Abstract
There is increasing evidence showing the involvement of CD4+ T cells in initiating and maintaining antitumor immune responses. NY-ESO-1 is expressed by various tumors but not normal tissues except testis. We conducted a cancer clinical trial by using full-length NY-ESO-1 protein formulated with ISCOMATRIX adjuvant and injected into patients intramuscularly. Autologous dendritic cells pulsed with NY-ESO-1 ISCOMATRIX in combination with overlapping synthetic peptides were used to identify immunodominant T cells from a vaccinated patient. We show here the identification and characterization of two novel CD4+ T cell epitopes. T cells specific to these epitopes not only recognized autologous dendritic cells loaded with NY-ESO-1 but also NY-ESO-1-expressing tumor cell lines treated with IFN-γ. One of the two responses identified was greater than the previously identified immunodominant HLA-DP4-restricted response and correlated with NY-ESO-1-specific CD8+ T cell induction after vaccination. This T cell response was vaccinated in most patients who expressed HLA-DR2. This study has systematically surveyed patients vaccinated with full-length tumor antigen for a vaccinated CD4 helper T cell response.
Keywords: CD4+ T cells, vaccine, tumor antigen
Activation of tumor-specific CD8+ T cells generally requires “help” from CD4+ T cells (1). The CD4+ T cells help CD8+ T cell priming by expressing CD40 ligand, which interacts with CD40 molecules expressed on dendritic cells (DCs), to “license” DCs (2). They may also help CD8+ T cells by providing general growth factors [such as IL-2 (3)] to promote activation and proliferation. More recently, CD4+ T cells were shown to be necessary in a memory response for CD8+ T cells to become fully activated (4), to sustain their functionality (5), and to expand efficiently (6). Since the early 1990s, many human tumor antigens recognized by CD8+ T cells have been characterized and used as antigens for peptide-based vaccines (see reviews in refs. 7 and 8). The majority of trials, however, have so far failed to reveal general practical strategies, and the clinical outcomes have been generally disappointing. One of the potential design flaws might be that not enough emphasis has been given to understanding the CD8+ and CD4+ T cell interaction. A successful vaccine will likely be one incorporating robust CD4+ helper epitopes (9).
NY-ESO-1 is one of the best characterized cancer testis antigens in terms of its immunogenicity, although little is known about its biological function. Patients who develop anti-NY-ESO-1 antibodies often have detectable CD8+ T cell responses (10, 11). More recently a similar observation has been extended to NY-ESO-1-specific CD4+ T cells (12).
We conducted a cancer clinical trial by using recombinant full-length NY-ESO-1 protein antigen formulated with ISCO-MATRIX adjuvant (IMX) (ISCOTEC AB, Parkville, Victoria, Australia) (13), particles composed of Quillaia saponins, cholesterol, and phospholipids (CSL Limited, Parkville, Victoria, Australia). The rationale was that IMX delivers formulated antigen to the cytosol of various cell types, including DCs (13), and that the full-length NY-ESO-1 protein should provide the immune system simultaneously with both CD4+“helper” and CD8+ T cell determinants to achieve enhanced immunity. Anti-NY-ESO-1 antibody, CD8+, and CD4+ T cell responses for a large number of NY-ESO-1 epitopes were observed (I.D.D. et al., unpublished work). Using monocyte-derived dendritic cells (MoDCs) loaded with NY-ESO-1 IMX, we report here the identification and fine characterization of two CD4+ T cell determinants from a vaccinated patient. We also show that most of the patients who were vaccinated with NY-ESO-1 IMX and who express HLA-DR2 molecule had detectable responses to the NY-ESO-186–99 epitope induced by vaccination.
Materials and Methods
Patient and Normal Blood Samples. All clinical trial subjects but subject 5 (30 μg) used in this study received 100 μg of NY-ESO-1 IMX vaccine three times at monthly intervals (Ludwig trial LUD99-008; ref. 21). The blood was collected with written informed consent. Normal blood samples were acquired from the Victorian Transplantation and Immunogenetics Service (Victoria, Australia). Peripheral blood mononuclear cells (PBMCs) from these individuals were isolated by Ficoll/Hypaque gradient and stored in liquid nitrogen until use.
Synthetic Peptides and Antibodies. NY-ESO-1 18-mer peptides overlapping by 12 aa were individually synthesized by Chiron Mimotopes (Clayton, Victoria, Australia). All other peptides were synthesized and purified (purity > 95%) by Auspep (Parkville, Victoria, Australia). The sequence of peptide 15 is NY-ESO-185–102 SRLLEFYLAMPFATPMEA, and the sequence of peptide 27 is NY-ESO-1157–174 SLLMWITQCFLPV-FLAQP. Anti-CD4 (phycoerythrin), anti-CD8 (Cy-Chrome), and anti-IFN-γ (FITC) were purchased from Becton Dickinson. Pan anti-DR (L243), anti-DP (B7/21), and anti-DQ (SPV-L3) (14) antibodies were used as culture supernatants.
Cell Culture. Epstein–Barr virus-transformed B lymphocyte cell lines (BLCLs) were established from autologous PBMCs by using standard techniques. Melanoma cell line LAR1 was established in our laboratory from a tumor biopsy. Melanoma cell line NW-MEL-38 is described in ref. 15. Epstein–Barr virus-transformed BLCLs 9080 and 9014 were made available from the 10th International HLA Workshop (New York), and T242 and T282 were made available by the Victorian Transplantation and Immunogenetics Service (a gift from B. Tait). All cells were cultured in RP-10 medium consisting of RPMI medium 1640 supplemented with 10% FCS (CSL, Melbourne), l-glutamine (2 mM), 2-ME (5 × 10-5 M), and antibiotics.
IFN-γ Treatment of Tumor Cell Lines. The NW-MEL-38 and LAR1 tumor cell lines were cultured in RP-10 medium plus 100 ng/ml recombinant human IFN-γ for 48 h before being used as antigen-presenting cells (APCs) or stained with anti-MHC antibodies.
Generation of DCs and Antigen-Specific T Cells. Monocytes were isolated from PBMCs by using anti-CD14-conjugated MACS beads (Miltenyi Biotec, Bergisch Gladbach, Germany) and cultured in RP-10 medium in the presence of granulocyte/macrophage colony-stimulating factor (20 ng/ml; Schering-Plough) and IL-4 (500 units/ml; Schering-Plough) for 7 days. The DCs were then pulsed with NY-ESO-1 IMX at a final concentration of ≈10–20 μg/ml at 37°C for 2 h followed by maturation induction with 20 ng/ml tumor necrosis factor α (R & D Systems), 1,000 units/ml IFN-α (RoferonA; Roche, Sydney), and 1 μM prostaglandin E2 (ICN) for 2 h. Antigen-loaded DCs were then used as APCs and cocultured with autologous CD14-depleted PBMCs at a ratio of 1:10 (DC:CD14 cells) in the presence of 10 units/ml human recombinant IL-2. Approximately 10–13 days later, the cells were screened against NY-ESO-1 18-mer peptides.
Generation of Antigen-Specific T Cell Lines. Briefly 5 × 106 PBMCs were pulsed in FCS containing medium with 10 μM NY-ESO-1 peptide as indicated for 30 min. Cells were then cultured in RP-10 medium containing IL-2 for the indicated time. The cultures that showed specific T cell activities were restimulated with the same antigen fortnightly to maintain them as T cell lines.
Intracellular Cytokine Staining. Autologous BLCLs were pulsed with peptide at a 10 μM concentration in the presence of 10% FCS at 37°C for 2 h to allow serum-mediated processing (16) and potential antigen uptake. In assays assessing restricting HLA alleles, free peptides were washed out after pulsing to prevent presentation from activated autologous T cells. T cells and Brefeldin A (10 μg/ml) were then added. Four hours later cells were stained with anti-CD4 and anti-CD8. The cells were then fixed with 1% paraformaldehyde and stained with anti-IFN-γ in the presence of 0.2% saponin. One hundred thousand cells were acquired on a FACSCalibur flow cytometer (Becton Dickinson) and analyzed with flowjo software (Tree Star, Ashland, OR).
Antibody-Blocking Assay. The target cells were pulsed with 10 μM peptide at 37°C for 1 h. The cells were washed, and 20 μl of anti-HLA-class II antibody supernatant was added for another hour before addition of T cells and Brefeldin A. T cell activation was measured by intracellular cytokine staining.
Results
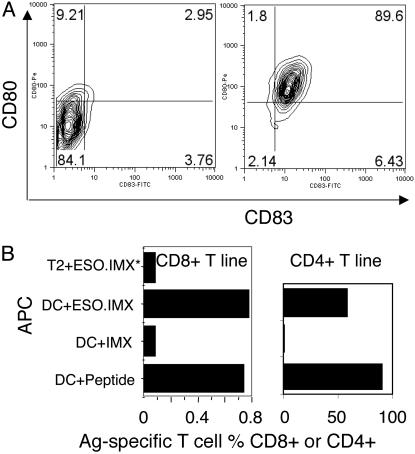
DCs Facilitated Identification of Immunodominant T Cell Determinants. It was rationalized that NY-ESO-1 IMX-loaded DCs should be able to process most of the naturally presented T cell epitopes from NY-ESO-1 to specific CD4+, as well as CD8+, and that T cells present in patient PBMC and the immunodominant T cells should be more abundant after stimulation. To prove the principle, T cell lines were established by using previously identified epitopes, including the HLA-A2-restricted CD8+ T cells specific to NY-ESO-1157–165 (17) and the HLA-DP4-restricted CD4+ T cells specific to NY-ESO-1157–170 (11). CD14+ monocytes from patient 7 were cultured for 7 days to derive MoDCs. As shown in Fig. 1A Left, the majority of MoDCs were of immature phenotype with limited CD80 and CD83 expression. These MoDCs were then loaded with 20 μg/ml NY-ESO-1 IMX and treated with tumor necrosis factor α, IFN-α, and prostaglandin E2. Upon maturation, the MoDCs all expressed high levels of CD80 and CD83 (Fig. 1A Right); they also expressed markedly higher CD86 and HLA-DR (data not shown). These NY-ESO-1 IMX-loaded MoDCs were able to activate NY-ESO-1157–165-specific CD8+ T cells as well as NY-ESO-1157–170-specific CD4+ T cells, raised from the same patient, to produce IFN-γ. Under the same conditions, the HLA-A2 expressing transporter associated with antigen processing (TAP)-deficient T2 cell line was not able to activate NY-ESO-1157–165-specific T cells (Fig. 1B), indicating that the antigen was processed endogenously.
Fig. 1.
MoDCs process T cell epitopes from NY-ESO-1 IMX. (A) The MoDCs from patient 7 were loaded with NY-ESO-1 IMX, matured, and stained for CD80 and CD83 (and also for CD86 and HLA-DR; data not shown). (B) Preestablished T cell lines specific to either NY-ESO-1157–165 or NY-ESO-1157–170 were used to detect antigen presentation after NY-ESO-1 IMX loading onto DC or T2 cells.
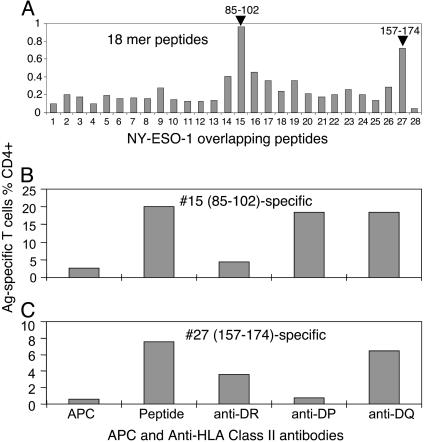
The antigen-loaded, matured MoDCs were cocultured with thawed CD14-depleted PBMCs for ≈10–13 days. The T cells were screened with a set of 18-mer NY-ESO-1 peptides by using autologous BLCLs as APCs. Two major responding clusters were identified around peptides 15 (NY-ESO-185–102) and 27 (NY-ESO-1157–174) for CD4+ T cells (Fig. 2A). Although there were clearly identifiable antigen-specific CD8+ T cells (data not shown), CD4+ T cell expansion tended to dominate the cultures.
Fig. 2.
Identification of previously uncharacterized CD4+ HLA-DR-restricted NY-ESO-1-specific T cells. (A) T cells stimulated as per Fig. 1 were assessed for their specificities against NY-ESO-1 18-mer overlapping peptides by using autologous BLCLs as APCs in the presence of FCS. (B and C) T cells used in A were stimulated one more time in vitro with either peptide 15 (NY-ESO-185–102)(B) or peptide 27 (NY-ESO-1157–174)(C). The cells were then assayed in the presence or absence of anti-class II antibodies in intracellular cytokine staining.
To gain information on restricting HLA molecules for the identified CD4+ T cells, an antibody-blocking assay was conducted. The anti-DR antibody efficiently blocked the T cell activation to peptide 15, whereas the anti-DP antibody blocked T cells specific to peptide 27. Anti-DQ did not block T cell responses to either peptide (Fig. 2 B and C). We later confirmed that the T cells that responded to peptide 27 recognized the previously reported, HLA-DP4-restricted NY-ESO-1157–170 epitope (11), which was entirely contained in our peptide 27. The T cells recognizing peptide 15 (NY-ESO-185–102) represented a previously uncharacterized finding.
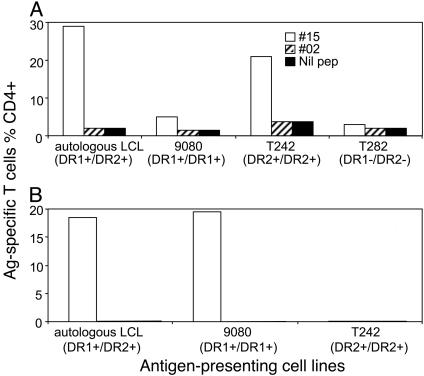
NY-ESO-185–102 Contains Multiple CD4+ T Cell Determinants. To further identify the restricting HLA-DR molecule, we obtained cell lines expressing homozygous DR alleles identical to those of the patient (Table 1). Bulk T cells originally stimulated by MoDCs loaded with NY-ESO-1 IMX were further stimulated with NY-ESO-185–102 and tested on various APCs pulsed with this peptide. The majority of NY-ESO-185–102-specific T cells were DR2-restricted (Fig. 3A). However, the homozygous DR1+ cell line (9080) also stimulated ≈10% of the total antigen-specific T cells, which could potentially explain the greater responses on autologous (heterozygous DR1+ and DR2+) versus homozygous DR2+ APCs (T242). To address the possibility of multiple CD4+ T cell determinants within the same peptide NY-ESO-185–102, we derived T cell sublines from the bulk T cell culture. As shown in Fig. 3B, one of the sublines was clearly DR1-restricted. Therefore, there were at least two CD4+ T cell determinants within NY-ESO-185–102.
Table 1. HLA details of the cell lines used in this work.
| Cell line ID | HLA-A | HLA-B | DR | DP | ||||
|---|---|---|---|---|---|---|---|---|
| Patient 7 | 0101 | 0201 | 0801 | 2705 | 0101 | 1501 | 0401 | |
| T242 | 2403 | 6801 | 0702 | 5106 | 1502 | 0402 | 26012 | |
| T282 | 0201 | 0301 | 4402 | 5701 | 1421 | 0701 | 0401 | |
| 9080 | 0301 | 3501 | 3503 | 0101 | ||||
| NW-Mel-38 | 0201 | 0101 | 0701 | |||||
| LAR-1 | 24 | 0201 | 7 | 70 | 1302 | 1501 | ||
Fig. 3.
Two DR-restricted CD4 determinants within peptide NY-ESO-185–102. BLCLs expressing homozygous HLA-DR molecule and a short-term T cell line (used in Fig. 2 A) and a long-term subline were used in these experiments. (A) The short-term line showed that the majority of T cells were restricted by DR2 and the minority of T cells were restricted by DR1. (B) Long-term subline derived from peptide 15-specific T cells showed exclusive DR1 restriction.
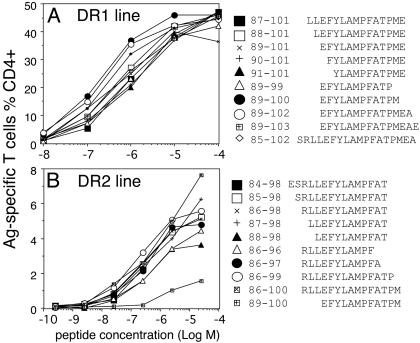
To further identify the minimum sequences, we first titrated 13-mer peptides with 2-aa shift within NY-ESO-185–102 and located the core sequences NY-ESO-185–97 and NY-ESO-189–101 by using either DR1- or DR2-restricted sublines (data not shown). We then synthesized extended and truncated peptides based on the 13-mer core sequences as shown in Fig. 4 A and B. These peptides were used to pulse homozygous PBMCs in the absence of FCS to avoid serum-mediated peptide processing (16). The minimum, yet most potent, sequence for the DR1-restricted determinant was NY-ESO-189–100 (Fig. 4A), and the minimum sequence for the DR2-restricted determinant was NY-ESO-186–99 (Fig. 4B). Although T cells specific for the latter determinant were more promiscuous in terms of peptide length, they did not recognize the DR1-restricted minimum peptide NY-ESO-189–100 (Fig. 4B) indicating that residue 88 was essential.
Fig. 4.
Fine mapping of the minimum T cell determinants. By using T cell lines [DR1-restricted (A) and DR2-restricted (B)], truncated or extended peptides surrounding the core sequences were tested by titration. Autologous PBMCs were pulsed with the peptides in the absence of FCS for 60 min. Excess peptides were washed out before addition of antigen-specific T cells.
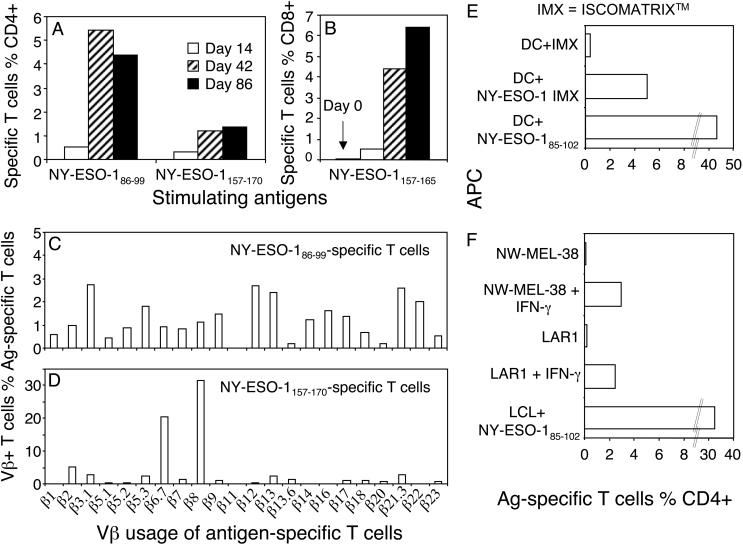
The Newly Identified DR2-Restricted CD4+ T Cell Determinant Was Immunodominant and Stimulated Polyclonal Responses. Having acquired the minimum sequences for the novel CD4+ T cell determinants, we wanted to investigate their vaccination status side by side with the DP4-restricted response. We stimulated PBMCs from patient 7, collected at various times after vaccination, with either the reported minimum DP4-restricted peptide (NY-ESO-1157–170) or the DR2-restricted peptide (NY-ESO-186–99). Under similar conditions, it was clear that the DR2-restricted T cell response was detected at the earliest time point after vaccination (on day 14, day-0 sample was not available) (Fig. 5A), matching the induced CD8+ responses to NY-ESO-1157–165 (Fig. 5B), which was not detectable before vaccination (day-0 sample). It was also clear that the DR2-restricted T cell response was greater than the DP4-restricted T cell response at each sampling point and had a broader T cell receptor usage (Fig. 5 A, C, and D).
Fig. 5.
The previously uncharacterized DR2-restricted T cells are more abundant and more polyclonal than the DP4-restricted T cells and recognize melanoma cell lines. (A) Multiple PBMC samples from patient 7 were thawed and stimulated with NY-ESO-186–99 or NY-ESO-1157–170. The T cell responses were analyzed on day 11. Note that the day-0 sample was not available for this assay. (B) An earlier analysis for NY-ESO-1157–165-specific CD8+ T cell response was performed including the day-0 sample. (C and D) T cells from day 86 after vaccination were stimulated with either NY-ESO-1157–170 or NY-ESO-186–99 and assessed with intracellular cytokine staining plus single Vβ antibodies. Vβ-positive and antigen-specific T cells were displayed as the percent of total antigen-specific T cells. (E and F) The NY-ESO-185–102-specific T cell line was used to read out antigen presentation of autologous MoDCs pulsed with NY-ESO-1 IMX (10 μg/ml, E) and DR1+ (NW-MEL-38) or DR2+ (LAR1) melanoma cell lines with or without IFN-γ treatment (F).
HLA-DR-Restricted T Cells Induced by NY-ESO-1 IMX Vaccine Recognize Naturally Presented Determinants. We confirmed that T cells from patient 7 stimulated with NY-ESO-185–102 were able to recognize autologous MoDCs loaded with NY-ESO-1 IMX (Fig. 5E). In addition, these T cells also recognized naturally presented NY-ESO-1 determinants on the NY-ESO-1-expressing DR1-positive melanoma line NW-MEL-38 and the DR2-positive melanoma line LAR1 (Table 1) after IFN-γ treatment in vitro (Fig. 5F). The CD4+ T cell line was not activated by either of the above cell lines without IFN-γ treatment (Fig. 5F) or tumor cells that did not express the appropriate DR allele even after IFN-γ treatment (data not shown).
Assessing the HLA-DR2-Restricted, NY-ESO-1-Specific T Cell Responses Among NY-ESO-1 IMX-Vaccinated Patients. It was reported that almost all patients who had serum antibody to NY-ESO-1 also expressed DP4 and exhibited the DP4-restricted NY-ESO-1157–170 T cell responses (11). Coincidentally, we had multiple subjects from the NY-ESO-1 IMX vaccine trial who expressed both HLA-DR2 and DP4. We sought to investigate which response might be more relevant to vaccination by screening the available samples for these antigen-specific T cells. We found that the DR2-restricted T cell response showed very good correlation to DR2 expression among our vaccine trial subjects (Table 2). All five DR2-positive samples showed the respected CD4+ T cell responses to NY-ESO-186–99, whereas of six DP4 positive samples, only one (patient 7) showed the DP4-restricted response. Among all of the detected DR2-restricted T cell responses, we were clearly able to demonstrate that three were due to vaccination because the same activities were not detected in the prevaccination samples under similar conditions. There were no prevaccination samples available to demonstrate the vaccination status for the other two subjects (Table 2, marked N/A). On the other hand, eight DR2-positive samples (six DP4-positive samples among them) from normal individuals were also tested the same way as described above, and no antigen specific T cell response was observed (data not shown).
Table 2. Survey of DR2-restricted T cell responses among NY-ESO-1 IMX-vaccinated patients.
| NY-ESO-1/DR2 responses
|
NY-ESO-1/DP4 responses
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Patient | Anti-ESO antibody | HLA-DR2 | T cells % CD4+ prevaccination | T cells % CD4+ postvaccination | HLA-DP4 | T cells % CD4+ postvaccination | |||
| 5 | + | + | N/A | + | 0.22 | — | — | 0.03 | |
| 7 | + | + | N/A | + | 2.46 | + | + | 1.07 | |
| 9 | + | — | — | 0 | — | 0.006 | — | NT | |
| 10 | + | + | — | 0.033 | + | 0.44 | + | +/— | 0.065 |
| 13 | + | — | +/— | 0.066 | + | 0.12 | + | — | 0.017 |
| 24 | + | + | — | 0.015 | + | 0.18 | + | — | 0.018 |
| 16 | + | — | +/— | 0.068 | NT | + | +/— | 0.062 | |
| 17 | + | + | — | 0.023 | + | 0.27 | — | — | 0.01 |
| 19 | + | — | NT | — | 0.008 | + | — | 0.016 | |
| 21 | + | — | — | 0.023 | — | 0.012 | — | — | 0 |
Postvaccination samples from listed patients were tested for the DR2-restricted (NY-ESO-186—99) and DP4-restricted (NY-ESO-1157—170) responses after a 13-day culture. The positive samples were then repeated side by side with prevaccination samples, if available. Eight DR2-positive samples (six of which were also DP4+) from normal individuals were also tested for the two T cell specificities, and the results were all negative (not listed). Intracellular cytokine staining values (shown as percentage of CD4+ cells) from all negative controls were <0.03% of total gated CD4+ cells. The definite positive results are in bold. +/— potential but weak responses. NT, not tested; N/A, sample not available.
Discussion
Whereas tumor-specific CD8+ T cell epitope identification has advanced rapidly, similar efforts for CD4+ T cell epitopes have lagged behind. It is increasingly evident that CD4+ T cells play important roles both as helpers as well as effectors in viral, tumor, and autoimmune responses (18, 19) and that a robust and lasting immune response hinges on a well balanced CD8+ and CD4+ T cell collaboration (4–6).
Using MoDCs loaded with full-length NY-ESO-1 formulated with IMX, we demonstrated two major CD4+ T cell responses from vaccinated patient 7. This method gives us the ability to potentially monitor or discover the most abundant T cell responses without necessarily knowing either the T cell determinants from a known tumor antigen or the MHC allelic details of a given individual. More importantly, the T cell determinants identified this way should only represent the ones naturally processed and presented because the patients were vaccinated with full-length NY-ESO-1 antigen and targeted to APCs efficiently. The T cells were most likely primed in vivo because we used only one round of antigen stimulation and a relatively short culture period before peptide screening, methods which do not lead to in vitro priming. Two HLA-DR-restricted specificities were discovered, and both determinants were presented by class II-expressing melanoma cell lines after IFN-γ treatment. These cell lines normally express relatively low-level surface class II molecules, and their expression was enhanced by 3- to 4-fold after IFN-γ treatment (data not shown), which resulted in detectable natural antigen presentation (Fig. 5F).
The natural CD8+ T cell responses to NY-ESO-1157–165 (17) and CD4+ T cell responses (11, 12) are tightly linked to antibody production to the same antigen. Previous work also suggested that anti-NY-ESO-1 antibody production might correlate directly with HLA-DP4 expression, hinting that the DP4-restricted and NY-ESO-1157–170-specific CD4+ T cells might be the dominant “helper,” although in the original survey a longer peptide was used and combined with multiple rounds of stimulation (11). Our data clearly confirmed some of these earlier observations in a vaccination setting. Many of the trial patients were DP4-positive, and most of them showed pronounced delayed-type hypersensitivity responses toward the vaccinating antigen and produced high titer anti-NY-ESO-1 antibodies (I.D.D., unpublished work) (Table 2). However, although we have confirmed the DP4-restricted NY-ESO-1157–170-specific response as one of the two immunodominant CD4+ T cell responses from patient 7 (Fig. 2 A), we have failed to detect significant T cell responses specific to the same epitope from any other DP4-expressing subjects (Table 2). In contrast, the newly identified DR2-restricted T cell response was more dominant than the DP4-restricted response in patient 7 samples when both were stimulated with the minimum peptides under similar conditions (Fig. 5A). That response was clearly induced by vaccination in all of the patients with available prevaccination samples (Table 2). These findings cast some doubt on the DP4-restricted determinant as the sole dominant helper determinant from NY-ESO-1. It is interesting to note that the same region containing our previously uncharacterized CD4+ determinants was reported recently to contain epitopes restricted by DR7 (NY-ESO-187–98) (12). In that report, multiple DR2-expressing (shown as DR15) patients did respond to NY-ESO-180–109, although the fine specificities were not further characterized (12). It might be possible that some of those responses represented the T cell responses to the immunodominant DR2 determinant, found in our study, caused by natural immunization. Various DR molecules may share binding motifs, and DR2 is normally expressed in ≈25% of the population of various ethnic backgrounds (20). It would not be surprising if the real responses to NY-ESO-186–99 were relatively larger because of broader DR binding, which might explain our positive results for NY-ESO-186–99 from patient 13 (Table 2), who is DR2- but DR1+ and DR11+. Further study on the DR2-restricted and NY-ESO-186–99 specific immune response from patients with a natural disease course will be helpful in confirming the above observation. The study on immunodominant CD4+ T cells and their determinants should help us to improve vaccine design.
Acknowledgments
We thank Tony Burgess, Andrew Scott, Brian Tait, Sharen Gibbs, Michael Rubira, Anna Kypridis, and Christine Millar, and Dr. Sacha Gnjatic for critical discussions. W.C. is supported by Wellcome Trust International Senior Research Fellow Fellowship 066646/Z/01/Z. I.D.D. is supported in part by an Australian National Health and Medical Research Council Career Development Award.
Abbreviations: APC, antigen-presenting cell; BLCL, B lymphocyte cell line; DC, dendritic cell; IMX, ISCOMATRIX adjuvant; MoDC, monocyte-derived dendritic cell; PBMC, peripheral blood mononuclear cell.
References
- 1.Cella, M., Scheidegger, D., Palmer-Lehmann, K., Lane, P., Lanzavecchia, A. & Alber, G. (1996) J. Exp. Med. 184, 747-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, S. R., Carbone, F. R., Karamalis, F., Flavell, R. A., Miller, J. F. & Heath, W. R. (1998) Nature 393, 478-480. [DOI] [PubMed] [Google Scholar]
- 3.Fearon, E. R., Pardoll, D. M., Itaya, T., Golumbek, P., Levitsky, H. I., Simons, J. W., Karasuyama, H., Vogelstein, B. & Frost, P. (1990) Cell 60, 397-403. [DOI] [PubMed] [Google Scholar]
- 4.Gao, F. G., Khammanivong, V., Liu, W. J., Leggatt, G. R., Frazer, I. H. & Fernando, G. J. (2002) Cancer Res. 62, 6438-6441. [PubMed] [Google Scholar]
- 5.Cardin, R. D., Brooks, J. W., Sarawar, S. R. & Doherty, P. C. (1996) J. Exp. Med. 184, 863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janssen, E. M., Lemmens, E. E., Wolfe, T., Christen, U., von Herrath, M. G. & Schoenberger, S. P. (2003) Nature 421, 852-856. [DOI] [PubMed] [Google Scholar]
- 7.Renkvist, N., Castelli, C., Robbins, P. F. & Parmiani, G. (2001) Cancer Immunol. Immunother. 50, 3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, I. D., Jefford, M., Parente, P. & Cebon, J. (2003) J. Leukocyte Biol. 73, 3-29. [DOI] [PubMed] [Google Scholar]
- 9.Zeng, G., Li, Y., El-Gamil, M., Sidney, J., Sette, A., Wang, R.-f., Rosenberg, S. A. & Robbins, P. F. (2002) Cancer Res. 62, 3630-3635. [PMC free article] [PubMed] [Google Scholar]
- 10.Jager, E., Nagata, Y., Gnjatic, S., Wada, H., Stockert, E., Karbach, J., Dunbar, P. R., Lee, S. Y., Jungbluth, A., Jager, D., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 4760-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng, G., Wang, X., Robbins, P. F., Rosenberg, S. A. & Wang, R. F. (2001) Proc. Natl. Acad. Sci. USA 98, 3964-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnjatic, S., Atanackovic, D., Jager, E., Matsuo, M., Selvakumar, A., Altorki, N. K., Maki, R. G., Dupont, B., Ritter, G., Chen, Y. T., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8862-8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ennis, F. A., Cruz, J., Jameson, J., Klein, M., Burt, D. & Thipphawong, J. (1999) Virology 259, 256-261. [DOI] [PubMed] [Google Scholar]
- 14.Robbins, P. A., Evans, E. L., Ding, A. H., Warner, N. L. & Brodsky, F. M. (1987) Hum. Immunol. 18, 301-313. [DOI] [PubMed] [Google Scholar]
- 15.Jager, E., Jager, D., Karbach, J., Chen, Y. T., Ritter, G., Nagata, Y., Gnjatic, S., Stockert, E., Arand, M., Old, L. J. & Knuth, A. (2000) J. Exp. Med. 191, 625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherman, L. A., Burke, T. A. & Biggs, J. A. (1992) J. Exp. Med. 175, 1221-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jager, E., Chen, Y. T., Drijfhout, J. W., Karbach, J., Ringhoffer, M., Jager, D., Arand, M., Wada, H., Noguchi, Y., Stockert, E., et al. (1998) J. Exp. Med. 187, 265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson, K. E., Stromnes, I., Messer, R., Hasenkrug, K. & Chesebro, B. (2002) J. Virol. 76, 7942-7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez, N. R., Augstein, P., Moustakas, A. K., Papadopoulos, G. K., Gregori, S., Adorini, L., Jackson, D. C. & Harrison, L. C. (2003) J. Clin. Invest. 111, 1365-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doolan, D. L., Southwood, S., Chesnut, R., Appella, E., Gomez, E., Richards, A., Higashimoto, Y. I., Maewal, A., Sidney, J., Gramzinski, R. A., et al. (2000) J. Immunol. 165, 1123-1137. [DOI] [PubMed] [Google Scholar]
- 21.Davis, I. D., Chen, W., Jackson, H., Parente, P., Shackleton, M., Hopkins, W., Chen, Q., Dimopoulos, N., Luke, T., Murphy, R., et al. (2004) Proc. Natl. Acad. Sci. USA 101, in press.