Abstract
Tumor hypoxia underlies treatment failure and yields more aggressive and metastatic cancer phenotypes. Although therapeutically targeting these hypoxic environments has been proposed for many years, to date no approaches have shown the therapeutic value to gain regulatory approval. Here, we demonstrated that a novel hypoxia-activated prodrug, Q6, exhibits potent antiproliferative efficacy under hypoxic conditions and induces caspase-dependent apoptosis in 2 hepatocellular carcinoma (HCC) cell lines, with no obvious toxicity being detected in 2 normal liver cell lines. Treatment with Q6 markedly downregulated HIF1A [hypoxia inducible factor 1, α subunit (basic helix-loop-helix transcription factor)] expression and transcription of the downstream target gene, VEGFA (vascular endothelial growth factor A). This dual hypoxia-targeted modulation mechanism leads to high potency in suppressing tumor growth and vascularization in 2 in vivo models. Intriguingly, it is the autophagy-dependent degradation pathway that plays a crucial role in Q6-induced attenuation of HIF1A expression, rather than the proteasome-dependent pathway, which is normally regarded as the predominant mechanism underlying posttranslational regulation of HIF1A. Inhibition of autophagy, either by short interfering RNA (siRNA) or by chemical inhibitors, blocked Q6-induced HIF1A degradation. Autophagic degradation of HIF1A was further confirmed by the observation that HIF1A coimmunoprecipitated with the ubiquitin-binding adaptor protein, SQSTM1, which is degraded through autophagy. Additionally, silencing of SQSTM1 inhibited Q6-induced HIF1A degradation. These findings suggest that the novel hypoxia-targeted agent, Q6, has potential clinical value in the therapy of HCC. Furthermore, the identification of autophagy as a crucial regulator of HIF1A provides new insights into hypoxia-related treatments.
Keywords: hypoxia-activated prodrug, dual modulation mechanisms, posttranslational regulation, autophagy, targeting treatment
Introduction
Many solid tumors, especially hepatocellular carcinoma (HCC), are characterized by regions of hypovascularity, as determined by diagnostic imaging. Direct intratumoral oxygen measurements using polagraphic electrode probes have also verified that hepatic tumors are significantly more hypoxic than adjacent normal tissues. 1 - 3 In liver cancer, the extent of tumor hypoxia seems to inversely correlate with patient prognosis and is often associated with resistance to conventional treatment modalities. 4 - 6 Thus, it also presents a unique opportunity for targeting treatment, thereby decreasing drug side effects and increasing antitumor efficacy.
Hypoxia-activated prodrugs (HAP) are a new class of drugs that act via a mechanism involving selective activation of a nontoxic prodrug within the hypoxic regions of tumors. The first promising drug, tirapazamine (TPZ), was a significant advance over previous ones and combination studies with fractionated radiation have demonstrated its ability to kill hypoxic cells in transplanted tumor models. However, this therapy only works in a subset of patients and is not curative, underlying an urgency to identify new compounds. 7 , 8 Many reports have shown that a major disadvantage of HAP therapy for hypoxia-targeted treatment is due to the heterogeneity of genetic abnormalities acquired during the course of carcinogenesis and microenvironmental changes. 9 Consequently, exploiting the unique molecular phenotype of hypoxic cells represents a viable strategy to supplement HAP therapy for improving patient outcomes.
The main oxygen-responsive signaling pathways that mediate adaptation to hypoxia are centered on hypoxia-inducible factor (HIF1), HIF1 is a heterodimeric protein composed of HIF1A and ARNT/HIF1B subunits. Whereas ARNT is constitutively expressed, HIF1A expression increases exponentially as O2 concentration declines. In order to respond rapidly to hypoxia, cells devote considerable energy to the continuous synthesis and degradation of HIF1A under nonhypoxic conditions. Under hypoxic conditions, the degradation of HIF1A is inhibited, resulting in accumulation of the protein, dimerization with ARNT, binding to hypoxia response elements (HREs) within target genes, and thereby modulating more than 100 target genes (e.g., VEGFA) involved in angiogenesis, cell survival, glucose metabolism, and resistance to conventional therapies. 10 - 12 Therefore, HIF1A has been identified as an attractive drug target, and a wide range of pharmacological approaches have been proposed to modulate its activity, particularly through decreasing its protein level. 7 , 8 , 13 A growing number of agents that can inhibit HIF1A activity have shown significant activity in arresting tumor xenograft growth. 14 , 15 Accordingly, we hypothesized that a therapeutic strategy aimed at both specifically killing hypoxic cells and inhibiting HIF1A expression may provide enhanced antitumor activity and clinical benefit.
In our laboratories, we have synthesized a series of 3-aryl-quinoxaline-2-carbonitrile 1, 4-Di-N-oxide analogs of TPZ, 16 several of which not only exerted antiproliferative activity and hypoxia selectivity to mutiple cell lines, but also displayed activity in inhibiting HIF1A expression and related signal transduction. Among these compounds, Q6 was outstanding with regard to its significantly antitumor activity both in vitro and in vivo. More importantly, we demonstrated that Q6-induced reduction in HIF1A expression was attributed, at least partially, to autophagy-mediated degradation, whereas the classic ubiquitin-proteasome pathway was not involved. In summary, these data provide strong support for the idea that, due to its targeting to hypoxic cells and its dual mechanisms of action, Q6 will be useful in the development of new types of HAP, and the novel role of autophagy in posttranslational regulation of HIF1A has also been unraveled.
Results
Q6 exerts potent antitumor activity and hypoxia-selectivity in 2 HCC cell lines
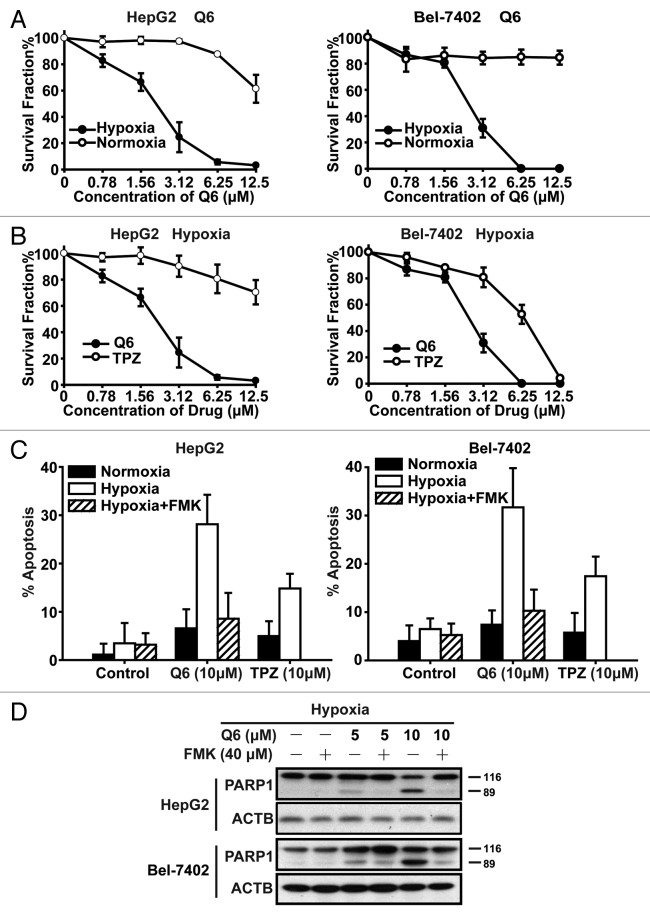
As a promising HAP, Q6 (Fig. S1) has shown broad antitumor activity and hypoxia-selectivity in a variety of cancer cell lines. 16 Thus, in the current study, we speculated that Q6 may inhibit HCC cell survival, especially under hypoxic conditions. To address this question, HepG2 and Bel-7402 cells were exposed to Q6/TPZ for 72 h under normoxia and hypoxia conditions, and cell viability was determined using the MTT assay. As expected, the survival rate of Q6-exposed cells under hypoxia was much lower than that of the cells under normoxia, with the average IC50 values of 1.99 ± 0.25 μM in hypoxia and 13.90 ± 5.15 μM in normoxia, a 7-fold selectivity (Fig. 1A). In addition, we compared the antiproliferative effects of Q6 and TPZ, in HepG2 and Bel-7402 cells under hypoxic conditions. The results showed that Q6 appeared to be more potent than TPZ with the IC50 values of 2.24 ± 0.18 and 1.74 ± 0.32 μM, respectively, compared with corresponding values of 24.92 ± 6.79 and 6.55 ± 1.78 μM for TPZ (Fig. 1B). In comparison to malignant cells, 2 normal human liver cell lines, HL-7702 and Chang liver cells, were resistant to both compounds (Fig. S2).
Figure 1. Q6 has potent antitumor efficacy and triggers caspase-dependent apoptosis in 2 HCC cell lines. (A and B) Two HCC cell lines, HepG2 (left) and Bel-7402 (right), were treated with Q6 or TPZ (0 to 12.5 μM) for 72 h under normoxia or hypoxia. Cell viability was determined by the MTT assay. Data are representative of 3 independent experiments and are expressed as the means ± SD (C and D) HepG2 and Bel-7402 cells were treated with Q6 or TPZ (0, 10 μM) or Q6 (0 to 10 μM) +Z-VAD-FMK (40 μM) for 24 h under normoxia or hypoxia. (C) Percentage of apoptosis was measured by flow cytometry using the ANXA5/PI apoptosis detection kit. Three independent experiments were performed and the values were expressed as the mean ± SD (D) Protein levels of PARP1 were detected by western blot analysis. ACTB was measured as the loading control. Data are representative of 3 independent experiments.
Q6 triggers caspase-dependent apoptosis
Increased apoptosis is an important mechanism by which HAPs kill tumor cells in hypoxia. Accordingly, 3 different approaches were used to investigate the effects of Q6 on apoptosis, and trypan blue exclusion analysis was also employed to investigate whether apoptosis is the main reason for Q6-induced cell death. Initially, the ANXA5/annexin V apoptosis assay was employed to detect levels of apoptosis. After treatment with Q6 or TPZ (0, 10 μM) for 24 h, we found that Q6 induced 28.14% apoptosis in HepG2 cells and 31.71% in Bel-7402 cells under hypoxic conditions, more potentially than that induced by TPZ (Fig. 1C). Moreover, in most cases, the process of apoptosis was accompanied by activation of the caspase cascade. Thus, we detected the expression of some caspase-related proteins, such as PARP1, CASP3, and cleaved-CASP3 by western blotting assay after Q6 treatment. We found that Q6 induced cleavage of PARP1 and CASP3 in a concentration and time-dependent manner in 2 HCC cell lines (Fig. S3B and S3C). Upon pretreatment of cells with a pan-caspase inhibitor, Z-VAD-FMK, Q6-induced apoptosis was significantly reduced from 28.14% to 8.55% in HepG2 cells, and from 31.71% to 12.81% in Bel-7402 cells (Fig. 1C), meanwhile Q6-induced cleavage of PARP1 was attenuated (Fig. 1D), implying the caspase-dependent pathway is involved in Q6-induced apoptosis.
Disruption of mitochondrial membrane potential (ΔΨm) is another important intracellular event that occurs during apoptosis. Using JC-1 analysis, we found that compared with corresponding controls, administration of Q6 for 24 h dramatically decreased ΔΨm in a concentration-dependent manner (Fig. S3D).
Finally, using trypan blue exclusion asssay, we found that Q6 (10 μM) induced 44% cell death in HepG2 cells and 51% in Bel-7402 cells. Upon pretreatment of cells with Z-VAD-FMK, Q6-induced cell death was significantly reduced from 44% to 15% in HepG2 cells, and from 51% to 18% in Bel-7402 cells (Fig. S3A), implying that increased apoptosis is an important mechanism by which Q6 killed tumor cells in hypoxia.
In summary, these experiments demonstrated that Q6 induces disruption of mitochondrial membrane potential, thereby triggering caspase-dependent apoptosis.
Q6 downregulates HIF1A expression and transcription activity
Previous studies have demonstrated the relationship between drug resistance and tumor progression with HIF1A overexpression; we observed that Q6 could significantly inhibit HIF1A and related signaling transduction. First, we investigated that treatment with Q6 (0 to 5 μM) for 6 h dramatically decreased hypoxia-induced HIF1A protein expression in HepG2 and Bel-7402 cells in a concentration-dependent manner. (Fig. 2A; Fig. S5A). Given that the inhibition of HIF1A accumulation in hypoxic cells could be correlated with Q6-induced cytotoxicity, parallel studies of cell viability were performed. The results showed that there was no significant alteration in cell viability after Q6 treatment (Fig. S4A), suggesting that Q6-induced reductions in HIF1A levels are not a result of its cytotoxic actions. Following this, we evaluated that Q6 could inhibit HIF1A-mediated transcriptional activity in a concentration-dependent manner in HepG2 and Bel-7402 cell lines, as determined using a hypoxia-responsive reporter construct, containing a luciferase gene under the control of HREs (Fig. 2B).
Figure 2. Q6 downregulates hypoxia-induced HIF1A protein expression and HIF1A-mediated signal transduction. (A and C) HepG2 (left) and Bel-7402 (right) cells were exposed to hypoxia or normoxia and treated with Q6 (0 to 5 μM) for 6 h. (A) Protein levels of HIF1A, EPAS1, and VEGFA were detected by western blot analysis. ACTB was analyzed as the loading control. Data are representative of 3 independent experiments. (B) An HRE-dependent reporter assay was used to determine the effect of Q6 on HIF1A transcriptional activity. Five independent experiments were performed and the values were expressed as the mean ± SD **P < 0.01 and ***P < 0.001, compared with untreated controls in hypoxia. (C) Total RNA was extracted and VEGFA mRNA expression was analyzed by RT-PCR, using GAPDH as a control gene. Five independent experiments were performed and the values were expressed as the mean ± SD **P < 0.01, compared with untreated controls in hypoxia.
Multiple studies have demonstrated that HIF1A-mediated VEGFA expression is considered the principal inducer of angiogenesis. 17 , 18 Therefore, in this study, we hypothesized that Q6 could inhibit VEGFA expression. As depicted in Figure 2A and C, Q6 significantly suppressed VEGFA protein expression and mRNA levels in a concentration-dependent manner under hypoxic conditions, further confirming that Q6 suppresses HIF1A-induced signal transduction.
Moreover, previous reports have shown that HIF1A and EPAS1/HIF2A are both particularly critical in mediating cellular responses to hypoxia, and are often regulated by the same mechanisms. 19 However, Q6 failed to exert an effect on EPAS1 protein levels in HepG2 and Bel-7402 cells (Fig. 2A), indicating that Q6-induced HIF1A suppression may occur through a mechanism that has not been previously reported.
Together, these results demonstrated that Q6 treatment suppresses expression and signaling transduction of HIF1A, but has no effect on EPAS1.
The autophagy–lysosome pathway participates in Q6-induced inhibition of HIF1A expression
In order to explore the mechanisms underlying Q6-induced HIF1A suppression, we first examined whether reduction of HIF1A by Q6 occurs at the transcriptional level. Real-time PCR analysis showed that HIF1A mRNA levels were not significantly altered after Q6 treatment in Bel-7402 and HepG2 cells (Fig. 3A). Furthermore, we found that Q6 had no effect on EGFR, PIK3CA-AKT1, or MAPK signaling pathways, which have been recently shown to control the protein synthesis of HIF1A (Fig. S4; Table S1). On the basis of these findings, we hypothesized that a degradative mechanism may be involved in Q6-induced reductions in HIF1A. To examine this possibility, cycloheximide (CHX, an inhibitor of protein synthesis) was used to prevent de novo protein synthesis; thus, changes in HIF1A levels would primarily reflect protein degradation. We exposed HepG2 and Bel-7402 cells to CHX under hypoxic conditions in the presence or absence of Q6 at different time points and measured expression of HIF1A. As shown in Figure 3B, although the intensity of the HIF1A signal was not obviously changed in Q6 untreated cells, the reduction of HIF1A protein levels were observed in Q6-treated cells in a time-dependent manner. Together, these results indicate that Q6 downregulates HIF1A protein expression through accelerating its degradation.
Figure 3. Q6 accelerates HIF1A protein degradation via the autophagy-lysosome pathway. (A) HepG2 (left) and Bel-7402 (right) cells were exposed to Q6 (0 to 5 μM) for 6 h in hypoxia. Total RNA was extracted and HIF1A mRNA expression was analyzed by RT-PCR, using GAPDH as a control gene. Five independent experiments were performed and the values were expressed as the mean ± SD (B) HepG2 and Bel-7402 cells exposed to hypoxia were treated with CHX in the presence or absence of Q6 (5 μM) for different times, and HIF1A protein levels were then measured by western blot analysis. ACTB was measured as the loading control. (C) HepG2 and Bel-7402 cells were pretreated with MG132 (a proteasome inhibitor) or 3-MA (an autophagy-lysosome inhibitor) for 30 min to allow functional inhibition of proteasomes and lysosomes. Cells were then exposed to hypoxia in the presence or absence of Q6 (5 μM) for 6 h, after which HIF1A protein levels were determined by western blot analysis. ACTB was measured as the loading control. (D) Ultrastructural features of HepG2 and Bel-7402 cells with or without Q6 treatment (5 μM) for 6 h were analyzed by electron microscopy. The typical images of autophagosomes (arrows) and autolysosomes (arrowheads) were shown at higher magnification. In the lower panel, the number of autophagosomes (AP) and autolysosomes (AL) were presented for HepG2 and Bel-7402 cells. Twenty cross sections were counted in each experiment. Data shown are means ± SD of 3 independent experiments. *P < 0.05 compared with HepG2 control group. **P < 0.01 compared with HepG2 control group. ## P < 0.01 compared with Bel-7402 control group. (E) HepG2 and Bel-7402 cells were treated with Q6 (0 to 5 μM) for 6 h and LC3B-I and LC3B-II protein levels were measured by western blot analysis. ACTB was measured as the loading control. Data are representative of 2 independent experiments.
Lysosomal and proteasomal degradation are the 2 major pathways for cellular protein turnover. Therefore, the 2 HCC cells lines were pretreated with MG132 (a specific proteasome inhibitor) or 3-MA (3-methyladenine, a nonspecific autophagy-lysosome inhibitor) before the addition of Q6. The results showed that 3-MA treatment abrogated Q6-induced reductions in HIF1A protein levels in both cell lines, while MG-132 exposure was without demonstrable effect (Fig. 3C). Consistent with this, pretreatment with another lysosome inhibitor CQ could also reverse Q6-induced suppression in HIF1A protein levels (Fig. S6A).
These findings indicate that Q6-induced reductions in HIF1A protein levels are mediated by the autophagy-lysosome pathway, whereas the classical proteasomal pathway is not involved.
Autophagy regulates Q6-induced degradation of HIF1A
Many previous studies have demonstrated that the major route for delivery of cellular proteins into lysosomes occurs through autophagy. 20 , 21 As mentioned above, we hypothesized that autophagy is induced by Q6. Two well-established methods were used to detect autophagosome formation. First, we investigated the number of autophagic vacuoles presenting in cells by ultrastructural analysis based on electron microscopy. As shown in Figure 3D, typical autophagic structures, including double-membrane structures containing cytoplasmic contents (autophagosome), as well as single-membrane structures containing cytoplasmic materials at different stage of degradation (autolysosome) were observed in Q6-treatment group. Next, we examined the conversion of MAP1LC3B (LC3B), ortholog of yeast Atg8, a widely accepted autophagy marker, to assess autophagy levels in Q6-treated cells. During autophagosome formation, cleaved LC3B-I is converted to lipidated LC3B-II, which represents a key event in the regulation of this process. As shown in Figure 3E and Figure S5A, Q6 stimulated significantly increased levels of LC3B-II protein expression, indicating induction of autophagy. In addition, because LC3B-II is continually degraded within autolysosomes during autophagy, it is recommended that cells be treated with lysosomal inhibitor before western blotting for LC3B-II to access autophagic flux. Here, in this study, 2 lysosomal inhibitors bafilomycin A1 and CQ were used to pretreat cells before addition of Q6. As shown in Figure S5B, Q6 could increase LC3B-II protein accumulation in a time-dependent manner, but a larger amount of LC3B-II accumulation was observed in bafilomycin A1/CQ pretreated cells, indicating induction of autophagic flux by Q6. However, treatment with other hypoxia-targeted agents, such as TPZ, had no apparent effect on inducing autophagy and autophagy-mediated degradation of HIF1A (Fig. S5C–S5E).
In addition, we characterized whether autophagy-related genes are involved in Q6-induced HIF1A reductions and RNA interference approaches were employed. Autophagosome formation requires 2 ubiquitin-like conjugation systems, the ATG12–ATG5 conjugate and the MAP1LC3 (LC3) systems, which are tightly associated with the expansion of autophagosomal membranes. Depletion of ATG5 and LC3B expression in these cells inhibited Q6-induced autophagy, and prolonged the half-life of HIF1A protein in a concentration and time-dependent manner (Fig. 4A–D; Fig. S6C). These data further suggest that autophagy regulates Q6-induced HIF1A turnover in HCC cells.
Figure 4. ATG5 and LC3B are required for Q6-induced autophagy and HIF1A degradation. (A and B) After transfection with specifically targeted siRNA (ATG5 or LC3B) for 36 h, HepG2 and Bel-7402 cells were treated with Q6 (0, 2.5, 5 μM) for 6 h, after which HIF1A, ATG5, and LC3B protein levels were measured by western blot analysis. ACTB was measured as the loading control. (C and D) After transfection with specifically targeted siRNA (ATG5 or LC3B) for 36 h, HepG2 and Bel-7402 cells were treated with Q6 (5 μM) for different times, after which HIF1A protein levels were measured by western blot analysis. ACTB was measured as the loading control. (E) HepG2 cells were treated with Q6 (0, 5 μM) for 6 h under hypoxic conditions, after which HIF1A protein was detected by immunoelectron microscopy analysis. Arrows indicate HIF1A. In the lower panel, the number of immuno-gold labeled HIF1A is presented for HepG2 cells. Twenty cross sections were counted in each experiment and a total of 40 autophagosomes were detected. Data shown are means ± SD of 3 independent experiments. **P < 0.01 compared with the control group.
Furthermore, to determine whether HIF1A could be engulfed in autophagosomes, HepG2 and Bel-7402 cells were exposed to bafilomycin A1 and/or Q6 and then subjected to following immunofluorescence analyses. Figure S6B revealed that HIF1A dots mostly colocalized with LC3B puncta in Q6-treated cells, whereas in Q6-untreated cells, HIF1A was localized in the nucleus with limited amounts being detected in the cytoplasm. Consistently, another supporting evidence was confirmed by immuno-transmission electron microscopy immuno-gold labeling: HIF1A (arrow) was sequestered in double-membrane structures after Q6 treatment. Together, these data indicate that Q6 activates autophagy and that HIF1A is engulfed in autophagosomes and then delivered to lysosomes for degradation.
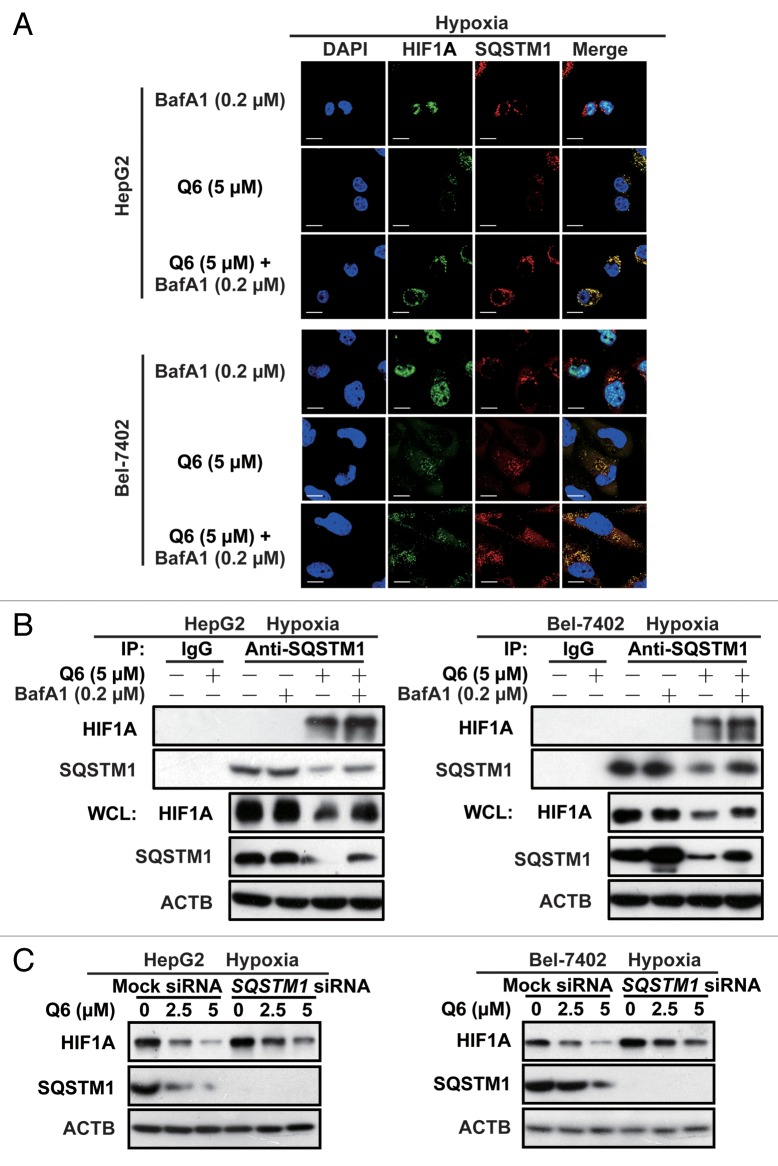
Accumulating studies have demonstrated that autophagy is an important mechanism to prevent the accumulation of polyubiquitinated protein aggregates, and recent studies have revealed that SQSTM1 recognizes polyubiquitinated protein aggregates and then binds to LC3B on autophagosomal membranes, thereby delivering the aggregates for degradation. 22 Therefore, we hypothesized that SQSTM1 could recognize the polyubiquitinated protein HIF1A. The first supporting evidence was provided by immunofluorescence analysis, where a significant increase in levels of HIF1A was found in the cytoplasm after Q6 treatment, and thus the colocalization between HIF1A and SQSTM1 was enhanced (Fig. 5A). The coimmunoprecipitation analysis using SQSTM1 and HIF1A antibodies further evaluated that under basal conditions there is no association between endogenous SQSTM1 and HIF1A, and this link is significantly increased after Q6 treatment. Particularly, cotreatment Q6 with bafilomycin A1, causes HIF1A and SQSTM1 to accumulate in autophagosomes and lysosomes, thus the more evident HIF1A band was observed. (Fig. 5B).
Figure 5. SQSTM1 regulates degradation of HIF1A. (A) HepG2 and Bel-7402 cells were exposed to Q6 (0, 5 μM) in the presence/absence of bafilomycin A1 (BafA1; 0.2 μM) for 6 h in hypoxia and then HIF1A and SQSTM1 protein levels were analyzed by confocal microscopy. Scale bar, 20 μm. (B) HepG2 (left) and Bel-7402 (right) cells were treated with Q6 (0, 5 μM) or Q6 (0, 5 μM) + bafilomycin A1 (0.2 μM) for 6 h and then assayed for protein expression levels as indicated by immunoprecipitation or western blotting. 10% of lysate used for IP was loaded for WCL. (C) After transfection with SQSTM1 siRNA for 36 h, HepG2 (left) and Bel-7402 (right) cells were treated with Q6 (0, 2.5, 5 μM) for 6 h, after which HIF1A protein levels were measured by western blot analysis. ACTB was measured as the loading control.
Furthermore, knockdown of SQSTM1 partially impaired Q6-induced degradation of HIF1A (Fig. 5C; Fig. S9), suggesting that SQSTM1 might play critical roles in Q6-triggered HIF1A degradation, by delivering HIF1A protein to autophagosomes for degradation.
Finally, all these observations collectively suggested that Q6-induced reduction in HIF1A protein levels were attributed to autophagy-mediated degradation and SQSTM1 was involved in the process.
Q6 arrests tumor growth in vivo
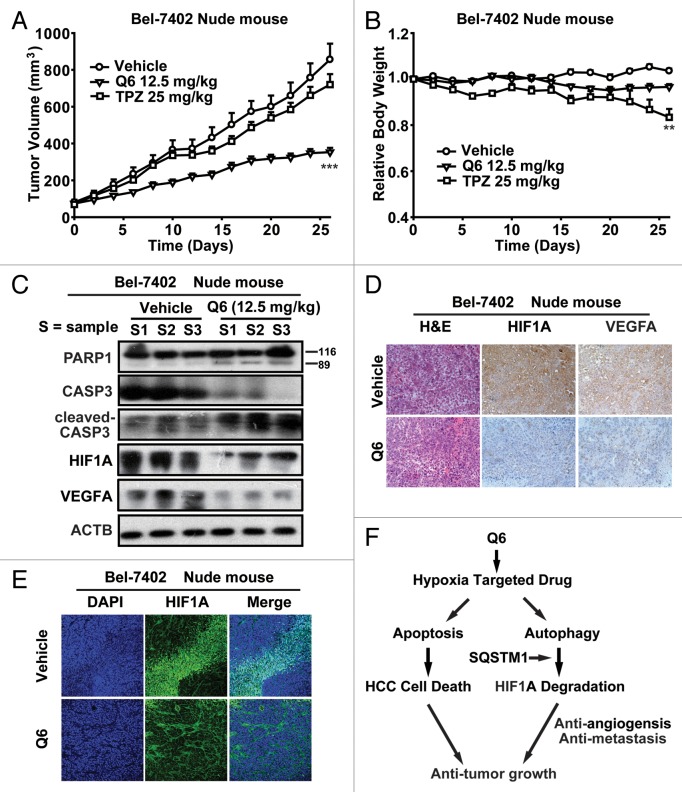
Given the superior antitumor activity of Q6 in vitro, we hypothesized that it should be capable of slowing tumor growth in vivo. As depicted in Figure 6A and Table S2, Q6 administration significantly inhibited growth of Bel-7402 xenograft tumors and H22 hepatoma. Furthermore, it is worth noting that Q6 treatment did not cause obvious weight loss in either of the 2 in vivo models (Fig. 6B; Table S2). Next, we performed western blot analysis of tumor xenografts and investigated that Q6 administration remarkably activated the caspase-dependent apoptosis pathway in Bel-7402 cell-derived tumors. Finally, western blot and immunohistochemical and immunofluoresence analyses verified that Q6 treatment inhibited HIF1A, VEGFA and CD31 expression in tumors from Q6-treated mice, indicating that Q6 suppresses HIF1A activity and related signaling mechanisms in vivo (Fig. 6D and E ; Fig. S7).
Figure 6. Q6 arrests tumor growth in vivo. (A–E) Nude mice bearing established Bel-7402 tumors were treated with TPZ or Q6 at 25 or 12.5 mg/kg by daily i.p. injection for 27 d. (A) Tumor volumes are expressed as the mean ± SEM ***P < 0.001 vs. vehicle-treated controls (n = 5 to 8 per group). (B) Relative body weights are expressed as the mean ± SEM **P < 0.01 vs. vehicle-treated controls (n = 5 to 8 per group). (C) Western blot analysis of Bel-7402 cell-derived tumors treated with Q6 (12.5 mg/kg) or vehicle for expression of PARP1, CASP3, cleaved-CASP3, HIF1A, and VEGFA. ACTB was measured as the loading control. (D and E) Effects of Q6 on expression levels of HIF1A and VEGFA in Bel-7402 cell-derived tumors were determined by hematoxylin and eosin staining and immunohistochemical and immunofluorescence analysis. (F) Schematic diagrams depicting the mechanism by which Q6 exerts its anticancer effects. On the one hand, Q6 triggers the caspase-dependent apoptosis pathway and induces HCC cell death both in vitro and in vivo. On the other hand, Q6 activates the ATG5 and LC3B-dependent autophagy pathway, which plays an essential role in HIF1A degradation and the anti-angiogenesis and anti-metastatic activities of Q6. Furthermore, interaction with SQSTM1 regulates the degradation of HIF1A. Thus, these data strongly support the conclusion that Q6 is a novel hypoxia-targeted drug for HCC therapy, and we propose that autophagy acts as a tumor suppressor by accelerating HIF1A degradation and arresting angiogenesis and metastatic processes.
Collectively, our results demonstrated that Q6 dramatically arrests tumor growth in vivo through dual hypoxia-targeted regulatory mechanisms.
Discussion
Given the association of hypoxia with treatment resistance and poor prognosis, the primary purpose of this study was to investigate responses of multiple HCC cell lines to a novel 3′-substituted TPZ analog, and the mechanism involved in mediating these responses. This study initially revealed the significant antitumor activity of the hypoxia-targeted drug, Q6, both in vitro and in vivo. More importantly, the present study demonstrated for the first time that autophagic degradation contributes to drug-induced proteolysis of HIF1A, thereby shedding light on a new role for autophagy as a regulator of HIF1A expression in hypoxia-targeted therapeutics.
The presence of hypoxic regions is a common feature of solid human tumors and some treatments for HCC, such as transarterial chemoembolization (TACE) can also cause hypoxia. 23 The most established approach to combat with hypoxia is the introduction of HAPs, which can specifically eliminate hypoxic tumor cells. The lead compound, TPZ, has shown selective toxicity toward hypoxic cells in cell culture and in preclinical tumor models. Nevertheless, randomized phase II and III clinical trials that have been completed to date have shown limited improvement in tumor control, especially a lack of efficient extravascular transport and substantially increased toxicity. 24 Therefore, new generation HAPs, such as AQ4N and TH-302, have been designed and potentially overcome, some of the recognized limitations of TPZ. In this study, we found that a 3′-substituted TPZ analog, Q6, exhibits 10-fold higher in vitro antiproliferative efficacy relative to TPZ against HCC cells. Moreover, like TPZ, the antitumor activity of Q6 is mediated, in part, through activation of the caspase-dependent apoptosis pathway. Pretreatment of the cells with a pan-caspase inhibitor, Z-VAD-FMK, attenuated Q6-induced cleavage of PARP1 and blocked the apoptosis process. It is not only efficacy, but also safety, that is an important factor when considering HAPs for cancer therapy. We evaluated the toxicity of Q6 in vitro and found that it exerts no apparent toxicity in 2 normal liver cell lines. Similarly, our primary toxicity studies showed that while 200 mg/kg TPZ was lethal, mice treated with 1000 mg/kg Q6 survived, indicating a reduced systemic toxicity of Q6 in vivo (unpublished observations). Collectively, these data strongly support the conclusion that Q6 is a potent hypoxia-selective prodrug, with limited toxicity.
The unique mechanism that likely underlies the high potency of the novel hypoxia-activated prodrug Q6 is that it takes advantage of the phenomenon of hypoxia-mediated gene expression. In recent clinical trials, many agents that suppress HIF1A expression and signal transduction, such as YC-1, sorafinib, and doxorubicin, had significantly enhanced activity when used in combination with HAPs, such as TPZ, PR-104, and TH-302. For example, it was reported that TH-302 enhanced the anticancer activity of doxorubicin in a broad panel of in vivo xenograft models including nonsmall cell lung cancer (NSCLC), colon cancer, prostate cancer, fibrosarcoma, melanoma, and pancreatic cancer. 25 Thus this increased understanding of hypoxia-targeted dual regulatory mechanisms may have good prospects for developing new approaches for cancer treatment. In addition, we demonstrated that Q6 suppresses HIF1-mediated VEGFA expression both in vitro and in vivo. Collectively, these observations strongly support the conclusion that Q6 is hypoxia-targeted drug with dual modulatory actions, which represents both an opportunity and a challenge for the design and synthesis of novel HAPs.
All eukaryotic cells use the ubiquitin-proteasome system and the autophagy-lysosome pathway for protein degradation and the proteasome is considered to be used for selective degradation of short-lived and abnormal/misfiled proteins, such as TP53/p53 or HIF1A. In this pathway, HIF1A is first hydroxylated by prolyl-hydroxylases and then scaffolded onto a ubiquitin E3 ligase complex that includes the product of the VHL (von Hippel-Lindau tumor suppressor, E3 ubiquitin protein ligase) gene. This eventually leads to rapid ubiquitination and proteasomal degradation. 13 Meanwhile, autophagy has been identified as a crucial cellular process that prevents accumulation of abnormal proteins and organelles in many physiological and pharmacological conditions. It has been demonstrated, using mice with a conditional, liver-specific knockout of Atg7, that loss of autophagy causes liver dysfunction accompanied by intracellular accumulation of ubiquitinated protein aggregates. 26 Recently, a paper has demonstrated that all-trans retinoic acid induces clearance of PML-RARA by autophagy to promote cell differentiation. 27 These reports suggest that basal constitutive autophagy is required in order to avoid accumulation of ubiquitinated protein aggregates and that drug-induced autophagy may play a positive role in downregulating crucial proteins in cancer therapies.
Several lines of evidence indicate that hypoxia-induced autophagy (or mitochondria autophagy) could be initiated by HIF1A and generally represents an adaptive mechanism that maintains cancer cell viability in hypoxic environments. Intriguingly, in this study, we found that autophagic flux is dramatically activated during the course of Q6 treatment in a HIF1A-independent manner (data not shown), and subsequently triggering autophagy-mediated degradation of HIF1A. Inhibition of Q6-induced autophagy would rescue the degradation of HIF1A, and simultaneously, attenuate the cell-killing abilities of Q6. Different from the hypoxia-induced and HIF1A-dependent protective autophagy reported previously, the autophagic cell death triggered by Q6 may represent a type of acute autophagy provoked by drug stimulation rather by the microenvironment and metabolism stresses. 28 , 29 The conclusion of autophagy-dependent lysosomal degradation of HIF1A by Q6 is based on several observations: 1) HIF1A was detected in autophagosome-like, double-membrane structures after Q6 treatment; 2) inhibition of autophagy by downregulating essential autophagy genes, such as ATG5 and LC3B, or by an autophagy inhibitor (3-MA), prevented Q6-induced degradation of HIF1A in a time- and concentration-dependent manner; and 3) pretreatment with the proteasome inhibitor, MG-132, could not reverse Q6-induced degradation of HIF1A in either of the HCC cell lines. Thus, the present study demonstrates for the first time that autophagic degradation contributes to drug-induced proteolysis of HIF1A, and also sheds light on a new role for autophagy as a regulator of HIF1A expression in hypoxia-targeted therapeutic approaches for liver cancer.
Furthermore, our results also demonstrate that autophagy promotes HIF1A degradation through an increased interaction with SQSTM1. It has been previously reported that SQSTM1 is a selective autophagy substrate, binding polyubiquitinated proteins (via its C-terminal ubiquitin-associated domain) and LC3 (via a newly identified LC3 interacting region), that acts as an adaptor by targeting ubiquitinated protein aggregates for degradation by autophagy. 30 In our study, without exposure to Q6, HIF1A was mainly located in the nucleus, whereas SQSTM1 was preferentially located in the cytoplasm. This quite different spatial distribution impedes the association between the 2 proteins, thus we were unable to detect the interaction between HIF1A and SQSTM1 in our experiment systems, although there is some constitutive autophagy occurring under hypoxia condition. In contrast, upon Q6 treatment, we observed a significant translocation of HIF1A from the nucleus to the cytoplasm, where HIF1A interacted with SQSTM1 and was subjected to lysosomal degradation. Furthermore, we also found that knockdown of SQSTM1 failed to completely reverse Q6-induced degradation of HIF1A, indicating that some other cargo proteins might involve in Q6-induced HIF1A lysosomal degradation. Further studies are required to confirm this hypothesis.
Based upon the work reported here and on previously published data, we propose a more nuanced conceptual model incorporating the promising hypoxia-targeted strategy for cancer therapy (Fig. 6F). In this model, Q6 triggers the caspase-dependent apoptosis pathway and induces HCC cell death. On the other hand, Q6 also activates the autophagy pathway, which plays an essential role in accelerating HIF1A degradation and suppression of its downstream target gene, VEGF. Furthermore, we determined that Q6-triggered apoptosis and autophagy were 2 mutually independent events, both of which contributed to the dual hypoxia-targeted regulatory mechanisms and might ultimately lead to the anticancer activity of Q6. (Fig. S8A and S8B). In summary, these data reveal a novel role for autophagy as a regulator of HIF1A expression and HIF1A-mediated signal transduction in hypoxia, and strongly support the conclusion that Q6 has potential clinical value in therapy of HCC.
Materials and Methods
Reagents and cell culture
Q6 was supplied by Professor Yong-zhou Hu (Zhejiang University, Hangzhou, China). TPZ (tirapazamine) was purchased from Topharman Shanghai Co. Ltd. 3-MA (M9281) was from Sigma-Aldrich. Primary antibodies to cleavage-CASP3 (9661), phospho-MAPK1/3 (9106), phospho-MAPK14 (9216), VEGFA (2463), ATG5 (8540), LC3B (2775), RARA (2554) and ACTB (4967) antibodies were obtained from Cell Signaling Technologies. Antibodies to CASP3 (sc-7272), PARP1 (sc-7150), and SQSTM1 (sc-25575) and HRP-labeled secondary anti-goat (sc-2354), anti-mouse (sc-2005), and anti-rabbit (sc-2004) antibodies were from Santa Cruz Biotechnology. Antibody to HIF1A (610959) was obtained from BD Biosciences. Antibody to CD31 (ab28364) was obtained from Abcam. Other anticancer agents and inhibitors were obtained from Sigma-Aldrich.
For the 2 HCC cell lines, Bel-7402 (TcHu10) was obtained from Cell Bank of China Science and HepG2 (HB8065) was obtained from the American Type Culture Collection (ATCC). Bel-7402/HepG2 cells normally cultured in RPMI 1640 medium (Gibco, 31800-022)/DMEM (Gibco, 12800-017) containing 10% fetal bovine serum (Hyclone, SH30088.03) and 1% antibiotics in a humidified atmosphere with 5% CO2 at 37 °C. Hypoxia treatment was performed by placing the cells in a CO2 Water Jacketed Incubator (Thermo Forma, 3110) filled with a mixture of 0.6% O2, 5% CO2, and 94.4% nitrogen.
Flow cytometric analysis of cell cycle, apoptosis, and mitochondrial membrane potential (ΔΨm)
Cells were treated with Q6 and/or the general caspase inhibitor, Z-VAD-FMK (Imgenex, IMI-2311). After harvesting and washing twice with cold PBS buffer, the ANXA5-FITC/PI apoptosis Detection Kit (Biovision, K101-100) was used for analysis of apoptotic cells. For JC-1 staining, cells were resuspended in PBS, containing 0.1 μΜ JC-1 (Sigma, T4069) and were incubated at 37 °C for 15 min in the dark. All samples were analyzed using a FACS-Calibur cytometer (Becton Dickinson).
Gene transfection and RNAi
Cells were seeded on 6-well plates and transfected 24 h later using Lipofectamine 2000 (Invitrogen, 11668-019), according to the manufacturer’s instructions. Human ATG5 siRNA, MAP1LC3B(LC3B) siRNA, HIF1A siRNA, SQSTM1/p62 siRNA, and control siRNA were obtained from GenePharma Co. Ltd.
Measurement of in vivo activity
Tumors were established by injection of Bel-7402 cells (5 × 106 cells per animal, subcutaneously into the armpit) to 5- to 6-wk-old BALB/c male athymic mice (National Rodent Laboratory Animal Resource). Treatments were initiated when tumors reached a mean group size of about 100 mm3. Tumor volume (mm3) was measured with calipers and calculated as (W2 × L)/2, where W is the width and L is the length. Athymic mice were intravenously administered with Q6 (12.5 mg/kg) and TPZ (25 mg/kg) dissolved in cremophor: ethanol: 0.9% sterile sodium chloride solution (1: 1: 8, volume) once daily. Mouse weight and tumor volumes were recorded every 2 d until the animals were sacrificed. Animal care was in accordance with institutional guidelines.
Statistical analyses
Results are expressed as the mean ± SD of at least 3 independent experiments. Differences between two means were analyzed by the Student t test and were considered statistically significant when P < 0.05.
A full description of additional Materials and Methods employed in these studies is provided in the supporting information.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 81072657 and No.81273535), Program for New Century Excellent Talents in University of Ministry of Education of China, and Zhejiang Provincial Program for The Cultivation of HigLevel Innovative Health Talents.
Glossary
Abbreviations:
- 3-MA
3-methyladenine
- ARNT/HIF1B
aryl hydrocarbon receptor nuclear translocator
- ATG
autophagy-related
- CASP3
caspase 3
- CHX
cycloheximide
- CQ
chloroquine
- DAPI
4',6-diamidino-2-phenylindole
- DMEM
Dulbecco’s modified Eagle’s medium
- EPAS1/HIF2A
endothelial PAS domain protein 1
- HAP
hypoxia-activated prodrug
- HIF1A
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
- HRE
hypoxia response element
- MAP1LC3B (LC3B)
microtubule-associated protein 1 light chain 3 beta
- MAPK1/3 (ERK1/2)
mitogen-activated protein kinase 1/3
- MAPK14 (p38 alpha)
mitogen-activated protein kinase 14
- PARP1
poly (ADP-ribose) polymerase 1
- siRNA
short interfering RNA
- SQSTM1
sequestosome 1
- TPZ
tirapazamine
- UBB
ubiquitin B
- VEGFA
vascular endothelial growth factor A
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/26838
References
- 1.American Cancer Society. Cancer Facts & Figures 2010. Atlanta, GA: American Cancer Society; 2010 [Google Scholar]
- 2. Lee YH, Andersen JB, Song HT, Judge AD, Seo D, Ishikawa T, Marquardt JU, Kitade M, Durkin ME, Raggi C, et al. . Definition of ubiquitination modulator COP1 as a novel therapeutic target in human hepatocellular carcinoma. Cancer Res 2010; 70:8264 - 9; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-0749; PMID: 20959491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yopp AC, Schwartz LH, Kemeny N, Gultekin DH, Gönen M, Bamboat Z, Shia J, Haviland D, D’Angelica MI, Fong Y, et al. . Antiangiogenic therapy for primary liver cancer: correlation of changes in dynamic contrast-enhanced magnetic resonance imaging with tissue hypoxia markers and clinical response. Ann Surg Oncol 2011; 18:2192 - 9; http://dx.doi.org/ 10.1245/s10434-011-1570-1; PMID: 21286939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matsumoto S, Yasui H, Mitchell JB, Krishna MC. . Imaging cycling tumor hypoxia. Cancer Res 2010; 70:10019 - 23; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-2821; PMID: 21159626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Semenza GL. . Targeting HIF-1 for cancer therapy. Nat Rev Cancer 2003; 3:721 - 32; http://dx.doi.org/ 10.1038/nrc1187; PMID: 13130303 [DOI] [PubMed] [Google Scholar]
- 6. Nath B, Szabo G. . Hypoxia and hypoxia inducible factors: diverse roles in liver diseases. Hepatology 2012; 55:622 - 33; http://dx.doi.org/ 10.1002/hep.25497; PMID: 22120903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown JM, Wilson WR. . Exploiting tumour hypoxia in cancer treatment. Nat Rev Cancer 2004; 4:437 - 47; http://dx.doi.org/ 10.1038/nrc1367; PMID: 15170446 [DOI] [PubMed] [Google Scholar]
- 8. Melillo G. . Targeting hypoxia cell signaling for cancer therapy. Cancer Metastasis Rev 2007; 26:341 - 52; http://dx.doi.org/ 10.1007/s10555-007-9059-x; PMID: 17415529 [DOI] [PubMed] [Google Scholar]
- 9. Moradpour D, Blum HE. . Pathogenesis of hepatocellular carcinoma. Eur J Gastroenterol Hepatol 2005; 17:477 - 83; http://dx.doi.org/ 10.1097/00042737-200505000-00002; PMID: 15827436 [DOI] [PubMed] [Google Scholar]
- 10. Finger EC, Giaccia AJ. . Hypoxia, inflammation, and the tumor microenvironment in metastatic disease. Cancer Metastasis Rev 2010; 29:285 - 93; http://dx.doi.org/ 10.1007/s10555-010-9224-5; PMID: 20393783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poon E, Harris AL, Ashcroft M. . Targeting the hypoxia-inducible factor (HIF) pathway in cancer. Expert Rev Mol Med 2009; 11:e26; http://dx.doi.org/ 10.1017/S1462399409001173; PMID: 19709449 [DOI] [PubMed] [Google Scholar]
- 12. Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. . Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 2005; 105:659 - 69; http://dx.doi.org/ 10.1182/blood-2004-07-2958; PMID: 15374877 [DOI] [PubMed] [Google Scholar]
- 13. Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. . Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001; 292:468 - 72; http://dx.doi.org/ 10.1126/science.1059796; PMID: 11292861 [DOI] [PubMed] [Google Scholar]
- 14. Semenza GL. . Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci 2012; 33:207 - 14; http://dx.doi.org/ 10.1016/j.tips.2012.01.005; PMID: 22398146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giaccia A, Siim BG, Johnson RS. . HIF-1 as a target for drug development. Nat Rev Drug Discov 2003; 2:803 - 11; http://dx.doi.org/ 10.1038/nrd1199; PMID: 14526383 [DOI] [PubMed] [Google Scholar]
- 16. Hu Y, Xia Q, Shangguan S, Liu X, Hu Y, Sheng R. . Synthesis and biological evaluation of 3-aryl-quinoxaline-2-carbonitrile 1,4-di-N-oxide derivatives as hypoxic selective anti-tumor agents. Molecules 2012; 17:9683 - 96; http://dx.doi.org/ 10.3390/molecules17089683; PMID: 22890172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xiang ZL, Zeng ZC, Fan J, Tang ZY, Zeng HY, Gao DM. . Gene expression profiling of fixed tissues identified hypoxia-inducible factor-1α, VEGF, and matrix metalloproteinase-2 as biomarkers of lymph node metastasis in hepatocellular carcinoma. Clin Cancer Res 2011; 17:5463 - 72; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-3096; PMID: 21712445 [DOI] [PubMed] [Google Scholar]
- 18. Yamaguchi R, Yano H, Iemura A, Ogasawara S, Haramaki M, Kojiro M. . Expression of vascular endothelial growth factor in human hepatocellular carcinoma. Hepatology 1998; 28:68 - 77; http://dx.doi.org/ 10.1002/hep.510280111; PMID: 9657098 [DOI] [PubMed] [Google Scholar]
- 19. Koh MY, Powis G. . Passing the baton: the HIF switch. Trends Biochem Sci 2012; 37:364 - 72; http://dx.doi.org/ 10.1016/j.tibs.2012.06.004; PMID: 22818162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levine B. . Cell biology: autophagy and cancer. Nature 2007; 446:745 - 7; http://dx.doi.org/ 10.1038/446745a; PMID: 17429391 [DOI] [PubMed] [Google Scholar]
- 21. Klionsky DJ, Emr SD. . Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717 - 21; http://dx.doi.org/ 10.1126/science.290.5497.1717; PMID: 11099404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. . Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res 2011; 17:654 - 66; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-2634; PMID: 21325294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Asghar U, Meyer T. . Are there opportunities for chemotherapy in the treatment of hepatocellular cancer?. J Hepatol 2012; 56:686 - 95; http://dx.doi.org/ 10.1016/j.jhep.2011.07.031; PMID: 21971559 [DOI] [PubMed] [Google Scholar]
- 24. Hicks KO, Siim BG, Jaiswal JK, Pruijn FB, Fraser AM, Patel R, Hogg A, Liyanage HD, Dorie MJ, Brown JM, et al. . Pharmacokinetic/pharmacodynamic modeling identifies SN30000 and SN29751 as tirapazamine analogues with improved tissue penetration and hypoxic cell killing in tumors. Clin Cancer Res 2010; 16:4946 - 57; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-1439; PMID: 20732963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Q, Sun JD, Wang J, Ahluwalia D, Baker AF, Cranmer LD, Ferraro D, Wang Y, Duan JX, Ammons WS, et al. . TH-302, a hypoxia-activated prodrug with broad in vivo preclinical combination therapy efficacy: optimization of dosing regimens and schedules. Cancer Chemother Pharmacol 2012; 69:1487 - 98; http://dx.doi.org/ 10.1007/s00280-012-1852-8; PMID: 22382881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. . Autophagy-deficient mice develop multiple liver tumors. Genes Dev 2011; 25:795 - 800; http://dx.doi.org/ 10.1101/gad.2016211; PMID: 21498569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Z, Cao L, Kang R, Yang M, Liu L, Zhao Y, Yu Y, Xie M, Yin X, Livesey KM, et al. . Autophagy regulates myeloid cell differentiation by p62/SQSTM1-mediated degradation of PML-RARα oncoprotein. Autophagy 2011; 7:401 - 11; http://dx.doi.org/ 10.4161/auto.7.4.14397; PMID: 21187718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. . Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 2008; 283:10892 - 903; http://dx.doi.org/ 10.1074/jbc.M800102200; PMID: 18281291 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29. Wilkinson S, O’Prey J, Fricker M, Ryan KM. . Hypoxia-selective macroautophagy and cell survival signaled by autocrine PDGFR activity. Genes Dev 2009; 23:1283 - 8; http://dx.doi.org/ 10.1101/gad.521709; PMID: 19487569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. . p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171:603 - 14; http://dx.doi.org/ 10.1083/jcb.200507002; PMID: 16286508 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.