Abstract
The genus Neisseria contains two pathogenic species of notable public health concern: Neisseria gonorrhoeae and Neisseria meningitidis. These pathogens display a notable ability to undergo frequent programmed recombination events. The recombination mediated pathways of transformation and pilin antigenic variation in the Neisseria are well studied systems that are critical for pathogenesis. Here we will detail the conserved and unique aspects of transformation and antigenic variation in the Neisseria. Transformation will be followed from initial DNA binding through recombination into the genome with consideration to the factors necessary at each step. Additional focus is paid to the unique type IV secretion system that mediates donation of transforming DNA in the pathogenic Neisseria. The pilin antigenic variation system uses programed recombinations to alter a major surface determinant which allows immune avoidance and promotes infection. We discuss the trans- and cis- acting factors which facilitate pilin antigenic variation and present the current understanding of the mechanisms involved in the process.
Introduction
The majority of species in the genus Neisseria are commensal bacteria that colonize mucosal surfaces. The two pathogenic species, Neisseria gonorrhoeae (the gonococcus) and Neisseria meningitidis (the meningococcus), are the causative agent of gonorrhea and the primary cause of bacterial meningitidis in young adults, respectively. Both organisms are strict human pathogens with no known environmental reservoirs that have evolved from commensal organisms within the human population (1). The study of the Neisseria is important for public health reasons, but also provides a defined system to study evolution of two highly related organisms that cause distinct diseases. One unique aspect of the pathogenic Neisseria is the presence of sophisticated genetic systems that contribute to pathogenesis. The processes of DNA transformation and pilin antigenic variation will be discussed in this Chapter.
Natural DNA Transformation
There is a diverse set of more than 80 identified naturally transformable bacterial species that are able to recognize free DNA in the environment, import it across the envelope and recombine exogenous DNA with resident DNA molecules (2). Unlike the majority of naturally competent species, Neisseria sp. are constitutively competent, capable of transformation at all phases of growth (3, 4). Natural transformation is the primary means of horizontal gene transfer (HGT) in Neisseria with a documented flow of information amongst both commensal and pathogenic members of the genus (5, 6). Similar to the vast majority of Gram-negative bacteria, transformation in Neisseria is dependent on a type IV pilus (Tfp) complex (7). Pathogenic Neisseria often undergo HGT with recombination occurring so frequently that there is a marked inability to establish stable clonal lineages (8). These genomic signatures suggest that mixed infections are common and this idea has been supported by some studies (9, 10). Although N. meningitidis maintains limited lineage structure, both species tend toward linkage equilibrium, and this frequent genetic exchange is thought to contribute to the rapid spread of antibiotic resistance among N. gonorrhoeae strains (11–14). This HGT has led to clinical isolation of N. gonorrhoeae strains with resistance to multiple antibiotics (15). Though no single strain has accumulated all of the resistance markers, there are resistant lineages to all currently recommended therapies (15). The threat of untreatable gonorrhea has earned N. gonorrhoeae a spot on the Center for Disease Control’s list of superbugs and its highest threat level reserved for only three organisms (16–18).
The Type IV Pilus
Tfp are a critical virulence factor for many pathogens and also promote interactions of nonpathogens with their environments. Tfp are long, thin fibers that undergo dynamic cycles of extension and retraction and mediate twitching motility, cellular adherence, microcolony formation, and natural transformation in both N. gonorrhoeae and N. meningitidis (19–23). The expression of Neisseria Tfp correlates directly with transformation efficiency, an observation that has been expanded to many Gram negative species (3). In the presence of excess DNA, highly piliated strains of N. gonorrhoeae can achieve transformation efficiencies more than a million times higher than strains lacking the major Tfp pilin PilE (24). There is a complex Tfp assembly apparatus present in the bacterial envelope that is responsible for pilus expression and associated functions (Figure 1). Many of the Tfp complex proteins are required for transformation. Among the Tfp complex proteins that have a defined function, PilD is a periplasmic protease responsible for processing PilE into the mature form that can be assembled into the pilus fiber (23). PilD is required for pilus expression and transformation competence. Mutations in Tfp structural proteins including the inner membrane protein PilG and the pore forming secretin PilQ abrogate transformation (25, 26). Additionally, transformation is dependent on the two cytoplasmic NTPases PilF and PilT (23, 27). PilF is thought to power pilus extension while PilT is required for pilus retraction and twitching motility. The requirement for many of the Tfp complex proteins in transformation has led to the generally accepted hypothesis that the pilus fiber mediates the binding and initial uptake of DNA into the periplasm as well as transport of DNA through the outer membrane during transformation (28–30). This has never been conclusively shown, and it is an open question whether Tfp or a pilus-like apparatus (pseudopilus) is actually responsible for DNA uptake across the outer membrane (31).
Figure 1. Type IV Pilus and DNA Uptake.
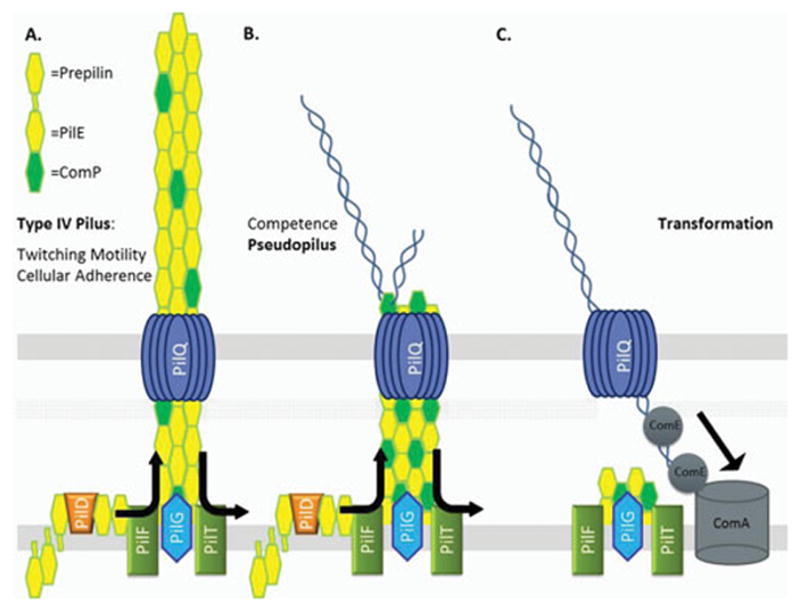
A. Type IV Pilus – The Tfp is a several micron long, 60 angstrom wide fiber anchored in the inner membrane by PilG that extends through the PilQ secretin pore. Composed mainly of the major pilin PilE (pilin), which is processed by a dedicated protease, PilD. The PilF and PilT NTPases mediate extension and retraction of the pilus through polymerization and depolymerization of the pilin subunits. B. Competence Pseudopilus – Hypothesized pseudopilus that could mediate transformation. Utilizes the type IV pilus complex including the PilQ pore but is not an extended fiber. Possible localization of ComP to the pseudopilus could mediate specific DNA binding. C. DNA Uptake Model - Retraction of the (pseudo)pilus mediated by PilT brings the initial length of DNA into the periplasm. DNA is then bound by a protein or protein complex possibly containing ComE which mediates import of the remaining length of DNA into the periplasm. The inner membrane protein ComA facilitates DNA entry into the cytoplasm.
Contributing to this uncertainty are the observations that small amounts and altered forms of pilin are sufficient for transformation. Pilin (pilE gene product) can exist in variant forms such as S-pilin (a short or secreted form) and L-pilin (long form). S-pilin results from a cleavage event and produces a soluble form that is secreted from the cell by an unknown mechanism (32). Some S-pilin variants display intermediate piliation phenotypes but wild-type levels of transformation (33, 34). L-pilin variants result from a duplication of coding sequences in pilE that produces an oversized pilin monomer that cannot be assembled into pilus fibers, yet only reduces transformation efficiencies 35-fold (32, 33). Alongside the pilin variants investigations, a 2003 study showed that when the level of pilin expression is reduced to the point where observable pili are extremely rare in a population of cells, the cells still exhibit considerable transformation efficiencies (24). The competence in these pilus-deficient gonococci was still dependent on PilT and PilQ, leading to the hypothesis that extended Tfp are not necessary for transformation; rather a pseudopilus apparatus, utilizing the type IV pilus complex of proteins, is sufficient for transformation (Figure 1).
DNA Uptake
Regardless of whether extended Tfp or pseudopili are responsible for specific binding of extracellular DNA, retraction of the Tfp or pseudopilus is likely not sufficient to account for the difficulties of transporting DNA across the outer membrane. The PilQ pore is only 6nm in width, the same width as the predicted Tfp structure (35). This leaves no room for concomitant transport of other substrates, much less the doubled up DNA structure that would result from binding at a mid-strand site. Taking into consideration that lengths of DNA several times longer than the cell are routinely transformed, a single pilus retraction event would not bring the entire DNA molecule into the periplasm (36). Therefore, DNA transport across the outer membrane requires a mechanism more complicated than pilus elaboration, DNA binding, and a single retraction event. Successive cycles of pilus extension and retraction could be responsible for pulling long DNA molecules across the outer membrane, but evidence from studies of the Gram-positive bacterium Bacillus subtilis suggests that the import of DNA is processive and occurs at a constant velocity (37). This could be explained by the cooperative retraction of several pili in succession but there is no experimental evidence to support this hypothesis (38).
An alternative hypothesis revolves around the gonococcal protein ComE. Although not surface localized, ComE is required for DNA uptake and transformation and binds DNA non-specifically (39). Four copies of ComE are encoded in the gonococcal genome and deletion of individual comE genes results in an additive negative effect on transformation. A recent study in V. cholerae showed that the ComE homolog of V. cholerae, ComEA, is required for DNA uptake into the periplasm and that ComEA binding of DNA can potentially prevent retrograde transport of DNA through the PilQ pore (40). The authors propose ComEA is responsible for pulling DNA into the periplasm through a ratcheting mechanism reliant on non-specific DNA binding and entropic forces similar to what has been proposed to drive double-stranded DNA (dsDNA) dsDNA transport through the nuclear pore complex in eukaryotic cells (41). While it remains to be seen if ComE plays a similar role in Neisseria, these observations form an alternative working model in which DNA is bound extracellularly by the (pseudo-)pilus (Figure 1). Retraction of the (pseudo-)pilus pulls the initial length of DNA into the periplasm. Non-specific binding of a complex possibly containing ComE then mediates import of the remaining length of DNA into the periplasm.
DNA Uptake Sequence
Although constitutively competent and lacking any apparent transformation regulation, Neisseria species preferentially transform self-DNA (42, 43). This is accomplished through repeat sequences spread across their genomes that aid in efficient uptake and transformation of self-DNA. The initially identified DNA uptake sequence (DUS) is a 10 base sequence (DUS10 5′-GCCGTCTGAA), but an extended 12-mer DUS (DUS12 5′-ATGCCGTCTGAA) occurs at greater than 75% of DUS10 locations and slightly increases transformation efficiencies over the 10-mer DUS (42, 44, 45). Remarkably, the 10 base sequence occurs about once every kb of the genome, a frequency a thousand times higher than chance predicts and is often located as an inverted repeat in putative Rho-independent transcriptional terminators (42, 45). It has been suggested that DUS sequences are more often found within DNA repair genes due to a role in genome maintenance, but this analysis does not take into account that DUS can act over several kb distances (46). It is possible that the enrichment of DUS sequences in core genes (genes shared among all isolates) may indicate slow accumulation of the sequences with the oldest (most essential) genes accumulating the highest proportion of DUS (47). The 10 base DUS sequence was identified for its ability to competitively inhibit transformation (42) and to enhance both DNA uptake and transformation when added to a previously untransformable plasmid (43). DUS mediated transformation enhancement is both strain and strand specific in N. gonorrhoeae; DUS containing DNA only enhances transformation 20-fold in strain FA1090 while it increases efficiencies 150-fold in strain MS11 (48). Because the DUS is non-palindromic, the two divergent single-stranded DUS sequences were investigated for their relative effect on transformation and given identifiers Watson (5′-ATGCCGTCTGAA) and Crick (5′-TTCAGACGGCAT). The single-stranded Watson DUS12 increased transformation efficiencies with single-stranded DNA (ssDNA) 180–470 fold while the single-stranded Crick DUS12 only enhanced transformation 7 fold over non-DUS containing ssDNA (49). Notably, even with the Watson DUS12, ssDNA was 2–24 fold less efficient than DUS12 containing dsDNA in transformation assays. These data suggest that there may be different uses for the double-stranded and single-stranded uptake sequences.
While the Tfp and its components were exhaustively investigated for a role in specific binding of the DUS, the mediator of self-DNA recognition was not identified until a landmark 2013 study by the Pelicic lab (39, 50–57). This study identified the type IV minor pilin ComP as the DUS receptor in N. meningitidis (54). The study investigated ComP due to its high level of sequence conservation amongst several Neisseria species (99%) suggesting an important function. They showed that ComP was the only known pilin component that bound dsDNA and had affinity for DUS-containing DNA. Through mutational analysis, it was also shown that an electronegative stripe on ComP that is predicted to be surface exposed on Tfp is responsible for the DNA binding ability of ComP (54). While the DUS sequence is conserved in both N. meningitidis and N. gonorrhoeae, different members of the Neisseria genus and the broader Neisseriaceae family have slightly different DUS sequences termed dialects (58). The efficiency of the DUS dialects for transformation of N. meningitidis was shown to be dependent on the expression of the cognate ComP protein (59). Notably, while the inner bases of the DUS are most critical for allowing high transformation efficiency and DNA binding by the ComP protein, not all residues that were important for full transformation efficiency were important for ComP binding (59). This result suggests that the DUS may act in a different step during transformation independent of ComP and could rely on the single stranded Watson DUS sequence shown to function in transformation (49).
Processing and Recombination
There are three gonococcal genes, comL, tpc, and comA; important for transformation competence that are proposed to act during the transport of DNA into the cytoplasm (60–62). ComL and Tpc are both localized to the periplasm and have been implicated in DNA transport across the peptidoglycan layer possibly through the creation of localized breaks in the cell wall (60, 61). ComA displays homology with ComEC of B. subtilis which is a polytopic membrane protein localized to the inner membrane that delivers ssDNA into the cytoplasm (62, 63). Whether ComA also transports dsDNA into the cell is an unanswered question.
Following import into the cell, DNA is subject to restriction modification (64). N. gonorrhoeae encodes a large array of methyltransferases and their corresponding endonucleases that form an effective restriction barrier to plasmid DNA, making plasmids 1,000 fold less transformable than chromosomal loci (65, 66). N. meningitidis is able to restrict transforming DNA through a unique CRISPR/Cas system that is not present in the sequenced gonococcal isolates (67). Clustered, regularly interspaced, short palindromic repeat (CRISPR) loci confer sequence specific adaptive immunity based on CRISPR-associated (Cas) protein complexes ability to cleave incoming DNA (68). While generally thought to be an adaptation to protect against phage invasion and foreign plasmid conjugation, the type II CRISPR/Cas system of N. meningitidis was the first system shown to naturally prevent transformation (67). The meningococcal CRISPR/Cas pathway is the most streamlined system characterized to date and encodes sequences that may be able to restrict the transfer of certain virulence determinants amongst cells of different lineages. This may explain the increased ability of the meningococcus to form semi-clonal lineages in comparison to the gonococcus which lacks a CRISPR/Cas system.
Following entry into the cytoplasm, transforming DNA is integrated into the chromosome through homologous recombination mediated by the major recombinase, RecA, which can bind ssDNA, find the complement strand in a homologous DNA duplex, and catalyze D-loop formation (69, 70). RecA activity is limited by RecX which facilitates more efficient recombination (71). Investigations into the role of DNA repair pathways in transformation showed the RecF-like pathway that mediates ssDNA gap repair in E. coli is not involved in transformation-mediated homologous recombination, but mutations in the RecBCD pathway show a 10 to 100-fold decrease in transformation efficiency (72, 73). The RecBCD pathway is involved in dsDNA break repair in E. coli and would be predicted to only act on dsDNA transformation substrates (74). It is likely that transformation proceeds mainly through ssDNA intermediates, and the observation that the majority of transformation is independent of the RecBCD pathway confirms the ssDNA dependence. However, the amount of transformation that is RecBCD-dependent suggests that either there is some dsDNA transported into the cytoplasm or that a portion of the transported ssDNA is converted to dsDNA and then used for recombination. A role for DprA nuclease, which is involved in DNA transformation in several bacterial species, has not been reported (2). Finally, mutation of PriA helicase, which helps restart stalled replication forks in E. coli, lowers transformation efficiency. The helicase may act directly on the D-loop produced by RecA mediated ssDNA invasion of the duplex, or alternatively, may process a different intermediate formed during the recombination process that requires replication restart (75, 76). There are no other naturally transformable organisms reported where PriA has been tested for a role in transformation processes, so it is unclear if this is a unique processing requirement in Neisseria. The details of DNA processing and recombination during transformation in the Neisseria is still open to further investigation.
Delivery of DNA for DNA Transformation - Gonococcal Genetic Island and Type IV Secretion
One of the major issues in genetic transfer is the source of transforming DNA. It has been argued that using DNA from dead bacteria may promote the acquisition of alleles that are at a selective disadvantage (77). This viewpoint ignores the fact that even if a genetic change occurs that causes death of a bacterial cell, the negative allele is only one gene in a chromosome of 500 – 8,000 genes that can be transferred. In addition, most competent bacteria exhibit a natural level of autolysis that provides DNA from cells that are not under negative selection (78). The gonococcus, however, has a unique way of providing donor DNA for transformation. Over 75% of investigated gonococcal strains carry a genomic island approximately 57kb in size encoding a type IV secretion system (T4SS) that secretes DNA into the extracellular environment (79, 80). While named the gonococcal genetic island (GGI) due to its discovery in N. gonorrhoeae, the GGI has also been found in 17.5% of meningococcal strains, although the functionality of the encoded T4SS varies amongst these strains (81, 82). The GGI displays many characteristics of a horizontally acquired genomic island; including a G + C content different than the rest of the chromosome and a paucity of DNA uptake sequences (DUS) relative to the rest of the genome (80). Like many other horizontally acquired genomic islands, the GGI has short direct repeats on both ends (80).
Based on the flanking direct repeat sequences difA, a consensus dif site, and difB, a dif site varying by 4 mismatches, it is thought that the acquisition of the GGI was mediated by the dif-recognizing gonococcal recombinase XerCD (80, 83). Under laboratory conditions, the GGI undergoes XerCD mediated excision once every 106 cells after 18 hours of growth (84). Notably, when the non-consensus difB site is mutated to restore the consensus dif sequence, the excision rate increases to once per 103 cells (85). These observations have incited speculation that the non-consensus sequence of the difB site acts to limit the rate of GGI excision (86). While the maintenance of the GGI and its encoded T4SS in 80% of gonococcal strains indicates a survival advantage, it is possible that infrequent excision of the GGI is maintained, perhaps to help propagate the GGI. The mutations in the difB site allow for this to happen, but also significantly decrease the frequency of GGI excision, which would favor maintenance. Interestingly, many N. meningitidis strains carry more mutations in the difB site than the gonococcal versions, which may explain why the GGI is maintained in the meningococcus despite carrying mutations that render the T4SS inactive (83).
The GGI consists of 62 open reading frames, 23 of which show significant homology to T4SS proteins (80). The encoded T4SS is similar to other F-like (named for the prototypical conjugative plasmid) T4SS, but in contrast to typical conjugative elements, the GGI secretes chromosomal DNA into the extracellular milieu in a contact-independent manner (87). Nuclease sensitivity experiments show that the secreted DNA is single-stranded and blocked at the 5′ end, presumably by a bound relaxase molecule (84). Unlike the prototypical T4SS, the secreted DNA is exposed to the extracellular environment rather than being transferred directly between cells (79). This is supported by the observation that the encoded TraA conjugative pilin subunit is not required for T4S (86). The secreted DNA, however, is competent for transformation and has been shown to be a better transformation substrate than DNA released by autolysis (79). This stands in contrast to the observation that ssDNA is a less efficient substrate than dsDNA for transformation (49). While protein secretion has yet to be shown, it has been postulated that the increased efficiency of transformation by T4S ssDNA over autolysis released dsDNA is due to proteins bound to the secreted DNA (86).
Although the gonococcal T4SS is non-conjugative, inferences can be made about the structure of the gonococcal T4SS based on the structure revealed by cryo-electron tomography for the conjugative plasmid pKM101 and gonococcal experimental data (88, 89) (Figure 2). In plasmid pKM101, the core complex spans both the inner and outer membrane with polymerized TraK forming the secretin in the outer membrane. Interacting with TraK in a one-to-one-to-one ratio are TraV, thought to stabilize the pore complex, and TraB (88, 89). Dillard et al. have established that TraK and TraV interact in gonococci and that TraK is surface exposed (86). In plasmid pKM101, TraB spans both the outer and inner membrane to connect the core complex and provide a channel for substrate translocation, and the same is predicted to be true in gonococci (88). The inner membrane complex is likely composed of the N-terminal domain of TraB, an ATPase TraC, a coupling protein TraD, and TraG (90, 91). While shown to stabilize mating pairs, aid in pilus production, and act in entry exclusion in homologous systems; TraG plays a novel, uncharacterized role in gonococcal T4S (91). Among sequenced N. gonorrhoeae strains, three different alleles of TraG are found (79). While the shortest allele was found to be nonfunctional for the secretion of DNA, it could still have a role in the secretion of proteins or other substrates (91). Like other T4SS, the GGI is dependent on lytic transglycosylases for secretion, presumably to produce localized breaks in the peptidoglycan to allow for secretion complex assembly (92). Unique to the gonococcal system is that two lytic transglycosylases, AtlA and LtgX, are required for T4S (92).
Figure 2. Type IV Secretion System Model.
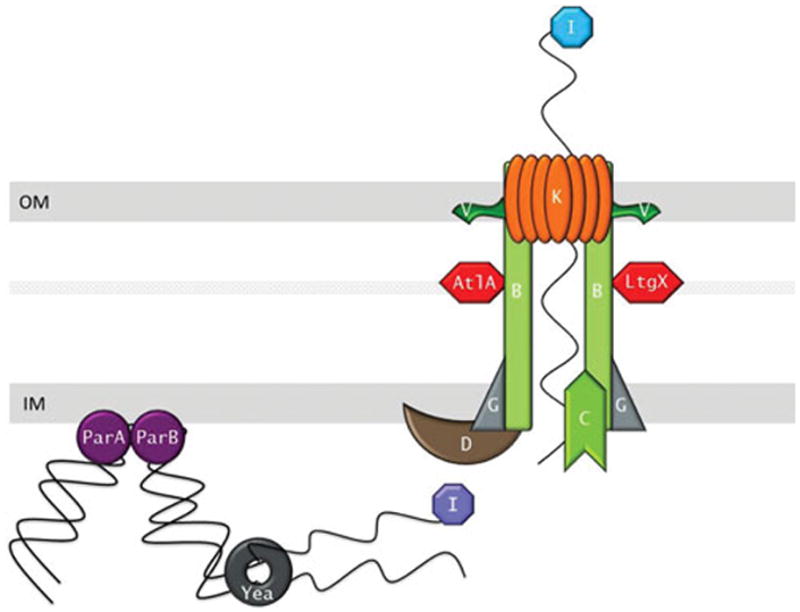
ParA and ParB recruit the chromosomal DNA to the type IV secretion system. TraI relaxase nicks the DNA at the oriT site and the DNA is unwound possiblibyl by the Yea helicase. The resulting single stranded DNA possibly still bound by TraI is then secreted through the type IV secretion complex into the extracellular mileu in a contact-independent manner. The inner membrane complex is predicted to consist of TraG, TraD and TraC with TraB spanning both the inner and outer membranes to form a channel for the DNA. The transglycosylases AtlA and LtgX create localized breaks in the peptidoglycan to allow the system to assemble. The outer membrane complex consists of TraB, TraK and TraV.
Besides the structural components of the T4SS, secretion of DNA is thought to occur through the action of ParA, ParB, TraI, Yea, and TraD (Figure 2). ParA and ParB are partitioning proteins responsible for segregation of chromosomes and plasmid DNA during replication (93). ParA, specifically the DNA binding Walker Box domain, has been shown to be required for DNA secretion leading to the hypothesis that ParA and ParB are responsible for recruitment of the DNA to the secretion complex and the processing enzymes (80). TraI is a GGI encoded relaxase, a class of proteins that nick DNA at a specific recognition site oriT (84, 94). TraI is required for DNA secretion and mutation of the putative catalytic tyrosine residues either blocks or severely hinders DNA secretion into the medium (84). While TraI does not appear to use the typical histidine-rich motif for metal coordination, an HD phosphohydrolase domain may substitute for metal ion coordination. Additionally, gonococcal TraI contains a predicted unique N-terminal amphiphatic helix which may allow association with the inner membrane. TraI interacts with the DNA at the single oriT site on the GGI. This site is an inverted repeat located near the traI gene and is required for DNA secretion. While the GGI encodes the only oriT site, the site can still support DNA secretion if moved to a distant location on the chromosome (84). This leads to the model that DNA secretion begins with a TraI induced nick at oriT and delivery of the nicked strand to the secretion complex by the required coupling protein TraD (95). Secretion then continues via unwinding of the chromosome by the GGI encoded helicase Yea. As the DNA is unwound for secretion, strand replacement synthesis is used to regenerate the chromosome and secretion proceeds along the length of the chromosome with the efficiency decreasing the further the DNA is from the original oriT.
Whether the GGI (and the encoded T4SS) has a direct role in gonococcal pathogenesis remains unclear. Certain versions of the GGI (containing atlA and the sac-4 allele of traG) are found more often in isolates that cause disseminated gonococcal infection (79). Additionally, the GGI T4SS is active during intracellular infection and can compensate for an inactive iron transport complex (96). These data suggest that the T4SS either acts as an import apparatus or exports proteins that can then be subsequently used to import iron into the bacterial cell. Finally, it has been established that secretion of ssDNA is required for initial stages of biofilm formation, which is presumed to help with colonization (97). While these studies establish possible roles for the GGI in N. gonorrhoeae virulence, the lack of an infection model makes it difficult to characterize the relative contribution of the system in causing disease. Perhaps the most interesting remaining question is what role the T4SS plays in protein secretion. The increased ability to transform with secreted DNA and the Ton-independent iron acquisition data suggest the possibility that the GGI mediates protein secretion, but this hypothesis still requires direct experimental support.
The fundamental question of what benefit HGT provides to any organism remains controversial (2). Several doubts have been raised about whether HGT is the evolutionary basis of competence due to the unpredictable results of genetic transfer with critics supporting a model where competence evolved as a nutrient acquisition system (77). An in-silico study, however, suggests that even if extracellular DNA comes from dead cells with more deleterious alleles than the recipient cell, HGT can actually allow a population to escape the predicted irreversible accumulation of harmful alleles (98). This in turn suggests that HGT may be important for long term genomic maintenance rather than having deleterious effects. Another possibility is that transformation competence allows a species to have a larger gene pool to draw upon (99). While this simulative study does not end the controversy, there is no doubt that natural transformation plays a significant role in genetic exchange and nutrient uptake and therefore is an important area for continued investigation. With the HGT-mediated spread of antibiotic resistance, the specter of untreatable gonorrheal infections is a looming reality.
Antigenic Variation
Introduction
As obligate human pathogens, N. gonorrhoeae and N. meningitidis are constantly under immune surveillance. One vital mechanism for immune avoidance is antigenic variation, by which a pathogenic organism constantly modifies surface-exposed immunogenic molecules. This strategy can result in prolonged colonization or allow for reinfection of a previously infected host who has a potentially effective immune response that is made ineffectual by the variation (reviewed in (100)). Phase variation is a related but distinct process of phenotypic variation where a cell switches between defined phases of expression, either between ON and OFF phases or between two variant forms. Both phase and antigenic variation processes generally occur at rates higher than the normal mutation rate of the organism. Phase variation systems have the ability to reversibly switch between the phases and are usually mediated by polynucleotide repeat variation, invertible elements, or differential methylation (reviewed in (101)). In contrast, antigenic variation systems have the ability to stochastically express many different forms of a gene product and can be based on multigene phase variation or a recombination-based diversity generation system (see next section). Both phase and antigenic variation systems can have functional as well as immune system consequences.
Both pathogenic Neisseria species express three antigenically or phase variable major surface determinants: the opacity (Opa) outer membrane proteins, which act as adhesins; lipooligosaccharide (LOS), which decorates the outer membrane and is also involved in host interactions; and Tfp (102). The commensal organisms express some of these molecules but do not undergo the variation processes. Both LOS and Opa proteins undergo antigenic variation through an ON/OFF phase variation mechanism, mediated by slipped-strand mispairing of tandem repeats in multiple genes (103–105). In the case of the Opa gene family, there are 4–13 individual genes that phase vary ON and OFF independently through changes in a pentamer repeat found in the signal sequence coding region of each gene (103). It is likely that the Opa protein antigenic variation process is mainly used to create functional variants to promote interactions with different human cellular receptors (106). Five of the LOS biosynthetic glycosyltransferases are phase variable due to polynucleotide repeats in promoter or coding regions that when altered turn each gene ON or OFF (105, 107). The LOS structure is then defined by which set of these five genes is expressed in combination with the eight invariant biosynthetic genes. Pilus antigenic variation, in contrast, uses a complex, programmed homologous recombination system to express antigenically distinct peptide sequences on the Tfp. While homologous recombination-based systems for antigenic variation are found in both prokaryotic and eukaryotic pathogens, the pilin antigenic variation in Neisseria has become a model system for these types of diversity generation systems.
Pilin Antigenic Variation
Pilin antigenic variation is mediated by non-reciprocal recombination events, where a sequence from a silent pilin copy is donated to the pilin expression locus but does not change in the reaction (Figure 3). Gonococci typically carry around 18 silent pilin copies in four to six pilS loci (108, 109). Lacking a promoter, ribosome binding site, and the coding sequence for the N-terminal alpha helix of pilin, the silent copies are not expressed (110–112). The pilE gene encodes a 5′ constant region followed by a semi-variable (SV) region, two highly conserved cys regions (containing the two-disulfide bond forming cysteines) sandwiched around the hypervariable loop, and finally the hypervariable tail. The amount of conservation and diversity in the different regions is due directly to the sequence variation in the silent copies (110). The hypervariable regions of pilE correspond to the coding regions that display the largest diversity amongst the silent copies. Highlighting the functional role of pilin variation in creating antigenic diversity, the hypervariable regions correspond to the surface and antibody-exposed areas of pilin in the pilus fiber (113, 114). During pilin antigenic variation a portion of a pilS copy is transferred into the expressed pilE. Because the amount of transferred sequence can range from a single nucleotide to the entire pilS copy and sequence can be donated from multiple pilS copies along the length of pilE, this process can result in a remarkably large set of expressed pilin sequences (Figure 3A–C) (115).
Figure 3. Molecular Description of Antigenic Variation.
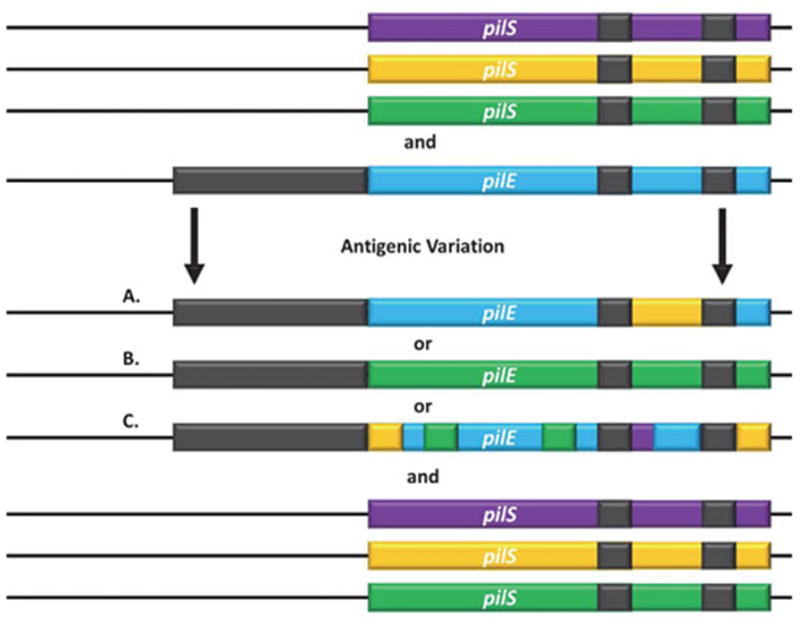
The pilE and pilS loci have regions of sequence microhomology (grey) and variability (colored). Sequence from a nonexpressed pilS loci copy is transferred into the expression locus with the pilS sequence not changing. Recombination can occur A. in just a section of the gene resulting in a pilE-pilS hybrid, B. across the entire pilS gene resulting in an entirely new variable region of pilE, or C. multiple times with different silent copies resulting in a new pilE sequence containing information from different silent copies throughout the variable regions.
The molecular process of antigenic variation results in a wide range of functional consequences. Noted as early as the 1960’s, gonococcal colonies can exhibit a visible phase variation due to expression or lack thereof of pili (116). While some pilus phase variation is due to ON/OFF switching of PilC or pilE deletion, antigenic variation events that introduce a premature stop codon from a silent copy or a non-favorable combination of silent copies can also prevent pilus expression (117–120). While not fully understood, it is likely that such nonfavorable combinations of silent copies result in a pilin molecule that is unable to efficiently assemble into a pilus fiber. This is supported by the observation that strains with different pilE coding sequences exhibit different levels of piliation (119). Although the lack of a suitable animal model means experimental data is lacking, it is likely that phase variation is critical for multiple reasons as all gonococcal isolates have this ability and multiple avenues to achieve it. Antigenic variation can also alter the sites of post-translational modification of the pilus which has been implicated in a variety of biological processes in N. gonorrhoeae including cellular adhesion and host-cell activation (121, 122). In N. meningitidis, changes in the glycosylation status of the pilus can enhance transit across epithelial barriers, a critical step in pathogenesis (123). In addition, variation of the exposed pilin residues has been implicated in controlling host cell response in N. meningitidis (124). Engineered pilin variants demonstrated that the C-terminal domain of pilin is critical for colonization promoting host cell interactions, as well as that different pilin sequences conferred different host cell specificities (124). These findings have direct implications on the pathogenesis of N. meningitidis and underscore the importance of the diversity generation systems of Neisseria to provide both functional and immune avoidance capabilities to the organism.
Trans-acting Factors Important for Pilin Antigenic variation
A series of broad and directed genetic screens have identified many factors important for pilin antigenic variation, although the majority of their roles have been inferred from orthologous proteins rather than direct biochemical characterization (125–127). The first protein identified as critical to antigenic variation is RecA, and mutations in the recA gene decreased pilus phase variation by 100–1000 fold (120). These results not only established RecA as a mediator of pilin antigenic variation but also demonstrated that antigenic variation is mediated by a homologous recombination-based process (120). E. coli RecA can complement a gonococcal RecA mutant and has similar biochemical properties (128, 129). Interestingly, expression of the E. coli RecA in N. gonorrhoeae resulted in increased antigenic variation and this increased frequency was due to the co-transcription of the E. coli RecX protein (130). This work led to the discovery that in the gonococcus, RecX is required for efficient pilin antigenic variation and that E. coli RecX is a negative regulator of RecA filamentation (71, 73, 130). Another protein that modulates RecA polymerization and activity named RdgC is also involved in promoting pilin antigenic variation presumably by also modulating RecA activity (131, 132).
The RecF-like recombination pathway has a central role in pilin antigenic variation (Neisseria do not encode a recF gene). In E. coli, the RecF pathway utilizes proteins both pathway specific (RecF, RecR, RecO, RecQ) and non-specific (RecA, RecN). The pathway is mainly responsible for repairing single-stranded gaps in DNA, although it can repair dsDNA breaks when the primary RecBCD pathway is inactive (133). In N. gonorrhoeae, mutational analysis of the pathway revealed that RecQ, and RecJ are required for some but not all antigenic variation events, but only the RecOR recombinase is required for all pilin antigenic variation (72, 134). RecQ involvement was shown to be dependent on the helicase activity of two of the three HRDC domains at the C-terminus (135). The Rep protein, a 3′–5′ helicase in E. coli, is also required for some events, and it is not known whether RecQ and Rep are partially redundant to one another (136, 137). RecJ is a 5′-3′ single-strand exonuclease whose role suggest that single-strand end resection is involved (134). RecR and RecO form a necessary recombinase that is the only known recombinase to act in the process of antigenic variation (126). RecN, whose role is not limited to the RecF pathway in E. coli, was not found to play a role in antigenic variation (126, 134). The identification of the RecF-like pathway as being necessary for pilin antigenic variation suggests that there is a gapped intermediate that is required for one step of the recombination process, but the molecular description of the intermediate has not been reported.
The RecBCD recombination pathway was reported to not to be involved in pilin antigenic variation, but subsequent studies reported that a recD mutant showed an increased frequency of antigenic variation and that a recB mutant was deficient for pilin antigenic variation in one strain of N. gonorrhoeae, but not another (72, 138, 139). All of these studies relied on assays that scored only a subset of potential variants and did not always account for the greatly reduced growth rate of the recB, C, and D mutants. A less biased sequencing assay, used to detect all pilin antigenic variation events, conclusively showed that insertional mutations in recB, recC and recD in both strains MS11 and FA1090 resulted in DNA repair phenotypes but not a pilin antigenic variation phenotype (140). This study confirmed that the impaired growth of the recB and recD mutants resulted in a shift in the frequency of some, but not all, donor silent copies—possibly explaining the reason for the contrasting conclusion of a role for the RecBCD pathway in pilin antigenic variation.
Both the RuvABC and RecG Holliday Junction processing pathways are required for pilin antigenic variation (126, 141). Mutations in either pathway prevent pilin antigenic variation, leading to the hypothesis that there are distinct substrates acted on by these pathways. Importantly, double mutants in recG and ruvA,B, or C created a partial synthetic lethality with an increase in pilE deletions in the surviving bacteria. This lethality could be rescued by a recA mutation as well as several other mutations that prevent antigenic variation (127, 141). This led to the hypothesis that antigenic variation involves two Holliday junctions, explaining the phenotype of the single mutations, and that the double recG and ruvA,B, or C mutations prevents the reversal of one or both of the Holliday junctions, thus locking the cell into a lethal antigenic variation intermediate. Preventing antigenic variation through processes such as recA mutation or pilE deletion prevents formation of the unresolvable lethal intermediate and rescues the synthetic lethality (141).
Required DNA Sequences and Structures
Several cis-acting sites have been identified as contributing to pilin antigenic variation. The 63 bp Sma/Cla repeat is located downstream of all pilin loci and often carries SmaI and ClaI restriction endonuclease sites (111). Because of the similarity of the Sma/Cla sequence to recombinase binding sites, the Sma/Cla site was investigated, and it was found that deletion of the site downstream of the expressed pilin locus in strain MS11 reduced antigenic variation using a semi-quantitative assay that only detected transfer from two donor silent copies (142). This observation has not been reported for other strains and remains unexplained. The cys2 region of pilin is another conserved sequence involved in antigenic variation. Two studies have implicated the importance of cys2 in antigenic variation as well as the spacing between the cys1 and cys2 region (143, 144). There are two likely roles of these conserved regions in antigenic variation. First, the sequence conservation of the cys2 region to that of the silent copies could serve as a shared region of homology to drive the recombination process. Secondly, the cys2 region might serve as a binding site for trans-acting factors required for antigenic variation. Whatever the reason, it is clear that conserved sequences have a role in pilin antigenic variation and more investigation is needed to identify the critical sequences.
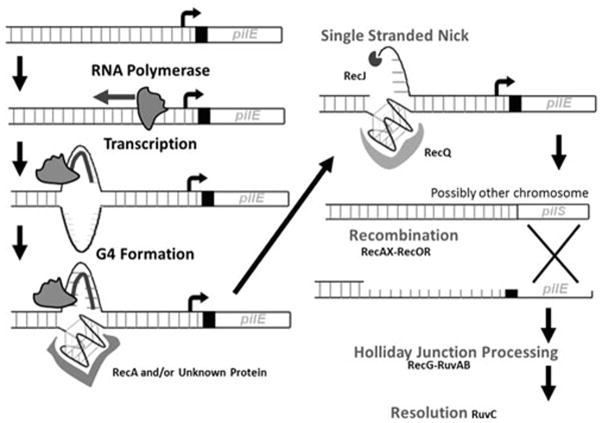
A breakthrough in the understanding of the mechanisms allowing pilin antigenic variation came from identification of a cis-acting DNA structure in the region of DNA immediately upstream of pilE. Transposon insertions isolated in a genetic screen prevented antigenic variation without disrupting pilin expression or an obvious open reading frame (126). A targeted mutagenesis of the genomic region carrying these transposon insertions identified twelve G-C base pairs that prevented antigenic variation when individually mutated (Figure 4A and 4B) (127). The sequence conforms to the definition of a guanine quadruplex (G4) forming motif (Figure 4B), which can adopt a four stranded, square planar structure utilizing non-traditional Hoogsteen base pairing (Figure 4C). Further investigation confirmed that the sequence forms a G4 structure in vitro and the individual mutations that abrogate antigenic variation also prevent G4 formation in vitro. Additionally, the point mutations that prevent G4 formation also decreased the detection of single-stranded nicks in both the G4 structure and in the C-rich strand opposite of the G4. These nicks are proposed to be required for the initiation of recombination and the subsequent gene conversion (127). NMR analysis defined the pilE G4 structure, showing the sequence forms a three-layer, all-parallel stranded monomeric G4 with single residue double-chain-reversal loops (Figure 4C) (145). Importantly, the pilE G4 structure (but not two other G4 forming sequences present in the N. gonorrhoeae genome) binds to RecA and does so with similar affinity as RecA for ssDNA (145). A G4 structure present on a ssDNA substrate can stimulate RecA-mediated strand exchange in vitro (145). Together these data suggest that the G4 serves to recruit RecA to the pilE locus and possibly serves as a nucleation site for RecA filamentation. Finally, the two extra HRDC domains of RecQ that were shown to be required for antigenic variation are also necessary to unwind the pilE G4 in vitro, suggesting that the effect of RecQ mutation on antigenic variation is due to the reduced capacity of the helicase to bind and unwind the G4 structure (135, 146).
Figure 4. The pilE Guanine Quartet (G4).
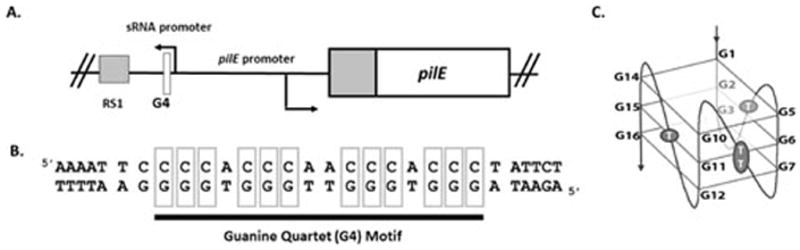
A. Gene map showing the location of the pilE-associated G4 forming sequence and the sRNA promoter required for antigenic variation at the pilE locus. B. The sequence upstream of pilE that forms a G4. Mutation of the boxed guanine residues lead to loss of antigenic variation implicating the G4 in antigenic variation. C. The parallel G4 structure of the pilE G4 as solved by NMR analysis.
Although the G4 structure was shown to be required for antigenic variation, it was unclear what initiated G4 formation since the dsDNA must be melted to allow the G4 structure to form. Directed mutational analysis identified a promoter downstream of the G4-forming sequence that is required for pilin antigenic variation (147). Transcription of a small RNA (sRNA) from this promoter was confirmed by 5′-RACE to start within the second set of Gs within the G4 forming sequence (Figure 4A). Expression of the sRNA at a distal chromosomal site did not restore antigenic variation in a promoter mutant showing that the sRNA was cis-acting. Together these data suggest that it is transcription of the sRNA at the G4 sequence and not some downstream role of the sRNA that initiates antigenic variation (147). Throughout the investigation of pilin antigenic variation, a variety of models have been proposed to explain the phenomenon. In light of the most recent data, the feasibility of each model can be re-evaluated.
Antigenic Variation Models
Pilin antigenic variation is a gene conversion event, i.e., an apparent nonreciprocal recombination process that has been mainly studied in eukaryotic cells that possess two copies of their chromosomes. It was therefore proposed that the efficient DNA transformation system of the Neisseria could be used to allow for gene conversion from pilS to pilE, if the donor molecule was from a different cell than the recipient (33, 148). While there are data supporting or refuting transformation as a way to allow pilin antigenic variation, it is now generally accepted that the majority of pilin antigenic variation events do not involve transformation. (33, 148–150). With transformation ruled out as the main source for antigenic variation, all remaining proposed models of antigenic variation involve intracellular recombination and necessitate at least two chromosomes. In most bacteria, two chromosomes only exist after replication, but it has been shown that both N. gonorrhoeae and N. meningitidis are polyploid, most likely diploid, and that Neisseria lactamica, which does not undergo pilin antigenic variation, has a single copy of its chromosome (151, 152). It has yet to be directly demonstrated that pilin antigenic variation relies on diploid homozygous chromosomes, but it remains a unique hypothesis.
One of the first models of antigenic variation proposed was the mini-cassette theory (110). It postulated that there were seven defined mini- cassettes of variable sequence interspersed among regions of homology which were used to affect the segmental recombination that defines pilin antigenic variation. This model was discounted by sequencing data that established that antigenic variation events can change as few as one nucleotide or incorporate an entire pilS copy and can occur anywhere in the variable sequences where microhomology occurs between the recombining silent and expressed gene (32, 153). There are three main models of pilin antigenic variation that have been proposed. The unequal crossing over model (or RecBCD-mediated double-chain-break repair model) (139, 154) (Figure 5A) proposes that a double strand break occurs in the pilE locus, and after RecBCD-mediated end resection, the single-stranded 3′ overhang invades the homologous pilS donor locus, presumably through RecA. The 3′ end of the invading strand is extended by polymerase while the displaced donor strand is used as a template to repair the non-invading strand. Holliday junction resolution results in the donor DNA sequence replacing the recipient pilE sequence. Although not conclusively ruled out, the data unambiguously showing RecBCD plays no role in antigenic variation make the unequal crossing-over model less likely (140).
Figure 5. Proposed Recombination Pathways.
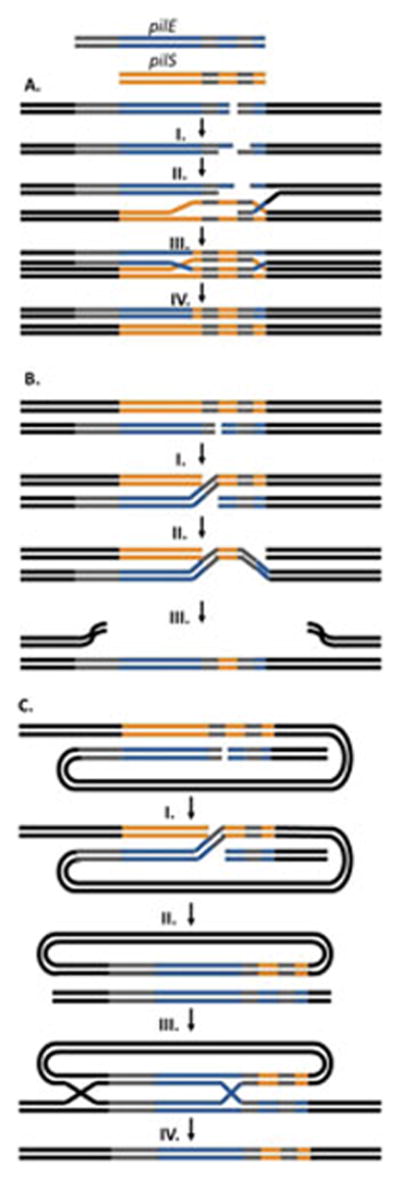
A. Unequal Crossing Over Model – A dsDNA break occurs at the pilE locus and I. the 5′ ends are resected by RecBCD to leave 3′ overhangs. II. A single 3′ end mediated by RecA, invades the pilS locus forming a D-loop. III. The 3′ ends are extended by DNA polymerase using the pilS gene as a template. IV. Resolution of the double Holliday junctions results in a new pilE sequence without altering the donor pilS sequence.
B. Successive Half Crossing Over Model – Recombination begins with a dsDNA break or single-stranded gap in pilE in a region of homology. I. A RecA and RecOR mediated half crossing over event occurs linking the pilE and a pilS locus on a sister chromosome. II. A second half crossing over event occurs in another region of microhomology downstream of the first event between the pilE:pilS hyrbid and the original pilE locus. III. This recombination event leads to a new sequence at the pilE locus and destruction of the donor chromosome.
C. Hybrid Intermediate Model – Similar to the half crossing over model, recombination initiates with a double stranded break or single-stranded gap at pilE and I. a half crossing over event with a donor pilS on the same chromosome. II. This results in a pilE:pilS hybrid intermediate and the loss of the donor chromosome. III. The hybrid intermediate then undergoes two recombination events with the recipient pilE on a different chromosome. The first recombination event would occur in the extensive region of homology upstream of the genes while the second even would utilize microhomology within the variable regions of the genes. IV. Resolution of the Holliday junction intermediates leads to a new pilE sequence on the recipient chromosome.
The successive half crossing-over model also proposes an initiating double-strand break at the recipient pilE locus, but could also be initiated by a gapped substrate (Figure 5B) (72, 154). RecJ catalyzed 3′ end resection at the pilE double-strand break or nick would provide a substrate for RecA, RecX and RecOR to mediate recombination with a donor pilS copy on a sister chromosome. This recombination event would create a pilE-pilS intermediate and link the sister chromosomes. A second half-crossing over event between the pilS region of the pilE-pilS 3′ intermediate and the original pilE locus would result in the original pilE locus containing a hybrid pilE-pilS sequence and the destruction of the donor pilS chromosome. As a result of the first half-crossing over event the original pilE locus will not be intact, requiring some sort of yet uncharacterized tethering mechanism to keep the loose pilE end of the dsDNA break in close proximity to the donor pilS locus. The hybrid intermediate model is a variation of the half crossing over model that evolved from experimental observations (Figure 5C) (143, 155). This model proposes that a recombination event occurs between pilE and pilS in a region of shared microhomology similar to that of the half-crossing over model but between genes located on the same chromosome. The crossover event would result in a circular pilE-pilS hybrid intermediate with the chromosomal sequences that existed between the two recombining loci also carried on the episomal circle and the resultant loss of the donor chromosome. The hybrid intermediate then requires two recombination events with a recipient pilE on a sister chromosome. One recombination event would occur in the extensive region of upstream homology and may utilize the homologous recombination factors while the second event occurs in a region of microhomology within the pilE coding sequence. Resolution of the resulting double Holliday junction would create a new pilE sequence without any changes outside of the variable regions of pilE. While pilE-pilS hybrids can be isolated from the gonococcus and have been shown to undergo recombination at the pilE locus more readily than pilS loci, a hybrid intermediate consistent with all experimental data has yet to be defined (155).
While the recombination event(s) that allow gene conversion in a bacterial chromosome remain undefined, the known antigenic variation proteins and the discovery of the G4 and cis-acting noncoding sRNA allows for the formation of a speculative, working model of the key events in antigenic variation (Figure 6). Initiation of antigenic variation occurs with transcription of the noncoding sRNA. The process of transcription melts the dsDNA and the occlusion of the C-rich strand by the formation of a DNA:RNA intermediate allows for the formation of the G4. The activation energy required for G4 formation may be lowered by a yet-unknown protein. The fact that G4 unwinding by RecQ (and possibly Rep) is required for pilin antigenic variation suggests that the G4 is resolved by RecQ either during or after antigenic variation. Formation of the G4 leads to local nicking of the DNA possibly induced by a stalled replication fork on the leading strand. The nicked substrate is likely processed by RecJ endonuclease and either the RecQ or Rep helicases. Based on RecA affinity for the pilE G4 structure, RecA may be recruited to the G4 structure to initiate RecA filamentation. A RecOR-assisted, RecA-mediated homologous pairing between the processed pilE and a pilS copy would create the half crossing over intermediate and the second half crossing over reaction. If the hybrid intermediate model is correct, these factors could be involved with the initial hybrid intermediate formation or recombination of the intermediate with the recipient pilE. The presence of microhomology at the ends of many pilin antigenic variation recombination tracts suggests there may be an annealing process involved, but the identity of the protein that promotes annealing is unknown. Regardless, the models need to account for the requirement for both RuvABC and RecG and an intermediate that cannot be resolved if both pathways are inactivated. Though the actual nature of the recombination events is still unclear, the proposed working model provides predictions for conducting future studies.
Figure 6. Proposed Antigenic Variation Initiation Pathway.
Transcription initiation at the sRNA upstream of pilE melts the DNA allowing the G4 structure to form. An unknown protein may bind the G4 to stabilize the structure. A single stranded nick may occur on the strand opposite the G4 due to a stalled replication fork. RecQ could unwind the G4 structure. RecJ resects the 5′ nicked end allowing RecA to mediate recombination, possibly enhanced by binding the G4 structure, with RecOR using regions of homology between pilE and the donor pilS, possibly through a recombination mechanism detailed in Figure 5. RecG and RuvABC then process and resolve the recombination intermediate.
Conclusions
The pathogenic Neisseria undergo transformation with remarkable frequency and efficiency, and this genetic exchange is critical to allow the pathogens to survive as human restricted organisms. Additionally, the system of antigenic variation allows for high frequency productive gene conversion events without adverse effects on genomic stability. Although pilin antigenic variation has undergone the most thorough investigation of any antigenic diversity generating system, implicating specific DNA sequences and structures, as well as a multitude of DNA repair proteins, the detailed mechanisms of recombination are largely unresolved. Continued exploration of the molecular mechanisms will provide understanding of how directed recombination can occur as well as increasing our knowledge of a system thought to allow N. gonorrhoeae to escape immune detection, re-infect core populations, and cause the failure of attempted gonococcal vaccines.
References
- 1.Virji M. Pathogenic Neisseriae: surface modulation, pathogenesis and infection control. Nature reviews Microbiology. 2009;7(4):274–86. doi: 10.1038/nrmicro2097. [DOI] [PubMed] [Google Scholar]
- 2.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. Bacterial transformation: distribution, shared mechanisms and divergent control. Nature reviews Microbiology. 2014;12(3):181–96. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 3.Sparling PF. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol. 1966;92(5):1364–71. doi: 10.1128/jb.92.5.1364-1371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas GD, Sox T, Blackman E, Sparling PF. Factors affecting genetic transformation of Neisseria gonorrhoeae. J Bacteriol. 1977;129:983–92. doi: 10.1128/jb.129.2.983-992.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koomey M. Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer. APMIS Suppl. 1998;84:56–61. doi: 10.1111/j.1600-0463.1998.tb05649.x. [DOI] [PubMed] [Google Scholar]
- 6.Sox TE, Mohammed W, Blackman E, Biswas G, Sparling PF. Conjugative plasmids in Neisseria gonorrhoeae. J Bacteriol. 1978;134(1):278–86. doi: 10.1128/jb.134.1.278-286.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nature reviews Microbiology. 2004;2(3):241–9. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 8.Smith JM, Smith NH, O’Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;90(10):4384–8. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin IM, Ison CA. Detection of mixed infection of Neisseria gonorrhoeae. Sex Transm Infect. 2003;79(1):56–8. doi: 10.1136/sti.79.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynn F, Hobbs MM, Zenilman JM, Behets FM, Van Damme K, Rasamindrakotroka A, et al. Genetic typing of the porin protein of Neisseria gonorrhoeae from clinical noncultured samples for strain characterization and identification of mixed gonococcal infections. J Clin Microbiol. 2005;43(1):368–75. doi: 10.1128/JCM.43.1.368-375.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs CP, Meyer TF. Genome plasticity in Neisseria gonorrhoeae. FEMS Microbiol Lett. 1996;145(2):173–9. doi: 10.1111/j.1574-6968.1996.tb08574.x. [DOI] [PubMed] [Google Scholar]
- 12.Hobbs MM, Seiler A, Achtman M, Cannon JG. Microevolution within a clonal population of pathogenic bacteria: recombination, gene duplication and horizontal genetic exchange in the opa gene family of Neisseria meningitidis. Mol Microbiol. 1994;12(2):171–80. doi: 10.1111/j.1365-2958.1994.tb01006.x. [DOI] [PubMed] [Google Scholar]
- 13.Snyder LA, Davies JK, Saunders NJ. Microarray genomotyping of key experimental strains of Neisseria gonorrhoeae reveals gene complement diversity and five new neisserial genes associated with Minimal Mobile Elements. BMC Genomics. 2004;5(1):23. doi: 10.1186/1471-2164-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckee CO, Jolley KA, Recker M, Penman B, Kriz P, Gupta S, et al. Role of selection in the emergence of lineages and the evolution of virulence in Neisseria meningitidis. Proc Natl Acad Sci U S A. 2008;105(39):15082–7. doi: 10.1073/pnas.0712019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goire N, Lahra MM, Chen M, Donovan B, Fairley CK, Guy R, et al. Molecular approaches to enhance surveillance of gonococcal antimicrobial resistance. Nature reviews Microbiology. 2014;12(3):223–9. doi: 10.1038/nrmicro3217. [DOI] [PubMed] [Google Scholar]
- 16.Kirkcaldy RD, Ballard RC, Dowell D. Gonococcal resistance: are cephalosporins next? Curr Infect Dis Rep. 2011;13(2):196–204. doi: 10.1007/s11908-011-0169-9. [DOI] [PubMed] [Google Scholar]
- 17.Lewis DA. The Gonococcus fights back: is this time a knock out? Sex Trans Inf. 2010;86(6):415–21. doi: 10.1136/sti.2010.042648. [DOI] [PubMed] [Google Scholar]
- 18.Prevention CfDCa. Antibiotic resistance threats in the United States, 2013. Atlanta: CDC; 2013. [Google Scholar]
- 19.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nature reviews Microbiology. 2004;2(5):363–78. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 20.Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407(6800):98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 21.Swanson J. Studies on gonococcus infection. IV. Pili: their role in attachment of gonococci to tissue culture cells. J Exp Med. 1973;137(3):571–89. doi: 10.1084/jem.137.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dietrich M, Bartfeld S, Munke R, Lange C, Ogilvie LA, Friedrich A, et al. Activation of NF-kappaB by Neisseria gonorrhoeae is associated with microcolony formation and type IV pilus retraction. Cell Microbiol. 2011;13(8):1168–82. doi: 10.1111/j.1462-5822.2011.01607.x. [DOI] [PubMed] [Google Scholar]
- 23.Freitag NE, Seifert HS, Koomey M. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol Microbiol. 1995;16(3):575–86. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 24.Long CD, Tobiason DM, Lazio MP, Kline KA, Seifert HS. Low-level pilin expression allows for substantial DNA transformation competence in Neisseria gonorrhoeae. Infect Immun. 2003;71(11):6279–91. doi: 10.1128/IAI.71.11.6279-6291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drake SL, Koomey M. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol Microbiol. 1995;18(5):975–86. doi: 10.1111/j.1365-2958.1995.18050975.x. [DOI] [PubMed] [Google Scholar]
- 26.Tonjum T, Freitag NE, Namork E, Koomey M. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol Microbiol. 1995;95(3):451–64. doi: 10.1111/j.1365-2958.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 27.Wolfgang M, Lauer P, Park HS, Brossay L, Hebert J, Koomey M. pilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29(1):321–30. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 28.Biswas GD, Lacks SA, Sparling PF. Transformation-deficient mutants of piliated Neisseria gonorrhoeae. J Bacteriol. 1989;171:657–64. doi: 10.1128/jb.171.2.657-664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aas FE, Wolfgang M, Frye S, Dunham S, Lovold C, Koomey M. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol Microbiol. 2002;46(3):749–60. doi: 10.1046/j.1365-2958.2002.03193.x. [DOI] [PubMed] [Google Scholar]
- 30.Berry JL, Cehovin A, McDowell MA, Lea SM, Pelicic V. Functional analysis of the interdependence between DNA uptake sequence and its cognate ComP receptor during natural transformation in Neisseria species. PLoS Genet. 2013;9(12):19. doi: 10.1371/journal.pgen.1004014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen I, Dubnau D. DNA transport during transformation. Front Biosci. 2003;8:s544–56. doi: 10.2741/1047. [DOI] [PubMed] [Google Scholar]
- 32.Haas R, Schwarz H, Meyer TF. Release of soluble pilin antigen coupled with gene conversion in Neisseria gonorrhoeae. Proc Natl Acad Sci U S A. 1987;84(24):9079–83. doi: 10.1073/pnas.84.24.9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibbs CP, Reimann BY, Schultz E, Kaufmann A, Haas R, Meyer TF. Reassortment of pilin genes in Neisseria gonorrhoeae occurs by two distinct mechanisms. Nature. 1989;338(6217):651–2. doi: 10.1038/338651a0. [DOI] [PubMed] [Google Scholar]
- 34.Long CD, Madraswala RN, Seifert HS. Comparisons between colony phase variation of Neisseria gonorrhoeae FA1090 and pilus, pilin, and S-pilin expression. Infect Immun. 1998;66(5):1918–27. doi: 10.1128/iai.66.5.1918-1927.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins RF, Davidsen L, Derrick JP, Ford RC, Tonjum T. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J Bacteriol. 2001;183(13):3825–32. doi: 10.1128/JB.183.13.3825-3832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamilton HL, Dillard JP. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol Microbiol. 2006;59(2):376–85. doi: 10.1111/j.1365-2958.2005.04964.x. [DOI] [PubMed] [Google Scholar]
- 37.Maier B, Chen I, Dubnau D, Sheetz MP. DNA transport into Bacillus subtilis requires proton motive force to generate large molecular forces. Nat Struct Mol Biol. 2004;11(7):643–9. doi: 10.1038/nsmb783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burton B, Dubnau D. Membrane-associated DNA transport machines. Cold Spring Harbor perspectives in biology. 2010;2(7):a000406. doi: 10.1101/cshperspect.a000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen I, Gotschlich EC. ComE, a competence protein from Neisseria gonorrhoeae with DNA-binding activity. J Bacteriol. 2001;183(10):3160–8. doi: 10.1128/JB.183.10.3160-3168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seitz P, Pezeshgi Modarres H, Borgeaud S, Bulushev RD, Steinbock LJ, Radenovic A, et al. ComEA Is Essential for the Transfer of External DNA into the Periplasm in Naturally Transformable Vibrio cholerae Cells. PLoS Genet. 2014;10(1):2. doi: 10.1371/journal.pgen.1004066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salman H, Zbaida D, Rabin Y, Chatenay D, Elbaum M. Kinetics and mechanism of DNA uptake into the cell nucleus. Proc Natl Acad Sci U S A. 2001;98(13):7247–52. doi: 10.1073/pnas.121067698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodman SD, Scocca JJ. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci U S A. 1988;85(18):6982–6. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elkins C, Thomas CE, Seifert HS, Sparling PF. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J Bacteriol. 1991;173(12):3911–3. doi: 10.1128/jb.173.12.3911-3913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ambur OH, Frye SA, Tonjum T. New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J Bacteriol. 2007;189(5):2077–85. doi: 10.1128/JB.01408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith HO, Gwinn ML, Salzberg SL. DNA uptake signal sequences in naturally transformable bacteria. Res Microbiol. 1999;150(9–10):603–16. doi: 10.1016/s0923-2508(99)00130-8. [DOI] [PubMed] [Google Scholar]
- 46.Davidsen T, Rodland EA, Lagesen K, Seeberg E, Rognes T, Tonjum T. Biased distribution of DNA uptake sequences towards genome maintenance genes. Nucleic Acids Res. 2004;32(3):1050–8. doi: 10.1093/nar/gkh255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Treangen TJ, Ambur OH, Tonjum T, Rocha EP. The impact of the neisserial DNA uptake sequences on genome evolution and stability. Genome Biol. 2008;9(3):2008–9. doi: 10.1186/gb-2008-9-3-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duffin PM, Seifert HS. DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent. J Bacteriol. 2010;192(17):4436–44. doi: 10.1128/JB.00442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duffin PM, Seifert HS. Genetic transformation of Neisseria gonorrhoeae shows a strand preference. FEMS Microbiol Lett. 2012;334(1):44–8. doi: 10.1111/j.1574-6968.2012.02612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mathis LS, Scocca JJ. On the role of pili in transformation of Neisseria gonorrhoeae. J Gen Microbiol. 1984;130:3165–73. doi: 10.1099/00221287-130-12-3165. [DOI] [PubMed] [Google Scholar]
- 51.Dorward DW, Garon CF. DNA-binding proteins in cells and membrane blebs of Neisseria gonorrhoeae. J Bacteriol. 1989;171:4196–201. doi: 10.1128/jb.171.8.4196-4201.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolfgang M, van Putten JP, Hayes SF, Koomey M. The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol Microbiol. 1999;31(5):1345–57. doi: 10.1046/j.1365-2958.1999.01269.x. [DOI] [PubMed] [Google Scholar]
- 53.Aas FE, Lovold C, Koomey M. An inhibitor of DNA binding and uptake events dictates the proficiency of genetic transformation in Neisseria gonorrhoeae: mechanism of action and links to Type IV pilus expression. Mol Microbiol. 2002;46(5):1441–50. doi: 10.1046/j.1365-2958.2002.03265.x. [DOI] [PubMed] [Google Scholar]
- 54.Cehovin A, Simpson PJ, McDowell MA, Brown DR, Noschese R, Pallett M, et al. Specific DNA recognition mediated by a type IV pilin. Proc Natl Acad Sci U S A. 2013;110(8):3065–70. doi: 10.1073/pnas.1218832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Assalkhou R, Balasingham S, Collins RF, Frye SA, Davidsen T, Benam AV, et al. The outer membrane secretin PilQ from Neisseria meningitidis binds DNA. Microbiology. 2007;153(Pt 5):1593–603. doi: 10.1099/mic.0.2006/004200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lang E, Haugen K, Fleckenstein B, Homberset H, Frye SA, Ambur OH, et al. Identification of neisserial DNA binding components. Microbiology. 2009;155(Pt 3):852–62. doi: 10.1099/mic.0.022640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benam AV, Lang E, Alfsnes K, Fleckenstein B, Rowe AD, Hovland E, et al. Structure-function relationships of the competence lipoprotein ComL and SSB in meningococcal transformation. Microbiology. 2011;157(Pt 5):1329–42. doi: 10.1099/mic.0.046896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frye SA, Nilsen M, Tonjum T, Ambur OH. Dialects of the DNA uptake sequence in Neisseriaceae. PLoS Genet. 2013;9(4):18. doi: 10.1371/journal.pgen.1003458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berry JL, Cehovin A, McDowell MA, Lea SM, Pelicic V. Functional Analysis of the Interdependence between DNA Uptake Sequence and Its Cognate ComP Receptor during Natural Transformation in Neisseria Species. PLoS Genet. 2013;9(12):e1004014. doi: 10.1371/journal.pgen.1004014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fussenegger M, Facius D, Meier J, Meyer TF. A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol Microbiol. 1996;19(5):1095–105. doi: 10.1046/j.1365-2958.1996.457984.x. [DOI] [PubMed] [Google Scholar]
- 61.Fussenegger M, Kahrs AF, Facius D, Meyer TF. Tetrapac (tpc), a novel genotype of Neisseria gonorrhoeae affecting epithelial cell invasion, natural transformation competence and cell separation. Mol Microbiol. 1996;19(6):1357–72. doi: 10.1111/j.1365-2958.1996.tb02479.x. [DOI] [PubMed] [Google Scholar]
- 62.Chaussee MS, Hill SA. Formation of single-stranded DNA during DNA transformation of Neisseria gonorrhoeae. J Bacteriol. 1998;180(19):5117–22. doi: 10.1128/jb.180.19.5117-5122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Draskovic I, Dubnau D. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol Microbiol. 2005;55(3):881–96. doi: 10.1111/j.1365-2958.2004.04430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sox TE, Mohammed W, Sparling PF. Transformation-derived Neisseria gonorrhoeae plasmids with altered structure and function. J Bacteriol. 1979;138:510–8. doi: 10.1128/jb.138.2.510-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stein DC, Gunn JS, Radlinska M, Piekarowicz A. Restriction and modification systems of Neisseria gonorrhoeae. Gene. 1995;157(1–2):19–22. doi: 10.1016/0378-1119(94)00649-d. [DOI] [PubMed] [Google Scholar]
- 66.Eisenstein BI, Sox T, Biswas G, Blackman E, Sparling PF. Conjugal transfer of the gonococcal penicillinase plasmid. Science. 1977;195:998–1000. doi: 10.1126/science.402693. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50(4):488–503. doi: 10.1016/j.molcel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barrangou R. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip Rev RNA. 2013;4(3):267–78. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- 69.Koomey JM, Falkow S. Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J Bacteriol. 1987;169(2):790–5. doi: 10.1128/jb.169.2.790-795.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453(7194):489–4. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 71.Gruenig MC, Stohl EA, Chitteni-Pattu S, Seifert HS, Cox MM. Less is more: Neisseria gonorrhoeae RecX protein stimulates recombination by inhibiting RecA. J Biol Chem. 2010;285(48):37188–97. doi: 10.1074/jbc.M110.171967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mehr IJ, Seifert HS. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol Microbiol. 1998;30(4):697–710. doi: 10.1046/j.1365-2958.1998.01089.x. [DOI] [PubMed] [Google Scholar]
- 73.Stohl EA, Seifert HS. The recX gene potentiates homologous recombination in Neisseria gonorrhoeae. Mol Microbiol. 2001;40(6):1301–10. doi: 10.1046/j.1365-2958.2001.02463.x. [DOI] [PubMed] [Google Scholar]
- 74.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72(4):642–71. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kline KA, Seifert HS. Mutation of the priA gene of Neisseria gonorrhoeae affects DNA transformation and DNA repair. J Bacteriol. 2005;187(15):5347–55. doi: 10.1128/JB.187.15.5347-5355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marians KJ. PriA-directed replication fork restart in Escherichia coli. Trends Biochem Sci. 2000;25(4):185–9. doi: 10.1016/s0968-0004(00)01565-6. [DOI] [PubMed] [Google Scholar]
- 77.Mell JC, Redfield RJ. Natural competence and the evolution of DNA uptake specificity. J Bacteriol. 2014;31:31. doi: 10.1128/JB.01293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewis K. Programmed death in bacteria. Microbiol Mol Biol Rev. 2000;64(3):503–14. doi: 10.1128/mmbr.64.3.503-514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol. 2001;41(1):263–77. doi: 10.1046/j.1365-2958.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- 80.Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol. 2005;55(6):1704–21. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- 81.Snyder LA, Jarvis SA, Saunders NJ. Complete and variant forms of the ‘gonococcal genetic island’ in Neisseria meningitidis. Microbiology. 2005;151(Pt 12):4005–13. doi: 10.1099/mic.0.27925-0. [DOI] [PubMed] [Google Scholar]
- 82.Woodhams KL, Benet ZL, Blonsky SE, Hackett KT, Dillard JP. Prevalence and detailed mapping of the gonococcal genetic island in Neisseria meningitidis. J Bacteriol. 2012;194(9):2275–85. doi: 10.1128/JB.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dominguez NM, Hackett KT, Dillard JP. XerCD-mediated site-specific recombination leads to loss of the 57-kilobase gonococcal genetic island. J Bacteriol. 2011;193(2):377–88. doi: 10.1128/JB.00948-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Salgado-Pabón W, Jain S, Turner N, Van Der Does C, Dillard JP. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol Microbiol. 2007;66(4):930–47. doi: 10.1111/j.1365-2958.2007.05966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Salgado-Pabon W, Jain S, Turner N, van der Does C, Dillard JP. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol Microbiol. 2007;66(4):930–47. doi: 10.1111/j.1365-2958.2007.05966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramsey ME, Woodhams KL, Dillard JP. The Gonococcal Genetic Island and Type IV Secretion in the Pathogenic Neisseria. Frontiers in microbiology. 2011;2(61):00061. doi: 10.3389/fmicb.2011.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bhatty M, Laverde Gomez JA, Christie PJ. The expanding bacterial type IV secretion lexicon. Res Microbiol. 2013;164(6):620–39. doi: 10.1016/j.resmic.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462(7276):1011–5. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G. Structure of a type IV secretion system core complex. Science. 2009;323(5911):266–8. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nature reviews Microbiology. 2009;7(10):703–14. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kohler PL, Chan YA, Hackett KT, Turner N, Hamilton HL, Cloud-Hansen KA, et al. Mating pair formation homologue TraG is a variable membrane protein essential for contact-independent type IV secretion of chromosomal DNA by Neisseria gonorrhoeae. J Bacteriol. 2013;195(8):1666–79. doi: 10.1128/JB.02098-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chan YA, Hackett KT, Dillard JP. The lytic transglycosylases of Neisseria gonorrhoeae. Microb Drug Resist. 2012;18(3):271–9. doi: 10.1089/mdr.2012.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Leonard TA, Moller-Jensen J, Lowe J. Towards understanding the molecular basis of bacterial DNA segregation. Philos Trans R Soc Lond B Biol Sci. 2005;360(1455):523–35. doi: 10.1098/rstb.2004.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grinter NJ. Analysis of chromosome mobilization using hybrids between plasmid RP4 and a fragment of bacteriophage lambda carrying IS1. Plasmid. 1981;5(3):267–76. doi: 10.1016/0147-619x(81)90004-4. [DOI] [PubMed] [Google Scholar]
- 95.Salgado-Pabon W, Du Y, Hackett KT, Lyons KM, Arvidson CG, Dillard JP. Increased expression of the type IV secretion system in piliated Neisseria gonorrhoeae variants. J Bacteriol. 2010;192(7):1912–20. doi: 10.1128/JB.01357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zola TA, Strange HR, Dominguez NM, Dillard JP, Cornelissen CN. Type IV secretion machinery promotes ton-independent intracellular survival of Neisseria gonorrhoeae within cervical epithelial cells. Infect Immun. 2010;78(6):2429–37. doi: 10.1128/IAI.00228-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zweig MA, Schork S, Koerdt A, Siewering K, Sternberg C, Thormann K, et al. Secreted single-stranded DNA is involved in the initial phase of biofilm formation by Neisseria gonorrhoeae. Environ Microbiol. 2013;3(10):1462–2920. doi: 10.1111/1462-2920.12291. [DOI] [PubMed] [Google Scholar]
- 98.Takeuchi N, Kaneko K, Koonin EV. Horizontal Gene Transfer Can Rescue Prokaryotes from Muller’s Ratchet: Benefit of DNA from Dead Cells and Population Subdivision. G3. 2013;17(113):009845. doi: 10.1534/g3.113.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baumdicker F, Hess WR, Pfaffelhuber P. The infinitely many genes model for the distributed genome of bacteria. Genome Biol Evol. 2012;4(4):443–56. doi: 10.1093/gbe/evs016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vink C, Rudenko G, Seifert HS. Microbial antigenic variation mediated by homologous DNA recombination. FEMS Microbiol Rev. 2011;25(10):1574–6976. doi: 10.1111/j.1574-6976.2011.00321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van der Woude MW. Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol. 2011;14(2):205–11. doi: 10.1016/j.mib.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 102.Kline KA, Sechman EV, Skaar EP, Seifert HS. Recombination, repair and replication in the pathogenic Neisseriae: the 3 R’s of molecular genetics of two human-specific bacterial pathogens. Mol Microbiol. 2003;50(1):3–13. doi: 10.1046/j.1365-2958.2003.03679.x. [DOI] [PubMed] [Google Scholar]
- 103.Stern A, Brown M, Nickel P, Meyer TF. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell. 1986;47(1):61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- 104.Danaher RJ, Levin JC, Arking D, Burch CL, Sandlin R, Stein DC. Genetic basis of Neisseria gonorrhoeae lipooligosaccharide antigenic variation. J Bacteriol. 1995;177(24):7275–9. doi: 10.1128/jb.177.24.7275-7279.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jennings MP, Hood DW, Peak IR, Virji M, Moxon ER. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol Microbiol. 1995;18(4):729–40. doi: 10.1111/j.1365-2958.1995.mmi_18040729.x. [DOI] [PubMed] [Google Scholar]
- 106.Bos MP, Hogan D, Belland RJ. Homologue scanning mutagenesis reveals CD66 receptor residues required for neisserial Opa protein binding. J Exp Med. 1999;190(3):331–40. doi: 10.1084/jem.190.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gotschlich EC. Genetic locus for the biosynthesis of the variable portion of Neisseria gonorrhoeae lipooligosaccharide. J Exp Med. 1994;180(6):2181–90. doi: 10.1084/jem.180.6.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Meyer TF, Mlawer N, So M. Pilus expression in Neisseria gonorrhoeae involves chromosomal rearrangement. Cell. 1982;30(1):45–52. doi: 10.1016/0092-8674(82)90010-1. [DOI] [PubMed] [Google Scholar]
- 109.Hamrick TS, Dempsey JA, Cohen MS, Cannon JG. Antigenic variation of gonococcal pilin expression in vivo: analysis of the strain FA1090 pilin repertoire and identification of the pilS gene copies recombining with pilE during experimental human infection. Microbiology. 2001;147(Pt 4):839–49. doi: 10.1099/00221287-147-4-839. [DOI] [PubMed] [Google Scholar]
- 110.Haas R, Meyer TF. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell. 1986;44(1):107–15. doi: 10.1016/0092-8674(86)90489-7. [DOI] [PubMed] [Google Scholar]
- 111.Haas R, Veit S, Meyer TF. Silent pilin genes of Neisseria gonorrhoeae MS11 and the occurrence of related hypervariant sequences among other gonococcal isolates. Mol Microbiol. 1992;6(2):197–208. doi: 10.1111/j.1365-2958.1992.tb02001.x. [DOI] [PubMed] [Google Scholar]
- 112.Segal E, Hagblom P, Seifert HS, So M. Antigenic variation of gonococcal pilus involves assembly of separated silent gene segments. Proc Natl Acad Sci U S A. 1986;83(7):2177–81. doi: 10.1073/pnas.83.7.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, et al. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell. 2006;23(5):651–62. doi: 10.1016/j.molcel.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 114.Forest KT, Bernstein SL, Getzoff ED, So M, Tribbick G, Geysen HMX, et al. Assembly and antigenicity of the Neisseria gonorrhoeae pilus mapped with antibodies. Infect Immun. 1996;64(2):644–52. doi: 10.1128/iai.64.2.644-652.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Criss AK, Kline KA, Seifert HS. The frequency and rate of pilin antigenic variation in Neisseria gonorrhoeae. Mol Microbiol. 2005;58(2):510–9. doi: 10.1111/j.1365-2958.2005.04838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kellogg DS, Jr, Peacock WL, Jr, Deacon WE, Brown L, Pirkle DI. Neisseria gonorrhoeae. I. Virulence Genetically Linked to Clonal Variation. J Bacteriol. 1963;85:1274–9. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jonsson AB, Nyberg G, Normark S. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J. 1991;10(2):477–88. doi: 10.1002/j.1460-2075.1991.tb07970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Segal E, Billyard E, So M, Storzbach S, Meyer TF. Role of chromosomal rearrangement in N. gonorrhoeae pilus phase variation. Cell. 1985;40(2):293–300. doi: 10.1016/0092-8674(85)90143-6. [DOI] [PubMed] [Google Scholar]
- 119.Hagblom P, Segal E, Billyard E, So M. Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae. Nature. 1985;315(6015):156–8. doi: 10.1038/315156a0. [DOI] [PubMed] [Google Scholar]
- 120.Koomey M, Gotschlich EC, Robbins K, Bergstrom S, Swanson J. Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics. 1987;117(3):391–8. doi: 10.1093/genetics/117.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jennings MP, Jen FE, Roddam LF, Apicella MA, Edwards JL. Neisseria gonorrhoeae pilin glycan contributes to CR3 activation during challenge of primary cervical epithelial cells. Cell Microbiol. 2011;13(6):885–96. doi: 10.1111/j.1462-5822.2011.01586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Marceau M, Forest K, Beretti JL, Tainer J, Nassif X. Consequences of the loss of O-linked glycosylation of meningococcal type IV pilin on piliation and pilus-mediated adhesion. Mol Microbiol. 1998;27(4):705–15. doi: 10.1046/j.1365-2958.1998.00706.x. [DOI] [PubMed] [Google Scholar]
- 123.Chamot-Rooke J, Mikaty G, Malosse C, Soyer M, Dumont A, Gault J, et al. Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science. 2011;331(6018):778–82. doi: 10.1126/science.1200729. [DOI] [PubMed] [Google Scholar]
- 124.Miller F, Phan G, Brissac T, Bouchiat C, Lioux G, Nassif X, et al. The Hypervariable Region of Meningococcal Major Pilin PilE Controls the Host Cell Response via Antigenic Variation. mBio. 2014;5(1):01024–13. doi: 10.1128/mBio.01024-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mehr IJ, Seifert HS. Random shuttle mutagenesis: gonococcal mutants deficient in pilin antigenic variation. Mol Microbiol. 1997;23(6):1121–31. doi: 10.1046/j.1365-2958.1997.2971660.x. [DOI] [PubMed] [Google Scholar]
- 126.Sechman EV, Rohrer MS, Seifert HS. A genetic screen identifies genes and sites involved in pilin antigenic variation in Neisseria gonorrhoeae. Mol Microbiol. 2005;57(2):468–83. doi: 10.1111/j.1365-2958.2005.04657.x. [DOI] [PubMed] [Google Scholar]
- 127.Cahoon LA, Seifert HS. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science. 2009;325(5941):764–7. doi: 10.1126/science.1175653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stohl EA, Blount L, Seifert HS. Differential cross-complementation patterns of Escherichia coli and Neisseria gonorrhoeae RecA proteins. Microbiology. 2002;148(Pt 6):1821–31. doi: 10.1099/00221287-148-6-1821. [DOI] [PubMed] [Google Scholar]
- 129.Stohl EA, Gruenig MC, Cox MM, Seifert HS. Purification and characterization of the RecA protein from Neisseria gonorrhoeae. PloS one. 2011;6(2):0017101. doi: 10.1371/journal.pone.0017101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stohl EA, Brockman JP, Burkle KL, Morimatsu K, Kowalczykowski SC, Seifert HS. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J Biol Chem. 2003;278(4):2278–85. doi: 10.1074/jbc.M210496200. [DOI] [PubMed] [Google Scholar]
- 131.Mehr IJ, Long CD, Serkin CD, Seifert HS. A homologue of the recombination-dependent growth gene, rdgC, is involved in gonococcal pilin antigenic variation. Genetics. 2000;154(2):523–32. doi: 10.1093/genetics/154.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Drees JC, Chitteni-Pattu S, McCaslin DR, Inman RB, Cox MM. Inhibition of RecA protein function by the RdgC protein from Escherichia coli. J Biol Chem. 2006;281(8):4708–17. doi: 10.1074/jbc.M513592200. [DOI] [PubMed] [Google Scholar]
- 133.Hiom K. DNA Repair: Common Approaches to Fixing Double-Strand Breaks. Curr Biol. 2009;19(13):R523–R5. doi: 10.1016/j.cub.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 134.Skaar EP, Lazio MP, Seifert HS. Roles of the recJ and recN genes in homologous recombination and DNA repair pathways of Neisseria gonorrhoeae. J Bacteriol. 2002;184(4):919–27. doi: 10.1128/jb.184.4.919-927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Killoran MP, Kohler PL, Dillard JP, Keck JL. RecQ DNA helicase HRDC domains are critical determinants in Neisseria gonorrhoeae pilin antigenic variation and DNA repair. Mol Microbiol. 2009;71(1):158–71. doi: 10.1111/j.1365-2958.2008.06513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lane HE, Denhardt DT. The rep mutation. III. Altered structure of the replicating Escherichia coli chromosome. J Bacteriol. 1974;120(2):805–14. doi: 10.1128/jb.120.2.805-814.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kline KA, Seifert HS. Role of the Rep helicase gene in homologous recombination in Neisseria gonorrhoeae. J Bacteriol. 2005;187(8):2903–7. doi: 10.1128/JB.187.8.2903-2907.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chaussee MS, Wilson J, Hill SA. Characterization of the recD gene of Neisseria gonorrhoeae MS11 and the effect of recD inactivation on pilin variation and DNA transformation. Microbiology. 1999;145(Pt 2):389–400. doi: 10.1099/13500872-145-2-389. [DOI] [PubMed] [Google Scholar]
- 139.Hill SA, Woodward T, Reger A, Baker R, Dinse T. Role for the RecBCD recombination pathway for pilE gene variation in repair-proficient Neisseria gonorrhoeae. J Bacteriol. 2007;189(22):7983–90. doi: 10.1128/JB.00980-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Helm RA, Seifert HS. Pilin antigenic variation occurs independently of the RecBCD pathway in Neisseria gonorrhoeae. J Bacteriol. 2009;191(18):5613–21. doi: 10.1128/JB.00535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sechman EV, Kline KA, Seifert HS. Loss of both Holliday junction processing pathways is synthetically lethal in the presence of gonococcal pilin antigenic variation. Mol Microbiol. 2006;61(1):185–93. doi: 10.1111/j.1365-2958.2006.05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wainwright LA, Pritchard KH, Seifert HS. A conserved DNA sequence is required for efficient gonococcal pilin antigenic variation. Mol Microbiol. 1994;13(1):75–87. doi: 10.1111/j.1365-2958.1994.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 143.Howell-Adams B, Wainwright LA, Seifert HS. The size and position of heterologous insertions in a silent locus differentially affect pilin recombination in Neisseria gonorrhoeae. Mol Microbiol. 1996;22(3):509–22. doi: 10.1046/j.1365-2958.1996.00128.x. [DOI] [PubMed] [Google Scholar]
- 144.Howell-Adams B, Seifert HS. Insertion mutations in pilE differentially alter gonococcal pilin antigenic variation. J Bacteriol. 1999;181(19):6133–41. doi: 10.1128/jb.181.19.6133-6141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kuryavyi V, Cahoon LA, Seifert HS, Patel DJ. RecA-binding pilE G4 sequence essential for pilin antigenic variation forms monomeric and 5′ end-stacked dimeric parallel G-quadruplexes. Structure. 2012;20(12):2090–102. doi: 10.1016/j.str.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cahoon LA, Manthei KA, Rotman E, Keck JL, Seifert HS. The Neisseria gonorrhoeae RecQ helicase HRDC domains are essential for efficient binding and unwinding of the pilE guanine quartet structure required for pilin Av. J Bacteriol. 2013 doi: 10.1128/JB.02217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cahoon LA, Seifert HS. Transcription of a cis-acting, noncoding, small RNA is required for pilin antigenic variation in Neisseria gonorrhoeae. PLoS Pathog. 2013;9(1):e1003074. doi: 10.1371/journal.ppat.1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Seifert HS, Ajioka RS, Marchal C, Sparling PF, So M. DNA transformation leads to pilin antigenic variation in Neisseria gonorrhoeae. Nature. 1988;336(6197):392–5. doi: 10.1038/336392a0. [DOI] [PubMed] [Google Scholar]
- 149.Swanson J, Morrison S, Barrera O, Hill S. Piliation changes in transformation-defective gonococci. J Exp Med. 1990;171(6):2131–9. doi: 10.1084/jem.171.6.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang QY, DeRyckere D, Lauer P, Koomey M. Gene conversion in Neisseria gonorrhoeae: evidence for its role in pilus antigenic variation. Proc Natl Acad Sci U S A. 1992;89(12):5366–70. doi: 10.1073/pnas.89.12.5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tobiason DM, Seifert HS. The obligate human pathogen, Neisseria gonorrhoeae, is polyploid. PLoS Biol. 2006;4(6) doi: 10.1371/journal.pbio.0040185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Stabler RA, Marsden GL, Witney AA, Li Y, Bentley SD, Tang CM, et al. Identification of pathogen-specific genes through microarray analysis of pathogenic and commensal Neisseria species. Microbiology. 2005;151(Pt 9):2907–22. doi: 10.1099/mic.0.28099-0. [DOI] [PubMed] [Google Scholar]
- 153.Seifert HS, Wright CJ, Jerse AE, Cohen MS, Cannon JG. Multiple gonococcal pilin antigenic variants are produced during experimental human infections. J Clin Invest. 1994;93(6):2744–9. doi: 10.1172/JCI117290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kobayashi I. Mechanisms for gene conversion and homologous recombination: the double-strand break repair model and the successive half crossing-over model. Adv Biophys. 1992;28:81–133. doi: 10.1016/0065-227x(92)90023-k. [DOI] [PubMed] [Google Scholar]
- 155.Howell-Adams B, Seifert HS. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol Microbiol. 2000;37(5):1146–58. doi: 10.1046/j.1365-2958.2000.02067.x. [DOI] [PubMed] [Google Scholar]