Abstract
Skeletal muscle adapts to different patterns of motor nerve activity by alterations in gene expression that match specialized properties of contraction, metabolism, and muscle mass to changing work demands (muscle plasticity). Calcineurin, a calcium/calmodulin-dependent, serine–threonine protein phosphatase, has been shown to control programs of gene expression in skeletal muscles, as in other cell types, through the transcription factor nuclear factor of activated T cells (NFAT). This study provides evidence that the function of NFAT as a transcriptional activator is regulated by neuromuscular stimulation in muscles of intact animals and that calcium influx from the transient receptor potential (TRPC3) channel is an important determinant of NFAT activity. Expression of TRPC3 channels in skeletal myocytes is up-regulated by neuromuscular activity in a calcineurin-dependent manner. These data suggest a mechanism for cellular memory in skeletal muscles whereby repeated bouts of contractile activity drive progressively greater remodeling events.
Skeletal muscles consist of a mosaic of multinucleated myofibers differing in size, metabolic capacity, mitochondrial content, and contractile properties. Determinants of specialized myofiber phenotypes are programmed during fetal development but are modulated in response to changing motor nerve input (1–3). This plasticity allows myofibers to match cellular capacities to different physiologic demands, as demonstrated in classic cross-innervation and chronic low-frequency stimulation experiments performed on adult muscles (4, 5). These adaptive responses are minimal after a single period of neuromuscular activity but become fully manifest only after repeated bouts of activity pursued over a period of days to weeks, as exemplified by the results of training in human athletes. Thus, the physiologically important phenomenon of muscle plasticity, familiar to muscle physiologists for decades (6, 7), includes a form of cellular memory, the molecular basis of which has been poorly understood.
We have reported that calcineurin, a serine–threonine phosphatase, functions as an effector for calcium-dependent signaling pathways in skeletal myofibers by inducing a program of gene expression associated with slow oxidative myofibers (8, 9). Calcineurin activates the transcription factors nuclear factor of activated T cells (NFAT) and myocyte enhancer factor 2, which in turn alter the expression of target genes (5, 9, 10). Constitutively active calcineurin expressed in myofibers of intact animals activates genes that encode contractile proteins, ion channels, and myoglobin that establish the slow oxidative myofiber phenotype (5).
How are changes in intracellular calcium associated with differing patterns of excitation–contraction coupling interpreted by calcineurin/NFAT to alter gene transcription? The present study used a transgenic mouse model to assess the functional state of NFAT proteins in the skeletal muscle of intact animals. In addition, we assessed the relative contributions of different intracellular pools of calcium to the control of NFAT function in cultured myocytes. These studies reveal a role for non-voltage-dependent calcium influx from the extracellular space in controlling the subcellular compartmentalization and transactivator function of NFAT. In particular, expression of TRPC3, a nonselective calcium influx channel, is increased in response to neurostimulation and activated calcineurin and is limiting to NFAT-dependent transactivation. We propose that TRPC3 channels participate in a positive feedback circuit, thereby conferring a form of cellular memory that is a prominent feature of adaptive plasticity of skeletal muscles in response to neurostimulation.
Methods
Construction of NRE-lacZ Reporter. The NFAT binding sites, flanked by HindIII restriction enzyme sites, were amplified by PCR by using the pNFAT plasmid (Stratagene). The construct used in this study contained four multimerized NFAT/AP-1 binding domains. The PCR product was ligated and cloned into the HindIII sites of the vector hsp68-lacZ (11).
Transgenic Mice. Transgenic mice were generated as previously described (5, 12). Genotyping was determined by Southern blotting as previously described, with a probe specific to the lacZ sequence. Of 16 founder animals examined, four independent lines carrying the reporter gene were established. Data presented here are from three transgenic lines representing different chromosomal insertion sites. All experiments with animals were reviewed and approved by the Institutional Animal Care and Research Advisory Committee.
Morphologic and Histologic Analysis. Dissected muscle were fixed in 2% paraformaldehyde and 0.2% glutaraldehyde in PBS on ice for 30 min and then stained histochemically for β-galactosidase as described in ref. 5.
Protein Lysates and Immunoblotting. Muscle protein homogenates were made as described (5). Individual skeletal muscles were dissected from each mouse and homogenized in a buffer containing PBS, 20% glycerol, 0.1% Triton X-100, 1 mM DTT, and protease inhibitors. Protein extracts were separated by using 10% SDS/PAGE and transferred to a membrane. Immunoblotting was performed with specific antibodies to galactosidase (1:1,000) from Promega, TRPC3 (1:2,000) from Alamone (Jerusalem), and ryanodine receptor (RYR), NFATc1, and tubulin (1:1,000) from Affinity Bioreagents (Golden, CO).
Cell Fractionation and Nuclear/Cytosolic Isolation. After stimulation with agonists, cells were washed twice in PBS and collected in lysis buffer (20 mM Hepes, pH 7.4/10 mM NaCl/1.5 mM MgCl2/20% glycerol/0.1% Triton X-100/1 mM DTT/protease inhibitors). Cell suspensions were centrifuged at 600 × g at 4°C for 1 min. Supernatants represented the cytosolic fraction, while pellets (nuclear fraction) were resuspended in lysis buffer supplemented with 500 mM NaCl and rotated for 1 h. The nuclear fraction was then centrifuged at 16,000 × g for 10 min at 4°C. Cytoplasmic and nuclear proteins were then analyzed by SDS/PAGE and immunoblotted to detect NFATc1 proteins with the monoclonal specific antibodies from Affinity Bioreagents. To quantify the NFAT translocation, immunoblots were imaged and densitometry was determined for individual bands, and results were expressed as fold translocation over basal state.
Voluntary Wheel Running and Chronic Low-Frequency Neurostimulation. Animals were maintained on a 12:12-h light and dark cycle, and running activity was monitored by the Dataquest Data Acquisition System (Data Sciences International, Arden Hills, MN). For chronic low-frequency stimulation, neurostimulators were implanted as previously described and attached to the sciatic nerve of one hind limb. Animals were treated with cyclosporin A (25 mg/kg) or vehicle for 3 days before pacing. For analysis of TRPC3 transcripts using semiquantitative PCR, animals were paced for 2 h, and plantaris muscles were harvested in Triazol to make cDNA. TRPC3 was amplified by using primers specified in ref. 5. TRPC3 transcripts from six animals (chronic low-frequency stimulation) were compared to six sham-operated animals. Data (mean ± SE) are expressed as the induction of TRPC3 relative to cyclophilin expression (TRPC3 intensity/cyclophilin intensity).
Cell Culture. C2C12 cells were grown in DMEM supplemented with 20% FBS, 100 IU/ml penicillin, and 100 μg/ml streptomycin. Cell differentiation into myotubes took place over 5 days while cultured in differentiating media (DM) containing DMEM containing 2% horse serum, 100 IU/ml penicillin, 100 μg/ml streptomycin, 10 μg/ml transferrin, 10 μg/ml regular insulin, and 50 mM Hepes buffer. Experiments involving C2C12 cells were performed after 5 days in DM. For Ca2+ measurement experiments, cells were plated on gelatin-coated glass coverslips and switched to DM for 5 days.
Ca2+ Imaging. Cells were grown on a glass coverslips and differentiated over 5 days. Cells were then loaded with 10 μM fura 2-acetoxymethyl ester for 60 min. The coverslips with fura 2-acetoxymethyl ester-loaded cells attached to them were assembled into the perfusion chamber and continuously perfused at a rate of 10 ml/min with a constant chamber volume of 200 μl. Application of agonists was made by inclusion in the perfusate. Loaded cells were illuminated by alternating 350/380 nm light delivered every 1–2 seconds, and fluorescence images were captured at an emission of 510 nm with a Delta-Scan fluorometric system. Ca2+ levels were derived from the change in ratio of fluorescence intensity at 340 and 380 nm as detailed in ref. 13.
Cloning of the TRPC3 Promoter. PCR primers were designed to amplify genomic sequences corresponding to the 1,800 bases upstream of the transcription start site for TRPC3. This PCR product was subcloned into PGL3-Luciferase obtained from Promega using the Xba restriction site.
Cell Transfection and Luciferase Assay. C2C12 myoblasts were plated at 105 cells per well of a six-well dish at least 12 h before transfection. Cells were transfected with 1.6 μg of NRE reporter or TRPC3 promoter along with CMV-LacZ constructs by using Lipofectamine 2000 (GIBCO/BRL). Cells were differentiated for 3 days in 2% horse serum and then assayed for luciferase and β-galactosidase from whole-cell extracts as described in ref. 5.
Results
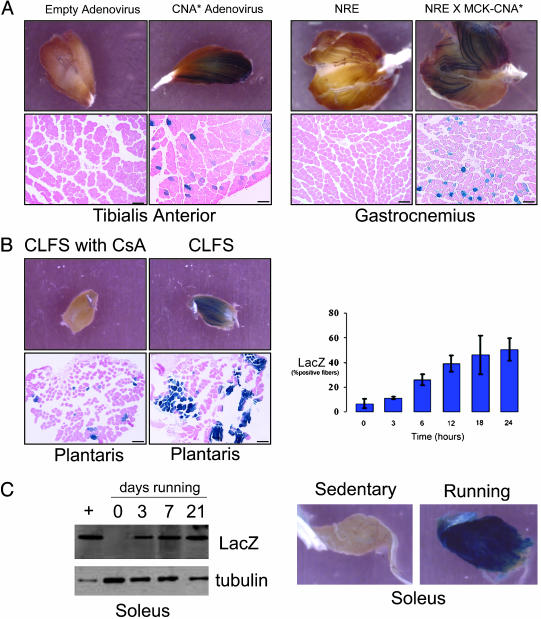
Transactivator Function of NFAT Proteins in Adult Skeletal Muscles. Transgenic mice were engineered to carry a lacZ reporter gene under the control of a three copies of a high-affinity NFAT binding sequence linked to a basal promoter (see Methods). Results were similar in each of three independent lines of mice representing different chromosomal insertion sites for the transgene. Expression of the NFAT-dependent lacZ transgene during embryonic and fetal life was consistent with known sites of NFAT-regulated developmental events (data not shown). Skeletal muscles of adult mice housed under standard conditions (sedentary) exhibited no detectable expression of lacZ, but the NFAT-dependent transgene was activated by expression of a constitutively active form of calcineurin, introduced either by an adenoviral vector or by genetic crosses of NFAT-lacZ mice to MCK-CnA animals (5, 14) (Fig. 1A). Likewise, NFAT-dependent transcription was induced in muscles of intact animals by neuromuscular activity, generated either by 18–24 h of direct electrical pacing (10 Hz) of the motor nerve (Fig. 1B) or by daily bouts of spontaneous wheel running for a 1-week period (Fig. 1C). This response to chronic low-frequency motor nerve stimulation was inhibited in animals that were pretreated with the calcineurin inhibitor cyclosporin A (25 mg/kg per day for 3 days) as compared with vehicle-treated animals (Fig. 1B). The response to running was limited to muscle groups (soleus, gastrocnemius) that power this form of locomotor activity (Fig. 1C) and were not observed in other hind limb muscles [extensor digitorum longus (EDL)] that are not recruited for this form of exercise. Induction of LacZ protein from the soleus and plantaris muscles in response to altered motor nerve activity (running or motor nerve stimulation) increased in a linear fashion, indicating a progressive training response.
Fig. 1.
NFAT indicator mice. Hind limb muscles dissected from NRE-LacZ transgenic mice were analyzed by histochemical staining for β-galactosidase activity after stimulation. (A) Expression of constitutively active calcineurin introduced by adenovirus or germ-line transgenesis. (Bar, 40 μmin Lower.) (B) Continuous low-frequency stimulation of the motor nerve at 10 Hz for 18 h in the presence or absence of cyclosporin A. (Bar, 40 μm.) (C) Spontaneous wheel running (sedentary, 3, 7, and 21 days).
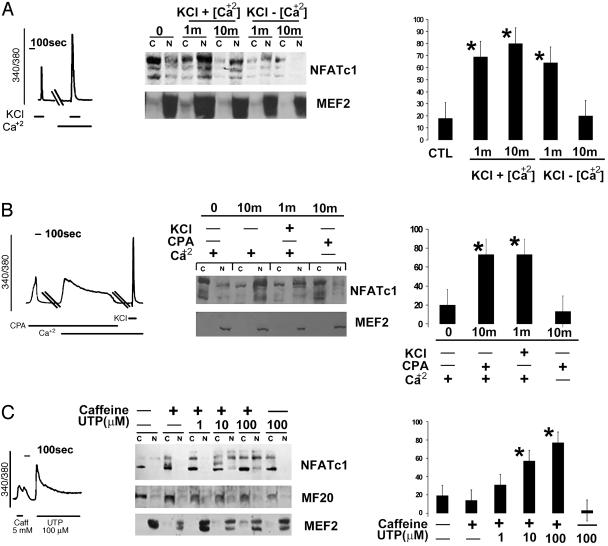
Extracellular Calcium Sustains NFAT in the Nucleus of Myotubes. C2C12 cells assemble components of two calcium signaling pathways including voltage-operated channels involved in excitation–contraction coupling (RYR and dihydropyridine receptor) and non-voltage-dependent calcium channels (IP3R and TRPC3) that are not required for muscle contraction but serve other signaling functions. The RYR Ca2+ pool contributes Ca2+ sufficient to initiate NFATc1 translocation to the nucleus, as was evident by stimulating myotubes with KCl in the absence of external Ca2+. However, with longer incubations of 10 min in a Ca2+-free bath, NFATc1 was depleted from the nuclear fraction, suggesting that Ca2+ influx from the extracellular space sustains NFATc1 in the nucleus (Fig. 2A). The specificity of the RYR pool for initiating NFATc1 translocation was supported by additional experiments with RYR activators (caffeine) and inhibitors (ruthenium red) (data not shown). When extracellular calcium is available, NFATc1 translocates and remains in the nucleus after cellular depolarization with KCl.
Fig. 2.
Calcium mobilized from different pools and NFAT translocation. Each panel includes calcium transients from fura 2-acetoxymethyl ester-labeled myotubes, immunoblots of cytoplasmic (C) and nuclear (N) fractions, and histograms presenting mean values (±SE) of duplicate determinations from three independent experiments (*, P < 0.01). (A) Release of the RYR pool of intracellular calcium by KCl promotes nuclear translocation of NFAT in C2C12 myotubes. However, in the absence of extracellular calcium, this response is short-lived and is blocked by ruthenium red, an inhibitor of RYR calcium release (data not shown). (B) Activation of non-voltage-dependent calcium channels by CPA translocates and sustains NFATc1 in the nucleus in the presence of, but not in the absence of, extracellular calcium. (C) Release of the RYR pool of calcium by a brief (1-min) exposure of cells to caffeine, followed by activation of non-voltage-dependent calcium channels with UTP, results in sustained maintenance of NFAT in the nuclear compartment.
To further characterize the calcium influx required to maintain NFATc1 in the nucleus, myotubes were stimulated with cyclopiazoic acid, an inhibitor of the sarcoplasmic reticulum sarco(endo)plasmic reticulum Ca2+ ATPase, in the presence and absence of external calcium. Used in this fashion, cyclopiazoic acid (CPA) activates non-voltage-dependent calcium influx (15). Even small calcium transients evoked by adding calcium to the media in the presence of CPA were sufficient to maintain NFATc1 translocation as efficiently as with KCl (Fig. 2B). Thus, activation of non-voltage-dependent calcium influx provides a signal sufficient to maintain NFATc1 in the nucleus of myotubes.
The finding that discharge of distinct Ca2+ pools regulates specific steps in NFATc1 translocation into the nucleus raised the possibility of cooperation between Ca2+ pools. Myotubes were stimulated with caffeine for 1 min to rapidly shuttle NFATc1 proteins to the nuclear fraction. NFATc1 proteins were depleted from the nuclear fraction within 9 min after removal of caffeine. Activation of Ca2+ entry by UTP, a purinergic agonist, was insufficient in itself to maintain NFATc1 proteins in the nuclear fraction of myotubes, but it sustained NFATc1 in nuclei of myotubes conditioned by a 1-min pulse of caffeine (Fig. 2C). Removal of Ca2+out during the stimulation with UTP depleted the NFATc1 proteins from the nuclear fraction (data not shown). These data suggest that enhanced Ca2+ influx through non-voltage-dependent calcium channels maintains NFATc1 in the nucleus.
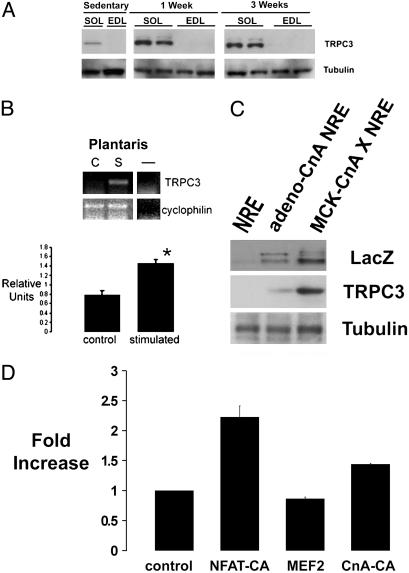
Increased TRPC3 Channel Expression by Neurostimulation and Calcineurin. TRPC channels encompass a family of nonselective calcium-influx channels that were first described in phototransduction in the Drosophila eye (16, 17). A role for TRPC channels in excitable cells has only recently been suggested (18–20). Here, we analyzed the expression of TRPC3 in skeletal muscles from adult mice. As assessed by immunoblotting experiments, TRPC3 expression is greater in muscles enriched in slow oxidative fibers (soleus) by comparison to muscles comprising mostly fast glycolytic fibers (EDL) (Fig. 3A). Animals that were provided access to free-wheel running, a stimulus that promotes mitochondrial biogenesis and myosin isoform switching (type IIb to IIa) in skeletal muscles (data not shown), demonstrated up-regulation of TRPC3 expression as compared with sedentary controls (Fig. 3A). As observed for NFAT transactivator function, little induction of TRPC3 channels was seen in EDL muscles that are not recruited during wheel running. Up-regulation of TRPC3 expression also occurs in fast muscles, as demonstrated by time-dependent accumulation of the NFAT-regulated transgene product (LacZ) in plantaris muscles subjected to 2 h of motor nerve stimulation at 10 Hz (Fig. 3B). TRPC3 expression was also increased in muscles expressing a constitutively active form of calcineurin introduced either by adenoviral delivery or by germ-line transgenesis (Fig. 3C). The effect of calcineurin to increase the abundance of TRPC3 in skeletal muscles is mediated, at least in part, by calcineurin-dependent induction of the TRPC3 gene promoter. An 1800-bp genomic fragment from the mouse TRPC3 gene was cloned upstream of a luciferase reporter gene and transfected into C2C12 myogenic cells. Expression of a constitutively active form of calcineurin or NFAT increased TRPC3 promoter activity (Fig. 3D). In contrast, expression of another calcineurin-regulated transcription factor, myocyte enhancer factor 2, failed to activate the TRPC3 promoter.
Fig. 3.
TRPC3 expression in skeletal muscles. TRPC3 expression is increased by slow oxidative [soleus (Sol)] compared with fast glycolytic (EDL) muscles, and by spontaneous wheel running for durations of 1 or 3 weeks (A), by chronic low-frequency neurostimulation at 10 Hz (S, n = 6) compared with control (C, n = 6) plantaris muscles (mean ± SE; *, P < 0.05) (B), and by constitutively active calcineurin introduced by adenovirus or germ-line transgenesis (C). (D) Transcriptional activity of the TRPC3 promoter (1800-bp genomic fragment linked to a luciferase reporter gene) is induced by constitutively active NFAT or calcineurin in C2C12 myogenic cells. *, P < 0.003.
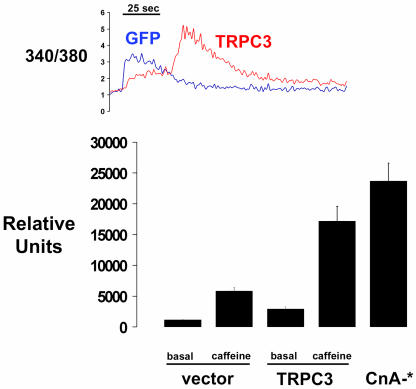
Expression of TRPC3 Channels Modulates Calcium Transients and Calcium-Regulated NFAT Function in Myocytes. Prolonged calcium influx was observed in calcium transients evoked by caffeine in myotubes transfected with a TRPC3 expression plasmid in comparison with control cells (Fig. 4 Upper). Consistent with established properties of TRPC3 channels in other cell types, CPA-induced calcium influx was not enhanced in cells expressing TRPC3 (13, 21–23). However, enhanced expression of TRPC3 channels influenced the transactivator function of NFAT. NFAT-dependent reporter gene activity was increased 2.5-fold in myotubes expressing TRPC3 compared with control cells, suggesting that increased expression of TRPC3 increased the regulatory pool of calcium that is pertinent to calcineurin/NFAT signaling (23), even under basal conditions. Prolonged exposure of cells to caffeine (120 min) induced a 7-fold increase in NFAT transactivation in myotubes expressing endogenous levels of TRPC3, but this response was approximately doubled (to 12-fold) in myotubes with increased expression of TRPC3 channels. Thus, TRPC3 channels control a calcium pool pertinent to NFAT transactivation, and the abundance of TRPC3 in skeletal myocytes is limiting both to the duration of calcium transients evoked by release of the RYR pool (and the ensuing Ca+2 influx) and to the magnitude of NFAT-dependent transcriptional responses.
Fig. 4.
TRPC3 expression is limiting to sustained calcium influx and NFAT transactivation in C2C12 myotubes. (Upper) Calcium transients in cells stimulated with 15 mM caffeine and expressing a CMV-TPRC3 construct or a control vector. Each trace represents the mean value of 5–10 cells from one experiment; similar results were obtained in two additional experiments. (Lower) NFAT-dependent reporter gene expression stimulated by caffeine in myocytes expressing a CMV-TPRC3 construct or a control vector. Data are corrected for variations in transfection efficiency by normalization of data to expression of a cotransfected pCMV-LacZ construct. *, P < 0.004.
Discussion
This study demonstrates that the calcineurin–NFAT signaling pathway is activated by neuromuscular stimulation in skeletal muscle of intact animals, and it provides evidence that TRPC3 channels serve as important regulators of this pathway. TRPC channels are expressed selectively in muscles undergoing sustained or repeated neurostimulation through a calcineurin-dependent mechanism. These findings provide a plausible explanation for the longstanding observation by physiologists that single periods of neurostimulation produce only minor and transient changes in muscle phenotype, while repeated bouts of contractile activity drive progressively greater and more stable remodeling events, a phenomenon we characterize as a form of cellular memory.
Calcineurin regulates the expression of several proteins involved in calcium homeostasis such as ion channels, transporters, and pumps (24–27). We also have recently shown that components of the NFAT–calcineurin pathway including NFATC2, calcsarcin 1, and latory calcineurin-interacting protein 1 are differentially expressed in slow oxidative muscles compared with modulatory fast glycolytic muscles (26). We propose that such profiles of gene expression represent a form of cellular memory that is important for maintaining skeletal muscle in the remodeled state evoked by changing patterns of neurostimulation. Studies of muscle plasticity going back over several decades (6, 7) show that the time course of responses evoked by neuromuscular stimulation requires days or weeks of sustained or repeated neuromuscular activity for conversion of fast glycolytic myofibers to the slow oxidative phenotype. Likewise, reversion of slow oxidative fibers back to the fast glycolytic phenotype when motor nerve input is reduced requires several weeks. The present report shows how activation of the calcineurin–NFAT pathway alters expression of important components of the signaling pathway itself, augmenting the gain to subsequent neurostimulation and stabilizing the slow oxidative phenotype.
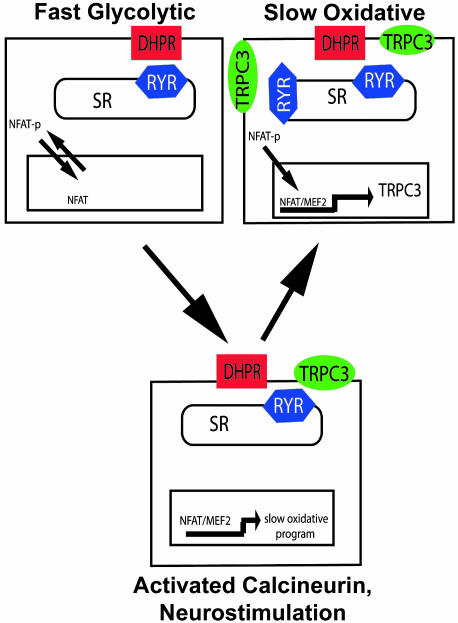
The present results also reveal features of calcium metabolism in muscle cells that discriminate, at least in part, pools of calcium required for muscle contraction from those required to drive calcium-regulated gene expression. Calcium released from RYR channels is sufficient to trigger actin–myosin cross-bridge cycling and muscle contraction without any requirement for calcium entry from the extracellular space (28). In contrast, the RYR pool of calcium is insufficient to maintain NFAT translocation to the nuclear compartment, and extracellular calcium entering through non-voltage-dependent Ca+2 channels, putatively represented by TRPC3, is required for NFAT-dependent transactivator function in skeletal myocytes. This model proposes that sustained contractile activity depletes sarcoplasmic reticulum calcium stores and opens TRPC channels in a manner capable of enhancing NFAT-dependent transactivation (Fig. 5).
Fig. 5.
Schematic model of the role of TRPC3 in calcium-dependent gene regulation in skeletal muscle. TRPC3 is expressed in greater amounts in myofibers after neuromuscular activity, a response mediated by the calcineurin–NFAT pathway. Increased expression of TRPC3 enhances calcineurin–NFAT signaling that results for subsequent periods of neuromuscular activity, thereby conferring cellular memory that stabilizes the slow oxidative phenotype.
Capacitative calcium entry and TRPC channel function have been found to be dysfunctional in mouse models of muscular dystrophy caused by mutations in dystrophin or mg29 (18, 20). In these studies no defects were found in excitation–contraction coupling. Interestingly, slow oxidative fibers are relatively spared from injury in muscular dystrophy (29), suggesting that increased expression of TRPC channels in slow oxidative fibers relative to fast glycolytic fibers perhaps has protective consequences. In support of a causal relationship among calcineurin–NFAT signaling, non-voltage-dependent Ca+2 channel function, and the pathobiology of muscular dystrophies, a series of patients with naturally occurring defects in non-voltage-dependent Ca+2 influx exhibited both impaired NFAT trafficking in lymphocytes and skeletal myopathy (30). These findings from human subjects are consistent with our proposal that TRPC3 channels are important to the processes that match contractile and metabolic properties of skeletal myocytes to changes in patterns of neurostimulation and work demand and that regulation of TRPC3 expression confers a form of cellular memory necessary for physiologically important muscle plasticity.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NFAT, nuclear factor of activated T cells; RYR, ryanodine receptor; CPA, cyclopiazoic acid; EDL, extensor digitorum longus.
References
- 1.Close, R. (1965) Nature 206, 718-719. [DOI] [PubMed] [Google Scholar]
- 2.Olson, E. N. & Williams, R. S. (2000) BioEssays 22, 510-519.10842305 [Google Scholar]
- 3.Olson, E. N. & Williams, R. S. (2000) Cell 101, 689-692. [DOI] [PubMed] [Google Scholar]
- 4.Close, R. (1965) Nature 206, 831-832. [DOI] [PubMed] [Google Scholar]
- 5.Wu, H., Rothermel, B., Kanatous, S., Rosenberg, P., Naya, F. J., Shelton, J. M., Hutcheson, K. A., DiMaio, J. M., Olson, E. N., Bassel-Duby, R. & Williams, R. S. (2001) EMBO J. 20, 6414-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.al-Amood, W. S., Buller, A. J. & Pope, R. (1973) Nature 244, 225-227. [DOI] [PubMed] [Google Scholar]
- 7.Buller, A. J. & Pope, R. (1977) Philos. Trans. R. Soc. Lond. B Biol. Sci. 278, 295-305. [DOI] [PubMed] [Google Scholar]
- 8.Chin, E. R., Olson, E. N., Richardson, J. A., Yang, Q., Humphries, C., Shelton, J. M., Wu, H., Zhu, W., Bassel-Duby, R. & Williams, R. S. (1998) Genes Dev. 12, 2499-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu, H., Naya, F. J., McKinsey, T. A., Mercer, B., Shelton, J. M., Chin, E. R., Simard, A. R., Michel, R. N., Bassel-Duby, R., Olson, E. N. & Williams, R. S. (2000) EMBO J. 19, 1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, Y., Cseresnyes, Z., Randall, W. R. & Schneider, M. F. (2001) J. Cell Biol. 155, 27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kothary, R., Perry, M. D., Moran, L. A. & Rossant, J. (1987) Dev. Biol. 121, 342-348. [DOI] [PubMed] [Google Scholar]
- 12.Zhou, B., Cron, R. Q., Wu, B., Genin, A., Wang, Z., Liu, S., Robson, P. & Baldwin, H. S. (2002) J. Biol. Chem. 277, 10704-10711. [DOI] [PubMed] [Google Scholar]
- 13.Kiselyov, K., Xu, X., Mozhayeva, G., Kuo, T., Pessah, I., Mignery, G., Zhu, X., Birnbaumer, L. & Muallem, S. (1998) Nature 396, 478-482. [DOI] [PubMed] [Google Scholar]
- 14.Naya, F. J., Mercer, B., Shelton, J., Richardson, J. A., Williams, R. S. & Olson, E. N. (2000) J. Biol. Chem. 275, 4545-4548. [DOI] [PubMed] [Google Scholar]
- 15.Xu, X., Star, R. A., Tortorici, G. & Muallem, S. (1994) J. Biol. Chem. 269, 12645-12653. [PubMed] [Google Scholar]
- 16.Montell, C., Birnbaumer, L. & Flockerzi, V. (2002) Cell 108, 595-598. [DOI] [PubMed] [Google Scholar]
- 17.Montell, C., Birnbaumer, L., Flockerzi, V., Bindels, R. J., Bruford, E. A., Caterina, M. J., Clapham, D. E., Harteneck, C., Heller, S., Julius, D., et al. (2002) Mol. Cell 9, 229-231. [DOI] [PubMed] [Google Scholar]
- 18.Vandebrouck, C., Martin, D., Colson-Van Schoor, M., Debaix, H. & Gailly, P. (2002) J. Cell Biol. 158, 1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, J. & Pan, Z. (2003) Front. Biosci. 8, D242-D255. [DOI] [PubMed] [Google Scholar]
- 20.Pan, Z., Yang, D., Nagaraj, R. Y., Nosek, T. A., Nishi, M., Takeshima, H., Cheng, H. & Ma, J. (2002) Nat. Cell. Biol. 4, 379-383. [DOI] [PubMed] [Google Scholar]
- 21.Vazquez, G., Wedel, B. J., Trebak, M., Bird, G. S. & Putney, J. W., Jr. (2003) J. Biol. Chem. 278, 21649-21654. [DOI] [PubMed] [Google Scholar]
- 22.Trebak, M., Bird, G. S., McKay, R. R. & Putney, J. W., Jr. (2002) J. Biol. Chem. 277, 21617-21623. [DOI] [PubMed] [Google Scholar]
- 23.Zitt, C., Obukhov, A. G., Strubing, C., Zobel, A., Kalkbrenner, F., Luckhoff, A. & Schultz, G. (1997) J. Cell Biol. 138, 1333-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genazzani, A. A., Carafoli, E. & Guerini, D. (1999) Proc. Natl. Acad. Sci. USA 96, 5797-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graef, I. A., Mermelstein, P. G., Stankunas, K., Neilson, J. R., Deisseroth, K., Tsien, R. W. & Crabtree, G. R. (1999) Nature 401, 703-708. [DOI] [PubMed] [Google Scholar]
- 26.Williams, R. S. & Rosenberg, P. (2002) Cold Spring Harbor Symp. Quant. Biol. 67, 339-343. [DOI] [PubMed] [Google Scholar]
- 27.Bonilla, M. & Cunningham, K. W. (2002) Sci. STKE 2002, PE17. [DOI] [PubMed] [Google Scholar]
- 28.Armstrong, C. M., Bezanilla, F. M. & Horowicz, P. (1972) Biochim. Biophys. Acta 267, 605-608. [DOI] [PubMed] [Google Scholar]
- 29.Lefaucheur, J. P., Pastoret, C. & Sebille, A. (1995) Anat. Rec. 242, 70-76. [DOI] [PubMed] [Google Scholar]
- 30.Feske, S., Draeger, R., Peter, H. H. & Rao, A. (2000) Immunobiology 202, 134-150. [DOI] [PubMed] [Google Scholar]