Abstract
Chromosomal instability (CIN) is defined as the perpetual missegregation of whole chromosomes during mitosis and represents a hallmark of human cancer. However, the mechanisms causing CIN and its consequences on tumor growth are largely unknown. We identify an increase in microtubule plus end assembly rates as a fundamental trigger for CIN in CRC cells. This trigger is mediated by overexpression of the oncogene AURKA or by loss of the tumor suppressor gene CHK2, a genetic constitution found in 73% of human colorectal cancers. Increased microtubule assembly rates are associated with transient abnormalities in mitotic spindle geometry promoting the generation of lagging chromosomes and resulting in CIN. Reconstitution of proper microtubule assembly rates by chemical or genetic means suppresses CIN and thereby, unexpectedly, accelerates tumor growth in vitro and in vivo. Thus, we identify a fundamental mechanism triggering CIN in cancer cells and reveal its adverse consequence on tumor growth.
INTRODUCTION
Colorectal cancer represents a prime example for a tumor entity exhibiting CIN, which is thought to contribute to tumorigenesis, tumor progression and therapy resistance1–4. However, despite being such an important phenotype the causes leading to and the direct consequences resulting from CIN are poorly understood. Several mechanisms including a weakened spindle checkpoint, chromatid cohesion defects and also replication stress have been proposed to contribute to CIN, but none of these mechanisms can account for the widespread appearance of CIN in human cancer4–6. However, a major pre-stage of chromosome missegregation in cancer cells are so-called lagging chromosomes, which are the consequence of erroneous attachments of a single kinetochore to microtubules emanating from opposing spindle poles7,8. In general, those erroneous kinetochore attachments also occur stochastically in the early phases of a normal mitosis, but are usually corrected before anaphase onset8. However, the details of the correction process are not well understood8–12 and it appears that error correction is rarely inactivated in human cancer. Hence, other mechanisms must exist that cause the generation of lagging chromosomes. It is conceivable that increased rates of kinetochore-microtubule mal-attachments might overwhelm the capacity of the cellular error correction machinery. Indeed, this has been demonstrated for a subset of cancer cells exhibiting supernumerary centrosomes. In these cases, transient multipolar spindle intermediates can facilitate the generation of erroneous kinetochore attachments and thus, promote the generation of lagging chromosomes despite the presence of error correction13,14. However, since the majority of cancer cells do not exhibit supernumerary centrosomes15, additional, yet unknown mechanisms promoting the generation of lagging chromosomes are likely to exist. Interestingly, in human cancer cells a number of established oncogenes and tumor suppressor genes are implicated in proper mitotic chromosome segregation16. For instance, Plk1 and Aurora-A are centrosomal kinases involved in mitotic spindle assembly17, but the consequences resulting from their overexpression, which is often detected in human cancer cells18,19, is unclear. Similarly, we and others previously reported on a requirement of the tumor suppressors CHK2 and BRCA1 for normal mitotic chromosome segregation20,21. The mechanism of how the overexpression of these oncogenes or the loss of these tumor suppressor genes causes CIN remained an open question.
RESULTS
Increased mitotic microtubule plus end assembly rates are characteristic for colorectal cancer cells exhibiting CIN
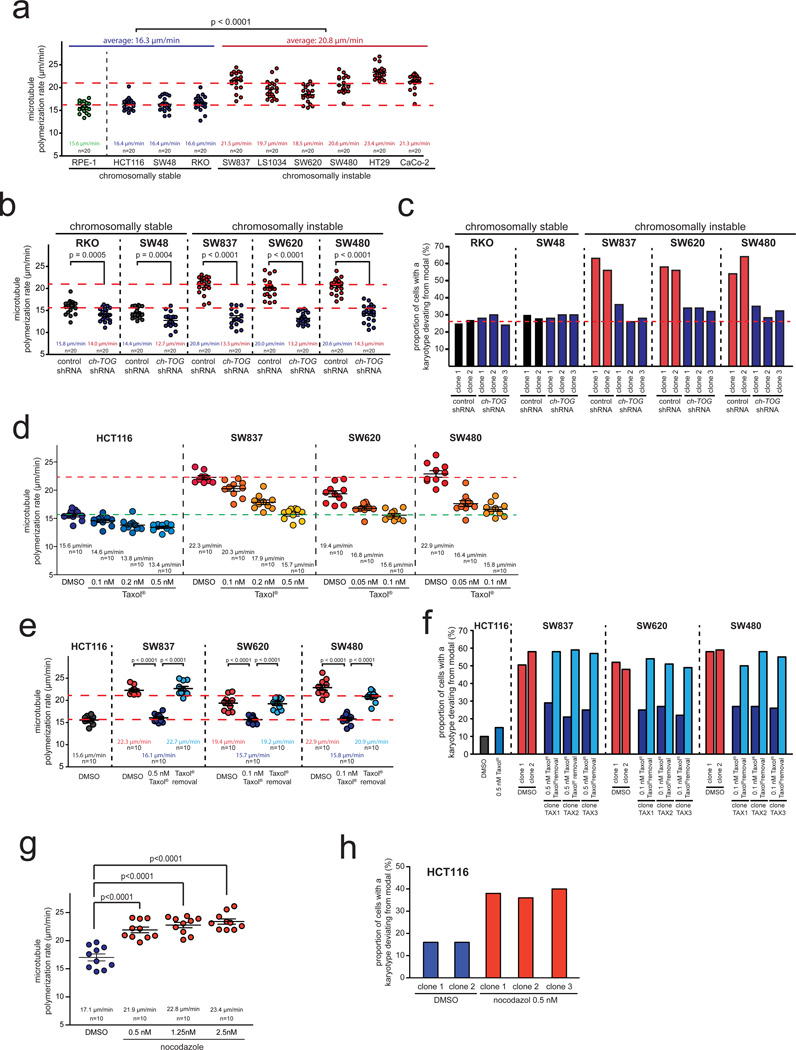
We addressed the role of microtubule assembly dynamics within mitotic spindles as a possible and yet unexplored cause for CIN in human cancer cells. To this end, we measured microtubule plus end assembly rates in live cells during mitosis by tracking the microtubule end binding protein EB3 fused to GFP22. We used a panel of colorectal cancer (CRC) cells, which can be categorized into chromosomally stable MIN/MSI cell lines with a near diploid karyotype (HCT116, SW48 and RKO) and cell lines exhibiting CIN (SW837, LS1034, SW620, SW480, HT29, CaCo-2). To ensure comparable measurements of the various cell lines, we synchronized cells in mitosis by using the small molecule inhibitor dimethylenastron (DME23) targeting the mitotic kinesin Eg5/Kif11, which resulted in the formation of monopolar spindles24. Neither this synchronization step nor the expression level of EB3-GFP influenced microtubule plus end assembly rates (Supplementary Fig. S1a, S1b, S2e). Intriguingly, we found that all CIN cell lines exhibited significantly increased microtubule assembly rates when compared to MIN/MSI cell lines or to non-transformed human RPE-1 cells (Fig. 1a) suggesting that abnormal microtubule plus end assembly rates might be linked to CIN.
Figure 1.
Increased mitotic microtubule assembly rates are a common characteristic of chromosomally instable CRC cells and mediate numerical chromosome instability. a, Measurement of mitotic microtubule plus end assembly rates in various CRC cell lines expressing EB3-GFP. Scatter dot plots show average assembly rates based on measurements of 20 microtubules per cell (mean +/− SEM, t-test, n=20 cells). b, Measurement of mitotic microtubule plus end assembly rates in stable CRC cell lines expressing control or shRNAs targeting CH-TOG/CKAP5. Scatter dot plots show average assembly rates (20 microtubules/cell, mean +/− SEM, t-test, n=20 cells). c, Determination of chromosome number variability in single cell clones shown in b. The proportion of cells showing chromosome numbers deviating from the modal were determined after 30 generations (n=50–103 cells). d, Measurement of mitotic microtubule plus end assembly rates after titration of sub-nanomolar doses of Taxol®. Scatter dot plots show average assembly rates (20 microtubules/cell; mean +/− SEM, n=10 cells). e, Measurement of mitotic microtubule plus end assembly rates of single cell clones derived from CRC cell lines and grown in the absence or presence of low doses of Taxol® or after subsequent removal of Taxol®. Scatter dot plots show average assembly rates (20 microtubules/cell, mean +/− SEM, t-test, n=10 cells). f, Determination of chromosome number variability in single cell clones generated as in e. The proportion of cells showing chromosome number deviations from the modal were determined after 30 generations (n=100–109 cells). g, Measurement of mitotic microtubule plus end assembly rates in chromosomally stable HCT116 cells after treatment with low doses of nocodazole. Scatter dot plots show average assembly rates (20 microtubules/cell, mean +/− SEM, t-test, n=10 cells). h, Determination of chromosome number variability in single cell clones generated in the absence or presence of low dose nocodazole. The proportion of cells showing chromosome number deviations from the modal were determined after 30 generations (n=50 cells).
Detailed data on karyotype analyses can be found in the Supplementary Table S1. Statistic source data for Figure 1 can be found in the Supplementary Table S2.
Increased mitotic microtubule plus end assembly rates cause CIN
To investigate the relationship between increased microtubule assembly rates and CIN we restored normal microtubule assembly rates in CIN cells by partially lowering the expression of the microtubule polymerase ch-TOG/CKAP5 (Supplementary Fig. S1c), which mediates the assembly of α/β-tubulin subunits at microtubule plus ends25,26. Live cell analyses of stable cell lines demonstrated that partial repression of CH-TOG/CKAP5 was sufficient to restore normal microtubule assembly rates in CIN cells to a level typically seen in chromosomally stable cells without affecting cell viability or normal cell cycle progression (Fig. 1b and data not shown). Most importantly, karyotype analyses using chromosome counting and interphase FISH revealed a significant reduction of karyotype variability and thus, of CIN after restoration of normal microtubule plus end assembly rates (Fig. 1c, Supplementary Fig. S1d, Supplementary Table S1). These results indicate that increased microtubule plus end assembly rates can trigger CIN in cancer cells.
Drug mediated alterations in mitotic microtubule plus end assembly rates affect karyotype stability
As another independent approach to restore normal microtubule assembly rates in CIN cells we used Taxol®, a microtubule binding drug known to suppress microtubule assembly, preferentially at the plus ends27–29. We identified sub-nanomolar concentrations of Taxol® that were sufficient to suppress the increased microtubule assembly rates in different CIN cell lines without affecting cell viability or normal cell cycle progression (Fig. 1d, Fig. 1e, Supplementary Fig. S1e). Most strikingly, low dose Taxol® treatment significantly suppressed CIN (Fig. 1f, Supplementary Fig. S1f, Supplementary Table S1). Remarkably, removal of Taxol® re-induced increased microtubule plus end assembly rates and CIN in the same single cell clones (Fig. 1e, Fig. 1f, Supplementary Table S1). In addition, we used sub-nanomolar concentrations of nocodazole, a microtubule binding drug known to have opposite effects on microtubule dynamics compared to Taxol®30, and detected an increase in microtubule assembly rates and an induction of CIN in otherwise chromosomally stable HCT116 cells (Fig. 1h, Supplementary Table S1). Together, these results indicate that subtle alterations in microtubule plus end assembly rates are sufficient to directly affect the numerical karyotype stability in cancer cells.
Overexpression of the oncogene AURKA or loss of the tumor suppressor gene CHK2 causes CIN by increasing mitotic microtubule assembly rates
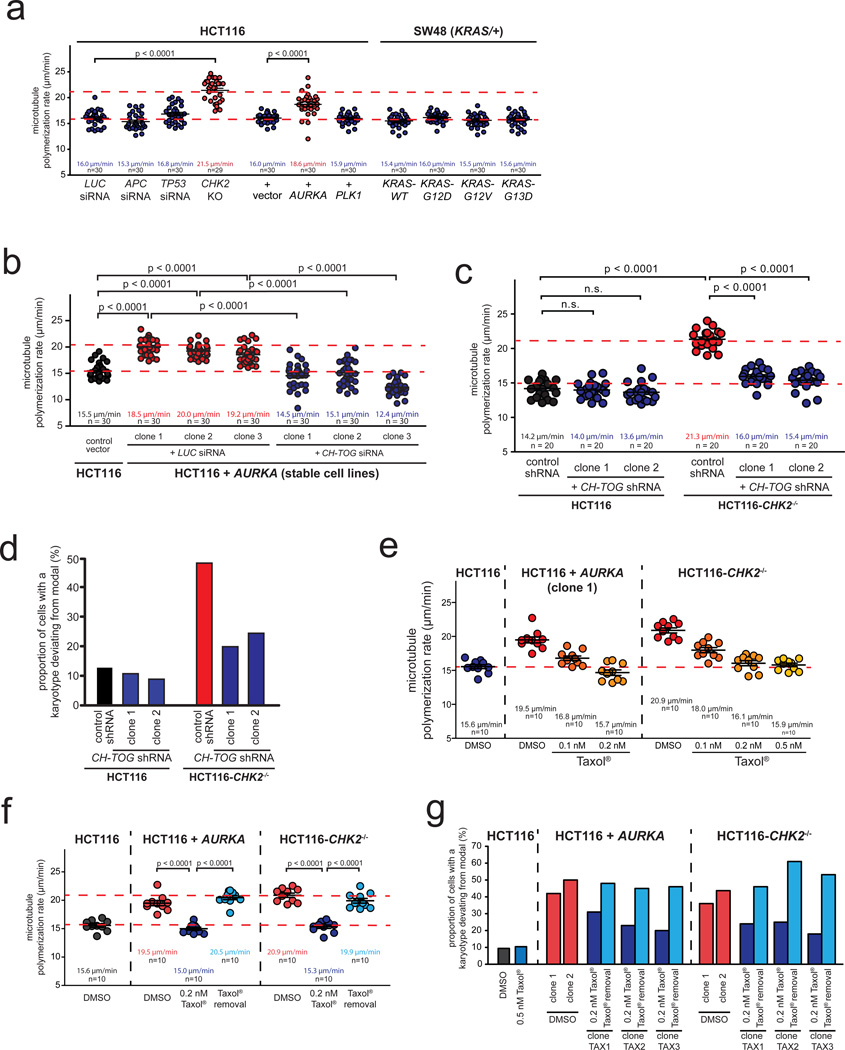
To identify cancer-relevant genetic lesions that confer increased microtubule assembly rates we investigated the role of the most frequent genetic alterations found in CRC (Supplementary Fig. S2a) previously implicated in mitotic processes 18,19,31–35. Live cell analyses of cells engineered to harbor these different genetic alterations (Supplementary Fig. S2b) showed that the overexpression of AURKA or loss of CHK2 increased microtubule assembly rates to a level typically found in chromosomally instable CRC cell lines (Fig. 2a). Moreover, single cell clones derived from stable cell lines exhibiting these genetic lesions evolved a high karyotype variability and thus, CIN (Fig. 2b, Fig 2c, Fig. 2d, Supplementary Fig. S2b–e, Supplementary Table S1). However, the increase in microtubule assembly rates were neither associated with supernumerary centrosomes (Supplementary Fig. S2f) nor with overt changes in other microtubule dynamics parameters such as overall dynamicity, frequency of catastrophe events or time spent paused (Supplementary Fig. 2g–i). Importantly, restoration of normal microtubule plus end assembly rates mediated by a partial repression of CH-TOG/CKAP5 (Fig. 2b, Fig. 2c, Supplementary Fig. S3a, S3b) or by treatment with sub-nanomolar doses of Taxol® (Fig. 2e, Fig. 2f) resulted in a significant suppression of CIN (Fig. 2d, Fig. 2g, Supplementary Fig. S3c–e, Supplementary Table S1). Thus, we demonstrate that overexpression of the oncogene AURKA or loss of the tumor suppressor gene CHK2 is sufficient to induce CIN in colorectal cancer cells by mediating increased microtubule assembly rates.
Figure 2.
Identification of AURKA overexpression and CHK2 loss as lesions triggering increased microtubule assembly rates in CRC cells. a, Measurement of mitotic microtubule plus end assembly rates in HCT116 cell harboring the indicated genetic alterations. Scatter dot plots show average assembly rates (20 microtubules/cell, mean +/− SEM, t-test, n=29–30 cells). b, Measurement of mitotic microtubule plus end assembly rates in cells stably overexpressing AURKA and transfected with control or CH-TOG/CKAP5 siRNAs. Scatter dot plots show average assembly rates based on measurements of 20 microtubules per cell (mean +/− SEM, t-test, n=30 cells). c, Measurement of mitotic microtubule plus end assembly rates in CHK2 deficient cells stably transfected with shRNAs targeting CH-TOG/CKAP5. Scatter dot plot show average assembly rates based on measurements of 20 microtubules per cell (mean +/−SEM, t-test, n=20 cells). d, Karyotype analyses of single cell clones derived from HCT116 and HCT116-CHK2−/− stably repressing CH-TOG/CKAP5. The proportion of cells showing chromosome number deviations from the modal were determined after 30 generations (n=50 cells). e, Measurement of mitotic microtubule plus end assembly rates of HCT116 cells overexpressing AURKA or deficient for CHK2 after titration of sub-nanomolar doses of Taxol®. Scatter dot plots show average assembly rates (20 microtubules/cell; mean +/− SEM, n=10 cells). f, Measurement of mitotic microtubule plus end assembly rates in single cell clones derived from HCT116 cells stably overexpressing AURKA or deficient for CHK2 and grown in the absence, presence or after removal of low dose Taxol®. Scatter dot plots show average growth rates (20 microtubules/cell, mean +/− SEM, t-test, n=10 cells). g, Determination of chromosome number variability in single cell clones shown in f. The proportion of cells showing chromosome numbers deviating from the modal were determined (n=100–103 cells).
Detailed data on karyotype analyses can be found in the Supplementary Table S1. Statistic source data for Figure 2 can be found in the Supplementary Table S2.
Increased microtubule assembly rates cause transient mitotic spindle geometry defects
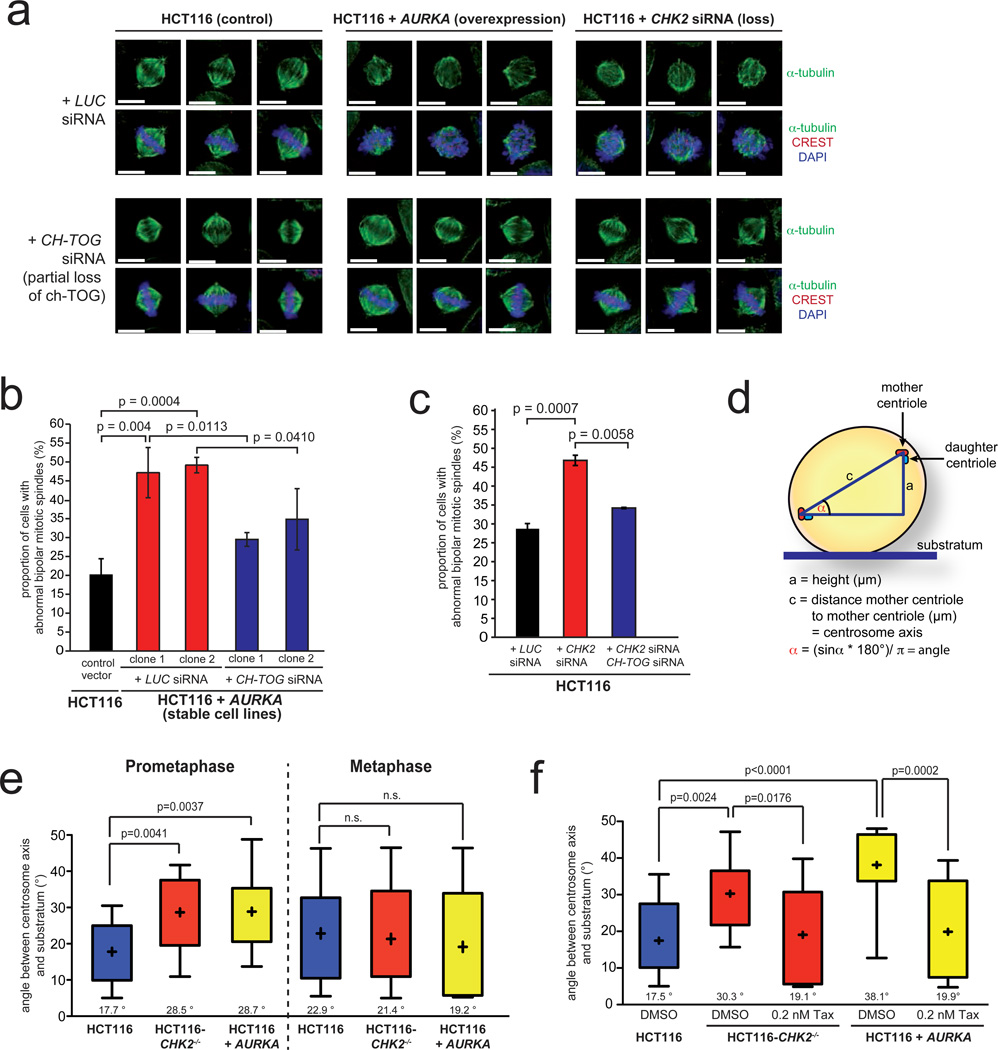
To investigate the link between increased microtubule assembly rates and chromosome missegregation we analyzed the structure of mitotic spindles in cell lines overexpressing AURKA or showing a loss of CHK2. Interestingly, we found abnormal mitotic spindle structures, which were suppressed when normal microtubule assembly rates were restored upon partial repression of CH-TOG/CKAP5 (Fig. 3a–c). Further live cell microscopy analyses revealed that the spindle structure abnormalities were transient and associated with a timely delay in chromosome alignment. Importantly, cells eventually achieved full alignment of chromosomes and initiated anaphase, albeit at the expense of chromosome missegregation (see example in: Movies S1 and S2). The transient nature of spindle abnormalities in those cells prompted us to analyze the dynamic positioning of the spindle axes by detecting GFP-centrin as a marker for centrosomes in live cells (Fig. 3d). In fact, during late prometaphase when the distance of the two centrosomes was maximal (Movies S3 and S4) we found significantly increased angles of spindle axes and thus, a misalignment of the mitotic spindle in cells exhibiting increased microtubule assembly rates (Fig. 3e). This spindle mispositioning was no longer apparent in metaphase (Fig. 3e) supporting our initial observation of transient spindle geometry abnormalities in fixed cells. Remarkably, restoration of normal microtubule assembly rates by low dose Taxol® treatment suppressed the spindle mispositioning (Fig. 3f) indicating that an increase in microtubule assembly rates can account for transient spindle geometry abnormalities during the early phases of mitosis.
Figure 3.
Increased microtubule assembly rates result in abnormal mitotic spindle geometry. a, Examples of metaphase spindle structures from cells overexpressing AURKA or deficient for CHK2 with or without partial repression of CH-TOG/CKAP5 (α-tubulin, green; CREST/kinetochores, red; DAPI, blue; scale bar: 10 µm). b, Quantification of abnormal bipolar metaphase spindle structures in HCT116 cells stably overexpressing AURKA before and after partial depletion of CH-TOG/CKAP5. The graphs show mean values +/− SD, t-test (n=240–1100 mitotic cells). c, Quantification of abnormal bipolar metaphase spindle structures in HCT116 cells after depletion of CHK2 and after concomitant repression of CH-TOG/CKAP5. The graphs show mean values +/−SD, t-test (n=1000–1300 mitotic cells). d, Schematic depiction of the experimental measurement of spindle axes positioning. e, Determination of angles of the spindle axes in cells overexpressing AURKA or deficient for CHK2 in prometaphase and metaphase cells. The box and whisker plots show the range, mean and quartil of the measurements (t-test, n=15 cells, prometaphase; n=9–11 cells, metaphase). f, Determination of angles of the spindle axes in prometaphase cells before and after reconstitution of normal microtubule assembly rates by low dose Taxol® treatment. The box and whisker plot shows the range, mean and quartil of the measurements (t-test, n=13–16 cells per group).
Statistic source data for Figure 3 can be found in the Supplementary Table S2.
Increased microtubule plus end assembly rates promote the generation of lagging chromosomes without interfering with error correction
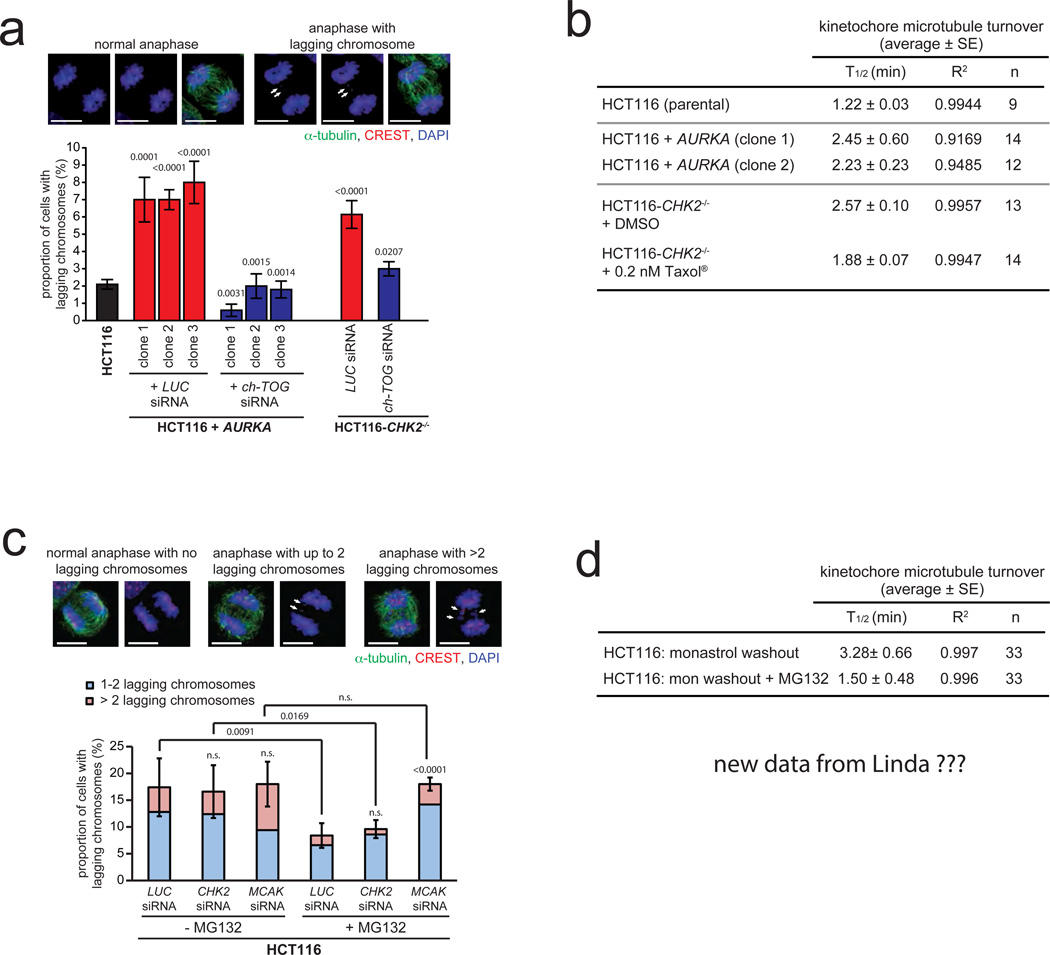
We investigated whether an increase in microtubule assembly associated with transient spindle geometry abnormalities might represent a cancer-relevant mechanism for the generation of lagging chromosomes. Indeed, the number of lagging chromosomes during anaphase was significantly increased in cells with increased microtubule assembly rates and suppressed when normal microtubule assembly rates were restored (Fig. 4a). Since lagging chromosomes can result from an impairment of error correction that is associated with increased microtubule-kinetochore attachment stability 8,10,36,37, we determined the microtubule-kinetochore attachment stability by measuring microtubule turnover in live cells (Supplementary Fig. S4a–c). Intriguingly, we found hyper-stable microtubule-kinetochore attachments in cells with increased microtubule assembly rates, which were suppressed by restoration of proper microtubule assembly (Fig. 4b). To test the possibility that an increased microtubule assembly rates might interfere with error correction, we induced erroneous kinetochore attachments by a monastrol washout procedure, followed by a release of these cells into medium containing the proteasome inhibitor MG132 in order to provide extra time for error correction by prolonging metaphase as shown previously38. Importantly, cells with increased microtubule assembly rates were able to efficiently reduce the number of lagging chromosomes during anaphase, whereas MCAK-depleted cells, which are impaired in error correction9–12, showed the persistence of lagging chromosomes even after prolonging the time for error correction (Fig. 4c, Supplementary Fig. S4d). Hence, despite the presence of hyper-stable microtubule-kinetochore attachments, the increase in microtubule plus end assembly rates as a trigger for CIN do not interfere with error correction. Vice versa, both, depletion of MCAK or APC, which reduces the turnover of kinetochore-microtubules10,37 or overexpression of MCAK in CIN cells, which promotes error correction10, caused no alteration in microtubule plus end assembly rates (Supplementary Fig. S4e–h). Thus, increased microtubule assembly rates and impaired error correction, both of which leading to lagging chromosomes and CIN are separable mechanisms. Since both conditions are associated with hyper-stable microtubule-kinetochore attachments we asked whether this might be associated with erroneous kinetochore attachments per se. To test this, we analyzed kinetochore-microtubule turnover in chromosomally stable HCT116 cells after release from a monastrol block in the presence or absence of MG132. Although the microtubule assembly rates were not changed (Supplementary Fig. S4i), the half-life of kinetochore microtubules was significantly increased upon induction of erroneous attachments and clearly suppressed after extended error correction (Fig. 4d, Supplementary Fig. S4j) suggesting that erroneous attachments might be associated with a higher stability per se. Thus, we conclude that increased microtubule assembly rates and impairment of error correction are separable mechanisms, both of which are associated with hyper-stability of erroneous microtubule-kinetochore attachments leading to the generation of lagging chromosomes.
Figure 4.
Increased microtubule assembly rates promote the generation of lagging chromosomes without interfering with error correction. a, Quantification of the proportion of cells exhibiting lagging chromosomes using HCT116 cells overexpressing AURKA or deficient for CHK2 and transfected with siRNAs targeting CH-TOG/CKAP5. Representative examples are given (scale bar, 10 µm). The graph shows mean values +/− SEM (t-test, n= 230–350 anaphase cells). b, Summary of the measurements of half-lifes of kinetochore-microtubule turnover in the indicated cell lines expressing photoactivatable GFP-tubulin (PA-GFP-tubulin). The addition of low concentrations of Taxol® reversed the observed hyper-stabilization of kinetochore microtubule attachments in CHK2 deficient cells (average +/− SE, n=9–14 cells as indicated). c, Quantification of the proportion of HCT116 cells showing lagging chromosomes after washout of monastrol and prolonging metaphase by MG132 treatment in the presence or absence of Chk2 or MCAK. The graph shows mean values +/− SD for cells exhibiting 1–2 and more than 2 lagging chromosomes per anaphase (t-test, n= 500 anaphase cells). d, Summary of kinetochore-microtubule turnover measurements in HCT116 cells expressing PA-GFP-tubulin immediately after establishing bipolar spindles upon release from a monastrol block and after release from monastrol into MG132 to prolong time for error correction (average +/− SE, n=33 cells for each condition).
Statistic source data for Figure 4 can be found in the Supplementary Table S2.
The CHK2-BRCA1 tumor suppressor pathway negatively regulates the oncogene AURKA to ensure proper microtubule assembly rates
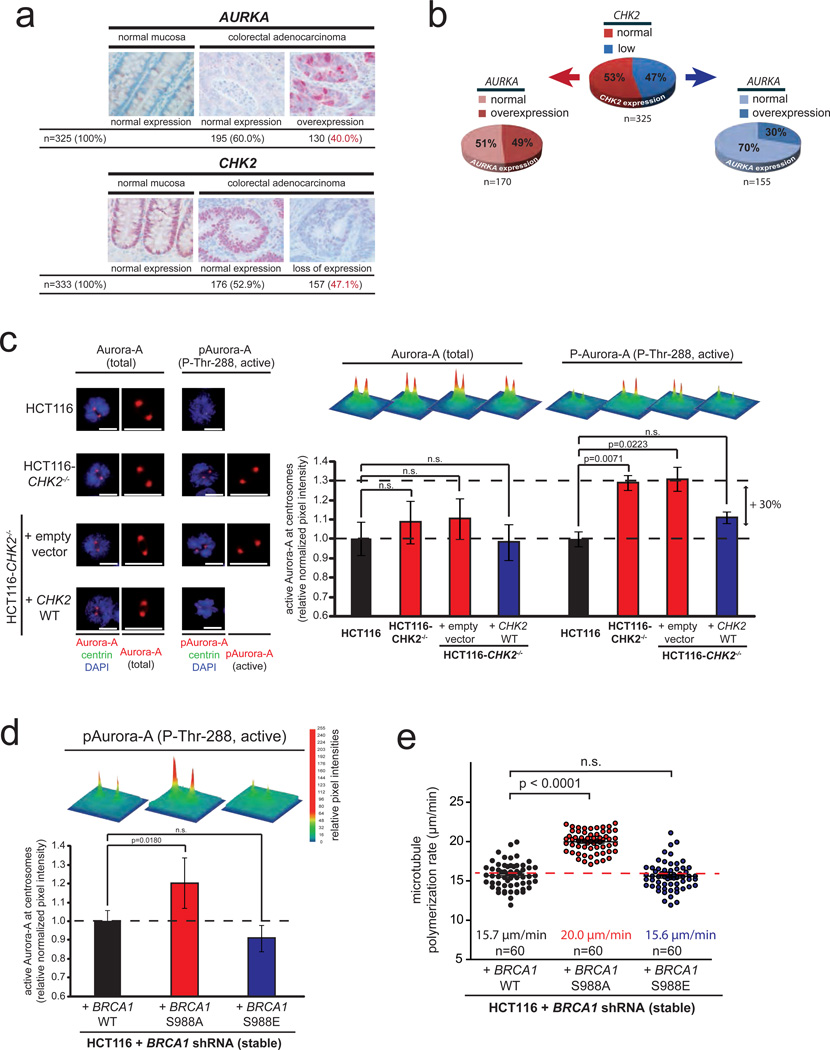
To investigate the cancer relevance of AURKA overexpression and loss of CHK2 as key mediators of increased microtubule assembly rates we determined their expression in tumor tissues from colorectal cancer (n=325–333 patients). We found a 40% frequency of AURKA overexpression and a 47% frequency for the loss of CHK2 (Fig. 5a), which ranges among the most frequent lesions detected so far in human colorectal cancer31. Based on 325 tumor samples, which could be used to simultaneously detect AURKA and CHK2 expression, we calculated a proportion of 73% of the cases, which exhibited at least one of the two lesions (Fig. 5b). Moreover, individual tumor specimens exhibiting a loss of CHK2 were found to be preferentially devoid of concomitant overexpression of AURKA (Fig. 5b). Similarly, in a panel of colorectal cancer cell lines increased microtubule assembly rates and CIN correlated with either an overexpression of AURKA or a loss of CHK2 (Supplementary Fig. S5a) suggesting possible redundant functions of these two genetic lesions. This prompted us to investigate a cross-talk of Aurora-A and Chk2. Not surprisingly, overexpression of AURKA resulted in increased levels of active and auto-phosphorylated Aurora-A kinase39 localized at mitotic centrosomes, which was associated with increased phosphorylation of the centrosomal Aurora-A target protein TACC340 (Supplementary Fig. S5b, S5c). Intriguingly, however, loss of CHK2 resulted in a very similar increase in the proportion of active Aurora-A at mitotic centrosomes and an increase in TACC3 phosphorylation without altering the overall level of Aurora-A (Fig. 5c, Supplementary Fig. S5d–g). Since the tumor suppressor Brca1 and more specifically, the Chk2-mediated phosphorylation of Brca1 (on serine-988) was shown to be crucial for proper mitotic progression20,21 we tested whether Chk2 might act on Aurora-A via Brca1. Indeed, cells expressing a non-phosphorylatable BRCA1 mutant20,41 (S988A; Supplementary Fig. S6a) showed an increase in active Aurora-A at mitotic centrosomes, which was accompanied with an increase in microtubule assembly rates (Fig. 5d, Fig. 5e). Moreover, in co-immunoprecipitation experiments we detected an interaction of active Aurora-A with Brca1, which was strongly enhanced upon loss of CHK2 or loss of the Chk2-mediated phosphorylation site on Brca1 while the interaction of Aurora-A with its co-factor TPX2 was not affected (Supplementary Fig. S6b–c). These results suggest that the tumor suppressor Chk2 restrains the activity of the oncogenic Aurora-A kinase during mitosis via the phosphorylation of the tumor suppressor protein Brca1 and indicate that the increase in Aurora-A activity might be a key trigger for increased microtubule plus end assembly rates and thus, for CIN in colorectal cancer cells.
Figure 5.
The CHK2-BRCA1 tumor suppressor pathway negatively regulates the oncogene AURKA to ensure proper microtubule plus end assembly rates. a, Detection of Chk2 and Aurora-A proteins by immunohistochemistry analyses in tissue sections from normal mucosa and from colorectal adenocarcinomas. Examples and overall quantifications are given. b, Depiction of the relationship of CHK2 and AURKA expression in CRC. Individual tumor samples were subdivided due to their CHK2 expression status and subsequently analyzed for their AURKA expression (n=325 tissue samples, t-test, p=0.001). c, Detection of total and active centrosomal Aurora-A (P-Thr-288) and centrin in prometaphase cells proficient or deficient for CHK2. Signal intensities were normalized to signals for centrosomal centrin and are depicted as 3D surface plots and were quantified (mean +/− SEM, t-test, n=49–50 cells for total Aurora-A and n=72–75 cells for P-Thr-288-Aurora-A). d, Detection of increased active centrosomal Aurora-A in mitotic HCT116 cells in which the endogenous Brca1 protein was replaced by the indicated BRCA1 mutants. Signal intensities for active Aurora-A (pThr-288) at mitotic centrosomes normalized to signals obtained for centrosomal centrin are depicted as 3D surface plots and were quantified (mean, +/− SEM, t-test, 55–57 cells). e, Measurement of mitotic microtubule plus end assembly rates in cells expressing either wild type or mutant BRCA1. Scatter dot plots show average growth rates based on measurements of 20 microtubules per cell (mean +/− SEM; t-test, n=60 cells).
Statistic source data for Figure 5 can be found in the Supplementary Table S2.
Increased Aurora-A kinase activity is a key trigger for increased microtubule assembly rates and CIN in colorectal cancer cells
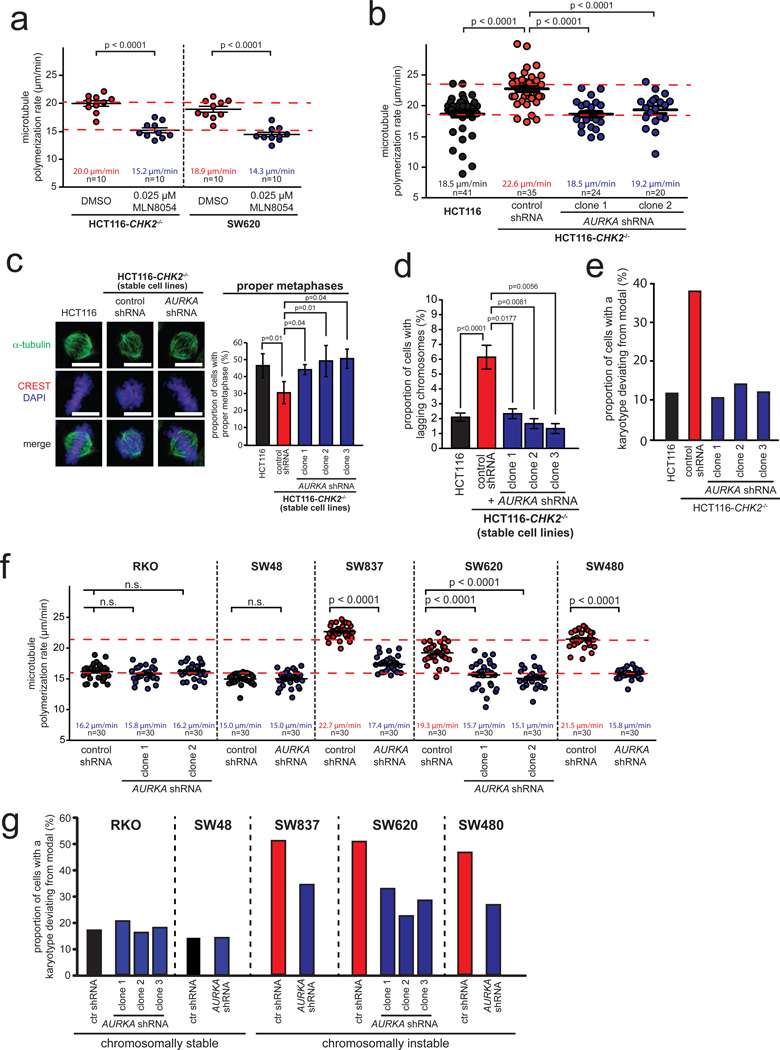
To test whether increased Aurora-A activity is a general trigger for increased microtubule assembly rates in CRC cells we partially inhibited Aurora-A in CHK2 deficient HCT116 and SW620 cells using the small molecule inhibitor MLN805442, which was indeed sufficient to suppress microtubule assembly rates (Fig. 6a). In addition, we generated stable cell lines derived from CHK2 deficient HCT116 cells that expressed reduced levels of AURKA. This partial loss of Aurora-A restorede its proper centrosomal activity level (Supplementary Fig. S7a–c), restored normal microtubule assembly rates (Fig. 6b), normal mitotic spindle geometry (Fig. 6c), timely mitotic progression (Supplementary Fig. S7d) and suppressed microtubule-kinetochore hyper-stability (Supplementary Fig. S7e). Consequently, the generation of lagging chromosomes was inhibited (Fig. 6d), which resulted in a striking suppression of CIN (Fig. 6e, Supplementary Fig. S7f, Supplementary Table S1). Importantly, partial repression of AURKA was also sufficient to restore normal microtubule assembly rates and to suppress CIN in various chromosomally instable CRC cell lines irrespective of their genetic constitution while not affecting non-CIN cells (Fig. 6f, Fig. 6g, Supplementary Fig. S7g, S7h, Supplementary Table S1). Thus, these findings demonstrate that increased microtubule growth rates resulting in CIN and commonly seen in chromosomally instable CRC cells are dependent on elevated Aurora-A kinase activity. Moreover, CIN in those cancer cells can be experimentally suppressed by different means including treatment with low dose Taxol®, partial loss of the microtubule polymerase ch-TOG and partial inhibition of Aurora-A.
Figure 6.
Increased Aurora-A kinase activity is a key trigger for increased microtubule assembly rates and CIN in CRC cells. a, Measurement of mitotic microtubule plus end assembly rates in CRC cells after partial inhibition of Aurora-A by low dose MLN8054 treatment. Scatter dot plots show average growth rates (20 microtubules/cell, mean +/− SEM, t-test, n=10 cells). b, Measurement of mitotic microtubule plus end assembly rates in CHK2 deficient cells after partial repression of AURKA. Scatter dot plots show average microtubule growth rates (20 microtubules/cell; mean +/− SEM, t-test, n=20–41 cells). c, Quantification of proper metaphase spindles and complete chromosome alignment in CHK2 deficient cells after restoration of proper microtubule assembly rates by partial repression of AURKA. Representative mitotic spindles are shown (scale bar, 10 µm) and proper metaphase spindles with completed chromosome alignment were quantified (mean +/− SD, t-test, n=200–2,500 mitotic cells). d, Detection of lagging chromosomes during anaphase in HCT116 and HCT116-CHK2−/− cells with or without partial repression of AURKA (mean +/− SD; t-test, n=300 anaphase cells). e, Determination of chromosome number variability in single cell clones derived from CHK2 deficient cells with partial repression of AURKA. The proportion of cells exhibiting chromosome numbers deviating from the modal were determined (n=91–100 cells). f, Measurement of mitotic microtubule plus end assembly rates in various chromosomally instable CRC cell lines stably expressing control or AURKA shRNAs. Average microtubule growth rates were determined (20 microtubules/cell, mean +/− SEM, t-test, n=30 cells). g, Chromosome number variability in single cell clones derived from various CRC cell lines and stably expressing control or AURKA shRNAs. The proportion of cells exhibiting chromosome numbers deviating from the modal were determined (n=81–104 cells).
Detailed data on karyotype analyses can be found in the Supplementary Table S1. Statistic source data for Figure 6 can be found in the Supplementary Table S2.
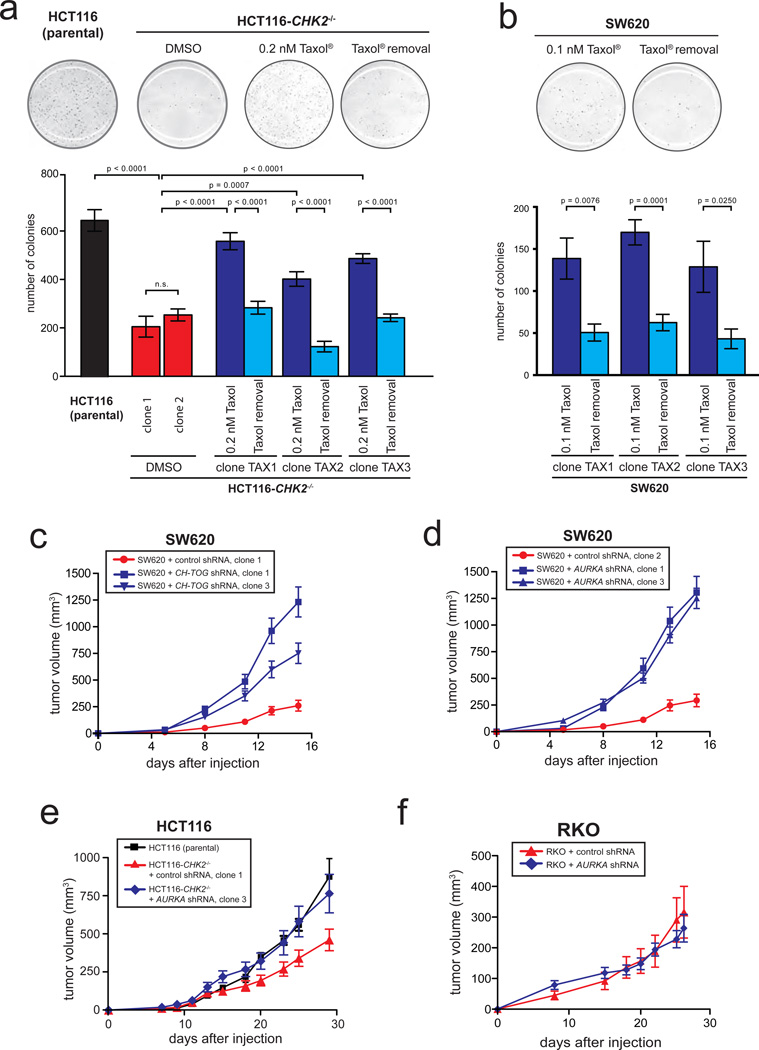
Restoring proper microtubule assembly rates associated with a suppression of CIN accelerates tumor growth in vitro and in vivo
The identification of increased microtubule plus end assembly rates as a trigger for CIN in CRC cells and the ability to suppress this trigger by various means enabled us to address the long-standing question for the impact of CIN on tumor growth. As a first step into this direction, we evaluated colony formation in soft agar, which reflects anchorage independent cell growth in tumors. Surprisingly, CHK2 deficient HCT116 and SW620 cells, in which increased microtubule assembly rates and CIN were suppressed either by treatment with low-dose Taxol®, by partial repression of CH-TOG/CKAP5 or by partial loss of AURKA showed a significantly higher rate of colony formation in soft agar when compared to the respective cells with high CIN (Fig. 7a, Fig. 7b, Supplementary Fig. S8a, S8b, S8c). Consequently, we investigated the consequence of CIN triggered by an increase in microtubule assembly rates on tumor growth in vivo. Astonishingly, xenograft tumors derived from SW620 cells with restored microtubule assembly rates and suppressed CIN, mediated either by partial repression of CH-TOG/CKAP5 or AURKA, exhibited an acceleration of tumor growth in vivo when compared to tumors showing increased microtubule assembly rates triggering CIN (Fig. 4c, 4d; Supplementary Fig. S8d, S8e). Analyses of cells re-isolated from these xenograft tumors showed the persisting maintenance of CIN suppression (Supplementary Fig. S8g–i). Moreover, direct comparison of xenograft tumors derived from isogenic HCT116 cells revealed that loss of CHK2, which is associated with increased microtubule assembly rates and CIN, caused reduced tumor growth when compared to tumor formation from parental cells (Fig. 7e). However, restoration of normal microtubule assembly rates and chromosomal stability in those cells accelerated tumor growth to a rate very similar to that of the chromosomally stable parental HCT116 cells (Fig. 4e, Supplementary Fig. S8f). In contrast, xenograft tumors derived from chromosomally stable RKO cells with a partial loss of AURKA not affecting their stable karyotype did not result in acceleration of tumor growth in vivo indicating that partial loss of AURKA does not alter the tumor growth of CRC cells per se (Fig. 7f). Thus, our data unexpectedly revealed a detrimental effect of increased microtubule assembly rates triggering CIN on tumor growth in vitro and in vivo.
Figure 7.
Suppression of CIN by restoring proper microtubule assembly rates accelerates tumor growth in vitro and in vivoa, Determination of colony formation in soft agar of HCT116 and HCT116-CHK2−/− cells in the presence or absence of low dose Taxol®. Single cell clones were generated and 5,000 cells were seeded onto soft agar. Colony numbers were quantified and the graphs show mean values +/− SEM, t-test (n=4–6 experiments). b, Determination of colony formation in soft agar of SW620 cells in the presence or absence of low dose Taxol®. Single cell clones were generated and 2,000 cells were seeded onto soft agar. Colony numbers were quantified and the graphs show mean values +/− SEM, t-test (n=6 experiments). c, Xenograft tumor growth in mice after s.c. injection of chromosomally instable SW620 expressing control or CH-TOG/CKAP5 shRNAs into both flanks of nude mice. Tumor growth was monitored and tumor volumes are shown as mean values +/− SEM (n=14–16 tumors for each group). d, Xenograft tumor growth in mice after s.c. injection of chromosomally instable SW620 expressing control or AURKA shRNAs into both flanks of nude mice. Tumor growth was monitored and tumor volumes are shown as mean values +/− SEM (n=14–16 tumors for each group). e, Xenograft tumor growth in mice after s.c. injection of HCT116 (chromosomally stable), HCT116-CHK2−/− (chromosomally instable) or CHK2 deficient cells stably expressing AURKA shRNAs (exhibiting restored normal microtubule assembly rates and suppressed CIN). Tumor volumes were measured over time and are shown as mean values +/− SEM (n=7–11 tumors for each group). f, Xenograft tumor growth in mice after s.c. injection of chromosomally stable RKO cells expressing control or AURKA shRNAs into both flanks of nude mice. Xenograft tumor growth was monitored and tumor volumes are shown as mean values +/−SEM (n=10 tumors for each group).
Statistic source data for Figure 7 can be found in the Supplementary Table S2.
DISCUSSION
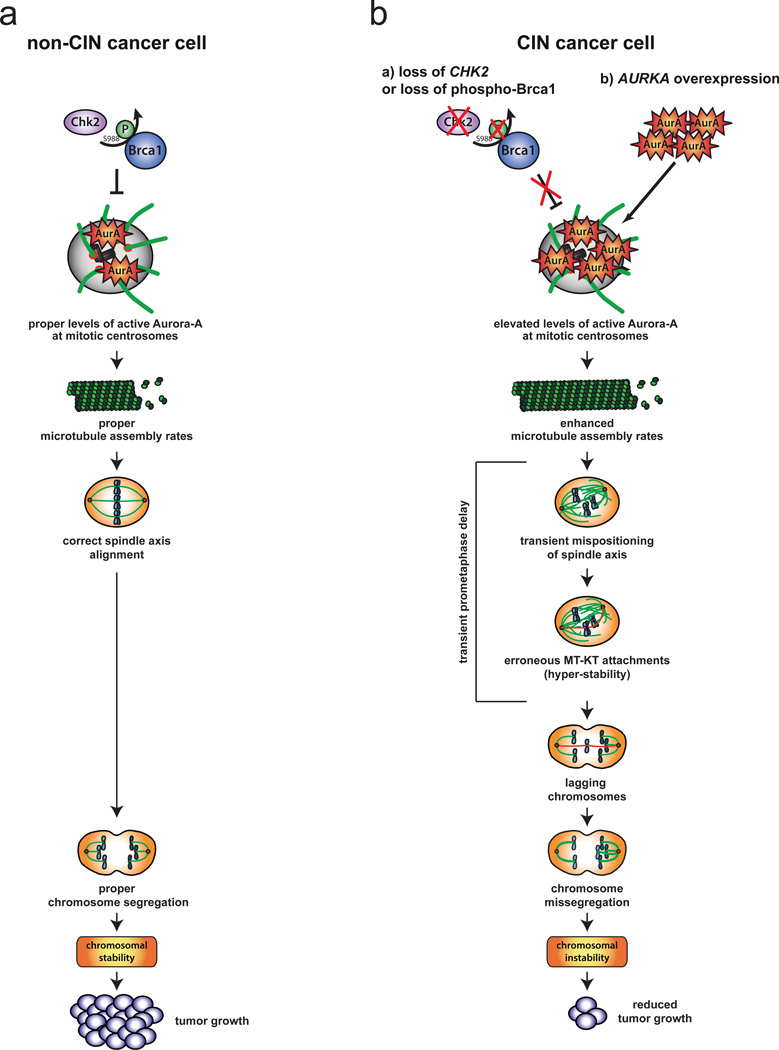
In cancer cells, CIN frequently correlates with the appearance of lagging chromosomes, which can persist either due to impairment of error correction or due to an increased rate of formation that might overwhelm the correction machinery8–14,36,37. So far, there is no evidence that error correction is frequently inactivated in human cancer cells(e.g. 43). However, our work now provides a mechanistic clue to the generation of lagging chromosomes as a source for CIN in colorectal cancer cells: an increase in microtubule plus end assembly rates within mitotic spindles is frequently found in CRC cells and can trigger transient spindle geometry abnormalities that facilitate the generation of erroneous kinetochore attachments resulting in lagging chromosomes and CIN (Fig. 8). This mechanism is reminiscent of a subset of cancer cells with supernumerary centrosomes15 that show transient multipolar spindle intermediates facilitating the generation of erroneous kinetochore attachments and leading to chromosome missegregation13,14. It is important to note that chromosome missegregation upon an increased rate of erroneous kinetochore attachments can occur in the presence of functional error correction. It is, nevertheless, associated with a decrease in kinetochore-microtubule turnover raising the possibility that hyper-stability of the kinetochore attachments might be related to the erroneous state of the attachment per se. In fact, increasing the number of erroneous attachments upon monastrol washout decreases kinetochore-microtubule turnover, while decreasing mal-attachments upon prolonging error correction increases the turnover of kinetochore-microtubules. It is not yet clear whether erroneous kinetochore attachments, possibly mediated by abnormal tension within the kinetochore7, are intrinsically more stable, but it would explain why conditions that induce erroneous kinetochore attachments without having an obvious function in error correction (e.g. APC) are also associated with an increase the stability of microtubule-kinetochore attachments37. An increase in microtubule assembly rates as a fundamental trigger for CIN is highly cancer-relevant. Genetic alterations triggering this phenotype including an overexpression of AURKA or a loss of CHK2 were found in about 70% of the cases of primary colorectal cancer and are also frequent in other types of cancer 18,20,44. Thus, an increase in microtubule assembly rates, together with cancer-associated centrosome amplification, which can also be driven by overexpression of AURKA in certain cell types45, might represent important mechanisms resulting in lagging chromosomes and CIN in human cancer cells. But how can elevated Aurora-A activity mediate an increase in microtubule plus end assembly rates? Intriguingly, increased microtubule assembly rates can be efficiently suppressed by repression of CH-TOG/CKAP5 indicating that this microtubule polymerase might be a target for Aurora-A. Interestingly, the recruitment of ch-TOG/CKAP5 to mitotic spindles is positively regulated by a Aurora-A mediated phosphorylation of the adaptor protein TACC340,46–48 and we detected an increase in this TACC3 phosphorylation in cells with increased microtubule assembly rates. Moreover, increased microtubule assembly rates in CIN cells can also be suppressed by partially depleting TACC3 (our unpublished results) indicating that Aurora-A might hyper-activate the TACC3-ch-TOG/CKAP5 axis. In this regard, it is of note that not only an overexpression of AURKA18, but also of TACC3 and CH-TOG have been linked to the etiology of various human cancers49,50 supporting a possible link between a general hyper-activity of this microtubule plus end assembly pathway and CIN in cancer cells.
Figure 8.
Model summarizing the main findings of this report. a, In chromosomally stable cancer cells the CHK2-BRCA1 tumor suppressor pathway is required to ensure proper levels of Aurora-A activity at mitotic centrosomes. This ensures proper microtubule plus end assembly rates as a prerequisite for normal progression of mitosis and proper chromosome segregation. b, Either the loss of the CHK2-BRCA1 tumor suppressor pathway or an overexpression of AURKA results in enhanced levels of Aurora-A at mitotic centrosomes, which triggers increased microtubule assembly rates. This, in turn, causes transient spindle geometry abnormalities facilitating the generation of erroneous microtubule-kinetochore attachments and lagging chromosomes as a source for chromosome missegregation. Thus, increased microtubule assembly rates represent a key trigger for perpetual chromosome missegregation, which is associated with reduced tumor growth in vitro and in vivo.
Giving the fact that we identified a key trigger for CIN in colorectal cancer cells that can be suppressed by various means we are now in a position to investigate the consequences of increased microtubule assembly rates and CIN on tumor growth. As a very first step into this important direction, we found that CIN cells with increased microtubule assembly rates exhibit a reduced tumor growth in vitro and in vivo when compared to tumors derived from cells with proper microtubule assembly rates and suppressed CIN. Although surprising at first glance, these results might reflect a detrimental effect of aneuploidy on cell proliferation, which might be mediated by altered metabolic properties and proteotoxic stress as shown previously for yeast and MEFs harboring defined trisomies51–53. This raises the question why cancer cells select for lesions such as AURKA overexpression or loss of CHK2 causing increased microtubule assembly rates and CIN. One plausible hypothesis might be that CIN can provide a high adaptation capability, which may be crucial for tumor initiation and progression 2,3,54. If so, then suppression of CIN by reverting a CIN trigger, as shown here, might be a useful strategy for anti-cancer treatment.
METHODS
Human Cell Lines
HCT116 and isogenic CHK2-deficient cells55 were a gift from Bert Vogelstein (John Hopkins University, Baltimore, MD, USA; colorectal cancer cell lines were from ATCC (Manassas, VA, USA). SW48 and derivative cell lines carrying KRAS mutations were from HORIZON (Cambridge, UK) and were generated by HORIZON’S proprietary adeno-associated virus (AAV) gene targeting technology GENESIS™. All cell lines were cultured at 37°C with 5% CO2 in RPMI1640 or DMEM medium containing 10% fetal calf serum, 1% Glutamine, 100 µg/ml streptomycin and 100 U/ml penicillin (Invitrogen, Netherlands).
cDNAs
HA-tagged BRCA1 cDNAs (wild type, S988A, S988E) cloned into pcDNA3 were provided by Jay Chung (NIH, Bethesda, MD, USA)41. Full-length human AURKA cDNA cloned into pcDNA3 (Invitrogen, Netherlands) was a gift from Martin Eilers (Würzburg, Germany). Reconstitution of CHK2 expression was carried out using pCEF-CHK2 and pcDNA3-CHK2 vectors as described previously20. HA-tagged human PLK1 cloned into pcDNA3 and pCMVflag-Plk4 were kind gifts from Ingrid Hoffmann (Heidelberg, Germany). For live-cell microscopy, pEGFP-EB322 was used to detect microtubule plus end tips. Plasmids for expression of GFP-tagged Centrin and RFP-tagged H2B were kind gifts from Stefan Duensing (Heidelberg, Germany) and Ody Sibon (Groningen, The Netherlands), respectively. Full-length human MCAK was cloned into pcDNA3 (Invitrogen, Netherlands).
shRNAs and siRNAs
The following short hairpin sequences were cloned into the pSuper-Retro vector carrying a puromycin resistance marker and were used to generate stable cell lines:
CHK2-#1: 5´-GCCUUAAGACACCCGUGGC-3´;
CHK2-#2: 5´-GAACCUGAGGACCAAGAAC-3´;
AURKA: 5´-GGCAACCAGUGUACCUCAU-3´;
CH-TOG/CKAP5: 5’-GAGCCCAGAGUGGUCCAAA-3’.
shRNA sequences targeting BRCA1 and the shRNA resistant mutants of BRCA1 and of CHK2 were previously described 20. The control scrambled shRNA sequence with no target in the human genome was: 5´- CAUAAGCUGAGAUACUUCA-3´. For transient siRNA transfections the following siRNA sequences were used:
CHK2: 5´-CCUUCAGGAUGGAUUUGCCAAUC-3´;
CH-TOG/CKAP5: 5´-GAGCCCAGAGUGGUCCAAA-3´;
TACC3: 5’-GUGGAUUACCUGGAGCAGU-3’;
APC: 5´-AAGACGUUGCGAGAAGUUGGA-3´
TP53: 5´-GUAAUCUACUGGGACGGAA-3´
MCAK: 5’-GCAGGCUAGCAGACAAAU-3’
LUCIFERASE: 5´-CUUACGCUGAGUACUUCGAUU-3´.
Transfections and generation of stable cell lines
Transient DNA transfections were carried out by using PEI (Sigma, Germany) or by electroporation using a BioRad electroporator (BioRad, USA) at 300 V and 500 µF (HCT116, SW48, SW620, SW837, LS1034), 220 V and 950 µF (HT29), and 200V and 950 µF (RKO). All siRNA transfections were carried out using Interferin transfection reagent (Polyplus, France) according to the manufacturer´s instructions.
Stable transfections were carried out using Metafectene (Biontex, Germany) using the protocol from the manufacturer followed by selection in medium containing 300 µg/ml G418 or 1 µg/ml puromycin. Single cell clones were isolated, expanded and further analyzed. The generation of stable HCT116 cell lines with low CHK2 expression (+ CHK2 shRNA) and cell lines in which CHK2 expression was reconstituted (CHK2 knowdown plus shRNA resistant CHK2 or CHK2−/− cells plus CHK2 wild type) and cell lines with reduced BRCA1 expression (HCT116 + BRCA1 shRNA) were described in detail previously 20. Stable colorectal carcinoma cell lines expressing shRNAs targeting CH-TOG/CKAP5 or AURKA were generated by stable transfection of cells with pSuper-Retro-CH-TOG/CKAP5 and with pSuper-Retro-AURKA, respectively and single cell clones were subsequently selected in medium containing 1 µg/ml puromycin. HCT116 cell lines stably expressing pcDNA-AURKA were selected in medium containing 300 µg/ml G418.
Cell treatments and cell cycle synchronization
To synchronize cells in G2 or in mitosis, cells were first synchronized at G1/S by a double thymidine block and released into fresh medium. 6–7 hours after the release cells were in G2 and 8–9 hours after release cells enter mitosis and various phases of mitosis could be observed. To block progression of mitosis beyond metaphase, 6 hours after release from the thymidine block cells were treated with 25 µM MG132 for additional 2.5 hours. To inhibit Aurora-A activity cells were treated with MLN8054 42 at concentrations up to 0.5 µM. To restore proper microtubule plus end growth rates low MLN8054 concentrations of up to 0.25 µM were used.
Immunofluorescence microscopy
To visualize mitotic spindles, kinetochores and centrosomes cells were fixed with 2% p-formaldehyde in PHEM (60 mM Pipes, 27 mM Hepes, 10 mM EGTA, 4 mM MgSO4, pH 7.0) followed by treatment with methanol at −20°C for 5 minutes. For detecting centrosomal Aurora-A, cells were fixed with methanol for 6 minutes. The following antibodies and dilutions were used for immunofluorescence microscopy experiments: anti-α-tubulin (1:700, B-5-1–2, Santa Cruz), anti-Centromere (CREST, 1:1000, Europa Bioproducts, UK), anti-Aurora-A (1:500, H130, Santa Cruz, USA), anti-phospho-Aurora-A (pT288, 1:200, Cell Signaling, USA), anti-γ-tubulin (1:500, GTU-88, Sigma Aldrich, USA) anti-centrin-2 (1:250, Abcam, UK). Secondary antibodies conjugated to Alexa Fluor-488/-594 (1:1000, Molecular Probes, USA) were used. Microscopy of fixed cells was performed on a Leica DM6000B microscope (Leica, Germany) equipped with a charged-coupled device camera (Orca-II-ERA, Hamamatsu, Japan) or using a DeltaVision-ELITE™ microscope (Applied Precision/GE Healthcare, Issaquah, WA, USA) equipped with a CoolSnap-HQ2 camera (Photometrics, USA). Images were recorded with a Z-optical spacing of 0.2 µm, deconvolved and analyzed using the Leica LAS-AF software (Leica) or the SoftWorx 5.0 (Applied Precision, USA) software. Pixel quantification of centrosomal signals were carried out using the Leica LAS-AF and ImagePro software packages (Leica, Germany; MediaCybernetics, USA) and normalized to signal intensities derived from centrosomal signals for centrin.
Measurement of microtubule plus end assembly rates
Microtubule plus end assembly rates were determined by tracking EB3-GFP protein in living cells 22,56. Cells were transfected with pEGFP-EB3 (provided by Linda Wordeman, USA), seeded onto glass bottom dishes (Ibidi, Germany) and after 48 hours cells were treated with the Eg5/Kif11 inhibitors monastrol (67 µM, Sigma, USA)24 or Dimethylenastron (2 µM; Calbiochem, Germany)23 for two hours or left untreated. This synchronization step was useful to ensure measurements of mitotic spindles in the same mitotic phase and did not affect the plus end growth rates per se. Four sections with a Z-optical spacing of 0.4 µm were taken every two seconds using a Deltavision ELITE™ microscope equipped with a Olympus 60X, 1.42 NA objective and a CoolSnap-HQ2 (Photometrics, USA) or a PCO Edge® sCMOS camera (PCO, Germany) at 37°C, 5% CO2. Images were deconvolved using the SoftWorx 5.0/6.0 software (Applied Precision, USA). Average assembly rates (µm/min) were calculated based on data retrieved for 20 individual microtubules per cell and a total of 10–60 cells were analyzed in 3–4 independent experiments.
Low dose Taxol® treatment
To identify low, sub-nanomolar concentrations of Taxol®, which are sufficient to reduce the increased microtubule growth rate of SW620, SW480, SW837, HCT116-CHK2−/− and of HCT116-AURKA cells to a level comparable with HCT116 cells without the enhanced microtubule growth phenotype, increasing, sub-nanomolar concentrations (0.05 – 0.5 nM) of Taxol® (Sigma, USA) were tested in EB3-GFP microtubule growth assays. The ascertained doses were used to generate single cell clones for karyotype analyses. Subsequently, these Taxol® treated cell clones were grown for additional 30 generations in the absence of Taxol® before the karyotypes were determined again.
Low dose Nocodazole treatment
To identify concentrations of nocodazol that increase microtubule assembly rates HCT116 cells were treated with various sub-nanomolar concentrations (0.5 – 2.5 nM) of nocodazol (Sigma, USA) before performing of EB3 tracking in live cells. For karyotype analyses, single cell clones derived from HCT116 cells were grown in 0.5 nM nocodazol for 30 generations.
Measurements of the half-life of kinetochore microtubule turnover
Cells were transfected with plasmids expressing PA-GFP-Tubulin and EB3-RFP to enable the laser to be focused on the kinetochore fibers prior to photoactivation. Late prometaphase cells were photoactivated on one side of the bipolar spindle with a 405nm laser on a microscope imaging system equipped with a charge-coupled device camera and a 60A 1.4 NA lens (Olympus, USA) and a QLM laser module (Personal Deltavision®; Applied Precision/GE Healthcare, Issaquah, WA, USA). Images were collected using adaptive frame rates for a period of five minutes post activation. Fluorescence intensity was measured and each time point was corrected for photobleaching (the decrease over time in the fluorescence of a photoactivated mark generated in Taxol®-treated cells). Each time point was also adjusted for incorporation of free photoactivated tubulin dimers using a reference point on the equivalent opposite side of the mitotic spindle20. Fluorescence values were normalized to the first time-point after photoactivation for each cell and the average intensity at each time point was fit to a two-phase exponential decay curve using Prism (GraphPad Software, Inc.) in which the normalized fluorescence (F) is described by: F=A1exp(-k1t) + A2exp(-k2t). A1 represents both the rapid turnover of non-kinetochore microtubules and instantaneous diffusion of photoactivated dimers and A2 represents the contribution of stable kinetochore fibers, k1 and k2 are their respective rate constants of turnover, and t is time after photoactivation. The turnover half-life for each process was calculated as ln2/k for each phase.
Determination of microtubule dynamicity
Cells were transfected with a plasmid expressing EB3-RFP and treated with 67 µM monastrol (Sigma, USA). Microtubule plus tips were automatically tracked using plusTipTracker software as described57 and overall dynamicity (collective displacement of all gap-containing tracks over their collective lifetimes), the percentage of time paused (total time all MTs spend in gap over the total time all tracks exist) and the catastrophe frequency (1/mean(T), where T is the lifetime (in minutes) of the growth sub-track just prior to the catastrophe) were determined. Rescue events could not be reliably calculated due to the interference from the high density of tips in the monasters.
Determination of mitotic timing
For live-cell analyses, cells were seeded on glass bottom dishes (Ibidi, Germany) and transfected with H2B–GFP expressing plasmids. Cells were followed by time-lapse microscopy at 37°C and 5% CO2 using a Leica DMI6000B (Leica, Germany). Image stacks were recorded every 2 minutes using a Hamamatsu EM-CCD camera (Hamamatsu, Japan) with a Z-optical spacing of 1 µm. Images were deconvolved and further processed using the Leica LAS-AF (Leica, Germany) or the SoftWorx 5.0/6.0 (Applied Precision, USA) software. The time from nuclear envelope breakdown (NEB=loss of the smooth appearance of the nucleus) until the beginning of anaphase (start of chromosome movement to the poles) was determined and box and whisker plots were calculated from at least 50 recorded cells using the Prizm software package, version 4 (GraphPad Software, Inc.).
Determination of spatial centrosome positioning
Cells were transfected with pcDNA-RFP-H2B (to detect chromosomes; a kind gift from Ody Sibon, Groningen, The Netherlands) and pEGFP-Centrin (to detect centrioles and thus, centrosomes; a kind gift from Stefan Duensing, Heidelberg, Germany) and seeded on glass bottom dishes (Ibidi, Germany). Asynchronously growing cells were followed by time-lapse microscopy at 37°C and 5% CO2 using a DeltaVision-ELITE™ microscope (Applied Precision/GE Healthcare, Issaquah, WA, USA). Image stacks were recorded every minute using a PCO Edge® sCMOS camera (PCO, Germany) with a Z-optical spacing of 1 µm. Images were deconvolved and further processed using the SoftWorx 6.0 (Applied Precision, USA) software. When the separation of the two mother centrioles was maximal (indicating maximal separation of the two centrosomes, see also: movies S3 and S4) the spatial positioning of the centrosomes was determined by measuring the angle between the centrosome axis and the growth surface (substratum) considering the distance between both mother centrioles and the height from the growth surface to the centrosome in Z-optical spacing planes following the mathematic formula ([α = (sinα * 180°)/ π]) (see also Figure 3d). Boxplots with mean values were calculated from 15 (prometaphase) and 9–11 (metaphase) recorded cells using the Prizm software package, version 4 (GraphPad Software, Inc.).
Detection of centrosome amplification
For quantification of centrosome amplification, asynchronously growing HCT116 parental cells, HCT116-CHK2−/− and AURKA overexpressing cells respectively, were fixed with 2% p-formaldehyde in PHEM (60 mM Pipes, 27 mM Hepes, 10 mM EGTA, 4 mM MgSO4, pH 7.0) followed by treatment with methanol at – 20°C for 5 minutes. Subsequently, the cells were stained for γ-Tubulin (1:500, GTU-88, Sigma-Aldrich, USA) to visualize centrosomes and with Hoechst (Hoechst 33342, 1:10000, Biomol) to identify interphase cells. As a positive control, HCT116 cells were transiently transfected with pCMVflag-Plk4 (kindly provided by Ingrid Hoffmann, Heidelberg, Germany) and 48 hours after transfection, interphase cells with more than two g-tubulin positive signals were evaluated.
Determination of lagging chromosomes
To detect lagging chromosomes in anaphase, cells were transfected with control or CH-TOG/CKAP5 siRNAs and subsequently synchronized in G1/S phase by a double Thymidin block. Cells were released into medium for 8–9 hours to accumulate cells in anaphase. Cells were analyzed by immunofluorescence microscopy detecting CREST-positive chromosomes. Only chromosomes that were clearly separated from the two pole-oriented chromosome masses were counted as “lagging chromosomes”. For the experiments, in which the time for error correction was prolonged, cells were transfected with siRNAs targeting CHK2, MCAK or luciferase. After 48 hours cells were treated with 2 µM DME or 67 µM monastrol for 3 hours, released from the block and fixed after 75 minutes when cells were mostly in anaphase. To prolong the time for error correction cells were released from the DME/monastrol block into medium containing 10 µM MG-132. After additional 2.5 hours MG-132 was washed out and cells were fixed 50 minutes later when they were mostly in anaphase.
Western blotting
Cells were lysed in lysis buffer (50 mM Tris-HCl, pH7.4, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1% (v/v) NP-40, 0.1% (w/v) SDS, 0.1% Sodium-Desoxycholate, protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Roche). Proteins were resolved on 7.5%, 10% or 12% SDS polyacrylamide gels and blotted onto nitrocellulose or PVDF membranes using semi-dry or tank-blot procedures. For western blot experiments the following antibodies and dilutions were used: anti-Chk2 (1:800, DCS-270, Santa Cruz, USA), anti-β-actin (1:40000, AC-15, Sigma, Germany), anti-Brca1 (1:500, C-20 or D-9, Santa Cruz), anti-Aurora-A (1:2000, H130, Santa Cruz), anti-phospho-Aurora-A (1:2000, pT288, Cell Signaling, USA), anti-phospho-Aurora-A, -B, -C (1:2000, pT288, pT232, pT198, Cell Signaling), anti-Tpx2 (1:1000; 18D5, Santa Cruz, USA), anti-α-tubulin (1:2000, B-5-1–2, Santa Cruz), anti-Plk1 (1:1000, F-8, Santa Cruz), anti-p53 (1:1000; Ab-2, Oncogene, USA), anti-MCAK (1:250; kindly provided by Linda Wordeman, Seattle, USA), anti-Axin-2 (used to verify the repression of APC; 1:1000; Cell Signaling), anti-TACC3 (1:1000; H300, Santa Cruz, USA), anti-phospho-TACC3 (Ser-558, 1:1000, D8H10, Cell Signaling, USA), anti-CH-TOG/CKAP5 (1:1000; H4, Santa Cruz, USA), secondary antibodies conjugated to horseradish peroxidase (1:10,000, Jackson ImmunoResearch, USA). Proteins were detected by enhanced chemoluminescence. Quantification of western blot bands was performed using the Image J software (NIH).
Immunoprecipitation
Cells were lysed in 50 mM Tris-HCl, pH7.4, 150 mM NaCl, 0.25% NP-40, 10% glycerol, 1 mM DTT, protease inhibitor and phosphatase inhibitor cocktails (Roche, Switzerland). 2 mg lysate were incubated with 1.5 µg anti-Brca1 (D-9, Santa Cruz, USA) or anti-TPX2 (18D5, Santa Cruz) antibodies and immunocomplexes were precipitated using protein-G sepharose beads (GE Healthscare, USA).
FACS analyses and determination of the mitotic index
Cells were fixed in 70% ethanol over night at 4°C and resuspended in propidium iodide (5 µg/ml) and RNaseA (1 µg/ml) in PBS. FACS analyses were performed on a BD FACSCANTO II (Becton Dickinson) and 10,000 events were counted. Data analyses was done using the BD FACSDIVA™ software (Becton Dickinson). The mitotic index was determined by staining of fixed cells with anti-MPM2 (1:1600, Millipore, USA) and secondary antibodies conjugated to Alexa Fluor-488 (1:2000, Molecular Probes) as described 20.
Karyotype analyses
Single cell clones were generated and chromosome spread analyses and chromosome counting in individual cells was performed after 30 generations as described 20 and the proportion of cells with chromosome numbers deviating from the modal were determined. The chromosome number variability generated in single cell clones within a defined time span and which showed deviation by up to 5 chromosomes was taken as a measure for CIN.
CEP-FISH analyses
Chromosome number variability was also determined by fluorescence in situ hybridization (FISH) using Aquarius® Satellite Enumeration probes (Cytocell, UK). Cells were washed with PBS and incubated in 40% (v/v) RPMI-1640 at RT for 15 min. Cells were then fixed in Carnoy`s solution (methanol – acetic acid (3:1)). FISH was performed using a-satellite probes specific for chromosomes 7 and 15 (Cytocell, UK) according to the manufacturer’s protocol. FISH images were acquired as 0.5 µm optical sections with the 60 × 1.4 NA objective and chromosome signals in at least 100 nuclei were determined.
Human tumor samples and immunohistochemistry analyses
Tissue samples from 333 primary human colorectal adenocarcinomas were investigated by using tissue microarrays. Microarray construction and immunohistochemical analyses were carried out as described 58 using a monoclonal anti-Chk2 antibody (DCS-273, 1:250, Sigma, Germany) and an anti-Aurora-A antibody (clone JLM28, 1:50, Novocastra Laboratories, UK). Staining was scored by two experienced pathologists (Al.S. and W.W.). Chk2 staining could be evaluated in all cases, for AURKA expression 325 cases were evaluable. Cases, which showed a moderate or strong expression in the majority of tumor cells were scored as positive.
Colony formation assays in soft agar
To analyze anchorage-independent colony formation 6-well culture dishes were coated with 1 ml culture medium containing 1% low-melt agarose (Sigma, Taufkirchen, Germany). Subsequently, 5,000 HCT116 and 2,000 SW620 cells were re-suspended in 1 ml of growth medium containing 0.4% low-melt agarose and plated on top of the solidified bottom layer. Soft agar was overlaid with medium containing Taxol® when indicated. The embedded cells were incubated at 37$°C and 5% CO2 until the arising colonies were stained with 0.001 % crystal violet. The growth area was scanned using a Quato IntelliScan 1600 (Quatographic Technology, Germany) and colonies were counted by using the Image-Pro Analyzer v7.0 Software (Media Cybernetics, Rockville, USA).
Mice xenografts
2 × 106 HCT116 cells in 150 µl PBS were injected s.c. into both flanks of 5-week old athymic nude mice (Crl:NU-Foxn1 nu, Charles River, Germany). For SW620 xenografts, 4 × 106 SW620 cells in 200 µl PBS were injected s.c. into both flanks of 6-week-old athymic mice (provided by the MEZ, University of Leipzig). Growth of two tumors per mouse was monitored regularly by determining the perpendicular diameters of the tumors. All xenograft experiments were performed according to and were approved by the national authorities.
Statistical analyses
All data are shown as mean ± standard deviations (SD), standard error (SE) or standard error of the mean (SEM). Where indicated students t-tests using the Prizm software package, version 4 were applied. For the signal intensities at centrosomes the 95% confidence interval (CI) was determined. Statistical analyses on CHK2 and AURKA expression patterns in human CRC tissue samples were performed using SPSS 19.0. and the significance of correlations was determined by applying two-sided Fisher’s exact tests. Quantification of mitotic spindles and lagging chromosomes are based on at least 3 independent experiments, in which at least 1500 mitotic figures or 300 anaphase cells, respectively, were evaluated. All karyotype analyses are based on quantification of individual chromosome numbers from 50–109 metaphase spreads (see Table S1).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Bert Vogelstein, Marian Grade, Martin Eilers, Stefan Duensing, Ingrid Hoffmann, Jay Chung, Ody Sibon and Michael Brandeis for cell lines and plasmids. We thank Frauke Alves and Joanna Napp for providing nude mice, Rovena Kampe and Bärbel Obst for excellent technical assistance and Tim Beiβbarth for the help with statistical analyses. We are grateful to Heike Krebber, Zuzana Storchova and all members of the Bastians lab for critically reading the manuscript. This work was supported by the Molecular Diagnostic Program of the German Consortium for Translational Cancer Research funded by the Germany Ministry of Education and Research (W.W.), the grant GM069429 from the National Institutes of Health (L.W.) and the DFG, the KFO179 and by a Heisenberg professorship awarded by the DFG to H.B.
Footnotes
SUPPLEMENTARY INFORMATION
Supplementary Information includes eight Supplementary Figures (Figure S1 – Figure S8), four Supplementary Movies (Movie S1 – Movie S4), one Supplementary Table (Table S1) and Statistical Source data (Table S2) and are available at …..
Contributions
N.E. and A.S. performed and analyzed most experiments; Al.S. and W.W. performed and analyzed the immunohistochemistry assessment on human tumor samples, S.K. P.B. and A.A. performed and analyzed the mouse xenograft experiments. L.W. performed and analyzed microtubule assembly and fluorescence dissipation assays. H.B. designed and coordinated the study, analyzed data and wrote the manuscript. All authors discussed the work and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests
REFERENCES
- 1.Rajagopalan H, Nowak MA, Vogelstein B, Lengauer C. The significance of unstable chromosomes in colorectal cancer. Nat Rev Cancer. 2003;3:695–701. doi: 10.1038/nrc1165. [DOI] [PubMed] [Google Scholar]
- 2.McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep. 2012;13:528–538. doi: 10.1038/embor.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfau SJ, Amon A. Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 2012;13:515–527. doi: 10.1038/embor.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 5.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrell RA, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregan J, Polakova S, Zhang L, Tolic-Norrelykke IM, Cimini D. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 2011;21:374–381. doi: 10.1016/j.tcb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol. 2007;179:869–879. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrews PD, et al. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 12.Lan W, et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 13.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghadimi BM, et al. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer. 2000;27:183–190. [PMC free article] [PubMed] [Google Scholar]
- 16.Orr B, Compton DA. A double-edged sword: how oncogenes and tumor suppressor genes can contribute to chromosomal instability. Front Oncol. 2013;3:164. doi: 10.3389/fonc.2013.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer. 2010;10:825–841. doi: 10.1038/nrc2964. [DOI] [PubMed] [Google Scholar]
- 18.Bischoff JR, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. Embo J. 1998;17:3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weichert W, et al. Polo-like kinase 1 expression is a prognostic factor in human colon cancer. World J Gastroenterol. 2005;11:5644–5650. doi: 10.3748/wjg.v11.i36.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stolz A, et al. The CHK2-BRCA1 tumour suppressor pathway ensures chromosomal stability in human somatic cells. Nat Cell Biol. 2010;12:492–499. doi: 10.1038/ncb2051. [DOI] [PubMed] [Google Scholar]
- 21.Joukov V, et al. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 22.Stepanova T, et al. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein) J Neurosci. 2003;23:2655–2664. doi: 10.1523/JNEUROSCI.23-07-02655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muller C, et al. Inhibitors of kinesin Eg5: antiproliferative activity of monastrol analogues against human glioblastoma cells. Cancer Chemother Pharmacol. 2007;59:157–164. doi: 10.1007/s00280-006-0254-1. [DOI] [PubMed] [Google Scholar]
- 24.Mayer TU, et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 25.Brouhard GJ, et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 28.Sironi L, et al. Automatic quantification of microtubule dynamics enables RNAi-screening of new mitotic spindle regulators. Cytoskeleton (Hoboken) 2011;68:266–278. doi: 10.1002/cm.20510. [DOI] [PubMed] [Google Scholar]
- 29.Derry WB, Wilson L, Jordan MA. Low potency of taxol at microtubule minus ends: implications for its antimitotic and therapeutic mechanism. Cancer Res. 1998;58:1177–1184. [PubMed] [Google Scholar]
- 30.Jordan MA, Thrower D, Wilson L. Effects of vinblastine, podophyllotoxin and nocodazole on mitotic spindles. Implications for the role of microtubule dynamics in mitosis. J Cell Sci. 1992;102(Pt 3):401–416. doi: 10.1242/jcs.102.3.401. [DOI] [PubMed] [Google Scholar]
- 31.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplan KB, et al. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat Cell Biol. 2001;3:429–432. doi: 10.1038/35070123. [DOI] [PubMed] [Google Scholar]
- 33.Vogel C, Kienitz A, Hofmann I, Muller R, Bastians H. Crosstalk of the mitotic spindle assembly checkpoint with p53 to prevent polyploidy. Oncogene. 2004;23:6845–6853. doi: 10.1038/sj.onc.1207860. [DOI] [PubMed] [Google Scholar]
- 34.Stolz A, Ertych N, Bastians H. Tumor suppressor CHK2: regulator of DNA damage response and mediator of chromosomal stability. Clin Cancer Res. 2011;17:401–405. doi: 10.1158/1078-0432.CCR-10-1215. [DOI] [PubMed] [Google Scholar]
- 35.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 37.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- 39.Littlepage LE, et al. Identification of phosphorylated residues that affect the activity of the mitotic kinase Aurora-A. Proc Natl Acad Sci U S A. 2002;99:15440–15445. doi: 10.1073/pnas.202606599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinoshita K, et al. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J Cell Biol. 2005;170:1047–1055. doi: 10.1083/jcb.200503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JS, Collins KM, Brown AL, Lee CH, Chung JH. hCds1-mediated phosphorylation of BRCA1 regulates the DNA damage response. Nature. 2000;404:201–204. doi: 10.1038/35004614. [DOI] [PubMed] [Google Scholar]
- 42.Manfredi MG, et al. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. Proc Natl Acad Sci U S A. 2007;104:4106–4111. doi: 10.1073/pnas.0608798104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 44.Zhou H, et al. Tumor amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Gen. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, et al. Overexpression of aurora kinase A in mouse mammary epithelium induces genetic instability preceding mammary tumor formation. Oncogene. 2006;25:7148–7158. doi: 10.1038/sj.onc.1209707. [DOI] [PubMed] [Google Scholar]
- 46.Gergely F, Draviam VM, Raff JW. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 2003;17:336–341. doi: 10.1101/gad.245603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin CH, Hu CK, Shih HM. Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. J Cell Biol. 2010;189:1097–1105. doi: 10.1083/jcb.200911120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LeRoy PJ, et al. Localization of human TACC3 to mitotic spindles is mediated by phosphorylation on Ser558 by Aurora A: a novel pharmacodynamic method for measuring Aurora A activity. Cancer Res. 2007;67:5362–5370. doi: 10.1158/0008-5472.CAN-07-0122. [DOI] [PubMed] [Google Scholar]
- 49.Charrasse S, et al. Characterization of the cDNA and pattern of expression of a new gene over-expressed in human hepatomas and colonic tumors. Eur J Biochem. 1995;234:406–413. doi: 10.1111/j.1432-1033.1995.406_b.x. [DOI] [PubMed] [Google Scholar]
- 50.Thakur HC, et al. Role of centrosomal adaptor proteins of the TACC family in the regulation of microtubule dynamics during mitotic cell division. Biol Chem. 2013 doi: 10.1515/hsz-2013-0184. [DOI] [PubMed] [Google Scholar]
- 51.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 52.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duijf PH, Benezra R. The cancer biology of whole-chromosome instability. Oncogene. 2013;32:4727–4736. doi: 10.1038/onc.2012.616. [DOI] [PubMed] [Google Scholar]
- 55.Jallepalli PV, Lengauer C, Vogelstein B, Bunz F. The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J Biol Chem. 2003;278:20475–20479. doi: 10.1074/jbc.M213159200. [DOI] [PubMed] [Google Scholar]
- 56.Piehl M, Tulu US, Wadsworth P, Cassimeris L. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc Natl Acad Sci U S A. 2004;101:1584–1588. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Applegate KT, et al. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol. 2011;176:168–184. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weichert W, et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: a retrospective analysis. Lancet Oncol. 2008;9:139–148. doi: 10.1016/S1470-2045(08)70004-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.