was randomly fragmented, tagged, and cloned into the retrophage vector (Fig. 3a). A protocol similar to the one described above using a full-length cDNA library was followed, with the slight modification that the RNA was isolated after target human osteosarcoma U2-OS cells had been subjected to a sublethal dose of radiation. As was done previously, the tags were rescued by RT-PCR, radiolabeled, and hybridized to two nylon replicas of the ATM retrophage library. Clones lost in irradiated vs. nonirradiated cells were isolated and individually confirmed by recloning into a tetracycline-regulated vector, followed by transduction into U2-OS cells, which were then tested for radiosensitivity (Fig. 3c). The positions of isolated clones in ATM cDNA and their functional validation are shown in Fig. 3b. Sense-oriented clones encode polypeptides spanning the functional domains of the protein. Some represent shorter versions of previously isolated dominant negative ATM mutant proteins (20, 21) with similar ability to stimulate p53-mediated transactivation, as determined by a CAT assay using p53-responsive reporter (Fig. 3c). When individually expressed in a tetracycline-regulated vector, they showed an inducible ability to sensitize U2-OS cells to γ radiation (Fig. 3d). These results provide a model for future applications of SSA to identify new gene targets for cancer treatment. SSA can be used to isolate genes whose suppression results in selective killing of cells or in their sensitization to chemical or radiation-induced damage.
Fig. 3.
Isolation of genetic suppressor elements against ATM. (a) The ATM cDNA was randomly fragmented by using DNaseI, and the subsequent fragments were ligated to two adaptors. One adaptor contains the start codon whereas the other contains stop codons in all three reading frames and the tag, an 80-bp random sequence. The tagged GSEs were PCR amplified by using primers specific for each adaptor and cloned into the retrophage vector. Selection was performed in U2-OS cells. (b) Several GSEs were isolated, including both antisense and dominant negative peptides in comparison with the full-length protein and previously published inhibitors (20, 21) (shown in the gray zone). The coordinates of isolated GSEs and the nature of their products are listed in Table 1. (c) Individual GSEs were transiently transfected with a p53-responsive CAT reporter to measure p53 transcriptional activity. Higher activity was observed in the presence of both antisense and sense GSEs in accordance with previously published results (20–23). (d) The radiosensitizing activity of ΔATMC2 was further confirmed by recloning into a tetracycline-regulated retroviral vector and infected in U2-OS. In the absence of doxycycline, the GSE is expressed and had no effect on cell growth under normal conditions of cultivation [compare colony sizes in +Dox (induction) and -Dox (no expression) plates]. However, activation of the GSE expression significantly sensitizes U2-OS to 5 Gy of γ radiation.
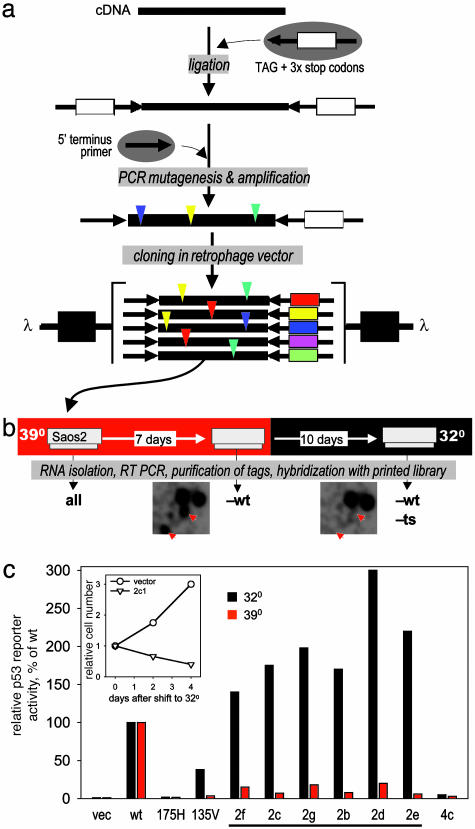
Temperature-sensitive (ts) mutants have been valuable genetic tools, especially in prokaryotes. Their use in mammalian genetics has been limited by lack of techniques that allow their systematic isolation. Here, we show that SSA can be used to isolate ts mutants by using the p53 gene (24) as a model. Previously, a ts mutant of p53 was isolated by serendipity (25, 26). To create a library of random mutations, p53 cDNA was ligated to 80-bp tags and then subjected to PCR-based mutagenesis (11, 12), resulting in the introduction of approximately two nucleotide substitutions per molecule. The tagged mutants were then cloned into the retrophage vector (Fig. 4a), and the resulting library was transduced into target p53-deficient Saos-2 cells, which are highly sensitive to wild-type p53. The first RNA isolation was done 48 hr after transduction of the library and then 7 days after cell growth at 39°C. By then, the cell population was presumably free of library clones with wild-type p53 activity, lethal to Saos-2 cells. The cells were then shifted to 32°C and propagated for an additional 10 days before the third RNA isolation was done (a longer time was used to reflect slower growth at the lower temperature). Tags were then isolated and compared (Fig. 4b) to identify clones that were maintained at high but lost at low temperature. The mutants isolated were analyzed individually in transactivation and colony growth assays (Fig. 4c) at 39°C and 32°C in comparison with the well known ts mutant, mouse p53Val-135, wild-type p53, and the tumor-derived p53 mutant p53175His, which lacks growth inhibitory and transactivation functions. As shown in Fig. 4c, the p53 mutants isolated by SSA (see Table 1 for their specific mutations) dramatically lose their function at the higher temperature, thus confirming that they are truly ts mutants. Therefore, SSA has allowed the direct isolation of multiple temperature-sensitive mutants from a complex library of random mutants of a single gene.
Fig. 4.
Isolation of temperature-sensitive p53 mutants. (a) Construction of library of random p53 mutants. The human p53 cDNA was blunt ended and ligated to the tag adaptor. PCR mutagenesis was performed in the presence of manganese and unequal dNTP pools (11, 12). A 5′ p53-specific primer and a primer specific for the 3′ terminus of the tag adaptor were used to control for cDNA orientation. The resulting tagged mutants were cloned into the retrophage vector, which was used to transduce Saos-2 at 39°C. (b) Selection scheme. Saos-2 were infected with the library at 39°C and passaged for 7 days before being shifted to 32°C for an additional 10 days. The tags were isolated by means of RT-PCR and hybridized. Similar fragments of autoradiograms resulting from hybridization with 32°C and 39°C probes are shown; positions of plaques corresponding to putative temperature-sensitive mutants are shown by red arrows. (c) Confirmation of biological activity for individual clones was assessed by transactivation of a p53 luciferase reporter and toxicity at both 32°C and 39°C as compared with wild type, p53R175H, and mouse p53A135V. Bars represent relative activity of p53-responsive luciferase reporter at two different temperatures calculated as a percentage of that of wild-type p53 cloned in the same vector and determined after transient transfection of Saos-2 cells. Underlined clones displayed predicted ts mutants. Clone 4c was picked as an example of a putative nonfunctional mutant (tag was not lost at any temperature). (Inset) Results of cell growth estimation of Saos-2 cells transduced with one of the ts mutants (2c) after temperature shift from 39° to 32°C. Mutations relative to wild-type human p53 are indicated in Table 1.
Table 1. Biologically active genetic elements isolated by SSA.
| Gene of origin | Name | Predicted product | Coordinates | Activity |
|---|---|---|---|---|
| ATM | GSE-A1 | Antisense RNAs | bp 155—403 | Radiosensitization |
| GSE-E1 | Antisense RNAs | bp 8231—8423 | Radiosensitization | |
| GSEF2 | Antisense RNAs | bp 8402—8772 | Radiosensitization | |
| GSEH2 | Antisense RNAs | bp 8550—8912 | Radiosensitization | |
| ΔATMC2 | Truncated proteins | aa 2945—3056 | Radiosensitization | |
| ΔATM-A1 | Truncated proteins | aa 894—1012 | Radiosensitization | |
| ΔATM-D3 | Truncated proteins | aa 1218—1302 | Radiosensitization | |
| p53 | 2f | Mutant proteins | E 221 D | ts GR&T |
| 2c | Mutant proteins | I 232 F | ts GR&T | |
| 2g | Mutant proteins | D 208 A | ts GR&T | |
| 2b | Mutant proteins | M 237 V | ts GR&T | |
| 2d | Mutant proteins | D 57 G V 272 L | ts GR&T | |
| 2e | Mutant proteins | T 256 A | ts GR&T | |
| 4c | Mutant proteins | I 254 F E 343 G | LOF mutant |
Radiosensitization, radiosensitization of U2 OS cells; ts GR&T, ts growth repression and transactivation; LOF, loss of function mutant.
In summary, we have demonstrated three applications illustrating the broad utility of SSA as a screening method. The functional isolation of p53 reveals gene discovery capability of SSA, making it possible to clone functionally new growth-inhibitory genes. Isolation of clones encoding dominant negative radiosensitizing mutant proteins from the library of gene fragments (combining SSA with the GSE technique) provides a straightforward approach to identify and validate molecular targets for selective killing of specific cells, a new possibility with obvious applications to the development of new cancer therapies. Even more powerful should be the combination of SSA with small interfering RNA (siRNA) libraries, a path that remains to be tested. Finally, direct and technically simple selection of growth-suppressing ts mutants from a library derived from a single gene opens new opportunities to generate effective tools for genes whose suppression is lethal, an impossible task with previous methods.
Acknowledgments
We thank Peter Chumakov and Alexey Ivanov for sharing their tetracycline-regulated retroviral vectors, Andrei Komarov for technical help, and George Stark for helpful discussions. This work was supported by Grants CA60730 and CA098374 from the National Institutes of Health and by a grant from Quark Biotech, Inc. (to A.V.G.).
Abbreviations: GSE, genetic suppressor element; SSA, selection–subtraction approach; ATM, ataxia telangiectasia mutated; ts, temperature-sensitive.
References
- 1.Stark, G. R. & Gudkov, A. V. (1999) Hum. Mol. Genet. 8, 1925-1938. [DOI] [PubMed] [Google Scholar]
- 2.Hannon, G. J., Sun, P., Carnero, A., Xie, L. Y., Maestro, R., Conklin, D. S. & Beach, D. (1999) Science 283, 1129-1130. [DOI] [PubMed] [Google Scholar]
- 3.Maestro, R., Dei Tos, A. P., Hamamori, Y., Krasnokutsky, S., Sartorelli, V., Kedes, L., Doglioni, C., Beach, D. H. & Hannon, G. J. (1999) Genes Dev. 13, 2207-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deiss, L. P. & Kimchi, A. (1991) Science 252, 117-120. [DOI] [PubMed] [Google Scholar]
- 5.Deiss, L. P., Feinstein, E., Berissi, H., Cohen, O. & Kimchi, A. (1995) Genes Dev. 9, 15-30. [DOI] [PubMed] [Google Scholar]
- 6.Ossovskaya, V. S., Mazo, I. A., Chernov, M. V., Chernova, O. B., Strezoska, Z., Kondratov, R., Stark, G. R., Chumakov, P. M. & Gudkov, A. V. (1996) Proc. Natl. Acad. Sci. USA 93, 10309-10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudkov, A. V., Kazarov, A. R., Thimmapaya, R., Axenovich, S. A., Mazo, I. A. & Roninson, I. B. (1994) Proc. Natl. Acad. Sci. USA 91, 3744-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudkov, A. V., Zelnick, C. R., Kazarov, A. R., Thimmapaya, R., Suttle, D. P., Beck, W. T. & Roninson, I. B. (1993) Proc. Natl. Acad. Sci. USA 90, 3231-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pestov, D. G. & Lau, L. F. (1994) Proc. Natl. Acad. Sci. USA 91, 12549-12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Primiano, T., Baig, M., Maliyekkel, A., Chang, B. D., Fellars, S., Sadhu, J., Axenovich, S. A., Holzmayer, T. A. & Roninson, I. B. (2003) Cancer Cell 4, 41-53. [DOI] [PubMed] [Google Scholar]
- 11.Cadwell, R. C. & Joyce, G. F. (1992) PCR Methods Appl. 2, 28-33. [DOI] [PubMed] [Google Scholar]
- 12.Cadwell, R. C. & Joyce, G. F. (1994) PCR Methods Appl. 3, S136-S140. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 3rd Ed., pp. 2.90-2.100.
- 14.Duguid, J. R. & Dinauer, M. C. (1990) Nucleic Acids Res. 18, 2789-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara, E., Kato, T., Nakada, S., Sekiya, L. & Oda, K. (1991) Nucleic Acids Res. 25, 7097-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedrick, S. M., Cohen, D. I., Nielsen, E. L. & Davis, M. M. (1984) Nature 308, 149-153. [DOI] [PubMed] [Google Scholar]
- 17.Feldhaus, M. J., Lualhati, M., Cardon, K., Roth, B. & Kamb, A. (2000) Nucleic Acids Res. 28, 534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kotlo, K. U., Yehiely, F., Efimova, E., Harasty, H., Hesabi, B., Shchors, K., Einat, P., Rozen, A., Berent, E. & Deiss, L. P. (2003) Oncogene 22, 797-806. [DOI] [PubMed] [Google Scholar]
- 19.Shiloh, Y. (2003) Nat. Rev. Cancer 3, 155-168. [DOI] [PubMed] [Google Scholar]
- 20.Morgan, S. E., Lovly, C., Pandita, T. K., Shiloh, Y. & Kastan, M. B. (1997) Mol. Cell. Biol. 17, 2020-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guha, C., Guha, U., Tribius, S., Alfieri, A., Casper, D., Chakravarty, P., Mellado, W., Pandita, T. K. & Vikram, B. (2000) Gene Ther. 7, 852-858. [DOI] [PubMed] [Google Scholar]
- 22.Xu, Y. & Baltimore, D. (1996) Genes Dev. 10, 2401-2410. [DOI] [PubMed] [Google Scholar]
- 23.Xu, Y., Yang, E. M., Brugarolas, J., Jacks, T. & Baltimore, D. (1998) Mol. Cell. Biol. 18, 4385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307-310. [DOI] [PubMed] [Google Scholar]
- 25.Michalovitz, D., Halevy, O. & Oren, M. (1990) Cell 24, 671-680. [DOI] [PubMed] [Google Scholar]
- 26.Martinez, J., Georgoff, I., Martinez, J. & Levine, A. J. (1991) Genes Dev. 5, 151-159. [DOI] [PubMed] [Google Scholar]