Abstract
MicroRNAs (miRNAs) are small single-stranded non-coding RNAs that have an important regulatory function in animal growth and developmental processes. However, the differential expression of miRNA and the role of these miRNAs in heat-stressed Holstein cows are still unknown. In this study, the profile of differentially expressed miRNAs and the target genes analysis in the serum of heat-stressed and normal Holstein cows were investigated by a Solexa deep-sequencing approach and bioinformatics. The data identified 52 differentially expressed miRNAs in 486 known miRNAs which were changed significantly between heat-stressed and normal Holstein cows (fold change >2, P < 0.001). Target genes analysis showed that at least 7 miRNAs (miR-19a, miR-19b, miR-146a, miR-30a-5p, miR-345-3p, miR-199a-3p, and miR-1246) were involved in the response to stress, oxidative stress, development of the immune system, and immune response among the identified 52 differentially expressed miRNAs. Five miRNAs (miR-27b, miR-181a, miR-181b, miR-26a, and miR-146b) were involved in stress and immune responses and the expression of five miRNAs was striking (P < 0.001). In addition, RT-qPCR and deep-sequencing methods showed that 8 miRNAs among the 12 selected miRNAs (miR-19a, miR-19b, miR-27b, miR-30a-5p, miR-181a, miR-181b, miR-345-3p, and miR-1246) were highly expressed in the serum of heat-stressed Holstein cows. GO and KEGG pathway analysis showed that these differentially expressed miRNAs were involved in a pathway that may differentially regulate the expression of stress response and immune response genes. Our study provides an overview of miRNAs expression profile and the interaction between miRNAs and their target genes, which will lead to further understanding of the important roles of miRNAs in heat-stressed Holstein cows.
Keywords: Heat stress, Serum, MicroRNAs, Target genes, Cows
Introduction
MicroRNAs (miRNAs) are a class of single-strand, endogenous, approximately 22-nt non-coding RNA molecules that negatively regulate gene expression at the post-transcriptional level (Llave et al. 2002; Bartel 2004). They are involved in almost every biological process, including cell growth and differentiation (McKenna et al. 2010), pathogenesis, and disease prevention. The mature miRNA strand is incorporated in the RNA-induced silencing complex (RISC), serving as a leading RNA to control the expression of cognate messenger RNA (mRNA) for degradation or translation repression (Zhang et al. 2007; McDaneld 2009). The first two miRNAs, lin-4 and let-7, were discovered in Caenorhabditis elegans (Lee et al. 1993; Reinhart et al. 2000). Subsequently, about 100 miRNAs were identified by cloning and Sanger sequencing (Friedländer et al. 2008). In cattle, miRNA expression profiles have been reported for fat tissue. The study identified 154 miRNA sequences from bovine fat tissue and mammary glands, 54 of which were determined to be fat tissue specific (Gu et al. 2007). Some studies found that miR-199b, miR-199a-5p, and miR-126 were expressed in the bovine mammary gland (Ogorevc et al. 2009). The role of miRNA in lipid metabolism was first reported in Drosophila, and deletion of miR-14 increased the accumulation of triacylglycerol and diacylglycerol (Xu et al. 2003). However, such approaches were limited in their ability to detect rare miRNAs, or tissue-specific miRNAs from tissues that are difficult to obtain. In mammals, temporal and spatial changes in miRNA expression have been well characterized to delineate the miRNA transcription of different tissues at different developmental stages (Li et al. 2010) or in different species (Li et al. 2012). At the same time, one study clearly demonstrated that levels of miRNAs in serum are stable, reproducible, and consistent among individuals of the same animal species (Chen et al. 2008).
Heat stress can be defined as a condition that occurs when an animal cannot adequately dissipate body heat in order to maintain thermal balance (Bernabucci et al. 2014). Virtually, the entire south of China is subject to extended periods of hot and dry conditions. Heat-tolerant breeds have been developed to deal with these conditions, in addition to nutritional management, to improve milk yields. Some studies have estimated the genetic components in both milk yield and reproductive traits under heat stress and have detected an unfavorable genetic relationship between the temperature humidity index (THI) and productive and reproductive traits (West 2003). Some studies suggest that the immune system is affected by heat stress in cows (Salak-Johnson and McGlone 2007). The study of miRNA expression profiling in the Holstein cow is meaningful because cows are the most important milk producers. However, the complete genome sequence, the number of miRNAs identified from the Holstein cow, is very limited as compared to other species. Furthermore, there has been no experimental validation of the related miRNA targets in the serum of heat-stressed Holstein cows.
In this study, we aimed to detect the differential expression of miRNAs in the serum of heat-stressed and normal Holstein cows by a Solexa deep-sequencing approach, and to analyze the target genes that may have an impact on stress response and immune function in cows.
Materials and methods
Animals
The experimental Holstein cows came from the dairy farm of Nanjing Agriculture University. The extraction of bovine venous blood experiments was approved by the Animals Ethics Committee of Nanjing Agriculture University. All experiments were carried out in accordance with the National Institutes of Health guidelines.
Samples and RNA preparation
The serum of heat-stressed and normal Holstein cows was collected separately from the same cows, but according to the two categories. There were six cattle in our study, all 3 years old, and all in the middle of the lactating period. Each cow had one calf. The same six experimental cows were tested both under normal physiological conditions in April and under heat-stressed conditions in August. The barn temperature and humidity index were used as heat-stress indicators, irrespective of whether the animals appeared heat stressed. The animal’s rectal temperature, breathing rate, milk yield, and food intake were recorded. A rectal temperature above 39 ° C, a respiratory rate of 90–120 times per minute, and decreased milk and dairy food intake were regarded as indicative of heat stress. The rectal temperature of heat-stressed cows was above 39 °C, and as high as 42 °C, the respiratory rate was 90~120 breaths per minute, and milk yield decreased significantly compared with the normal. Rectal temperature and breathing rate data showed that the cattle were under moderate heat stress. Blood was collected at 2:00 p.m. in 3 days. Samples of serum were frozen in liquid nitrogen immediately after 4,000 rpm centrifugation (4 °C, 15 min), and then stored at −80 °C until total RNA isolation. Serum total RNA was isolated separately from a 3-mL sample with total a RNA isolation Kit (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. The quality and concentration of total RNA were determined by routine agarose gel analysis and spectrophotometry (ND-1,000, Thermo Scientific, Waltham, MA, USA), respectively.
Solexa deep-sequencing array
An miRNA Solexa deep-sequencing assay was performed by the service provider (LC-Biotech, USA). Ten micrograms of total RNA were used for sequencing by the Genome Analyzer GA-I (Illumina, San Diego, USA) following protocols. The RNA fractions with the length of 10–40 nt were isolated by 15 % denaturing polyacrylamide gel electrophoresis. After connecting with 5′ and 3′ adaptors, the obtained short RNAs were reversely transcribed to complementary DNA (cDNA) according to the Illumina protocol. The raw sequences were first processed by Illumina’s Genome Analyzer Pipeline software to filter out the adapter sequences, low-quality and low-copy sequences, and then, the extracted small RNA sequences with 15–26 nt in length were subjected to mRNA, RFam, and Repbase filter. Finally, the remaining unique sequences were compared to the miRNA database, miRBase 20.0 by BLASTn search to identify the conserved miRNAs in bovine (http://www.mirbase.org/). In identifying potential miRNA precursor sequences, a maximum of three mismatches was allowed between identified short miRNAs and known animal miRNAs. All identified mature miRNA sequences were further BLASTed against the draft genome sequences which were downloaded from a bovine genome database (http://www.ncbi.nlm.nih.gov/genome/82) and predicated for the hairpin RNA structures for their flanking sequences by RNAfold software (http://mfold.rit.albany.edu/?q=mfold/RNA-Folding-Form).
Non-coding sequences, which met previously described criteria, were then considered to be a potential miRNA precursor. Specifically, (1) the identified miRNAs were located in the arms of a stem–loop structure. (2) No large loop or break occurred in the identified miRNA sequence. (3) A maximum of six mismatches were allowed between the identified miRNAs and the opposite miRNA sequence (miRNA*). (4) The potential miRNA precursor must have a higher negative minimal folding energy (MFE) and minimal free folding energy indexes (MFEI) to distinguish it from other small RNAs. To identify potential novel miRNAs in genome, the remaining unmapped soluble RNA (sRNA) sequences were also BLASTed against the genome as described above, and their folded secondary structure was predicted. Only the non-coding sequences that formed a perfect stem–loop structure and met the criteria for miRNAs prediction were considered to be a potentially novel miRNA candidate.
Quantitative real-time PCR
The candidate differentially expressed miRNAs were selected based on their pattern of expression as seen in the deep sequencing array results (fold change >2, P < 0.001). Real-time quantitative PCR (RT-qPCR) was performed using the ABI PRISM® 7,300 Real-Time PCR System. Briefly, 2 μg of miRNA were reverse transcribed using the One Step PrimeScript® miRNA cDNA Synthesis Kit (Takara Biotechnology Co., Ltd., Japan, D350A). The reverse transcription reaction system included 10 μL of 2 × miRNA reaction buffer, 2 μL of 0.1 % BSA, 2 μL of miRNA PrimeScript® RT Enzyme Mix, 2 μg of total RNA, and RNA-free dH2O to a final volume of 20 μL. The RT-PCR program was set to 37 °C for 60 min followed by 85 °C for 5 s. The cDNA products were stored at −20 °C. The relative real-time quantitative PCR was performed with SYBR® Premix Ex Taq™ (Takara Biotechnology Co., Ltd., Japan, DRR081A). The reaction solution was prepared on ice and comprised 10 μL of 2× SYBR® Premix Ex Taq™, 0.8 μL of PCR forward primer (10 μM), 0.8 μL of Uni-miR qPCR Primer (10 μM), 0.4 μL of 50× ROX reference dye, 2 μL of cDNA (100 ng/μL), and dH2O to a final volume of 20 μL. The reaction mixtures were incubated in a 96-well plate at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 30 s. All reactions were performed in triplicate. The primers for the miRNA had the same sequences as the Bos taurus miRNA with appropriate adjustments at their 5′ (Table 1). The △△Ct method was used to determine the miRNAs expression level of serum between heat-stressed and normal Holstein cows. The housekeeping gene beta-5S RNA was used as an endogenous control.
Table 1.
Primer sequences for RT-qPCR
| Assay name | Forward Primer (5′ to 3′) | Length |
|---|---|---|
| bta-5sRNA | CTCGTCTGATCTCGGAAGCTAA | 22 |
| miR-19a | CGGCGGTGTGCAAATCTAT | 19 |
| miR-19b | TGTGCAAATCCATGCAAAACTG | 22 |
| miR-26a | GGTTCAAGTAATCCAGGATAGGCT | 24 |
| miR-27b | CGGCTTCACAGTGGCTAAGTTCT | 23 |
| miR-30a-5p | CGGTGTAAACATCCTCGACTGG | 22 |
| miR-146a | GCGGCGGTGAGAACTGAAT | 19 |
| miR-146b | CGCCGGTGAGAACTGAAT | 18 |
| miR-181a | GAACATTCAACGCTGTCGGTG | 21 |
| miR-181b | AACATTCATTGCTGTCGGTGG | 21 |
| miR-199a-3p | CGGACAGTAGTCTGCACATTGG | 22 |
| miR-345-3p | GCCTGAACTAGGGGTCTGGAG | 21 |
| miR-1246 | GGAATGGATTTTTGGAGCAGG | 21 |
miRNAs Target genes prediction and functional analysis by bioinformatics
To understand the molecular function of the differentially expressed miRNAs in serum of heat-stressed and normal Holstein cows, we used the TargetScan (Lewis et al. 2003) to predict their target mRNA. In cases where the tested Bos taurus miRNA was not included in the current version of TargetScan, the target genes were predicted by a custom search using the seed sequences. This approach was based on the assumption that miRNA target sites in the 3′ UTR of the genes are more highly conserved among closely related animals than in more distant species (Chen and Rajewsky 2006; Rajewsky 2006). With the use of DAVID (Dennis et al. 2003) bioinformatics resources, the genes were classified according to KEGG functional annotations to identify pathways that were actively regulated by miRNA in serum of heat-stressed and normal Holstein cows. Enriched GO terms in the target genes were identified with the GO: Term Finder Perl module using the Term Enrichment tool (Consortium 2004).
Statistical analysis
The data were obtained from one independent experiment carried out in triplicate. Interactive and main effects were analyzed by one-way ANOVA using SPSS 18.0 software. When justified by one-way ANOVA, differences between individual group means were analyzed by Fisher’s PLSD test. Differences were considered statistically significant at P < 0.001.
Results
miRNAs Deep sequencing result
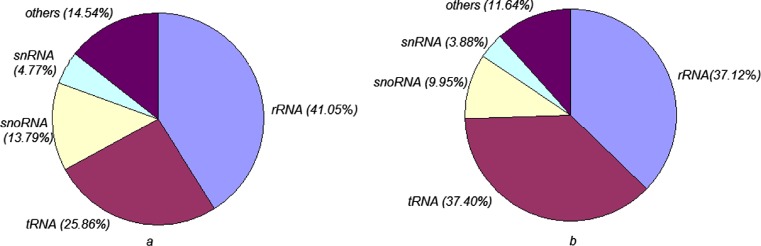
To identify miRNAs in Holstein cows, two independent sRNA libraries were constructed from the serum of normal and heat-stressed Holstein cows by Solexa deep sequencing. There were 13,912,941 raw reads in the normal cows and 9,668,506 raw reads in the heat-stressed cows. Filtering out the adapter sequences and low-quality sequences revealed 11901654 sequences with a length of 16–32 nt in the serum of normal cows and 5278893 sequences of the same length in the serum of the heat-stressed cows. As shown in Table 2, most of the unique sequences were mapped to the bovine genome; 55.52 % of the unique sequences in normal cows were mapped and 53.17 % of the unique sequences in heat-stressed cows were mapped. Analysis of the fragments of ribosomal RNA (rRNA), transfer RNA (tRNA), snRNA, snoRNA, and others in the sRNA libraries (Fig. 1) were revealed. The majority of the fragments in the RNA libraries were identified as rRNA and tRNA. In the serum RNA library of the normal cows, rRNA and tRNA accounted for 41.05 and 25.86 %, respectively. In the heat-stressed cows, rRNA and tRNA accounted for 37.12 and 37.40 %, respectively.
Table 2.
Statistics of small RNA sequences from cow’s serum
| Category | Heat-stress | Normal | ||
|---|---|---|---|---|
| Sequences | Unique sequences | Sequences | Unique sequences | |
| Raw reads | 9668506 | 495021 | 13912941 | 545772 |
| Low-quality reads removed | 11026 | 3040 | 13931 | 1656 |
| 3ADT and length filtera | 1202522 | 164358 | 581309 | 159157 |
| mRNA, RFam, Repbase, matches removed | 5005388 | 72864 | 1704071 | 61936 |
| Mappable sequencesb | 5278893 | 263210 | 11901654 | 303021 |
a3ADT and length filter: reads removed due to 3ADT
bMappable sequences: The raw reads were passed through a series of digital filters by Illumina’s Genome Analyzer Pipeline software and ACGT101-miR program, and the resulting sequences were called “mappable sequences”
Fig. 1.
The distribution of fragments of rRNA, tRNA, snRNA, snoRNA, and others in the small RNA libraries. a The serum of normal Holstein cows. b The serum of heat-stressed Holstein cows. The distribution of these fragments was focused on rRNA and tRNA in the serum of Holstein cows
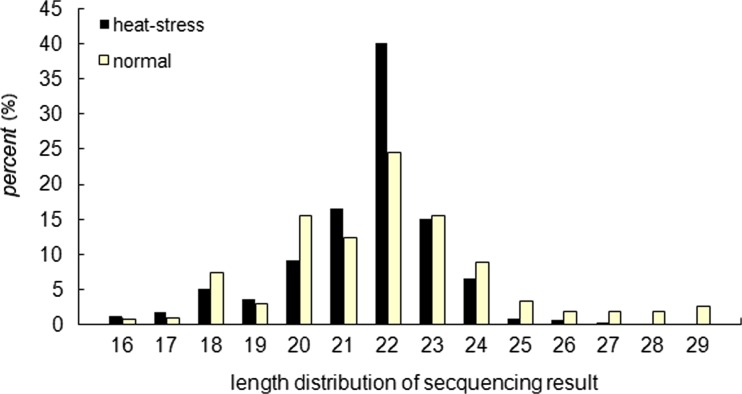
To assess the sequencing quality, we analyzed the distribution and length of the sRNA based on both total abundance and distinct sequences. In both libraries, the majority of the sRNAs were 20–24 nt in size. The top four abundant sizes were 20, 21, 22, and 23, which accounted for 15.57, 12.36, 24.48, and 15.40 % of the total reads in normal cows and 9.04, 16.42, 39.98, and 14.94 % in heat-stressed cows, respectively (Fig. 2). These results were consistent with the typical sRNA distribution of mammals.
Fig. 2.
The size distribution of the small RNAs in the serum of heat-stressed and normal Holstein cows. In both libraries, the majority of the small RNAs were 20~24 nt, and 22 nt in size having the highest abundance
Based on the sequence similarity, the mappable sRNA sequences were first compared to the currently known bovine miRNAs in miRBase v20 database. Four hundred eighty-six miRNAs sequence showed similarity to the known bovine miRNAs (date not shown), which means that 486 miRNAs were identified in the serum of Holstein cows. These identified miRNAs could be grouped into several miRNA families. The families of miR-29, miR-30, miR-2284, and miR-2285 were abundant in two libraries. The expression profiles of miRNAs were analyzed and compared in two libraries. We identified 52 differentially expressed miRNAs in 486 known miRNAs which were changed significantly between heat-stressed and normal Holstein cows (fold change >2, P < 0.001) (Table 3).
Table 3.
The differential expression of miRNAs in serum of heat-stressed and normal Holstein cows by sequencing method (P < 0.001)
| miRNA | Normal (N) | Heat stress (HS) | Fold change [Log2 (HS/N)] | P value |
|---|---|---|---|---|
| Lowly expressed miRNA in HS [Log2(N/HS) >2] | ||||
| bta-miR-96 | 408 | 68 | 2.58 | 5.77E-05 |
| bta-miR-99b | 24,874 | 2,556 | 3.28 | 2.21E-09 |
| bta-miR-126-5p | 46,523 | 46,523 | 2.34 | 1.36E-08 |
| bta-miR-134 | 147 | 36 | 2.04 | 7.35E-02 |
| bta-miR-146a | 94,687 | 20,502 | 2.21 | 3.74E-02 |
| bta-miR-150 | 158,552 | 36,283 | 2.13 | 3.74E-02 |
| bta-miR-151-5p | 81,993 | 19,084 | 2.10 | 1.25E-06 |
| bta-miR-199a-3p | 22,653 | 4,238 | 2.42 | 1.23E-04 |
| bta-miR-223 | 52,160 | 11,808 | 2.14 | 1.57E-03 |
| bta-miR-331-3p | 1,566 | 352 | 2.15 | 2.12E-08 |
| bta-miR-376b | 29 | 1.89 | 3.94 | 1.58E-07 |
| bta-miR-376a | 128 | 1.89 | 6.08 | 2.14E-02 |
| bta-miR-379 | 505 | 96 | 2.39 | 1.79E-04 |
| bta-miR-381 | 1,452 | 218 | 2.74 | 5.29E-02 |
| bta-miR-410 | 1,029 | 96 | 3.41 | 3.67E-01 |
| bta-miR-431 | 157 | 7 | 4.38 | 5.54E-06 |
| bta-miR-493 | 153 | 26 | 2.54 | 3.55E-05 |
| bta-miR-654 | 1972 | 201 | 3.29 | 1.86E-08 |
| bta-miR-758 | 470 | 81 | 2.53 | 3.64E-03 |
| bta-miR-873 | 456 | 45 | 3.33 | 2.51E-07 |
| bta-miR-2478 | 11,379 | 1,877 | 2.59 | 5.29E-02 |
| bta-miR-4286 | 393 | 73 | 2.43 | 2.19E-09 |
| Highly expressed miRNA in HS [Log2(N/HS) ≤2] | ||||
| bta-miR-19b | 17,57 | 11,927 | −2.76 | 9.07E-04 |
| bta-miR-19a | 537 | 6256 | −3.54 | 6.93E-06 |
| bta-miR-30a-5p | 2,739 | 25,140 | −3.19 | 9.36E-06 |
| bta-miR-99a-3p | 4 | 182 | −5.26 | 7.98E-04 |
| bta-miR-122 | 2,413 | 59,724 | −4.63 | 1.54E-03 |
| bta-miR-130b | 532 | 2,473 | −2.22 | 1.75E-08 |
| bta-miR-141 | 403 | 2,604. | −2.69 | 1.13E-03 |
| bta-miR-147 | 40 | 252 | −2.65 | 8.58E-03 |
| bta-miR-192 | 11,798 | 84,839 | −2.85 | 1.27E-08 |
| bta-miR-193b | 97 | 762 | −2.96 | 2.14E-05 |
| bta-miR-193a-5p | 215 | 931 | −2.11 | 1.75E-08 |
| bta-miR-200a | 286 | 4,421 | −3.95 | 1.27E-03 |
| bta-miR-204 | 14 | 290 | −4.32 | 5.94E-09 |
| bta-miR-205 | 79 | 1,323 | −4.06 | 3.49E-08 |
| bta-miR-211 | 0.26 | 24 | −6.55 | 4.74E-06 |
| bta-miR-214 | 18 | 168 | −3.15 | 7.11E-04 |
| bta-miR-215 | 1,562 | 34,823 | −4.48 | 1.18 E-03 |
| bta-miR-296-3p | 156 | 4,579 | −4.87 | 3.56E-06 |
| bta-miR-301a | 1,004 | 5,466 | −2.44 | 1.33E-06 |
| bta-miR-320a | 5,355 | 24,525 | −2.19 | 2.14E-07 |
| bta-miR-345-3p | 3 | 1,754 | −9.12 | 9.07E-06 |
| bta-miR-378b | 23 | 1,25 | −2.41 | 3.83E-04 |
| bta-miR-380-3p | 9 | 70 | −2.89 | 4.39E-02 |
| bta-miR-421 | 446 | 4,427 | −3.31 | 9.03E-06 |
| bta-miR-423-5p | 19,199 | 229,399 | −3.58 | 1.22E-06 |
| bta-miR-429 | 6 | 161 | −4.56 | 1.27E-08 |
| bta-miR-497 | 23 | 170 | −2.85 | 4.19E-02 |
| bta-miR-499 | 30 | 426 | −3.82 | 1.69E-06 |
| bta-miR-1246 | 394 | 7,196 | −4.19 | 1.74E-02 |
| bta-miR-1343-3p | 36 | 276 | −2.93 | 9.58E-06 |
Validation of the selected miRNA by RT-qPCR
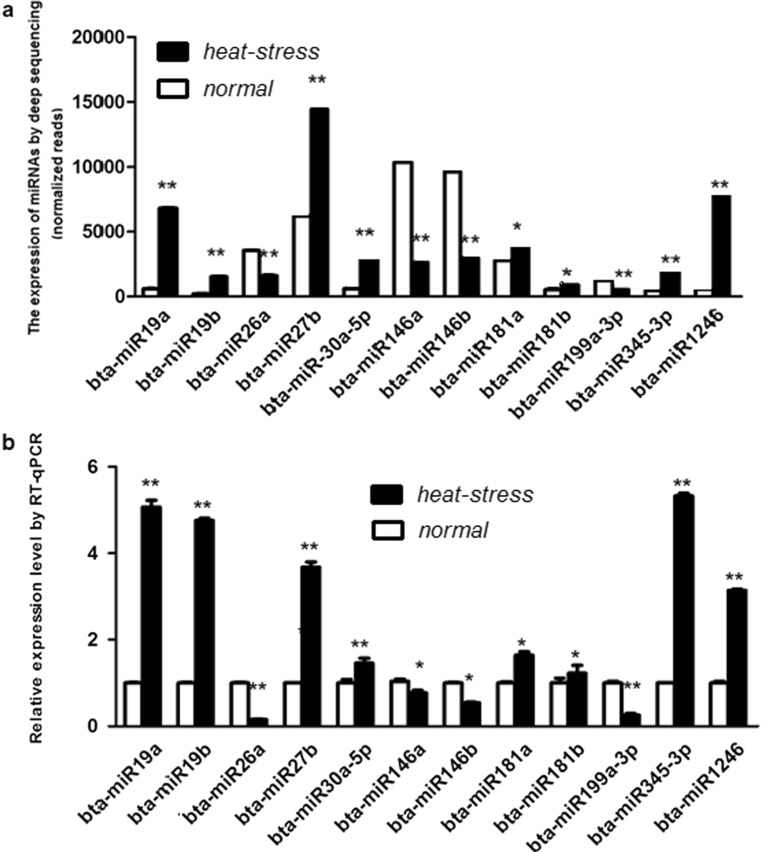
To validate whether the deep sequencing and real-time quantitative PCR (RT-qPCR) data were identical, RT-qPCR was conducted to measure the expression levels of eight selected up-regulated miRNAs (miR-19a, miR-19b, miR-27b, miR-30a-5p, miR-181a, miR-181b, miR-345-3p, and miR-1246) and four down-regulated miRNAs (miR-26a, miR-146a, miR-146b, and miR-199a-3p) in heat-stressed serum. The results showed that the expression level of these selected miRNA by RT-qPCR was in concordance with the normalized sequencing data (Fig. 3).
Fig. 3.
Analysis the selected 12 differential expressed miRNAs in serum of heat-stressed and normal Holstein cows by sequencing and RT-qPCR methods. a The expression of miRNAs by Solexa deep sequencing (normalized reads). b The relative expression of miRNAs tested RT-qPCR. The results showed that expression of these selected miRNAs by RT-qPCR were in agreement with the normalized sequencing data
Prediction of target genes and functional bioinformatics analysis
Target gene analysis showed that 12 of the 52 differentially expressed miRNAs were involved in the response to heat, oxidative stress, and immune system responses among the identified 52 differentially expressed significant miRNAs. The analysis showed that the 12 selected miRNAs had 1,210 putative target genes (data not shown). Among the 1,210 putative target genes, according to the enrichment results, 58 target genes were related to biological functions associated with the response to stress, oxidative stress, development of the immune system, and immune response (Table 4 and Table 5). The number of potential target genes varied among miRNAs with a high of 13 predicted genes such as miR-181b, and a low of 2 predicted genes such as miR-199a-3p.
Table 4.
Predicted miRNAs target genes with function related to response to stress, oxidative stress, and immune response
| miRNA | Count | Target genes |
|---|---|---|
| miR-19a | 3 | HSPBAP1, DNAJB1, HPX |
| miR-19b | 5 | HSPBAP1, DNAJB1, HPX, APOA2, NOD2 |
| miR-26a | 3 | HSPA8, CD38, PRKCB |
| miR-27b | 7 | TP53, PLA2R1, HSPCB, PSEN2, FYN, THY1, BCL10 |
| miR-30a-5p | 2 | PLA2R1, PICEN |
| miR-146a | 3 | TLR4, CD3E, PLEKHA2 |
| miR-146b | 3 | TLR4, CD3E, PLEKHA2 |
| miR-181a | 9 | HSPA1L, IKBKB, HPX, APOA2, TNF, NOD2, PSEN2, THEMIS, CD38, BCL10 |
| miR-181b | 11 | PPP1R15A, HSPA1L, IKBKB, HPX, APOA2, TNF, HPX, NOD2, PSEN2, THEMIS, CD38, BCL10 |
| miR-199a-3p | 2 | TP53, PSEN2 |
| miR-345-3p | 2 | NOD2, THY1 |
Table 5.
Functional analysis of gene targets involved in stress, oxidative stress, and immune response
| GO Term | Count | Target genes |
|---|---|---|
| GO:0090403~oxidative stress induced premature senescence | 2 | TP53, PLA2R1 |
| GO:0006950~response to stress | 22 | PYCR1, PRKRA, PP1R15A, HSPB8, HPSH1, HSPBAP1, HSPCB, ERP44, MAPK13, MAPK1, HSPA8, PPP3CA, HSF2BP, HSPB6, PARK7, HSPB3, AHA1, HSPAIL, DNAJB1, UCHl1, UCN3, PICEN |
| GO:000863 ~induction of apoptosis by oxidative stress | 1 | DIABLO |
| GO:0001776~leukocyte homeostasis | 3 | MENI, BAX, NKX2-6 |
| GO:0001782~B cell homeostasis | 3 | IKBKAPI, IKBKB, BAX |
| GO:0002218~activation of innate immune response | 2 | TMEM173, TLR4 |
| GO:0002286~T cell activation involved in immune response | 2 | PSEN1, PSEN2 |
| GO:0002309~T cell proliferation involved in immune response | 1 | TP53 |
| GO:0002313~mature B cell differentiation involved in immune response | 1 | EBI2 |
| GO:0002426~immunoglobulin production in mucosal tissue | 1 | GCNT3 |
| GO:0002639~positive regulation of immunoglobulin production | 3 | SASH1, HPX, FOXP1 |
| GO:0002740~negative regulation of cytokine secretion involved in immune response | 3 | IL-10, APOA2, TNF |
| GO:0002830~positive regulation of type2 immune response | 2 | NOD2, II33 |
| GO:0002925~positive regulation of humoral immune response mediated by circulating immunoglobulin | 4 | LTA, HPX, NOD2, TNF |
| GO:0050852~T cell receptor signaling pathway | 12 | PSEN1, FYN, CD247, PSEN2, BCL10, THEMIS, CD3E, TRAF6, RBCK1, GATA3, THY1, UBE2N |
| GO:0050853~B cell receptor signaling pathway | 6 | CD79A, PLEKHA2, SYK, CD38, PRKCB, LCK |
| GO:0050870~positive regulation of T cell activation | 4 | CCL2, BCL10, THY1, LCK |
| GO:0050871~positive regulation of B cell activation | 2 | IL-6, NOD2 |
Using DAVID bioinformatics resources, these predicted target genes were classified according to KEGG functional annotations (Table 6). Interestingly, most of the 12 miRNAs differentially expressed in the serum of the heat-stressed cows and normal cows seemed to be involved in three pathways: mitogen-activated protein kinase (MAPK) signaling, cancer, and regulation of the cytoskeleton. Although the predicted target genes need to be validated experimentally in subsequent experiments, collectively, the findings illustrate some possible pathways and roles of differentially expressed miRNAs in cows exposed to heat stress.
Table 6.
KEGG Pathways enriched for targets of the 12 miRNAs that are differentially expressed in the serum of normal and heat-stressed cows
| Term | Gene number | P value |
|---|---|---|
| bta04722: Neurotrophin signaling pathway | 30 | 3.4E-03 |
| bta05211: Renal cell carcinoma | 19 | 5.3E-03 |
| bta04666:Fc gamma R-mediated phagocytosis | 23 | 6.2E-03 |
| bta05218: Melanoma | 19 | 6.3E-03 |
| bta04130: SNARE interactions in vesicular transport | 13 | 7.0E-03 |
| bta04810: Regulation of actin cytoskeleton | 41 | 1.2 E-02 |
| bta05200: Pathways in cancer | 60 | 1.5 E-02 |
| bta04010: MAPK signaling pathway | 52 | 1.6 E-02 |
| bta04660:T cell receptor signaling pathway | 25 | 1.8 E-02 |
| bta04110: Cell cycle | 28 | 2.1 E-02 |
| bta04142: Lysosome | 26 | 2.4 E-02 |
| bta05214: Glioma | 16 | 2.7 E-02 |
| bta05216: Thyroid cancer | 9 | 3.0 E-02 |
| bta04115:p53 signaling pathway | 16 | 3.0 E-02 |
| bta04150:mTOR signaling pathway | 14 | 3.2 E-02 |
| bta04620: Toll-like receptor signaling pathway | 22 | 3.3 E-02 |
| bta04670: Leukocyte transendothelial migration | 24 | 6.3 E-02 |
| bta05212: Pancreatic cancer | 16 | 6.4 E-02 |
| bta00601: Glycosphingolipid biosynthesis | 8 | 6.7 E-02 |
| bta04664:Fc epsilon RI signaling pathway | 17 | 6.7 E-02 |
| bta00510:N-Glycan biosynthesis | 11 | 7.3 E-02 |
| bta03040: Spliceosome | 25 | 8.7 E-02 |
| bta05219: Bladder cancer | 10 | 8.8 E-02 |
| bta04210: Apoptosis | 18 | 9.3E-02 |
| bta03018: RNA degradation | 13 | 9.6E-02 |
Discussion
The innate immune system of animals provides an immediate defense against infection of pathogens in a non-specific manner, which is the first line of defense found in vertebrates and invertebrates. At the same time, there is an important relationship between miRNA and innate immunity in invertebrates (Yang et al. 2012). Apoptosis, activation of the innate immune response, and the T/B cell receptor signaling pathway are considered to be the most important responses in innate immunity, with the responses regulated by complex systems mainly involving the control of gene expression. Identification of miRNAs and their targets genes are the basis for understanding the physiological functions of miRNAs. Many animal miRNAs have been deposited to miRBase and their physiological functions have also been studied. Many studies on the related miRNAs of cows focus on lipid metabolism (Cui et al. 2014) or mastitis (Li et al. 2014). Research on the connection between heat-stress and miRNAs showed that miRNAs were an emerging layer of novel regulators in the mammalian heat-shock response (Islam et al. 2013; Place and Noonan 2013). But no studies to date have been made about the related miRNAs in heat-stressed Holstein cows.
In this study, we assessed the expression of miRNAs by Solexa deep sequencing. We found many differences in the expression of miRNAs between the serum of normal and heat-stressed Holstein cows. In total, 486 miRNAs were detected and 52 miRNAs changed significantly, suggesting that miRNAs may play an important role when cows are exposed to heat stress. Target genes analysis showed that at least 12 miRNAs were involved with heat stress, oxidative stress, and immune system responses. The number of target genes may be related to the different levels at which miRNAs regulate the immune and stress responses. The number of potential target genes varied among the selected miRNAs, as a result of the different functions of the different miRNAs. We also analyzed the KEGG pathway and GO-term enrichment of the predicted target genes of the 12 different expressed miRNAs using the DAVID bioinformatics resources and the Term enrichment tool.
The 12 selected miRNAs and their target genes have been involved in apoptosis, and in activation of the innate immune response and T/B cell receptor signaling pathway. Our study presents the first comprehensive view of miRNAs associated with innate immunity and heat-stress response in heat-stressed Holstein cows. As revealed in this study, miR-181a is significantly up-regulated in heat-stressed serum (P < 0.001). Other studies indicate that miR-181a also plays an important role in T cell differentiation (Chen et al. 2004) and T cell receptor (TCR) signaling sensitivity (Li et al. 2007). The target genes of miR-181a were associated with T cell activation, immune response, and the B cell receptor signaling pathway. Among several genes of the toll-like receptor (TLR) pathway up-regulated during intramammary infections (IMI) challenge (Moyes et al. 2009), the increased expression of the target gene of miR181a (FOS) after IMI supported its involvement in inflammation and apoptosis during tissue remodeling following inflammation (Clarkson et al. 2004). More importantly, the target gene TLR4 (miR-146a/b) involved in non-specific immunity (innate immunity) is not only an important class of protein molecules, but is also the connection between non-specific and specific immunity, when microorganisms break through the body’s physical barriers, such as skin or mucous membranes. It has been reported that miR-146a has an anti-inflammatory function and is up-regulated after lipopolysaccharide (LPS) challenge on monocytes (Dilda et al. 2012). This finding does not agree with the results of the present study. The discord might be due to heat stress not yet causing inflammation. Both the physiological state and the response were different after heat stress or LPS. The toll-like receptor can activate the immune response when the body subjected to high temperature or the stimulation of LPS. Our study also showed that miR-1246 and TLR are closely related. As an important miRNA target gene, miR-1246 has an influence on the immune system. Other target genes of the selected miRNAs were also closely connected with innate immunity. The interactions of NOD2 and LPS may be spurring inflammation in the ileum. A previous work reported that a CD38 polymorphism was related to leukemia and that tumor necrosis factor (TNF) was associated with cytokine production and inflammatory reactions. At the same time, functional analysis of gene targets reveals that several target genes of selected miRNAs are involved in stress response, and oxidative stress such as TP53, PLA2R1, PYCR1, PRKRA, PP1R15A, HSPB8, HPSH1, HSPBAP1, and HSPCB. Numerous studies indicate that there is an important relationship between the family of heat-shock proteins and the heat-stress response (Song et al. 2013; McConnell et al. 2014). Our study provides an important theoretical basis for further research on the link between heat-shock proteins and miRNAs (such as miR-19a, miR-19b, and miR-27a).
In conclusion, we carried out this experiment to study miRNAs expression profile in the serum of heat-stressed Holstein cows, and to understand the relationship of these miRNAs to stress and immune responses in farm animals. We have provided useful knowledge on the regulation mechanism of miRNAs in cows exposed to heat-stress and on the relationship between miRNAs and regulation of the immune function/stress response. Meanwhile, a significant difference in the expression of miRNAs can be used as a biomarker to determine the extent of heat stress and stress response. For example, miR345-3p is significantly up-regulated in heat-stress serum (Fold change = −9.12, P < 0.001). Such knowledge may also apply to other animals or to people, especially in terms of regulation of stress and immune system.
Acknowledgments
This work was supported by the Natural Science Foundation of Jiangsu Province (no. SBK201241530), the National Supporting Projects for Science and Techniques (no. 2011BAD28B02, 2012BAD12B10), and the Fundamental Research Funds for the Central Universities (no. KYZ201413).
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bernabucci U, Biffani S, Buggiotti L, Vitali A, Lacetera N, Nardone A. The effects of heat stress in Italian Holstein dairy cattle. J Dairy Sci. 2014;97(1):471–486. doi: 10.3168/jds.2013-6611. [DOI] [PubMed] [Google Scholar]
- Chen C-Z, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. Deep conservation of microRNA-target relationships and 3′ UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb Symp Quant Biol. 2006;71:149–156. doi: 10.1101/sqb.2006.71.039. [DOI] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Clarkson R, Wayland MT, Lee J, Freeman T, Watson CJ. Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression. Breast Cancer Res. 2004;6(2):R92–R109. doi: 10.1186/bcr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GO. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(1):D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Wang Y, Sun B, Xiao Z, Ye L, Zhang X. MiR-205 modulates abnormal lipid metabolism of hepatoma cells via targeting acyl-CoA synthetase long-chain family member 1 (ACSL1) mRNA. Biochem Biophys Res Commun. 2014;444:270–275. doi: 10.1016/j.bbrc.2014.01.051. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Dilda F, Gioia G, Pisani L, Restelli L, Lecchi C, Albonico F, Ceciliani F. Escherichia coli lipopolysaccharides and Taphylococcus aureus enterotoxin B differentially modulate inflammatory microRNAs in bovine monocytes. Vet J. 2012;192(3):514–516. doi: 10.1016/j.tvjl.2011.08.018. [DOI] [PubMed] [Google Scholar]
- Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26(4):407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Gu Z, Eleswarapu S, Jiang H. Identification and characterization of microRNAs from the bovine adipose tissue and mammary gland. FEBS Lett. 2007;581(5):981–988. doi: 10.1016/j.febslet.2007.01.081. [DOI] [PubMed] [Google Scholar]
- Islam A, Deuster PA, Devaney JM, et al. An exploration of heat tolerance in mice utilizing mRNA and microRNA expression analysis. PLoS One. 2013;8(8):e72258. doi: 10.1371/journal.pone.0072258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih I-h, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li M, Xia Y, Gu Y, Zhang K, Lang Q, Chen L, Guan J, Luo Z, Chen H, Li Y. MicroRNAome of porcine pre-and postnatal development. PLoS One. 2010;5(7):e11541. doi: 10.1371/journal.pone.0011541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q-J, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Li R, Sun Q, Jia Y, Cong R, Ni Y, Yang X, Jiang Z, Zhao R. Coordinated miRNA/mRNA expression profiles for understanding breed-specific metabolic characters of liver between Erhualian and large white pigs. PLoS One. 2012;7(6):e38716. doi: 10.1371/journal.pone.0038716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang H, Chen L, Wang L, Liu X, Ru C, Song A. Identification and characterization of novel and differentially expressed microRNAs in peripheral blood from healthy and mastitis Holstein cattle by deep sequencing. Anim Genet. 2014;45(1):20–27. doi: 10.1111/age.12096. [DOI] [PubMed] [Google Scholar]
- Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14(7):1605–1619. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Alexander LA, McAlpine SR. A heat shock protein 90 inhibitor that modulates the immunophilins and regulates hormone receptors without inducing the heat shock response. Bioorg Med Chem Lett. 2014;24:661–666. doi: 10.1016/j.bmcl.2013.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaneld T. MicroRNA: mechanism of gene regulation and application to livestock. J Anim Sci. 2009;87(14 suppl):E21–E28. doi: 10.2527/jas.2008-1303. [DOI] [PubMed] [Google Scholar]
- McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, Kaestner KH. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. 2010;139(5):1654–1664. doi: 10.1053/j.gastro.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes K, Drackley J, Morin D, Bionaz M, Rodriguez-Zas S, Everts R, Lewin H, Loor J. Gene network and pathway analysis of bovine mammary tissue challenged with Streptococcus uberis reveals induction of cell proliferation and inhibition of PPARγ signaling as potential mechanism for the negative relationships between immune response and lipid metabolism. BMC Genomics. 2009;10(1):542. doi: 10.1186/1471-2164-10-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogorevc J, Kunej T, Dovc A, Razpet P. Database of cattle candidate genes and genetic markers for milk production and mastitis. Anim Genet. 2009;40(6):832–851. doi: 10.1111/j.1365-2052.2009.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Noonan EJ (2013) Non-coding RNAs turn up the heat: an emerging layer of novel regulators in the mammalian heat shock response. Cell Stress Chaperones 1–14 [DOI] [PMC free article] [PubMed]
- Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Salak-Johnson J, McGlone J. Making sense of apparently conflicting data: stress and immunity in swine and cattle. J Anim Sci. 2007;85(13 suppl):E81–E88. doi: 10.2527/jas.2006-538. [DOI] [PubMed] [Google Scholar]
- Song H-M, Mu X-D, Gu D-E, Luo D, Yang Y-X, Xu M, Luo J-R, Zhang J-E, Hu Y-C (2013) Molecular characteristics of the HSP70 gene and its differential expression in female and male golden apple snails (Pomacea canaliculata) under temperature stimulation. Cell Stress Chaperones 1–11 [DOI] [PMC free article] [PubMed]
- West J. Effects of heat-stress on production in dairy cattle. J Dairy Sci. 2003;86(6):2131–2144. doi: 10.3168/jds.S0022-0302(03)73803-X. [DOI] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13(9):790–795. doi: 10.1016/S0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- Yang G, Yang L, Zhao Z, Wang J, Zhang X. Signature miRNAs involved in the innate immunity of invertebrates. PLoS One. 2012;7(6):e39015. doi: 10.1371/journal.pone.0039015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210(2):279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]