Abstract
Complex interactions between a series of environmental factors and genes result in progression to clinical type 1 diabetes in genetically susceptible individuals. Despite several decades of research in the area, these interactions remain poorly understood. Several studies have yielded associations of certain foods, infections, and immunizations with the onset and progression of diabetes autoimmunity, but most findings are still inconclusive. Environmental triggers are difficult to identify mainly due to (i) large number and complex nature of environmental exposures, including bacteria, viruses, dietary factors, and environmental pollutants, (ii) reliance on low throughput technology, (iii) less efforts in quantifying host response, (iv) long silent period between the exposure and clinical onset of T1D which may lead to loss of the exposure fingerprints, and (v) limited sample sets. Recent development in multiplex technologies has enabled systematic evaluation of different classes of molecules or macroparticles in a high throughput manner. However, the use of multiplex assays in type 1 diabetes research is limited to cytokine assays. In this review, we will discuss the potential use of multiplex high throughput technologies in identification of environmental triggers and host response in type 1 diabetes.
1. Introduction
Type 1 diabetes (T1D) results from complex yet poorly defined interactions between environmental agents, the immune system, and genetic factors (Figure 1). T1D is a chronic T-cell mediated disease, characterized by selective loss of insulin-producing β-cells in the pancreatic islets [1]. There is an annual average of 3% increase in T1D incidence worldwide and the incidence rates are also increasing in the countries with no previous record of having T1D [2, 3]. It is believed that genetic susceptibility is a prerequisite for the development of T1D; however, not all genetically predisposed individuals develop clinical disease and subjects with low risk or protective genes also have been found to develop T1D. These observations suggest that apart from genetic susceptibility additional factors trigger the process of β-cell autoimmunity and subsequent clinical disease.
Figure 1.
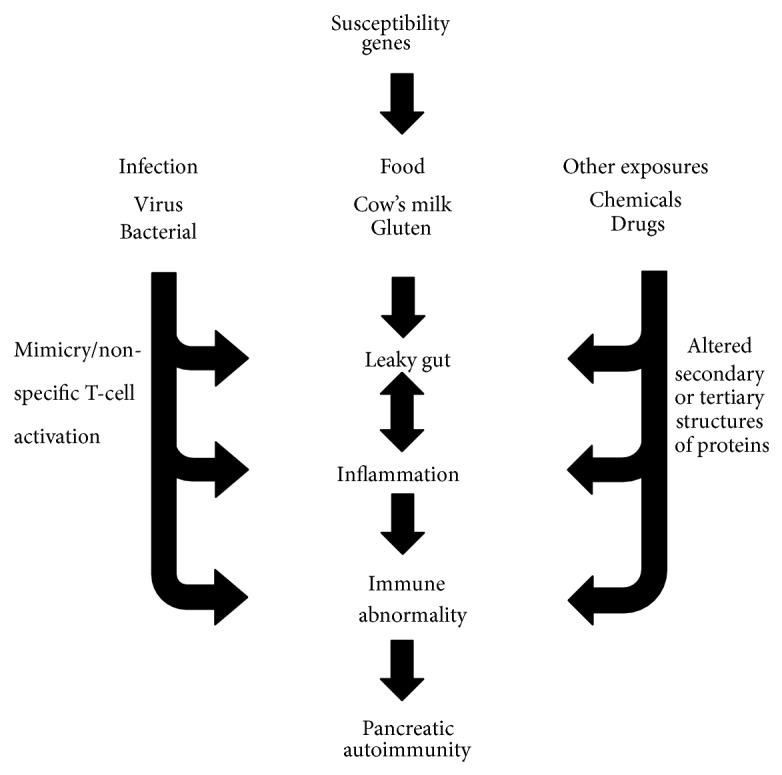
Susceptibility genes and environmental triggers in development and progression of type 1 diabetes. In genetically susceptible individuals, different classes of environmental exposures such as diet, infection, and pollutants lead to increase in peripheral and mucosal inflammation causing leaky gut and aberrant immune reaction towards pancreatic β-cells.
If these environmental triggers are known, change in life style is likely to offer the most powerful strategy for effective prevention of T1D. If successful, this approach can target the whole population or at least the population with increased genetic susceptibility. In pilot studies, dietary interventions have been successfully tested to manipulate appearance of β-cell autoimmunity in high risk children [4, 5]. However, there has been little progress in this area partly due to nonavailability of technologies to measure different types of environmental exposures and host response in large sample sets. The recently developed multiplex technologies have enabled the measurement of greater number of analytes in a high throughput manner.
2. Environmental Triggers of T1D
Several studies have observed seasonal patterns in the presence of serum antibody titers (incidence being more common during cold), in part due to the role of recent infections in the development of β-cell auto-antibodies [6]. Viral infections have been suggested to be responsible for T1D autoimmunity for a century, but recent studies have provided stronger data [7]. A number of viruses have been shown to be associated with T1D autoimmunity including Enterovirus, rubella, mumps, and rotavirus [8]. Taking into account the timing and profiles of the autoantibody peaks observed in several studies, Enterovirus infections appear to be the most probable trigger of β-cell autoimmunity [9]. Despite significant amount of evidence, the role of viruses in the development of T1D autoimmunity is not conclusive. The long silent period between the infection and clinical onset of T1D may lead to loss of the viral signatures in serum [9, 10]. Some authors believe that infections may protect from development of T1D autoimmunity. The proponents of the “hygiene hypothesis” suggest that children experiencing more infection in childhood are more protected; however there has been no consensus among researchers [11–13]. On the other hand the “trigger-booster hypothesis” claims that progression to clinical type 1 diabetes typically requires the unfortunate combination of genetic disease susceptibility, a diabetogenic trigger, and a high exposure to a driving antigen [14].
A number of dietary factors have also been found to be associated with development of T1D, including cow's milk, wheat gluten, and vitamin D deficiency [15, 16]. Some studies have shown the protective role of breast feeding and other nutrients [17, 18]. Several other studies have shown positive association of β-cell autoantibodies with introduction of milk based or wheat based formula early in life [19]. The results of these studies have always been mixed with no consensus on specific dietary factor or nutrient being conclusively responsible for development of T1D [20].
Recent studies suggest that β-cell autoantibodies are preceded by active inflammation [21]. Viral infections, dietary factors, and changes in gut microbiome lead to intestinal inflammation and may contribute to the increased permeability of the gut [16]. Vaarala et al. showed that the complex interactions between gut microbiome, intestinal permeability, and mucosal immunity contribute to the pathogenesis of T1D [22, 23]. These authors suggested that leaky gut allows entry to certain proteins present in cow's milk and wheat and as such leads to T1D autoimmunity in at risk subjects [22].
3. Measurement of Environmental Exposures and Host Response in T1D
Elucidation of the environmental exposure in T1D has been a highly contentious issue. Although studies have postulated a role of several environmental agents in T1D, progress in this area has been slow. This at least in part is attributed to the complex nature of the environmental exposures. A large number of environmental exposures need to be explored from different classes, including bacteria, viruses, dietary factors, and pollutants and the measurement involves different classes of molecules including DNA, RNA, proteins, metabolites, small molecules, and antibodies.
Also, due to a huge lag time between the time of exposure and the onset of disease, sometimes it is difficult to identify the environmental trigger itself. However, such an exposure may leave a signature or fingerprint which may be present for a longer time (host response). Thus measuring this host response may provide additional information correlated with the environmental triggers. For example, circulating levels of IgG and IgM against viruses have been shown in T1D patients [9, 24–26]. Similarly, measurement of cytokines, chemokines, and other plasma proteins could provide us a hint on the class of environmental exposures [27]. Measuring the host response may provide us unique fingerprints which may be used as additional markers for disease progression.
4. Current Approaches
Currently, many available “omics” technologies are being used to study the environmental exposures and host response in T1D. To identify molecular and cellular signatures, we have measured several classes of biomolecules in our laboratory [28–32]. ELISA assays are being used for detection of Saccharomyces cerevisiae [33], wheat protein Glb-3 [34, 35], gluten [36], gliadin, cow milk proteins in T1D, and celiac disease [37]. Giongo et al. used pyrosequencing approach to show that the intestinal microbiome of the children progressing to clinical disease was less diverse than healthy children [38]. PCR based typing was utilized to identify the Enterovirus DNA circulating in the serum of newly diagnosed T1D patients [16, 39]. However, the studies focusing on measurement of single environmental agent provide a skewed view on the environmental exposure. This can be remedied by measuring several types of environmental agents and host response to obtain a fingerprint of overall environmental exposure in a high throughput manner.
5. Multiplex Technologies
To measure several hundreds of the environmental triggers and the host response in larger sample sets economically, high throughput technologies are needed [28–32]. ELISAs or radioimmunoassays have been the preferred technologies for the measurement of low abundance agents in the serum. Recently, multiplex assays have been developed from traditional ELISA assays with the purpose of measuring multiple analytes in the same sample at the same time. Multiplex assays are available in several different formats based on the utilization of flow cytometry, chemiluminescence, and array technology (Table 1). Compared with traditional ELISA, multiplex arrays have a number of advantages including (i) high throughput multiplex analysis, (ii) less sample volume requirements, (iii) efficiency in terms of time and cost, (iv) ability to evaluate the levels of given analyte in the context of multiple others, (v) ability to perform repeated measures of the multiplex panels in the same experimental assay conditions, and (vi) ability to reliably detect analytes across a broad dynamic range of concentrations [40].
Table 1.
Characteristics of currently available array based high throughput technologies.
| Technology/manufacturer | Maximum number of analytes | Maximum number of samples | Volume of sample | Dynamic range |
|---|---|---|---|---|
| Luminex/Luminexcorp | 500 | 96 or 384 | 1–5 µL | >4.5 logs |
| SimOa/Quanterix | 10 | 384 | 1–10 µL | >4 logs |
| Flow Cytomix/Afffymetrix | 20 | 96 | 25 µL | — |
| Cytometric Bead assay/BD biosciences | 30 | 96 | 25–50 µL | — |
| Barcoded Magnetic Beads/Applied Biocode | 128 | 96 | 1–5 µL | 5 logs |
| Antibody arrays/Quansys Bioscience* | 25 | 96 | 30–50 µL | — |
| Antibody arrays/Meso Scale discovery* | 10 | 96 or 384 | 30–50 µL | >4 logs |
| Antigen Arrays/Thermo Scientific | 10 | 96 | 40 µL | — |
*Detection is based on chemiluminescence or electrochemiluminescence.
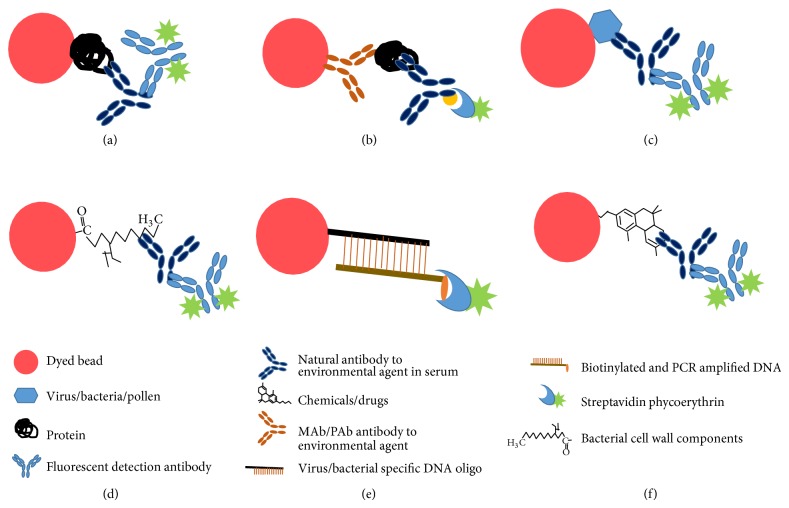
Bead-based multiplex assays represent probably the most commonly used format developed by several companies. Multianalyte profiling (xMAP) technology from Luminex (http://www.luminexcorp.com/) and several other companies employ proprietary bead sets which are distinguishable under flow cytometry (Figure 2). The platform is a suspension array where capture moieties are covalently coupled with internally dyed microspheres, and phycoerythrin-labeled anti-human antibodies bind to the specific antigen-antibody complex on the bead set. Response is thus recognized and measured by the differences in both bead sets, with fluorogenic emissions detected using red (bead set) and green (detection of entities) lasers. The flexibility of the system allows covalent coupling and detection of several different classes of molecules or macroparticles.
Figure 2.
Luminex bead arrays could be used to detect different classes of environmental triggers. (a) Protein(s), (b) monoclonal (MAb) or polyclonal (PAb) antibodies, (c) viruses, (d) bacterial cell wall components, (e) DNA from virus/bacteria, and (f) chemicals/drugs can be covalently coupled to the beads. Coupled entities can be detected using fluorescently labeled appropriate detection agents.
6. Use of Multiplex Assays in Other Research Areas
Multiplex assays have been at the forefront for epidemic monitoring by health agencies in USA and abroad. Developments in the PCR technology and discrimination methods combined with the multiplex assays have improved the detection of coinfections with reduced cost and sample volumes required for analysis [40]. The most active research areas using the multiplex immunoassays are allergy, asthma, infectious disease, autoimmunity, and toxicology (Table 2). Extensive research efforts have been taken to test the feasibility of the Luminex xMAP technology to detect the autoantibodies to autoantigens, IgE response to grass and tree pollen, virus and bacterial serotypes, and weaponized microbial agents. Researchers in the fields of vaccine development and epidemiology have extensively documented the use of multiplexed assays to identify targets using antibody-based capture or DNA fragments specific to each serotype of bacteria or viruses. Using monoclonal antibodies to the individual serotypes, Yu et al. used Luminex technology to detect 26 different serotypes for Streptococcus pneumoniae in serum [41]. Other investigators have used antibody-based multiplex assays to characterize microbial pathogens and agents [42–44]. Conventional and real-time RT-PCR, combined with Luminex bead array, were used in detection of multiple viruses to identity the microbial agents in disease individuals [45, 46]. All these reports provide sufficient feasibility regarding the development of multiplex assays to identify the environmental triggers in T1D.
Table 2.
Research areas and available assays for Luminex platform.
| Research Areas | Manufacturers | Species |
|---|---|---|
| Immunology/inflammation/apoptosis/tissue remodeling markers | Millipore, RnD Systems, Life Technologies, Luminex, Biorad | Hu/Ms/Rt/Ca/Mo |
| Phosphoproteins, signal transduction proteins | Millipore, Life Technologies, Biorad | Hu/MS |
| Cancer markers | Millipore, Biorad | Hu/Ms |
| Metabolic markers | Millipore, Life Technologies | Hu |
| Cardiovascular markers | Millipore, Life Technologies, Biorad | Hu |
| Toxicity markers | Millipore | Hu |
| Neuroscience | Millipore, Biorad | Hu |
| Antibody isotyping | Biorad | Hu/Ms |
| Auto-antibody measurement | One Lambda, Origene | Hu |
| Genotyping, epigenetics, and gene expression profiling | Affymetrix, One Lambda, Origene, Active Motiff | Hu |
| HLA typing | One Lambda | Hu |
| Environmental agents/allergens/food | Thermo Scientific | Hu |
| Bacterial/virus serotyping | Biovet/Luminex | Hu |
| Drugs/chemical | Under development in research laboratories | Hu |
| Vaccine testing | Luminex | Hu |
Ca: cat, Hu: human, Mo: monkey, Ms: mouse, Rt: rat.
7. Technological Considerations for Developing Multiplex Assays for Environmental Triggers of T1D
Although multiplex technologies offer several advantages over ELISA approaches, caution must be exercised for developing assays. In this section some of the critical issues are discussed and possible solutions are offered for development of these assays.
Multiplex bead assays, by their very nature, involve measurement of several potential analytes in a single well. Therefore, cross-interactions between different capture antibodies and antigens in the sample/assay solution are inherently possible. Cross-reactivity of antibodies should be tested first and the lowest amount should be used to minimize such cross-reactions. The individual panels should be designed using the analytes with minimal cross-reactivity among the analytes and detection reagents. Secondly, multiplex assays are performed using a common binding and wash buffers, and these may not be the optimum conditions for all the analytes. This can be solved by creating custom multiplex assays having similar binding and washing conditions.
In the bead-based multiplex arrays, the reactions take place among capture entities and analytes which are freely mobile in solution, providing more sensitivity to measure circulating levels of analytes. However, abundant proteins present in bodily fluids, such as serum, may affect multiplex results. The abundant proteins serve both as reservoir and as carrier of small molecules such as cytokines and metabolites. The bound complexes may not get captured in the solution phase or the detecting reagent may not be able to access the required binding site. This interference from abundant proteins may require development of additional processing steps prior to multiplex assay.
The commercially available Luminex kits can measure up to 60 cytokines, chemokines, and ligands. However the linear range of the standard curve and the levels in the samples limits the number of analytes which can be measured simultaneously. This issue can be solved by performing pilot experiments to select the analytes having similar dynamic range (customized panels). For a particular value of sample dilution, the analyses will be selected in the same panel if their median fluorescence intensities fall in the linear range of the standard curve.
A defined set of principles are required to establish good laboratory practices and must be followed in the planning, performing, monitoring, recording, reporting, and archiving of all laboratory measurements. To prevent quality problems a good quality assurance policy must be established. The variations in high throughput measurements emerge from many sources. To reduce plate to plate variation and to produce consistent results over time, a dilution series of pooled control serum on each plate should be included for the normalization.
The bulk of our knowledge about T1D pathogenesis comes from studies of animal models. Data from human subjects are scarce and difficult to replicate for many reasons including, but not limited to, large variations of the studied phenotypes at the individual and population levels and differences in study design (insufficient sample size, poor matching of patients and controls, case/control versus prospective, and so forth). Important considerations in the proper design of human studies include prospective studies, which minimize many of the drawbacks associated with cross sectional comparisons that are commonly used in human studies.
8. Conclusions
Multiplex technologies offer opportunities to examine the different “classes” of environmental triggers of T1D in a time- and cost-efficient high throughput manner. While the use of such technologies is still at early stage, recent reports from other research areas highlight their usefulness and feasibility to evaluate the environmental exposure and host response in T1D pathogenesis. Also, multiplex technologies offer substantial sample savings over traditional ELISA measurements. Despite potential advantages of this new technology, expertise and experience are required for new assay development. We have used this technology in examining several classes of serum proteins in T1D [47–49]. In our view, multiplex technology could be successfully used for the evaluation of different classes of environmental exposures and host responses in T1D pathogenesis.
Acknowledgments
This work is partly supported by NIH Grants to Jin-Xiong She (U01DK063865 and RO1HD37800). Sharad Purohit was supported by JDRF Grants (10-2006-792 and 2-2011-153).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Achenbach P., Bonifacio E., Koczwara K., Ziegler A.-G. Natural history of type 1 diabetes. Diabetes. 2005;54(supplement 2):S25–S31. doi: 10.2337/diabetes.54.suppl_2.S25. [DOI] [PubMed] [Google Scholar]
- 2.Egro F. M. Why is type 1 diabetes increasing? Journal of Molecular Endocrinology. 2013;51(1):R1–R13. doi: 10.1530/JME-13-0067. [DOI] [PubMed] [Google Scholar]
- 3.Harjutsalo V., Podar T., Tuomilehto J. Cumulative incidence of type 1 diabetes in 10,168 siblings of finnish young-onset type 1 diabetic patients. Diabetes. 2005;54(2):563–569. doi: 10.2337/diabetes.54.2.563. [DOI] [PubMed] [Google Scholar]
- 4.Åkerblom H. K., Virtanen S. M., Ilonen J., et al. Dietary manipulation of beta cell autoimmunity in infants at increased risk of type 1 diabetes: a pilot study. Diabetologia. 2005;48(5):829–837. doi: 10.1007/s00125-005-1733-3. [DOI] [PubMed] [Google Scholar]
- 5.Knip M., Virtanen S. M., Seppä K., et al. Dietary intervention in infancy and later signs of beta-cell autoimmunity. The New England Journal of Medicine. 2010;363(20):1900–1908. doi: 10.1056/NEJMoa1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghazarian L., Diana J., Simoni Y., Beaudoin L., Lehuen A. Prevention or acceleration of type 1 diabetes by viruses. Cellular and Molecular Life Sciences. 2013;70(2):239–255. doi: 10.1007/s00018-012-1042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galleri L., Sebastiani G., Vendrame F., Grieco F. A., Spagnuolo I., Dotta F. Viral infections and diabetes. Advances in Experimental Medicine and Biology. 2012;771:252–271. doi: 10.1007/978-1-4614-5441-0_20. [DOI] [PubMed] [Google Scholar]
- 8.Yoon J.-W., Jun H.-S. Role of viruses in the pathogenesis of type 1 diabetes mellitus. In: LeRoith D., Taylor S. I., Olefsky J. M., editors. Diabetes Mellitus: A Fundamental and Clinical Text. Philadephia, Pa, USA: Lippincott Williams & Wilkins; 2004. [Google Scholar]
- 9.van der Werf N., Kroese F. G. M., Rozing J., Hillebrands J.-L. Viral infections as potential triggers of type 1 diabetes. Diabetes/Metabolism Research and Reviews. 2007;23(3):169–183. doi: 10.1002/dmrr.695. [DOI] [PubMed] [Google Scholar]
- 10.Mercalli A., Lampasona V., Klingel K., et al. No evidence of enteroviruses in the intestine of patients with type 1 diabetes. Diabetologia. 2012;55(9):2479–2488. doi: 10.1007/s00125-012-2591-4. [DOI] [PubMed] [Google Scholar]
- 11.Tracy S., Drescher K. M., Jackson J. D., Kim K., Kono K. Enteroviruses, type 1 diabetes and hygiene: a complex relationship. Reviews in Medical Virology. 2010;20(2):106–116. doi: 10.1002/rmv.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooke A. Review series on helminths, immune modulation and the hygiene hypothesis: how might infection modulate the onset of type 1 diabetes? Immunology. 2009;126(1):12–17. doi: 10.1111/j.1365-2567.2008.03009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludvigsson J., Holmqvist B.-M., Samuelsson U. Does modern high standard life style cause type 1 diabetes in children? Diabetes/Metabolism Research and Reviews. 2013;29(2):161–165. doi: 10.1002/dmrr.2377. [DOI] [PubMed] [Google Scholar]
- 14.Knip M., Veijola R., Virtanen S. M., Hyöty H., Vaarala O., Åkerblom H. K. Environmental triggers and determinants of type 1 diabetes. Diabetes. 2005;54(2):S125–S136. doi: 10.2337/diabetes.54.suppl_2.S125. [DOI] [PubMed] [Google Scholar]
- 15.Harinarayan C. V. Vitamin D and diabetes mellitus. Hormones. 2014;13(2):163–181. doi: 10.1007/BF03401332. [DOI] [PubMed] [Google Scholar]
- 16.Knip M., Simell O. Environmental triggers of type 1 diabetes. Cold Spring Harbor Perspectives in Medicine. 2012;2(7) doi: 10.1101/cshperspect.a007690.a007690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alves J. G. B., Figueiroa J. N., Meneses J., Alves G. V. Breastfeeding protects against type 1 diabetes mellitus: a case-sibling study. Breastfeeding Medicine. 2012;7(1):25–28. doi: 10.1089/bfm.2011.0009. [DOI] [PubMed] [Google Scholar]
- 18.Pereira P. F., Alfenas R. D. C. G., Araújo R. M. A. Does breastfeeding influence the risk of developing diabetes mellitus in children? A review of current evidence. Jornal de Pediatria. 2014;90(1):7–15. doi: 10.1016/j.jped.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 19.Thorsdottir I., Thorisdottir A. V. Whole cow's milk in early life. Nestle Nutr Workshop Ser Pediatr Program. 2011;67:29–40. doi: 10.1159/000325573. [DOI] [PubMed] [Google Scholar]
- 20.Norris J. M. Infant and childhood diet and type 1 diabetes risk: recent advances and prospects. Current Diabetes Reports. 2010;10(5):345–349. doi: 10.1007/s11892-010-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pflueger M., Seppänen-Laakso T., Suortti T., et al. Age- and islet autoimmunity-associated differences in amino acid and lipid metabolites in children at risk for type 1 diabetes. Diabetes. 2011;60(11):2740–2747. doi: 10.2337/db10-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaarala O. Leaking gut in type 1 diabetes. Current Opinion in Gastroenterology. 2008;24(6):701–706. doi: 10.1097/MOG.0b013e32830e6d98. [DOI] [PubMed] [Google Scholar]
- 23.Vaarala O., Atkinson M. A., Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57(10):2555–2562. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tracy S., Drescher K. M., Chapman N. M. Enteroviruses and type 1 diabetes. Diabetes/Metabolism Research and Reviews. 2011;27(8):820–823. doi: 10.1002/dmrr.1255. [DOI] [PubMed] [Google Scholar]
- 25.Filippi C. M., von Herrath M. G. Viral trigger for type 1 diabetes: pros and cons. Diabetes. 2008;57(11):2863–2871. doi: 10.2337/db07-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bason C., Lorini R., Lunardi C., et al. In type 1 diabetes a subset of anti-coxsackievirus B4 antibodies recognize autoantigens and induce apoptosis of pancreatic beta cells. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0057729.e57729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dezayee Z. M. I. The status of serum γ-interferonand antiviral antibodies in patients with type I and type 2 diabetes: a comparative study. Journal of Research in Medical Sciences. 2012;17(9):855–858. [PMC free article] [PubMed] [Google Scholar]
- 28.Carey C., Purohit S., She J.-X. Advances and challenges in biomarker development for type 1 diabetes prediction and prevention using “omic” technologies. Expert Opinion on Medical Diagnostics. 2010;4(5):397–410. doi: 10.1517/17530059.2010.508492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins C. D., Purohit S., Podolsky R. H., et al. The application of genomic and proteomic technologies in predictive, preventive and personalized medicine. Vascular Pharmacology. 2006;45(5):258–267. doi: 10.1016/j.vph.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Purohit S., Podolsky R., Schatz D., et al. Assessing the utility of SELDI-TOF and model averaging for serum proteomic biomarker discovery. Proteomics. 2006;6(24):6405–6415. doi: 10.1002/pmic.200600420. [DOI] [PubMed] [Google Scholar]
- 31.Purohit S., She J. X. Biomarkers for type 1 diabetes. International Journal of Clinical and Experimental Medicine. 2008;1(2):98–116. [PMC free article] [PubMed] [Google Scholar]
- 32.Zhi W., Purohit S., Carey C., Wang M., She J.-X. Proteomic technologies for the discovery of type 1 diabetes biomarkers. Journal of Diabetes Science and Technology. 2010;4(4):993–1002. doi: 10.1177/193229681000400431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakly W., Mankaï A., Sakly N., et al. Anti-Saccharomyces cerevisiae antibodies are frequent in type 1 diabetes. Endocrine Pathology. 2010;21(2):108–114. doi: 10.1007/s12022-010-9118-7. [DOI] [PubMed] [Google Scholar]
- 34.Simpson M., Mojibian M., Barriga K., et al. An exploration of Glo-3A antibody levels in children at increased risk for type 1 diabetes mellitus. Pediatric Diabetes. 2009;10(8):563–572. doi: 10.1111/j.1399-5448.2009.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacFarlane A. J., Burghardt K. M., Kelly J., et al. A type 1 diabetes-related protein from wheat (Triticum aestivum): cDNA clone of a wheat storage globulin, Glb1, linked to islet damage. The Journal of Biological Chemistry. 2003;278(1):54–63. doi: 10.1074/jbc.M210636200. [DOI] [PubMed] [Google Scholar]
- 36.Troncone R., Franzese A., Mazzarella G., et al. Gluten sensitivity in a subset of children with insulin dependent diabetes mellitus. The American Journal of Gastroenterology. 2003;98(3):590–595. doi: 10.1111/j.1572-0241.2003.07301.x. [DOI] [PubMed] [Google Scholar]
- 37.Adler K., Mueller D. B., Achenbach P., et al. Insulin autoantibodies with high affinity to the bovine milk protein alpha casein. Clinical and Experimental Immunology. 2011;164(1):42–49. doi: 10.1111/j.1365-2249.2011.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giongo A., Gano K. A., Crabb D. B., et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME Journal. 2011;5(1):82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salminen K., Sadeharju K., Lönnrot M., et al. Enterovirus infections are associated with the induction of β-cell autoimmunity in a prospective birth cohort study. Journal of Medical Virology. 2003;69(1):91–98. doi: 10.1002/jmv.10260. [DOI] [PubMed] [Google Scholar]
- 40.Leng S. X., McElhaney J. E., Walston J. D., Xie D., Fedarko N. S., Kuchel G. A. ELISA and multiplex technologies for cytokine measurement in inflammation and aging research. Journals of Gerontology A: Biological Sciences and Medical Sciences. 2008;63(8):879–884. doi: 10.1093/gerona/63.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu J., Lin J., Kim K.-H., Benjamin W. H., Jr., Nahm M. H. Development of an automated and multiplexed serotyping assay for Streptococcus pneumoniae . Clinical and Vaccine Immunology. 2011;18(11):1900–1907. doi: 10.1128/CVI.05312-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croft H., Malinowski T., Krizbai L., et al. Use of Luminex xMAP-derived Bio-Plex bead-based suspension array for specific detection of PPV W and characterization of epitopes on the coat protein of the virus. Journal of Virological Methods. 2008;153(2):203–213. doi: 10.1016/j.jviromet.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 43.Anderson S., Wakeley P., Wibberley G., Webster K., Sawyer J. Development and evaluation of a Luminex multiplex serology assay to detect antibodies to bovine herpes virus 1, parainfluenza 3 virus, bovine viral diarrhoea virus, and bovine respiratory syncytial virus, with comparison to existing ELISA detection methods. Journal of Immunological Methods. 2011;366(1-2):79–88. doi: 10.1016/j.jim.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 44.Opalka D., Lachman C. E., MacMullen S. A., et al. Simultaneous quantitation of antibodies to neutralizing epitopes on virus-like particles for human papillomavirus types 6, 11, 16, and 18 by a multiplexed luminex assay. Clinical and Diagnostic Laboratory Immunology. 2003;10(1):108–115. doi: 10.1128/CDLI.10.1.108-115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y., Xu Z. Q., Zhang Q., et al. Simultaneous detection of seven enteric viruses associated with acute gastroenteritis by a multiplexed luminex-based assay. Journal of Clinical Microbiology. 2012;50(7):2384–2389. doi: 10.1128/JCM.06790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takao S., Hara M., Okazaki T., Suzuki K. Simultaneous multiple assay (Luminex xTAG respiratory viral panel FAST assay) efficacy in human respiratory virus detection. Kansenshōgaku Zasshi. 2011;85(1):31–36. doi: 10.11150/kansenshogakuzasshi.85.31. [DOI] [PubMed] [Google Scholar]
- 47.Zhi W., Sharma A., Purohit S., et al. Discovery and validation of serum protein changes in type 1 diabetes patients using high throughput two dimensional liquid chromatography-mass spectrometry and immunoassays. Molecular and Cellular Proteomics. 2011;10(11) doi: 10.1074/mcp.M111.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guan R., Purohit S., Wang H., et al. Chemokine (c-c motif) ligand 2 (ccl2) in sera of patients with type 1 diabetes and diabetic complications. PLoS ONE. 2011;6(4) doi: 10.1371/journal.pone.0017822.e17822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu S., Purohit S., Sharma A., et al. Serum insulin-like growth factor binding protein 6 (IGFBP6) is increased in patients with type 1 diabetes and its complications. International Journal of Clinical and Experimental Medicine. 2012;5(3):229–237. [PMC free article] [PubMed] [Google Scholar]