Abstract
Mammalian hibernators survive low body temperatures, ischemia–reperfusion, and restricted nutritional resources via global reductions in energy-expensive cellular processes and selective increases in stress pathways. Consequently, studies that analyze hibernation uncover mechanisms which balance metabolism and support survival by enhancing stress tolerance. We hypothesized processing factors that influence messenger ribonucleic acid (mRNA) maturation and translation may play significant roles in hibernation. We characterized the amino acid sequences of three RNA processing proteins (T cell intracellular antigen 1 (TIA-1), TIA1-related (TIAR), and poly(A)-binding proteins (PABP-1)) from thirteen-lined ground squirrels (Ictidomys tridecemlineatus), which all displayed a high degree of sequence identity with other mammals. Alternate Tia-1 and TiaR gene variants were found in the liver with higher expression of isoform b versus a in both cases. The localization of RNA-binding proteins to subnuclear structures was assessed by immunohistochemistry and confirmed by subcellular fractionation; TIA-1 was identified as a major component of subnuclear structures with up to a sevenfold increase in relative protein levels in the nucleus during hibernation. By contrast, there was no significant difference in the relative protein levels of TIARa/TIARb in the nucleus, and a decrease was observed for TIAR isoforms in cytoplasmic fractions of torpid animals. Finally, we used solubility tests to analyze the formation of reversible aggregates that are associated with TIA-1/R function during stress; a shift towards the soluble fraction (TIA-1a, TIA-1b) was observed during hibernation suggesting enhanced protein aggregation was not present during torpor. The present study identifies novel posttranscriptional regulatory mechanisms that may play a role in reducing translational rates and/or mRNA processing under unfavorable environmental conditions.
Keywords: Hibernation, Stress response, RNA-binding proteins, Nucleus, Transcription, Translation, mRNA processing
Introduction
Living organisms inhabit all corners of the earth, surviving anywhere from the dry and arid conditions of the desert to the frigid ice-locked arctic. Each environment offers both resources and challenges, which are reflected in the strategies the resident organisms use to survive and reproduce. A common organismal response to environmental stress is metabolic depression. Entry into a hypometabolic state (termed hibernation, torpor, dormancy, or diapause) minimizes energy expenditures and preserves life until the stress passes. The ability to reorganize metabolic activity, yet retain full viability upon restoration of the aroused state, could have numerous medical applications including the treatment of strokes, heart attacks, or traumatic battlefield injuries as well as increasing the “shelf life” of organs removed for transplantation. Consequently, there is great interest in highlighting specific networks that help to transition an organism from periods of growth and proliferation to those of conservation and preservation. The mechanisms utilized to limit energy expenditure involve profound behavioral and physiological changes that can be traced to important molecular “switches” that shut down metabolism and aid in meeting challenges associated with hypometabolism (Storey 2010; Bouma et al. 2012). A prime example is hibernation, where mammals limit activity in their burrows and settle into a deep torpor. This behavior is matched with the strong downregulation of ATP-costly intracellular processes including gene transcription and protein synthesis (Wang and Lee 1996; Storey and Storey 2004). A secondary, yet equally important component to the hypometabolic state, is the selective upregulation of multiple preservation mechanisms that counteract the otherwise deleterious aspects of surviving hypothermia, ischemia–reperfusion, and restricted ATP stores (Storey and Storey 2004, 2010; Storey 2010).
During the winter season, hibernating mammals settle into prolonged periods (days or weeks) of torpor during which their core body temperature (Tb) can fall to near ambient temperatures. A process of regulated metabolic suppression triggers a fall in Tb. This lower Tb further slows metabolism allowing some torpid species to have a Tb that approach 0 °C, with metabolic rates of <1 % of their resting euthermic rate (Geiser 2004; Heldmaier et al. 2004; Chung et al. 2011). Low Tb is not the only challenge hibernators must overcome; in ground squirrels, heart rate, respiration rate, and cerebral blood flow all fall to approximately 1, 3, and 10 % of their normal levels, respectively, resulting in a dramatic reduction in oxygen demand in order for tissues to maintain normoxic status during torpor (Zatzman 1984; McArthur and Milsom 1991; Martin et al. 1999; Dave et al. 2012). These and other physiological parameters are rapidly returned to euthermic levels during intermittent arousals, as a 10–20-fold surge in oxygen consumption occurs within minutes of the animal rewarming to 37 °C (Storey 2003; Staples 2011). This surge in oxygen consumption increases the production of reactive oxygen species (ROS) necessitating most organisms that use hypometabolism to possess increased antioxidant defenses, which protect macromolecules from damage by ROS both during extended periods of torpor and arousal (Storey 1996). Since these physiological changes are poorly tolerated in nonhibernating species, hibernators represent an excellent model system for understanding the intricacies of the stress response and the biochemical adaptations that mitigate stress.
The genes that support the hibernating phenotype appear to be common among all mammals, suggesting that regulatory changes determine why some mammals can enter torpor and survive the associated stresses while others cannot. Such regulatory changes may include altered controls over messenger ribonucleic acid (mRNA) transcripts. From the moment these molecules are synthesized in the nucleus to their demise following translation in the cytoplasm, eukaryotic mRNAs are subject to an intricate array of regulatory events that mediate every step of their life cycle (Moore 2005). A host of RNA-binding proteins act as mRNA chaperones and, through these protein adaptors, individual mRNAs can respond to a multitude of inputs. T cell intracellular antigen 1 (TIA-1), TIA1-related (TIAR), and poly(A)-binding proteins (PABP-1) are stress-responsive DNA/RNA-binding proteins that exert control over mRNA fate during transcription (Suswam et al. 2005), pre-mRNA splicing (Minvielle-Sebastia et al. 1997; Suswam et al. 2005), translation (Sachs and Davis 1989; Kedersha and Anderson 2007), mRNA stabilization (Decker and Parker 1993), and decay (Caponigro and Parker 1995). TIA-1 and TIAR are composed of three N-terminal RNA recognition motifs (RRM1–3) and a C-terminal glutamine-rich motif (Suswam et al. 2005). The glutamine-rich motifs of TIA-1 and TIAR are structurally related to prion proteins (prion-like domains (PRD)) and have the capacity to form reversible aggregates (Gilks et al. 2004). TIA-1/TIAR are expressed as two major isoforms (TIA-1a/TIA-1b, TIARa/TIARb), which regulate alternative pre-mRNA splicing of various genes (e.g., FGFR-2, msl-2, Fas) and display distinct functional properties (Izquierdo and Valcárcel 2007). PABP-1 contains four RNA-binding domains (RBDs) and a proline-rich C-terminal domain which associates with the 3′ poly(A) tail of mRNA (Burd et al. 1991).
In the present study, we endeavored to identify the role played by RNA-binding proteins in the posttranscriptional regulatory control of mRNA transcripts during hibernation in the liver of thirteen-lined ground squirrels (Ictidomys tridecemlineatus). Since RNA-binding proteins such as TIA-1/R and PABP-1 influence transcription and mRNA splicing as well as the localization, stability, and association of transcripts with the translation machinery, we hypothesized that these factors could play a significant role in the global suppression of translation during hypometabolism. Further to this, we propose that this response would be especially vital to the hibernating liver since it plays an established role in metabolism and in the maintenance of homeostasis over cycles of torpor and arousal. To address this hypothesis, we characterized the amino acid sequence of ground squirrel TIA-1, TIAR, and PABP-1 and identified the presence of TIA-1 and TIAR splice variants in the liver. The generation of alternative splice variants and their differential expression have the capacity to modify protein function, yet their involvement remains a largely unexplored concept during mammalian hibernation. Additionally, we utilized fluorescence microscopy, protein fractionation, and Western blotting to assess the localization and relative expression of the RNA-binding proteins. Finally, we used solubility tests to assess the formation of reversible aggregates that are associated with TIA-1/R function during stress. The data presented here demonstrates the presence of two major TIA-1 and TIAR isoforms, which possessed a high degree of sequence similarity among higher mammals. Surprisingly, the shorter variants of both TIA-1 and TIAR (isoform b) displayed consistently higher expression across experimental conditions. Subnuclear foci were observed during hibernation using immunofluorescence microscopy, complementing the observation of a relative increase of TIA-1a and TIA-1b in the nuclear fractions of torpid ground squirrels. The solubility of TIARa/b, TIA-1 p15, and PABP-1 did not change, but a relative increase in the ratio of soluble/insoluble fractions was observed for TIA-1a and TIA-1b during hibernation. The data revealed that RNA-binding proteins play a role in mRNA processing during hibernation, and these proteins could provide a framework for the global reductions in translation and selective regulation of key cellular networks as well as facilitate the rapid reversal of the hibernating phenotype.
Materials and methods
Animals
Thirteen-lined ground squirrels (I. tridecemlineatus; formerly Spermophilus tridecemlineatus) were wild-captured by a licensed trapper (TLS Research, Bloomingdale, IL) and transported to the Animal Hibernation Facility of the National Institute of Neurological Disorders and Stroke (NIH, Bethesda, MD). Hibernation experiments were conducted by the laboratory of Dr. J.M. Hallenbeck, as described by McMullen and Hallenbeck (2010). Animals were fitted with a subcutaneous sensor chip (IPTT-300; Bio Medic Data Systems) while anesthetized with 5 % isoflurane and transferred to a holding room in individually housed shoebox cages (ambient temperature, Ta, of 21 °C). Ground squirrels were fed standard rodent diet and water ad libitum until they gained sufficient lipid stores to enter hibernation. To induce hibernation, the animals were transferred to an environmental chamber (Ta of 5 °C and 60 % humidity) in cages containing wood shavings. The hibernaculum was kept in constant darkness, except for a photographic red safe light (3–5 lx), and could be entered only through a darkened anteroom. Body temperature, time, and respiration rate was monitored and used to determine the stage of torpor–arousal cycle. Experiments were done during the winter months of December–March; thus, all animals had been through a series of torpor–arousal bouts prior to sampling. Animals were sampled as in McMullen and Hallenbeck (2010), and excised tissues were then shipped to Carleton University on dry ice (stored at −80 °C until use). EC designates euthermic, cold room; these euthermic squirrels had a stable Tb (∼37 °C) for at least 3 days. EC animals were capable of entering torpor but had not done so in the past 72 h. LT designates late torpor; animals were in deep torpor for 5 days (Tb = 5–8 °C).
Computational analysis
The full sequences for ground squirrel (I. tridecemlineatus) TIA-1, TIAR, and PABP-1 were deduced using whole genome shotgun (wgs) reads. A wgs BLAST (NCBI) search using the human and mouse sequences allowed overlapping contigs to be assembled into the full coding sequences for the ground squirrel. Human sequences used for TIA-1, TIAR, and PABP-1 wgs BLAST (NCBI) searches had the accession numbers NM_022173.2, NM_001033925.1, and NM_002568.3, respectively. Mouse sequences used for TIA-1, TIAR, and PABP-1 wgs BLAST (NCBI) searches had the accession numbers NM_011585.4, NM_009383.2, and NM_008774.3, respectively. A minimum of two overlapping contigs were used to construct the full ground squirrel sequence. The predicted ground squirrel mRNA coding sequence was translated into amino acids and aligned with human (Homo sapiens), Rhesus macaque (Mucaca mulatta), bovine (Bos taurus), rat (Rattus norvegicus), mouse (Mus musculus), and African clawed frog (Xenopus laevis) using Geneious (Biomatters Ltd) in order to determine percent homology. The domains/motifs were predicted using the Eukaryotic Linear Motif resource for Functional Sites in Proteins (ELM, http://elm.eu.org/) and compared to those predicted using the ScanProsite tool (http://prosite.expasy.org/scanprosite/).
Primer design and synthesis
Primers for Tia-1 and TiaR were designed using alignments created by Geneious (Biomatters Ltd) and online OligoAnalyzer software (Integrated DNA Technologies, Inc.) based on the deduced ground squirrel coding sequence and consensus sequences for the genes derived from several mammalian species (mouse, rat, cow, human). The Tia-1 primer pairs span exon 5 (i.e., amplify Tia-1 splice variants a and b), while the TiaR primer pairs span an alternate in-frame splice site in the coding region (i.e., amplify TiaR splice variants a and b). The predicted transcript size of Tia-1a amplified from F1R1 and F2R1 is 314 and 319 bp, respectively, and the predicted transcript size of Tia-1b amplified from the same primer pairs is 281 and 286, respectively. The predicted transcript size of TiaRa amplified from F1R1 and F1R2 is 290 and 338 bp, respectively, and the predicted transcript size of TiaRb amplified from the same primer pairs is 239 and 287, respectively. The primer sequences (purchased from Integrated DNA Technologies, Inc.) were as follows:
Tia-1 forward 1 (Tia-1 F1) 5′GATAATGGGTAAGGAAGTCA–3′
Tia-1 forward 2 (Tia-1 F2) 5′–CGGAAGATAATGGGTAAGGA–3′
Tia-1 reverse 1 (Tia-1 R1) 5′–CCAGTTAGTTCTGATTTGTC–3′
TiaR forward 1 (TiaR F1) 5′–GGACCCTGTAAAAGCTGTAA–3′
TiaR reverse 1 (TiaR R1) 5′–GGGCAAATGCTGATTTGATA–3′
TiaR reverse 2 (TiaR R2) 5′–GTTGCCATGTCTTTAACTAC–3′
RNA isolation, cDNA synthesis, and PCR amplification
Total RNA was extracted from the liver (∼100 mg) of n = 4 individual animals from control (EC) and LT groups. Samples were homogenized using a Polytron homogenizer in 1 mL TRIzolTM reagent (Invitrogen), according to manufacturer’s instructions and as previously described (Tessier and Storey 2010). In brief, following homogenization, chloroform was added to each sample and upper aqueous layers were removed, RNA was precipitated with isopropanol, and, finally, washed with 70 % ethanol. RNA concentrations were determined by reading absorbance at 260 nm on a GeneQuant Pro spectrophotometer (Pharmacia) using the ratio of absorbance at 260/280 nm as an indicator of RNA purity (A260/280 ratio = 1.8–2). RNA quality was confirmed by native gel electrophoresis with ethidium bromide staining to check the integrity of 18S and 28S ribosomal RNA (rRNA) bands (28S rRNA band was about twice as intense as the 18S rRNA band). All samples were standardized to 1 μg/μl with diethylpyrocarbonate (DEPC)-treated water.
First strand cDNA synthesis used 3 μg aliquots of total RNA from the liver, according to manufacturer’s instructions and as previously described (Tessier and Storey 2010). RNA aliquots were combined with 7 μl of DEPC-treated water and 1 μl of oligo-dT (Sigma Genosys; 200 ng/μl). Following a 5-min incubation at 65 °C in a thermocycler (iCycler, BioRad) and rapid chilling on ice, the following components were added to each sample: 4 μl 5× first strand buffer, 2 μl of 0.1 M dithiothreitol (DTT), 1 μl of 10 mM dNTPs, and 1 μl Superscript II reverse transcriptase (all reagents from Invitrogen). The final mixture (19 μl) was incubated at 42 °C for 1 h and then held at 4 °C. Dilutions of cDNA were prepared (10−1) in DEPC water and were used to amplify Tia-1 and TiaR splice variants in ground squirrel liver.
PCR was performed by mixing 5 μl of cDNA (10−1) with 1.25 μl of primer mixture (0.3 nmol/μl forward and 0.3 nmol/μl reverse), 13 μl of DEPC-treated water, 2.5 μl of 10× PCR buffer (Invitrogen), 1.75 μl of 50 mM MgCl2, 0.5 μl of 10 mM dNTPs, and 1 μl of Taq Polymerase (Invitrogen) for a total volume of 25 μl. The cycles performed for amplification consisted of an initial step of 7 min at 95 °C, followed by 35 cycles at 95 °C for 1 min, annealing at 53–64.6 °C for 1 min, and 72 °C for 1 min; the final step was 72 °C for 10 min. Annealing temperature was 53 °C for Tia-1a/b primers (i.e., F1R1 and F2R1) and 64.6 °C for TiaRa/b primers (i.e., F1R1 and F1R2). PCR products containing 2 μg aliquots of xylene blue loading dye and 1× SYBR Green 1 (Invitrogen, Cat. no. S7563) were separated on 2.0 % agarose gels (80 min, 130 V).
Immunofluorescence and fluorescence microscopy
Liver cryosections were prepared by the Morphology Unit (Department of Pathology and Laboratory Medicine, University of Ottawa) and were stored at −80 °C until use. Sections were immediately fixed in ice-cold methanol (5 min), transferred to ice-cold acetone (5 min), and washed twice with 1× phosphate-buffered saline (PBS). The sections were then incubated with Hoeschst 33342 (Invitrogen). Sections were blocked for 1 h in goat serum (10 % v/v made up in 1× PBS) and washed twice in 1× PBS at room temperature. All primary antibodies were diluted 1:100 v/v in 10 % goat serum (1× PBS) and incubated overnight at 37 °C (with 5 % CO2). Antibodies for TIA-1/TIAR were purchased from Santa Cruz (D-9, sc-48371), while PABP-1 antibody was purchased from Upstate (Cat. no. 05-847). Before secondary antibody application, slides were washed four times in 1× PBS. Secondary Alexa 488-conjugated antibodies (Invitrogen) were used at 1:200 v/v in 10 % goat serum (1× PBS) and incubated for 1 h and 40 min at 37 °C. Glass coverslips were mounted in Fluoromount G, sealed with nail polish, and visualized on a Zeiss AxioImager.Z1 microscope. A ×100 oil immersion lens with a 1.3 NA was controlled by Zeiss AxioVision v4.8.2 software, and images were captured on an AxioCam HRm camera. Thirty nuclei stained for TIA-1/TIAR and PABP-1 were counted for the presence or absence of signal localization in four separate fields (n = 120 nuclei/antibody/experimental condition). Images were prepared using Adobe Creative Suite CS4.
Cytoplasmic/nuclear extract preparation
Samples of frozen liver from n = 4 individuals from EC and LT experimental conditions were used to prepare cytoplasmic and nuclear fractions as previously described by Tessier and Storey (2010). Samples of liver (∼200 mg) were homogenized using a Dounce homogenizer with two piston strokes and 400 μl of homogenization buffer (10 mM 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), pH 7.9, 10 mM KCl, 10 mM EDTA, 20 mM β-glycerophosphate) with 4 μl of 100 mM DTT and 4 μl protease inhibitor cocktail (BioShop) added immediately before homogenization. Samples were centrifuged (10,000 rpm, 10 min, 4 °C) and the supernatants were removed as cytoplasmic fractions. Pellets were resuspended in 147 μl of extraction buffer (20 mM HEPES, pH 7.9, 400 mM NaCl, 1 mM EDTA, 10 % v/v glycerol, 20 mM β-glycerophosphate) with 1.5 μl of 100 mM DTT and 1.5 μl protease inhibitor cocktail added (a total of 150 μl/g starting material). After 1 hour incubation, samples were centrifuged (10,000 rpm, 10 min, 4 °C) and the supernatants were collected as nuclear fractions. Protein concentration was quantified by the Coomassie blue dye-binding method using the BioRad reagent (BioRad Laboratories, Hercules, CA) at 595 nm on an MR5000 microplate reader. Samples were then adjusted to 10 μg/μl and aliquots were combined 1:1 v/v with 2× sodium dodecyl sulfate (SDS) loading buffer (100 mM Tris base pH 6.8, 4 % w/v SDS, 20 % v/v glycerol, 0.2 % w/v bromophenol blue, 10 % v/v 2-mercaptoethanol), boiled, and final protein samples (5 μg/μl) were stored at −40 °C until use. Identical protein amounts of each sample (15–25 μg) were loaded onto SDS-polyacrylamide gels or 15 % Tris-Tricine gels to enhance resolving power, as described in the Western blotting section below.
Soluble–insoluble fractionation of lysates
Samples of frozen liver from n = 4 individuals from each experimental condition were separately extracted in order to assess total protein solubility (protocol adapted from Ripaud et al. 2003). The basic method involved tissue homogenization using mechanical/physical cell disruption thereby releasing proteins from both cytoplasmic and nuclear compartments as well as separating soluble and insoluble proteins using a suspension (i.e., aqueous) and solubilizing (i.e., containing detergent) buffer, respectively. Samples were quickly weighed, crushed into small pieces under liquid nitrogen, and then homogenized 1:5 w/v using a Polytron PT10 in ice-cold suspension buffer (1× PBS pH 7.4, 100 mM NaCl, 0.2 % Triton-X) with 10 μl/mL protease inhibitor cocktail (Bioshop) added immediately before homogenization. Homogenates were incubated with gentle agitation for 20 min at room temperature and then centrifuged at 11,000 rpm (20 min, 4 °C). The supernatant was transferred to a clean Eppendorf tube (soluble fraction), and the pellet was washed twice with water (500 μl/wash) to minimize cross-contamination. Using equivalent volumes to the suspension buffer, the pellet was then resuspended in SDS solubilization buffer (1× PBS pH 7.4, 300 mM NaCl, 2 % v/v SDS, 1 % v/v Triton-X) with 2 mM DTT and 10 μl/mL protease inhibitor cocktail. Samples were then centrifuged at 11,000 rpm (30 min, room temperature), and the supernatant was collected as the insoluble fraction. Total protein control samples were used in order to ensure that the composition of total protein was preserved in each experimental condition. Total protein control samples were prepared using 1:10 v/v solubilization buffer (1× PBS pH 7.4, 300 mM NaCl, 2 % v/v SDS, 1 % v/v Triton-X) with 2 mM DTT and 10 μl/mL protease inhibitor cocktail. Control homogenates were incubated with gentle agitation for 1 h at room temperature and then centrifuged at 11,000 rpm (30 min, room temperature). The supernatant was removed, transferred to a clean Eppendorf tube, and saved as total protein controls. All samples (control and soluble/insoluble fractions) were combined 1:1 v/v with 2× SDS loading buffer, boiled, and stored at −20 °C until use. Identical volumes of each sample (3 μl of soluble/insoluble fractions and controls) were loaded onto SDS-polyacrylamide gels or 15 % Tris-Tricine gels as described in the Western blotting section below.
Western blotting
Equal amounts from each sample were loaded onto 10–12 % SDS-polyacrylamide gels and run at 180 V for 45–60 min (except for TIA-1 p15 which was run on a 15 % Tris-Tricine gel) as previously described by Rouble et al. (2013). Proteins were then transferred to polyvinylidene difluoride (PVDF) membrane (0.45 μm PVDF for SDS-PAGE, 0.2 μm PVDF for Tris-Tricine gels) by electroblotting at 160 mA for 1–1.5 h. Membranes were blocked with milk and made up in TBST (20 mM Tris base, pH 7.6, 140 mM NaCl, 0.05 % v/v Tween-20) to prevent nonspecific binding of antibodies. Milk was prepared 2–5 % (w/v) in TBST and incubated with the membrane on a rocker for 20 min. Membranes were probed with specific primary antibodies and diluted in TBST, at 4 °C for 12 h. Antibodies specific for mammalian TIA-1 (C-20, sc-1751) and TIAR (C-18, sc-1749) were purchased from Santa Cruz Biotechnologies, TIA-1 p15 was purchased from Abcam (ab2712), and PABP-1 was purchased from GeneTex (GTX101515). Antibodies for TIA-1a/TIA-1b and TIARa/TIARb cross-reacted with bands on the immunoblots at molecular masses of 43/41 and 41/40 kDa, respectively. Antibodies for TIA-1 p15 and PABP-1 cross-reacted with bands on the immunoblots at molecular masses of 15 and 70 kDa, respectively. Antibodies were used at 1:1,000 v/v dilution in TBST. Membranes that had been probed with TIA-1 p15 were probed with horseradish peroxidase (HRP)-linked anti-mouse IgG secondary antibody (1:4,000 v/v dilution), PABP-1 were probed with HRP-linked anti-rabbit IgG secondary antibody (1:5,000 v/v dilution), and TIA-1 and TIAR were probed with HRP-linked anti-goat IgG secondary antibody (1:10,000 v/v dilution). All membranes were washed three times between incubation periods in 1× TBST for ∼10 min/wash. Bands were visualized by enhanced chemiluminescence (H2O2 and Luminol), and then, blots were restained using Coomassie blue (0.25 % w/v Coomassie brilliant blue, 7.5 % v/v acetic acid, 50 % methanol) to visualize all protein bands.
Quantification and statistics
Band densities on chemiluminescent immunoblots were visualized using a Chemi-Genius BioImaging system (Syngene, Frederick, MD) and quantified using the associated Gene Tools software. Immunoblot band density in each lane was standardized against the summed intensity of a group of Coomassie-stained protein bands in the same lane (except for solubility data); these were chosen because they did not show variation between different experimental states and were not located close to the protein bands of interest. Solubility data are expressed as the ratio of soluble/insoluble fractions. Data are expressed as means ± SEM; n = 4 independent samples from different animals. Statistical testing of standardized band intensities used the Student’s t test (Figs. 2, 3, 4, and 5).
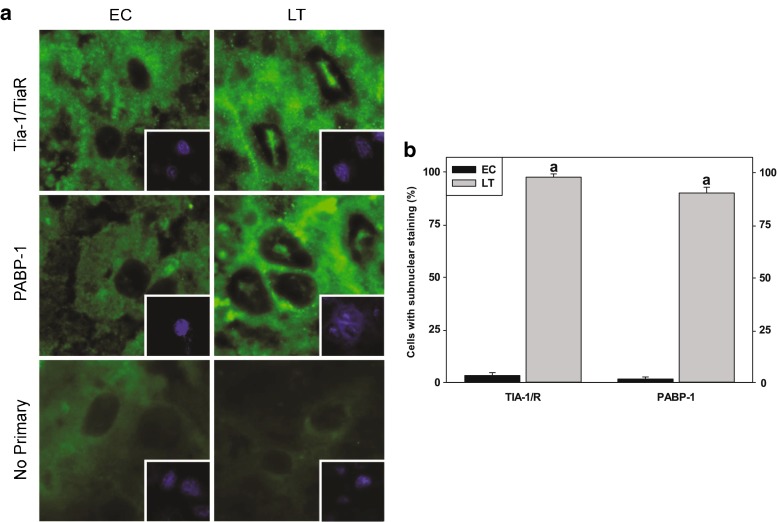
Fig. 2.
Localization of RNA-binding proteins to the nucleus in the liver of torpid thirteen-lined ground squirrels (I. tridecemlineatus). a Immunostaining of endogenous TIA-1/R and PABP-1. Fluorescent signals from TIA-1/R and PABP-1 were found to be distributed in the cytoplasm and nucleus during EC but displayed distinct subnuclear foci during LT. Shown are representative cryosections immunostained with TIA-1/R and PABP-1 as well as DAPI-stained nuclei (depicted in the bottom right quadrant) from EC and LT liver samples. b Number of cells possessing subnuclear foci averaged across four fields comparing euthermic control (EC) and late torpor (LT) conditions. Data were analyzed using the Student’s t test; a denotes values are significantly different from EC, p < 0.05
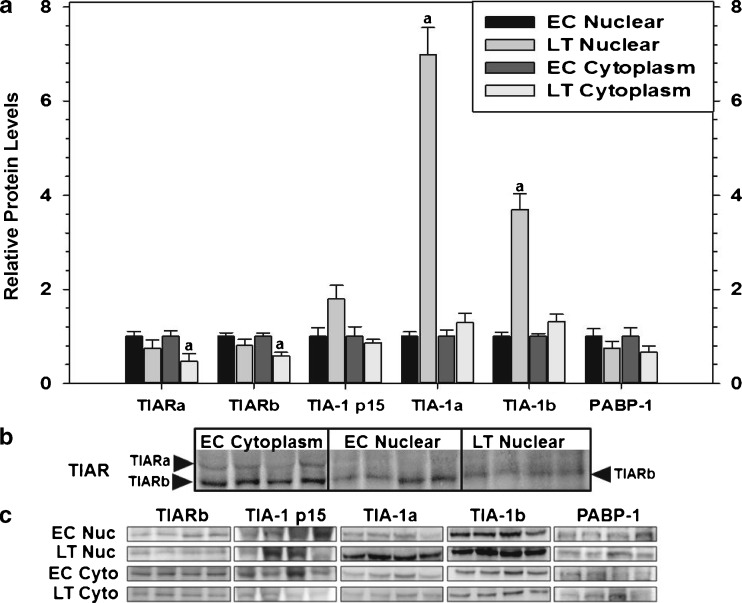
Fig. 3.
Subcellular distribution of RNA-binding proteins in cytoplasmic and nuclear fractions sampled from the liver of thirteen-lined ground squirrels (I. tridecemlineatus) comparing euthermic control (EC) and late torpor (LT) conditions. a Histogram showing mean relative expression of TIARa/b, TIA-1 p15, TIA-1a/b, and PABP-1 (± SEM, n = 4 independent protein isolations from different animals). b Representative Western blots are shown of the cytoplasmic and nuclear distribution of TIAR with the subcellular fraction and experimental conditions labeled at the top of the gel and the TIAR isoform labeled to the left of the gel. c Representative Western blots are shown with the experimental conditions labeled to the left of the gel and the protein target labeled at the top of the gel. Data were analyzed using the Student’s t test; a denotes values are significantly different from EC, p < 0.05
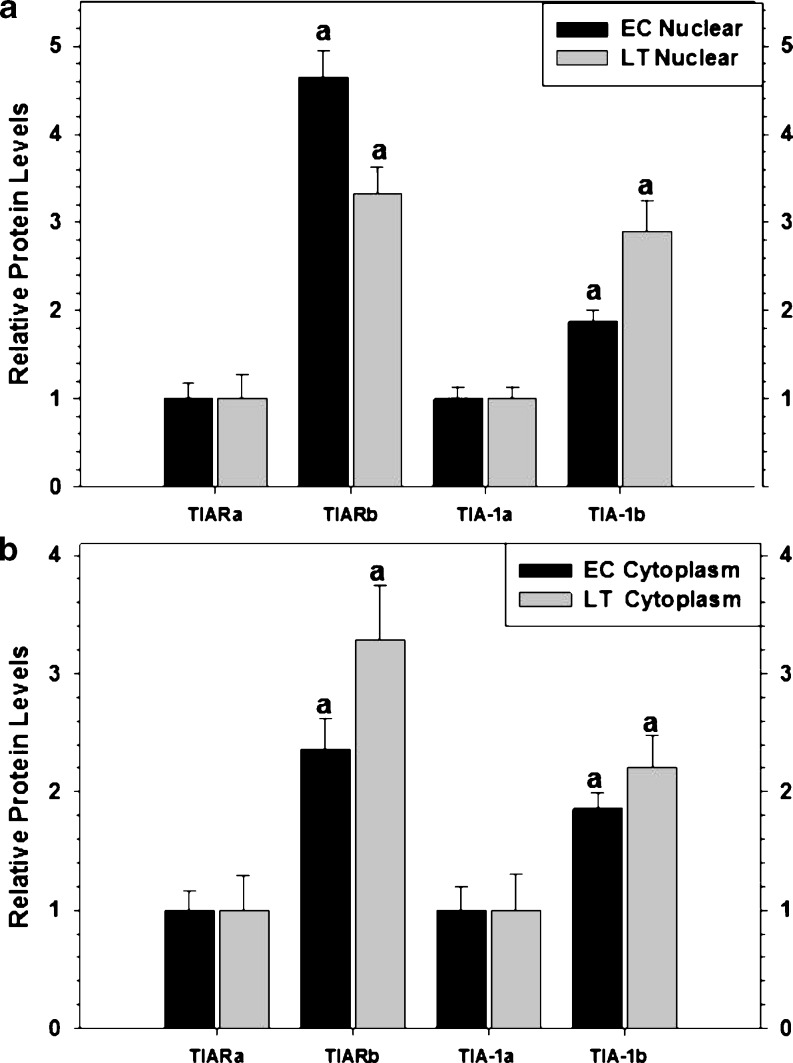
Fig. 4.
The expression levels of RNA-binding proteins isoform a relative to isoform b sampled from the liver of thirteen-lined ground squirrels (I. tridecemlineatus) comparing euthermic control (EC) and late torpor (LT) conditions as well as their subcellular distribution. a Histogram showing mean relative expression levels of TIARa versus TIARb and TIA-1a versus TIA-1b in nuclear fractions (±SEM, n = 4 independent protein isolations from different animals). b Histogram showing mean relative expression of TIARa versus TIARb and TIA-1a versus TIA-1b in cytoplasmic fractions (±SEM, n = 4 independent protein isolations from different animals). Other information as in Fig. 3
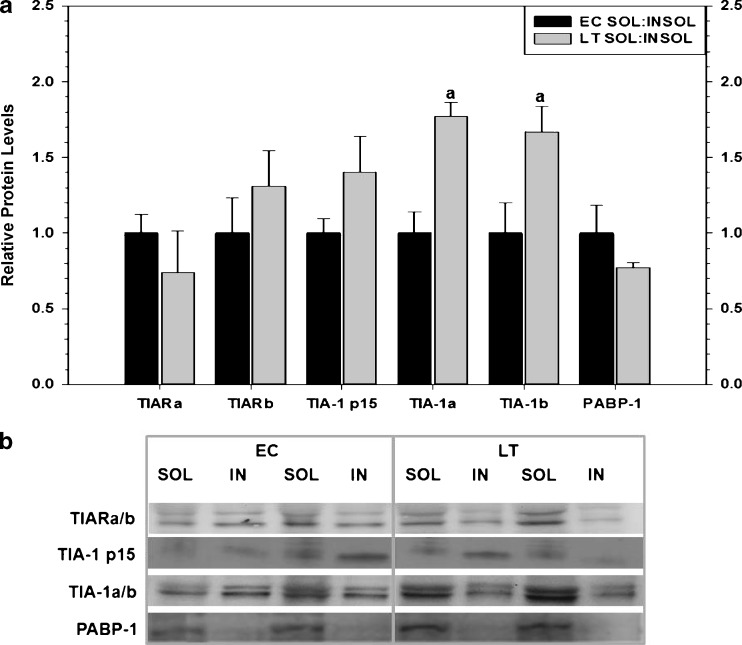
Fig. 5.
The relative expression of soluble and insoluble protein fractions of RNA-binding proteins sampled from the liver of thirteen-lined ground squirrels (I. tridecemlineatus) comparing euthermic control (EC) and late torpor (LT) conditions. a Histogram showing mean relative protein levels in soluble versus insoluble fractions for TIARa/b, TIA-1 p15, TIA-1a/b, and PABP (±SEM, n = 4 independent protein isolations from different animals). b Representative Western blots are shown with the protein targets labeled to the left of the gel and the experimental conditions labeled at the top of the gel. Other information as in Fig. 3
Results
Characterization of thirteen-lined ground squirrel TIA-1, TIAR, and PABP-1
Human and mouse mRNA coding sequences were used in whole genome shotgun NCBI BLAST analysis to deduce the I. tridecemlineatus. TIA-1, TIAR, and PABP-1 mRNA sequences. The encoded ground squirrel RNA-binding proteins were translated from the in silico-derived open reading frames, and motifs were identified using online software (ELM and ScanProsite) (Fig. 1a and Table 1). As expected, protein alignment scores revealed that the ground squirrel amino acid sequences were highly conserved when compared to other species: (1) TIA-1 was 99, 97, 96, and 85 %, (2) TIAR was 99, 93, 98, and 68 %, (3) PABP-1 was 100, 99, 99, and 93 % identical to human, bovine, mouse, and African clawed frog sequences, respectively (Table 2). RT-PCR with primers designed to amplify both isoforms of Tia-1 and TiaR confirmed the presence of the alternative splice variants in ground squirrel liver (Fig. 1b). These splicing variants were also detected as mature proteins via Western blotting, with antibodies targeting TIA-1 identifying two bands in the 42-kDa range, TIA-1a (top) and TIA-1b (bottom), while TIAR antibodies detect two bands in the 41-kDa range, TIARa (top) and TIARb (bottom).
Fig. 1.
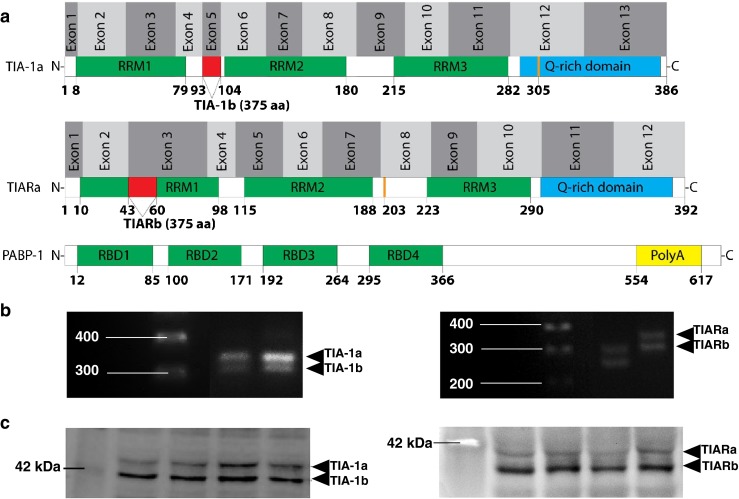
Alternative gene variants of RNA-binding proteins in the liver of hibernating thirteen-lined ground squirrel (I. tridecemlineatus). a Illustration of alternative splice sites. Patterns of alternative splicing are represented by a line and a red box, exons are denoted by alternating light and dark gray boxes, RNA recognition motifs (RRMs) are represented by green boxes, the Q-rich domain is represented by a blue box, the Poly-A-tail binding domain is represented by a yellow box, the location of ground squirrel amino acid substitutions (as compared to the human sequence) are represented by thick orange lines, and numbers indicate amino acid residues. b Representative bands on agarose gels are shown for Tia-1 and TiaR gene variants amplified by primers spanning exon 5 and an alternate in-frame splice site in the coding region, respectively. The DNA ladder is labeled to the left and the isoform to the right of the gel. c Representative Western blots are shown for TIA-1 and TIAR protein isoforms with the protein ladder labeled to the left and the isoform to the right of the gel
Table 1.
The predicted thirteen-lined ground squirrel mRNA coding sequence for TIA-1, TIAR, and PABP-1 were translated into amino acids, and the domains/motifs were predicted using the Eukaryotic Linear Motif resource for Functional Sites in Proteins (ELM) and compared to those predicted using the ScanProsite tool
| 13LGS TIA-1a | ELM | ScanProsite |
| RRM1 | 8–79 | 7–83 |
| RRM2 | 107–180 | 106–184 |
| RRM3 | 215–282 | 214–286 |
| Q-rich domain | 295–366 | |
| 13LGS TIARa | ELM | ScanProsite |
| RRM1 | 10–98 | 9–102 |
| RRM2 | 115–188 | 114–192 |
| RRM3 | 223–290 | 222–294 |
| 13LGS PABP-1 | ELM | ScanProsite |
| RRM1 | 12–85 | 11–89 |
| RRM2 | 100–171 | 99–175 |
| RRM3 | 192–264 | 191–268 |
| RRM4 | 295–366 | 294–370 |
| PolyA | 554–617 | 542–619 |
Table 2.
Alignment scores comparing TIA-1, TIAR, and PABP-1 protein sequences of thirteen-lined ground squirrel to human, bovine, mouse, and African clawed frog. NCBI accession numbers are included
| Human (Homo sapiens) | Bovine (Bos taurus) | Mouse (Mus musculus) | African clawed frog (Xenopus lavis) | |
|---|---|---|---|---|
| TIA-1 | 99 % (NP_071505.2) | 97 % (NP_001069577.1) | 96 % (NP_035715.1) | 85 % (NP_001087561.1) |
| TIAR | 99 % (NP_001029097.1) | 93 % (NP_001179985.1) | 98 % (NP_033409.1) | 68 % (NP_001167497.1) |
| PABP-1 | 100 % (NP_002559.2) | 99 % (NP_776993.1) | 99 % (NP_032800.2) | 93 % (NP_001080204.1) |
Relative expression and nuclear localization of RNA-binding proteins
To better understand the functions of the mRNA processing factors during hibernation, we employed immunohistochemistry to visualize their subcellular distribution in euthermic control (EC) and torpor (LT) animals (Fig. 2). While a large pool of TIA-1/TIAR and PABP-1 was found in the cytoplasm, a population of these molecules was also present in subnuclear bodies within the nuclei of LT ground squirrels (Fig. 2a). These large (∼48 μm) structures appear as a single foci rather than the multiple, speckled domains as is sometimes observed for other types of nuclear bodies. Targeting of TIA-1/TIAR and PABP-1 to nuclear foci was observed in approximately 98 and 90 % of LT liver nuclei, respectively (Fig. 2b and Table 3). Conversely, EC ground squirrels possessed subnuclear TIA-1/TIAR and PABP-1 in less than 3 % of liver nuclei visualized (Fig. 2b).
Table 3.
The number of cells possessing subnuclear foci viewed by immunofluorescence of cryosections obtained from thirteen-lined ground squirrel liver stained for TIA-1/R and PABP-1. Across 4 separate fields (30 nuclei/field), a total of 120 nuclei were counted for either the presence or absence of signal localization comparing euthermic control (EC) and late torpor (LT) conditions
| Field | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| TIA-1/R | EC | 1 | 1 | 0 | 2 |
| LT | 30 | 30 | 28 | 29 | |
| PABP-1 | EC | 1 | 0 | 0 | 1 |
| LT | 27 | 29 | 27 | 25 | |
| No 1 AB | EC | 0 | 0 | 0 | 0 |
| LT | 0 | 0 | 0 | 0 | |
| No. of nuclei counted/antibody | 30 | 30 | 30 | 30 | |
We also utilized subcellular fractionation and immunoblotting to determine the relative expression of TIARa/b, TIA-1 p15, TIA-1a/b, and PABP-1 in the cytoplasmic and nuclear fractions of ground squirrel liver, during EC and LT experimental conditions (Fig. 3). No change was observed in the protein levels of TIAR isoforms when comparing nuclear fractions across experimental conditions (Fig. 3a); however, TIARa and TIARb decreased significantly in the cytoplasm during torpor to levels that were 46 and 59 %, respectively, of EC values. As shown in Fig. 3b, TIARa was predominantly localized in the cytoplasm with negligible amounts in the nuclei under both EC and LT conditions, whereas TIARb was present in both fractions. TIA-1a and TIA-1b showed parallel, statistically significant changes in relative protein expression levels whereby 7- and 3.7-fold increases were observed in the nuclear fractions from torpid ground squirrels as compared to EC, respectively (Fig. 3a). This increased nuclear population was in agreement with the immunohistochemistry results observed in Fig. 2. In contrast, neither TIA-1 isoform changed in cytoplasmic fractions between EC and LT. TIA-1 p15 and PABP-1 showed no significant changes in either cytoplasmic or nuclear fractions across experimental conditions. Representative Western blots corresponding to those quantified to produce histograms depicted in Fig. 3a are shown in Fig. 3c.
Relative expression of TIA-1a/b and TIARa/b isoform expression
Next, we wanted to determine the relative abundance of each TIA-1/TIAR isoform in EC and LT ground squirrels. We compared the expression levels of the b isoform relative to the a isoform from both the nuclear and cytoplasmic fractions (Fig. 4). Interestingly, when assessing the relative expression of TIARa versus TIARb in the nucleus, consistently higher levels of TIARb were observed in both EC (4.7-fold) and LT (3.3-fold) livers (Fig. 4a). TIA-1b expression was also consistently higher than TIA-1a in the nucleus of both EC (1.9-fold) and LT (2.9-fold) (Fig. 4a) animals. When comparing the relative levels of TIARa/b and TIA-1a/b in the cytoplasmic fractions, the b isoform of both molecules was regularly higher than the a isoform: (1) TIAR EC (2.4-fold) and LT (3.3-fold) and (2) TIA-1 EC (1.9-fold) and LT (2.2-fold) (Fig. 4b). Together, this data suggests that the b isoform of both proteins is the predominant splice variant.
Detergent solubility test of RNA-binding proteins
Soluble and insoluble protein fractions were collected through classic methods used to isolate pathological protein aggregates present in brain tissues of subjects with neurodegenerative diseases (Wolozin 2012); however, interchangeable forms in this cellular context are associated with TIA protein function during stress (Gilks et al. 2004). The present study tested for differences in solubility of RNA-binding proteins, particularly those with the glutamine-rich PRD such as TIA-1 and TIAR (Fig. 5). The solubility of TIARa/b, TIA-1 p15, and PABP-1 did not change when comparing the ratio of soluble/insoluble fractions between euthermic and torpid conditions. In contrast, a relative increase in the ratio of soluble/insoluble fractions was observed for TIA-1a and TIA-1b during torpor (LT was 1.8- and 1.7-fold higher than EC for TIA-1a and TIA-1b, respectively). As such, relatively more TIA-1a and TIA-1b proteins were recovered in the soluble fractions during torpor as compared to euthermic controls.
Discussion
The present study investigates the presence of alternative isoforms, localization, and solubility of protein targets involved in mRNA processing in the liver of control (EC) and hibernating (LT) thirteen-lined ground squirrel. Hibernating ground squirrels are an excellent model system for uncovering mechanisms involved in mRNA processing in response to stress since they demonstrate a marked global decrease in transcription (Morin and Storey 2006) and translation (Frerichs et al. 1998) during torpor yet show increases in the levels of selected mRNA/protein species (Pan and van Breukelen 2011). While a suite of transcriptional and posttranscriptional controls vital to achieving this molecular restructuring has been described (Hittel and Storey 2002; Storey and Storey 2004; Morin and Storey 2009; Kornfeld et al. 2012; Wu and Storey 2012), many regulatory mechanisms remain to be described during hibernation. In order to further understand the mechanisms that direct mRNA fate, we analyzed the DNA/RNA-binding proteins that are stress-responsive and are involved in guiding gene transcription, mRNA processing, and the formation of stress granules.
It has been previously established that RNA-binding proteins have different roles when present in the cytoplasm versus nucleus (Suswam et al. 2005; Zhang et al. 2005); therefore, we sought to compare the subcellular distribution and localization of TIA-1/TIAR and PABP-1 in euthermic and torpid animals. In the cytoplasm, RNA-binding proteins such as TIA-1/R and PABP-1 are localized to stress granules, which store stalled translational pre-initiation complexes in response to various stresses (Zhang et al. 2005; Kedersha and Anderson 2007). While an attractive candidate for involvement in the hibernating liver, our current study did not reveal evidence of cytoplasmic stress granule formation using indirect fluorescence microscopy (Fig. 2a). Furthermore, the formation of cytoplasmic stress granules tend to be concentration-dependent (Gilks et al. 2004); however, in the present study, the relative expression of cytoplasmic TIA-1 and TIAR did not change or decreased significantly during LT, respectively. Nonetheless, recent evidence suggests that RNA-binding proteins such as TIAR may play a more generalized role in translational repression which is not exclusively linked with the presence of cytoplasmic stress granules (Mazan-Mamczarz et al. 2006). Additionally, when ribosomal fractions from ground squirrel kidney were separated on sucrose gradients, TIA-1 was restricted to monosome fractions (i.e., translationally silent) during control and torpor (Hittel and Storey 2002). Taken together, this data suggests cytoplasmic RNA binding proteins may play a generalized role in translational suppression in thirteen-lined ground squirrels.
In contrast to the diffuse signal observed for cytoplasmic pools of RNA-binding proteins, protein targets visualized by fluorescence microscopy during LT including antibodies directed towards TIA-1/TIAR and PABP-1 cross-reacted with proteins that are localized to relatively large (∼48 μm) subnuclear foci (up to 98 % of LT liver cells contained nuclear foci, Fig. 2b and Table 3). The presence of nuclei containing structural components in response to the hibernating phenotype has also been observed in the hazel dormouse (Muscardinus avellanarius) and the edible dormouse (Glis glis) (Malatesta et al. 1994, 1999, 2001, 2008); however, the ground squirrel subnuclear structures presently described appear as single foci/nucleus with distinct characteristics, including size. Since nuclear TIA-1 and TIAR perform a range of functions including rendering mRNA translationally silent, modulating the rates of gene transcription, and regulating constitutive and alternative pre-mRNA splicing (Förch et al. 2001; López de Silanes et al. 2005; Suswam et al. 2005), further studies were performed in order to better understand the function of subnuclear foci during torpor.
Using subcellular fractionation and Western blotting, up to a 7-fold increase in relative protein expression of TIA-1a and TIA-1b was observed in the nucleus during LT, compared with EC (Fig. 3). By contrast, the TIARa isoform was largely restricted to the cytoplasmic fraction, and there was no relative change in the expression of the TIARb isoform and PABP-1 in the nucleus between EC and LT. These data suggests that TIA-1 isoforms are major components of the nuclear foci observed by fluorescence microscopy during the hibernating phenotype. It has been shown using by both experimental and bioinformatic approaches that approximately 3,000 mRNAs interact with TIA-1 spanning a range of cellular processes and, when bound by TIA-1, these mRNAs are translationally repressed (Piecyk et al. 2000; López de Silanes et al. 2005). Consequently, it is possible that, during hibernation, nuclear TIA-1 associate with target mRNAs destined for translational suppression, while the mRNA is still in the nucleus. It should also be noted that while the localization of TIA-1 into subnuclear structures was correlated with significant increases in relative protein levels during torpor, this was not the case for TIAR and PABP-1 whereby signal localization occurred despite no observed changes in nuclear protein expression levels. This data suggests TIAR/PABP-1 proteins undergo a reorganization of the nuclear pool during LT, and protein localization to subnuclear structures is not necessarily dose-dependent.
While a case has been made for the role TIA-1 in ground squirrel subnuclear foci during LT, proposed roles for nuclear TIAR and PABP-1 may also be gleaned from the present data. In the nucleus, TIAR binds single-stranded DNA (ssDNA), and ssDNA conformations occur in chromatin undergoing active transcription (Suswam et al. 2005). Given that (1) the TIARa isoform occurred mostly in the cytoplasm (Fig. 3), (2) there was no change in relative expression of TIARb in the nucleus across experimental conditions, and (3) that a state of enhanced transcription does not agree with the general energy conservation strategy of the hypometabolic state (Morin and Storey 2006), it is unlikely that TIAR associates with ssDNA during hibernation, but further evidence is required in order to confirm. Instead, TIAR in the nucleus during hibernation may be related to other tasks such as regulating mRNA processing and/or enhancing mRNA stability, which are also functions associated with TIA-1 and PABP-1 activity (Minvielle-Sebastia et al. 1997; Afonina et al. 1998; Förch et al. 2001; Suswam et al. 2005). In this capacity, nuclear TIA proteins and PABP-1 may represent the inhibition of mRNA processing and, in hibernating hazel dormice, the accumulation of pre-mRNAs at the splicing/cleavage stage has been demonstrated (Malatesta et al. 2008). It should be noted, however, that some level of active pre-mRNA processing may be critical during torpor since a small subset of genes are selectively activated in order to support the torpid phenotype. Consequently, while repression of global mRNA processing may be occurring during torpor, this generalized response would parallel selective increases in a subpopulation of mRNA species essential for mitigating stresses associated with hibernation. In summary, the nuclear functions of RNA-binding proteins described above would be highly advantageous during torpor since mRNA could be held in highly organized storage centers, selectively processed when needed, and possibly gain benefits of enhanced stability. Indeed, all of these proposed functions would support an overall suppression of translation during torpor, complement the proposed role of microRNAs as stabilizing molecules during hypometabolism (Biggar and Storey, 2011), while also promoting a rapid resumption of protein synthesis when animals arouse back to euthermia.
Further to containing distinct functional roles in the cytoplasm versus nucleus, structural transitions mediated by the prion-like domain of TIA-1 and TIAR have been associated with their function during stress (Gilks et al. 2004). TIA-1/R proteins are composed of an RRM and a glutamine-rich PRD, which have been shown to be important for RNA recruitment and assembly of stress granules, respectively (Anderson and Kedersha 2002). Similar to infectious prion proteins, the PRD of TIA-1 seemingly displays conformation-dependent changes of function which are related to differences in solubility (Gilks et al. 2004). Furthermore, the soluble form of TIA proteins has been suggested to function as a translational silencer, while aggregated forms promote stress-induced translational arrest via stress granule formation (Gilks et al. 2004). As such, the solubility of RNA-binding proteins was assessed in the liver of ground squirrels (Fig. 5) in order to assess if these interchangeable forms were responsive to torpor. During LT, TIA-1a/TIA-1b showed a shift towards the soluble fraction and all other targets showed no change, suggesting that enhanced protein aggregation was not present during torpor. While the functional consequences of this shift towards the soluble fraction are not clear, the data supports the role of RNA-binding proteins, especially TIA-1, as translational suppressors during torpor.
To characterize ground squirrel RNA-binding proteins, the amino acid sequences of TIA-1, TIAR, and PABP-1 were deduced using sequencing information available as whole genome shotgun reads (Fig. 1a). All RNA-binding proteins analyzed were highly conserved in comparison to the human sequence (Table 2) with TIA-1 and TIAR containing only one amino acid substitution located within the Q-rich domain (residue 305) of TIA-1 or the linker region between RRM2 and RRM3 (residue 203) in TIAR. The full length TIA-1 protein is proteolytically cleaved in order to derive the C-terminus TIA-1 p15 (Kawakami et al. 1994; Taupin et al. 1995) and, in human, TIA-1a and TIA-1b are generated by alternative splicing surrounding exon 5 (Izquierdo and Valcárcel 2007). Exon 5 is located between RRM1 and RRM2 and its inclusion generates the longer TIA-1 isoform a, whereas exon skipping generates the shorter TIA-1 isoform b (Fig, 1a). The full length TIAR protein (TIARa) uses an alternate in-frame splice site in the coding region, compared to TIARb, resulting in the longer protein. The TIA-1 and TIAR isoforms differ by 11 and 17 amino acids, respectively. The present study detected the presence of the two TIA-1 and TIAR isoforms in the liver of ground squirrels, which was confirmed by RT-PCR and SDS-PAGE (Fig. 1b, c). The presence of alternate isoforms not only agrees with data obtained by Izquierdo and Valcárcel (2007) but also supports the possibility that active alternative splicing may indeed be occurring in ground squirrel liver and/or play a role during hibernation.
Izquierdo and Valcárcel (2007) observed that the relative expression of TIA isoforms differed in human tissues and cell lines. For example, the overall protein levels of TIA-1 were higher in HeLa than NRK cells, with isoform a being dominant in HeLa cells and isoform b being the only detectable species in NRK cells. Additionally, TIAR was expressed at roughly the same level in the two cell lines; however, while HeLa cells expressed both TIARa and b equally, TIARb was the dominant isoform in NRK cells. In thirteen-lined ground squirrel liver the relative expression of isoforms a and b were considered for cytoplasmic and nuclear fractions (Fig. 4). When comparing the relative expression of TIA-1a to TIA-1b and TIARa to TIARb, a consistently higher expression level of isoform b was observed for both proteins across experimental conditions in both cytoplasmic and nuclear fractions (Fig. 4). The TIA-1 isoforms appear to have overlapping roles yet exhibit functional differences in splicing activity. For example, TIA-1 isoforms exhibit identical subcellular distribution and RNA-binding properties; however, TIA-1b displayed enhanced splicing activity (Izquierdo and Valcárcel 2007). Previous results have shown that TIAR can regulate the levels of mRNA isoforms of TIA-1a versus TIA-1b; thus, a reduction in the levels of TIAR resulted in increases in the TIA-1 isoform b in comparison to isoform a (Izquierdo and Valcárcel 2007). While stable levels of TIAR were observed in nuclear fractions across experimental conditions in the present study, the selective localization of TIARa to cytoplasmic fractions may have influenced the relative expression of TIA-1b in the nucleus. Nonetheless, more evidence will be required to further elucidate the relationship between TIAR and TIA-1 isoforms in ground squirrel liver.
In summary, the present study provides insight into the regulation of RNA-binding proteins in the liver of hibernating thirteen-lined ground squirrels. Torpor-specific subnuclear structures were observed by fluorescence microscopy, and RNA-binding proteins such as TIA-1, TIAR, and PABP-1 show selective increases in relative expression in the nuclear compartment as evidenced by subcellular fractionation and Western blotting. These data suggest a role for these proteins in reducing translational rates, regulating mRNA processing, and/or enhancing mRNA stability during torpor. Additionally, a case has been made for the presence of alternate splice variants of TIA-1 and TIAR in ground squirrel liver, opening up the field to further studies that would elucidate a role for alternative gene variants during hibernation. An important future milestone would involve delineating the dynamic (i.e., playing an active role in mRNA processing/splicing) versus static (i.e., acting primarily as storage/sequestering molecules) nature of subnuclear structures during torpor. This may be achieved by characterizing the interactions of RNA-binding proteins with specific mRNA species and tracking their movement between subcellular compartments. Since selective increases in gene/protein expression must be balanced with an overall reduction in transcriptional and translational rates, the hibernating ground squirrel provides a unique opportunity to study nuclear dynamics and posttranscriptional control under changing environmental conditions.
Acknowledgments
We would like to thank Dr. J.M. Hallenbeck and Dr. D.C. McMullen (NINDS, NIH, Bethesda) for providing the tissue samples for this study. Thanks also to J.M. Storey for editorial review of the manuscript. Research was supported by a grant from the Canadian Institute of Health Research (CIHR) to SL, a discovery grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada to KBS, and the Canada Research Chairs program. SNT held an NSERC PGSD scholarship.
References
- Afonina E, Stauber R, Pavlakis GN. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:VSTROE>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar KK, Storey KB. The emerging roles of microRNAs in the molecular responses of metabolic rate depression. J Mol Cell Biol. 2011;3:167–175. doi: 10.1093/jmcb/mjq045. [DOI] [PubMed] [Google Scholar]
- Bouma HR, Verhaag EM, Otis JP, Heldmaier G, Swoap SJ, Strijkstra AM, Henning RH, Carey HV. Induction of torpor: mimicking natural metabolic suppression for biomedical applications. J Cell Physiol. 2012;227:1285–1290. doi: 10.1002/jcp.22850. [DOI] [PubMed] [Google Scholar]
- Burd CG, Matunis EL, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro G, Parker R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- Chung D, Lloyd GP, Thomas RH, Guglielmo CG, Staples JF. Mitochondrial respiration and succinate dehydrogenase are suppressed early during entrance into a hibernation bout, but membrane remodeling is only transient. J Comp Physiol B. 2011;181:699–711. doi: 10.1007/s00360-010-0547-x. [DOI] [PubMed] [Google Scholar]
- Dave KR, Christian SL, Perez-Pinzon MA, Drew KL. Neuroprotection: lessons from hibernators. Comp Biochem Physiol B Biochem Mol Biol. 2012;162:1–9. doi: 10.1016/j.cbpb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Förch P, Puig O, Kedersha N, Martínez C, Granneman S, Séraphin B, Anderson P, Valcárcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell. 2001;6:1089–1098. doi: 10.1016/S1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Frerichs KU, Smith CB, Brenner M, DeGracia DJ, Krause GS, Marrone L, Dever TE, Hallenbeck JM. Suppression of protein synthesis in brain during hibernation involves inhibition of protein initiation and elongation. Proc Natl Acad Sci U S A. 1998;95:14511–14516. doi: 10.1073/pnas.95.24.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser F. Metabolic rate and body temperature reduction during hibernation and daily torpor. Annu Rev Physiol. 2004;66:239–274. doi: 10.1146/annurev.physiol.66.032102.115105. [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldmaier G, Ortmann S, Elvert R. Natural hypometabolism during hibernation and daily torpor in mammals. Respir Physiol Neurobiol. 2004;141:317–329. doi: 10.1016/j.resp.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Hittel D, Storey KB. The translation state of differentially expressed mRNAs in the hibernating thirteen-lined ground squirrel (Spermophilus tridecemlineatus) Arch Biochem Biophys. 2002;401:244–254. doi: 10.1016/S0003-9861(02)00048-6. [DOI] [PubMed] [Google Scholar]
- Izquierdo JM, Valcárcel J. Two isoforms of the T-cell intracellular antigen 1 (TIA-1) splicing factor display distinct splicing regulation activities. Control of TIA-1 isoform ratio by TIA-1-related protein. J Biol Chem. 2007;282:19410–19417. doi: 10.1074/jbc.M700688200. [DOI] [PubMed] [Google Scholar]
- Kawakami A, Tian Q, Streuli M, Poe M, Edelhoff S, Disteche CM, Anderson P. Intron-exon organization and chromosomal localization of the human TIA-1 gene. J Immunol. 1994;152:4937–4945. [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- Kornfeld SF, Biggar KK, Storey KB. Differential expression of mature microRNAs involved in muscle maintenance of hibernating little brown bats, Myotis lucifugus: a model of muscle atrophy resistance. Genomics Proteomics Bioinforma. 2012;10:295–301. doi: 10.1016/j.gpb.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Silanes I, Galbán S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta M, Zancanaro C, Martin TE, Chan EKL, Almaric F, Luhrmann R, Vogel P, Fakan S. Cytochemical and immunocytochemical characterization of nuclear bodies during hibernation. Eur J Cell Biol. 1994;65:82–93. [PubMed] [Google Scholar]
- Malatesta M, Cardinali A, Battistelli S, Zancanaro C, Martin TE, Fakan S, Gazzanelli G. Nuclear bodies are usual constituents in tissues of hibernating dormice. Anat Rec. 1999;254:389–395. doi: 10.1002/(SICI)1097-0185(19990301)254:3<389::AID-AR10>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Luchetti F, Marcheggiani F, Fakan S, Gazzanelli G. Disassembly of nuclear bodies during arousal from hibernation: an in vitro study. Chromosoma. 2001;110:471–477. doi: 10.1007/s004120100166. [DOI] [PubMed] [Google Scholar]
- Malatesta M, Biggiogera M, Baldelli B, Barabino SM, Martin TE, Zancanaro C. Hibernation as a far-reaching program for the modulation of RNA transcription. Microsc Res Tech. 2008;71:564–572. doi: 10.1002/jemt.20587. [DOI] [PubMed] [Google Scholar]
- Martin SL, Maniero GD, Carey C, Hand SC. Reversible depression of oxygen consumption in isolated liver mitochondria during hibernation. Physiol Biochem Zool. 1999;72:255–264. doi: 10.1086/316667. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Ashish L, Martindale JL, Kawai T, Gorospe M. Translational repression by RNA-binding protein TIAR. Mol Cell Biol. 2006;26:2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur MD, Milsom WK. Changes in ventilation and respiratory sensitivity associated with hibernation in Columbian (Spermophilus columbianus) and golden-mantled (Spermophilis lateralis) ground squirrels. Physiol Zool. 1991;64:940–959. [Google Scholar]
- McMullen DC, Hallenbeck JM. Regulation of Akt during torpor in the hibernating ground squirrel, Ictidomys tridecemlineatus. J Comp Physiol B. 2010;180:927–934. doi: 10.1007/s00360-010-0468-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Preker PJ, Wiederkehr T, Strahm Y, Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc Natl Acad Sci U S A. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Morin P, Jr, Storey KB. Evidence for a reduced transcriptional state during hibernation in ground squirrels. Cryobiology. 2006;53:310–318. doi: 10.1016/j.cryobiol.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Morin P, Jr, Storey KB. Mammalian hibernation: differential gene expression and novel application of epigenetic controls. Int J Dev Biol. 2009;53:433–442. doi: 10.1387/ijdb.082643pm. [DOI] [PubMed] [Google Scholar]
- Pan P, van Breukelen F. Preference of IRES-mediated initiation of translation during hibernation in golden-mantled ground squirrels, Spermophilus lateralis. Am J Physiol Regul Integr Comp Physiol. 2011;301:R370–R377. doi: 10.1152/ajpregu.00748.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M, Wax S, Beck ARP, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripaud L, Maillet L, Cullin C. The mechanisms of [URE3] prion elimination demonstrate that large aggregates of Ure2p are dead-end products. EMBO J. 2003;22:5251–5259. doi: 10.1093/emboj/cdg488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouble AN, Helfer J, Mamady H, Storey KB, Tessier SN. Anti-apoptotic signaling as a cytoprotective mechanism in mammalian hibernation. Peer J. 2013;1:e29. doi: 10.7717/peerj.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Davis RW. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- Staples JF. Maintaining metabolic balance in mammalian hibernation and daily torpor. In: Nowakowska A, Caputa M, editors. Hypometabolism: strategies of survival in vertebrates and invertebrates. Kerala: Research Signpost; 2011. pp. 95–115. [Google Scholar]
- Storey KB. Oxidative stress: animal adaptations in nature. Braz J Med Biol Res. 1996;29:1715–1733. [PubMed] [Google Scholar]
- Storey KB. Mammalian hibernation: transcriptional and translational control. In: Roach RC, Wagner PD, Hackett PH, editors. Hypoxia: through the lifecycle. New York: Kluwer Academic/Plenum Publishers; 2003. pp. 7–24. [Google Scholar]
- Storey KB. Out cold: biochemical regulation of mammalian hibernation—a mini-review. Gerontology. 2010;56:220–230. doi: 10.1159/000228829. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM. Metabolic rate depression in animals: transcriptional and translational controls. Biol Rev Camb Philos Soc. 2004;79:207–233. doi: 10.1017/S1464793103006195. [DOI] [PubMed] [Google Scholar]
- Storey KB, Storey JM. Metabolic rate depression: the biochemistry of mammalian hibernation. In: Makowski GS, editor. Advances in clinical chemistry. Burlington: Academic; 2010. pp. 77–108. [PubMed] [Google Scholar]
- Suswam EA, Li YY, Mahtani H, King PH. Novel DNA-binding properties of the RNA-binding protein TIAR. Nucleic Acids Res. 2005;33:4507–4518. doi: 10.1093/nar/gki763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taupin JL, Tian Q, Kedersha N, Robertson M, Anderson P. The RNA-binding protein TIAR is translocated from the nucleus to the cytoplasm during Fas-mediated apoptotic cell death. Proc Natl Acad Sci U S A. 1995;92:1629–1633. doi: 10.1073/pnas.92.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier SN, Storey KB. Expression of myocyte enhancer factor-2 and downstream genes in ground squirrel skeletal muscle during hibernation. Mol Cell Biochem. 2010;344:151–162. doi: 10.1007/s11010-010-0538-y. [DOI] [PubMed] [Google Scholar]
- Wang LCH, Lee TF. Torpor and hibernation in mammals: metabolic, physiological, and biochemical adaptations. In: Fregley MJ, Blatteis CM, editors. Handbook of physiology: environmental physiology. New York: Oxford University Press; 1996. pp. 507–532. [Google Scholar]
- Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CW, Storey KB. Regulation of the mTOR signaling network in hibernating thirteen-lined ground squirrels. J Exp Biol. 2012;215:1720–1727. doi: 10.1242/jeb.066225. [DOI] [PubMed] [Google Scholar]
- Zatzman ML. Renal and cardiovascular effects of hibernation and hypothermia. Cryobiology. 1984;21:593–614. doi: 10.1016/0011-2240(84)90220-7. [DOI] [PubMed] [Google Scholar]
- Zhang T, Delestienne N, Huez G, Kruys V, Gueydan C. Identification of the sequence determinants mediating the nucleo-cytoplasmic shuttling of TIAR and TIA-1 RNA-binding proteins. J Cell Sci. 2005;118:5453–5463. doi: 10.1242/jcs.02669. [DOI] [PubMed] [Google Scholar]