Abstract
Removal of ubiquitinated targets by autophagosomes can be mediated by receptor molecules, like SQSTM1, in a mechanism referred to as selective autophagy. While cytoplasmic protein aggregates, mitochondria, and bacteria are the best-known targets of selective autophagy, their role in the turnover of membrane receptors is scarce. We here showed that fasting-induced wasting of skeletal muscle involves remodeling of the neuromuscular junction (NMJ) by increasing the turnover of muscle-type CHRN (cholinergic receptor, nicotinic/nicotinic acetylcholine receptor) in a TRIM63-dependent manner. Notably, this process implied enhanced production of endo/lysosomal carriers of CHRN, which also contained the membrane remodeler SH3GLB1, the E3 ubiquitin ligase, TRIM63, and the selective autophagy receptor SQSTM1. Furthermore, these vesicles were surrounded by the autophagic marker MAP1LC3A in an ATG7-dependent fashion, and some of them were also positive for the lysosomal marker, LAMP1. While the amount of vesicles containing endocytosed CHRN strongly augmented in the absence of ATG7 as well as upon denervation as a model for long-term atrophy, denervation-induced increase in autophagic CHRN vesicles was completely blunted in the absence of TRIM63. On a similar note, in trim63−/− mice denervation-induced upregulation of SQSTM1 and LC3-II was abolished and endogenous SQSTM1 did not colocalize with CHRN vesicles as it did in the wild type. SQSTM1 and LC3-II coprecipitated with surface-labeled/endocytosed CHRN and SQSTM1 overexpression significantly induced CHRN vesicle formation. Taken together, our data suggested that selective autophagy regulates the basal and atrophy-induced turnover of the pentameric transmembrane protein, CHRN, and that TRIM63, together with SH3GLB1 and SQSTM1 regulate this process.
Keywords: acetylcholine receptor, atrophy, Bif-1, endophilin B1, LC3, MuRF1, neuromuscular junction, selective autophagy, SQSTM1/p62
Introduction
Macroautophagy (hereafter short autophagy) refers to a membrane-engulfment process leading to degradation of bulk material in response to various stress stimuli, including starvation. Selective autophagy is a process by which ubiquitinated target structures, such as cytosolic protein aggregates and organelles, are engulfed and degraded via autophagy (for recent review, see 1 ). Sequestration and transport of ubiquitinated targets to newly forming autophagosomes is typically mediated by selective autophagy receptors, like SQSTM1 2 and NBR1 3 that interact with both the polyubiquitin chain of the target and the autophagic marker protein, MAP1LC3A. 4 While most targets for selective autophagy described so far include cytosolic protein aggregates, 5 damaged mitochondria 6 and bacteria, 7 information regarding the degradation of plasma membrane-bound transmembrane receptor molecules via selective autophagy is emerging recently. 8
CHRN is a pentameric ligand-gated ion-channel present in the central nervous system and in skeletal muscle. 9 In the latter it mediates nerve-induced muscle contraction and exhibits extremely high densities of about 10,000 molecules per square micron in the postsynaptic membrane of the nerve-muscle synapse, called the neuromuscular junction. 10 For simplicity, in the following text we refer to muscle-type CHRN, which is composed of the 5 subunits CHNRA1 (2 subunits), CHRNB1, CHRND, and CHRNE (in adult muscle) or CHRNG (in developing/regenerating muscle). CHRN are bona fide transmembrane proteins and are composed of 5 subunits that are synthesized and assembled at the level of the endoplasmic reticulum, before they reach the postsynapstic membrane via Golgi apparatus and exocytic vesicles. 11 , 12 After their arrival at the membrane, CHRN can either persist there or get endocytosed in vesicular carriers 13 , 14 from where it might be either recycled back to the membrane or get degraded 15 - 17 in lysosomes. 18 While the expression of CHRN displays strong upregulation under muscle wasting conditions, 19 , 20 such as immobilization and denervation, its metabolic stability decreases at the same time, 10 , 21 - 23 implicating post-transcriptional regulation in CHRN turnover under stress. Indeed, CHRN is subject to a wealth of regulatory post-translational modifications ranging from phosphorylation 24 , 25 to ubiquitination. 26 , 27 We have recently reported that the E3-ubiquitin ligase TRIM63 plays an important role in the turnover of CHRN. 28 TRIM63 was found to be highly enriched in the perisynaptic region and to coprecipitate with CHRN that was in vivo-labeled with α-bungarotoxin (BGT)-biotin. Furthermore, in vivo imaging showed that TRIM63-GFP is almost exclusively localized to endo/lysosomal structures containing endocytosed CHRN, and metabolic destabilization of CHRN protein upon denervation-induced muscle atrophy was significantly halted in trim63−/− mice. Since yeast 2-hybrid screens previously indicated direct interaction between TRIM63 and SQSTM1, 29 we tested the hypothesis that CHRN, is directed to selective autophagy via TRIM63.
Results
Starvation destabilizes CHRN
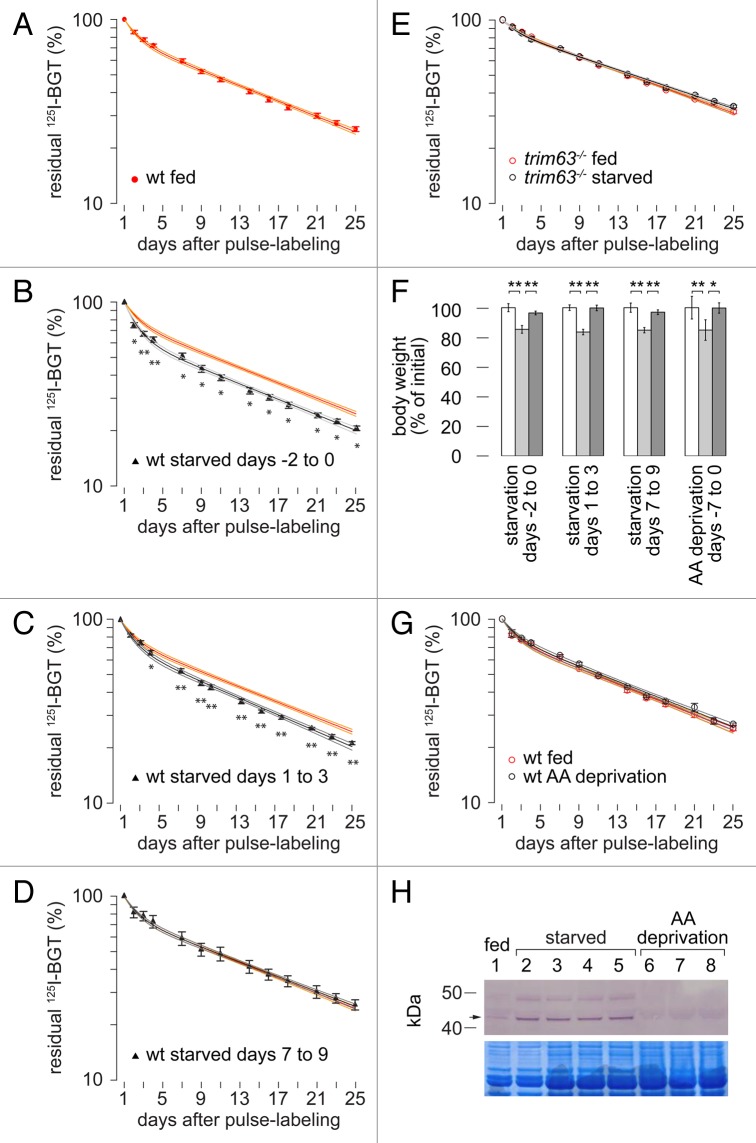
A wealth of previous reports describes a strong negative impact of skeletal muscle denervation on the metabolic stability of CHRN. 10 , 21 - 23 However, it is unclear if this effect is due to a generalized atrophic signaling or specific to the lack of nerve-derived input. To address this point, we used a recently established in vivo-labeling approach that measures the metabolic stability of CHRN. 23 This assay is based on the irreversible binding to CHRN of 125I-labeled BGT (125I-BGT), an 8 kDa, non-cell-penetrating snake venom (for scheme, see Fig. 1). The total amount of radioiodine that is present in the muscle is determined at several time points without discriminating between different locations (e.g., on the cell surface, in endocytic compartments, etc.). In fact, radioiodine emission is only lost if CHRN and with it bound 125I-BGT is degraded and the 125I, incorporated in tyrosine residue 54 of BGT, is shuttled out of the muscle into the blood stream (Fig. 1D). As previously described, in denervated muscles an increase in size of a short-lived pool of CHRN (half-life of about 1 d) and a reduction in half-life of a long-lived receptor pool from 13 to 8 d had been observed. 23 Now, we applied the approach to fasted mice. Animals were sorted into 4 different groups. While CHRN in the hind legs of all animals were pulse labeled with 125I-BGT at day 0, 3 groups were kept without food for 48 h. This happened either immediately before pulse labeling (days -2 to 0, group B), or 1 (days 1 to 3, group C) or 7 d after pulse labeling (days 7 to 9, group D). A control group (group A) was fed ad libitum for the entire course of the experiment. 125I-emission from the hind limbs was then recorded at shown intervals for the next 4 weeks and was normalized to the values measured on day 1. This showed that compared with control (Fig. 2A) CHRN lifetime was mildly but significantly reduced in mice starved immediately before and immediately after pulse labeling (Fig. 2B and C). In both cases starvation led to an increase in size of the short-lived receptor pool. The half-life of the long-lived pool was either unaffected or even slightly higher. Notably, CHRN lifetime was completely unaffected when fasting was executed 7 d after pulse labeling (Fig. 2D). These data show that the metabolic status of mice directly and immediately affects the stability of CHRN. This effect is transient and acts primarily on the ratio between short- and long-lived CHRN.
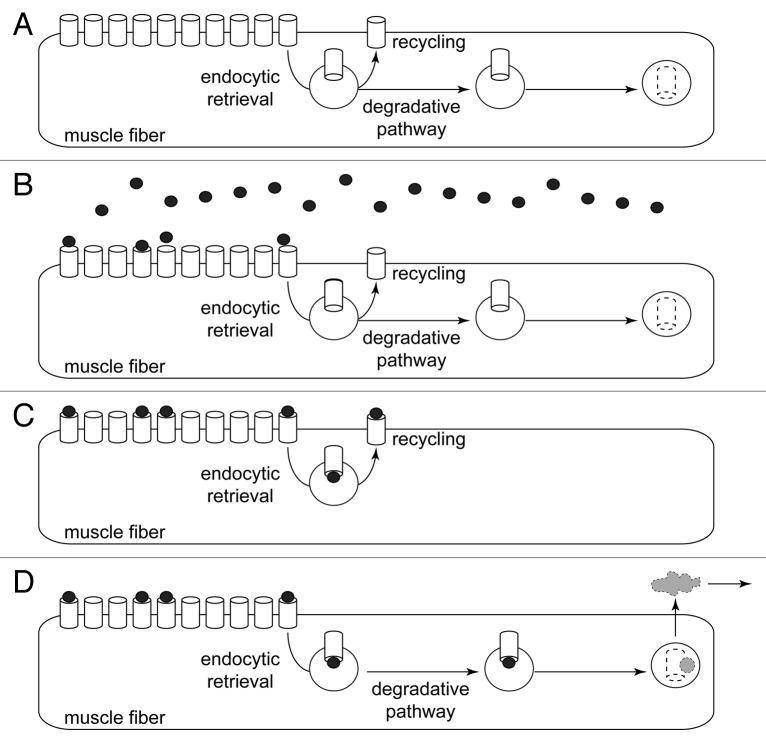
Figure 1. Radioiodine labeling detects total amount of 125I contained in muscle. Scheme illustrating the working principle of the radioiodine approach. (A) Pre-pulse situation. CHRN channels (white barrels) are present at high amounts in the postsynaptic membranes of muscle fibers. Small amounts of CHRN are constantly internalized by endocytic retrieval. From there, CHRN might undergo recycling back to the plasma membrane or follow a degradative pathway. (B) Pulse labeling with 125I-BGT (black dots). About 10% of the 125I-BGT binds to CHRN in the injected hind limb, the rest of the 25I-BGT is washed out and binds to other muscles or is excreted. (C and D) CHRN labeled with 125I-BGT undergo either recycling (C) or are degraded (D). Only in the latter case, 125I is removed from the muscle (gray cloud), i.e., the radioiodine approach measures the rate of 125I removal from the muscle as a consequence of CHRN degradation.
Figure 2. Fasting affects CHRN turnover only of newly formed receptors in a TRIM63-dependent manner. (A–E) Tibialis anterior muscles of WT or trim63−/− mice were labeled with 125I-BGT (“pulse-labeling”) on day 0 and 125I-emission from hindlegs was then repetitively monitored at indicated time points. Animals were either fed ad libitum (A) or starved for 48 h 2 d before (B and E), 1 d (C) or 7 d after pulse-labeling (D). Graphs depict residual 125I-emission normalized to the values measured on day 1. Symbols and red or black lines represent measured values (mean ± SEM, n = 32, n = 15, n = 14, and n = 4 muscles in A–D, respectively) and 2-term exponential fits, respectively. Orange and gray lines show 95% confidence intervals. The ” WT fed” graph is repeated in (A–D) for better comparison. Significance was tested between measurement values at each time point: *P < 0.05; **P < 0.01 according to the Welch test. Note significant CHRN destabilization in (B and C), but not in (D and E). (F–H) WT mice were either fasted for 48 h or fed ad libitum with amino acid-free chow for 7 d. Animals were weighed immediately before and after treatment and one more time 3 to 4 weeks later. After 7 d of amino acid deprivation, tibialis anterior muscles of amino acid-deprived animals were labeled with 125I-BGT (“pulse-labeling”) on day 0 and 125I-emission from hindlegs was then repetitively monitored at the indicated time points. (F) Body weights of animals before (white columns), after (light gray columns), and 3 to 4 weeks after fasting (dark gray columns). Mean ± SEM (n ≥ 3 mice for each condition). *P < 0.05; **P < 0.01 according to the Welch test. (G) Graphs depict residual 125I-emission normalized to the values measured on day 1. Symbols and black/red lines show measured values (mean ± SEM, n = 3 mice for each condition) and 2-term exponential fits, respectively. Orange and gray lines show 95% confidence intervals. Significance was tested between measurement values at each time point. (H) Images depict western blot signals (upper) and Coomassie-stained corresponding SDS-PAGE (lower). Lanes, loading of muscle lysates from animals that were either control fed (lane 1), starved for 48 h (lanes 2 to 5), or amino acid-deprived for five days (lanes 6 to 8). Arrow indicates height of TRIM63.
Starvation-induced destabilization of CHRN is absent in trim63−/− mice
Given the recently described role of the atrogene and E3 ubiquitin ligase TRIM63, in regulating the turnover of CHRN 28 and the known importance of TRIM63 in fasting-induced muscle remodeling, 19 , 30 , 31 we then asked if this atrogene also participates in starvation-induced regulation of CHRN turnover. Therefore, trim63−/− mice were starved for 48 h and the radioiodine experiment was performed as before. This revealed indistinguishable CHRN lifetime curves for starved and unstarved trim63−/− mice (Fig. 2E), indicating a central role of TRIM63 in this process.
Amino acid deprivation-induced muscle loss is not sufficient to destabilize CHRN
Muscle wasting is due to an imbalance between protein synthesis and degradation. 32 Although both amino acid deprivation as well as total fasting lead to muscle atrophy, the underlying mechanisms triggering the metabolic response are likely to be different. 33 To address specific reasons for the destabilization observed upon total starvation, we thus switched to an amino acid-deprivation paradigm. As expected, the rate of weight loss was smaller under this condition, but after 7 d of amino acid deprivation the total weight loss was comparable to that upon 2 d of complete starvation (Fig. 2F). Thus, the 125I-BGT metabolic-labeling approach was repeated with animals that were either fed with full diet [wild type (WT) fed] or with amino acid-free chow for 7 d preceding the 125I-BGT-pulse (WT AA deprivation). This treatment was insufficient to induce any detectable change in CHRN lifetime (Fig. 2G). Western blot analysis showed that compared with normally fed mice TRIM63 protein was more abundant in muscles from completely fasted but not amino acid-deprived animals (Fig. 2H). This is consistent with the notion that TRIM63 is not universally connected to atrophic conditions and together with the radioiodine experiments in the presence (Fig. 2A to D) and absence of TRIM63 (Fig. 2E) this finding further supports a role of TRIM63 in CHRN stabilization.
Internalization of synaptic CHRN is enhanced upon fasting
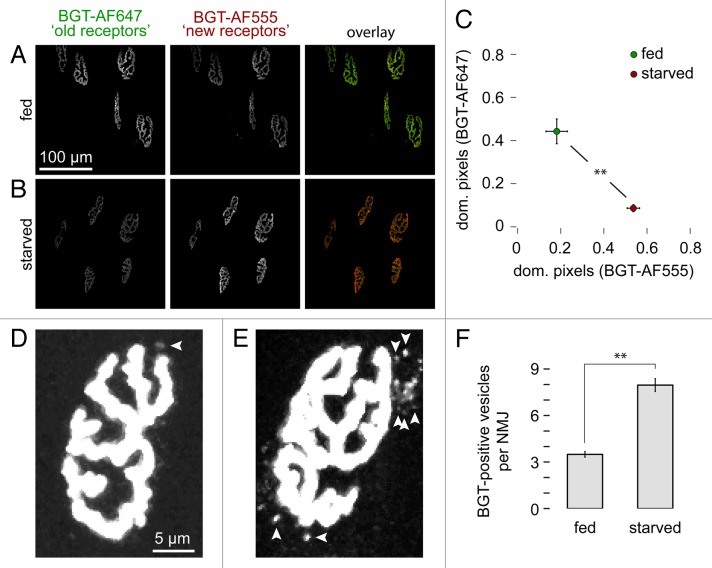
A logical consequence of the increased turnover of CHRN upon total fasting would be enhanced endocytic retrieval in endosomes and subsequent degradation of CHRN. Since the radioiodine assay is incapable of discriminating between the different subcellular locations of CHRN after their arrival at the plasma membrane (see Fig. 1), we next addressed this issue with in vivo imaging as described previously. 25 In brief, a first pool of CHRN (“old receptors”) was fluorescently labeled with BGT-AlexaFluor 647 (BGT-AF647, infrared dye) after 48 h of fasting. Ten days after this first mark, newly arrived CHRN (“new receptors”) were stained with BGT-AlexaFluor 555 (BGT-AF555, red dye) and animals were imaged shortly afterwards. The ratio of red to infrared label intensity, thus, indicated the turnover of CHRN. Furthermore, the imaging also provided insights into CHRN distribution. This approach clearly confirmed an increased turnover of CHRN. Compared with unstarved controls (Fig. 3A), where the “old receptor” label was maintained indicating low receptor turnover, fasted muscles exhibited much more “new receptor” labels (Fig. 3B). This was also evident from the quantitative analysis performed in a semiautomated manner (Fig. 3C). In addition, the in vivo imaging revealed the presence of 2 to 3 times more BGT-positive, i.e., endocytosed CHRN-containing vesicular structures per NMJ in fasted vs. nonfasted muscles (Fig. 4D–F).
Figure 3. Starvation leads to increased CHRN turnover and enhances the amount of endo/lysosomal carriers containing CHRN at the NMJ. CHRNs in tibialis anterior muscles were labeled with BGT-AF647 (“old receptors”) after 48 h of starvation and 10 d later muscles were reinjected with BGT-AF555 (“new receptors”). Then, in vivo microscopy was performed and an automated algorithm determined the relative amount of pixels dominant for either “new receptor” or “old receptor” signals. The number of endo/lysosomal carriers containing CHRN was counted manually after electronically enhancing the signal using ImageJ. (A and B) Maximum z-projections of confocal stacks showing automatically segmented NMJs from fed (A) and starved (B) mice. In the overlay panels (right) old receptor and new receptor signals are shown in green and red, respectively. (C) Quantitative analysis. Graph depicts mean ± SEM of the relative amounts of pixels dominant for either new receptor or old receptor signals in each NMJ (n = 12 [293 NMJs] and n = 4 [106 NMJs] muscles for fed and starved conditions, respectively). **P < 0.01 according to the Welch test. (D and E) Maximum z-projections of NMJs from fed (D) and fasted muscles (E). Arrowheads indicate BGT-AF positive endo/lysosomal carriers. (F) Quantitative analysis. Graph depicts the average amount of BGT-positive vesicles per NMJ. Mean ± SEM (n = 6 and n = 3 muscles for fed and fasted conditions, respectively. 50 NMJs analyzed for each condition). **P < 0.01 according to the Welch test.
Figure 4. Endo/lysosomal carriers containing CHRN colocalize with TRIM63 and are capped by MAP1LC3A. Mouse tibialis anterior muscles were either transfected with GFP-MAP1LC3A (A–F) or cotransfected with GFP-MAP1LC3A and TRIM63-mCherry (G–J). After 7 d, CHRNs were marked with BGT-AF555 (A–F) or with BGT-AF647 (G–J) and 24 h later, in vivo imaging was performed. (A–C) Single optical slice of a GFP-MAP1LC3A-positive fiber showing the NMJ as pretzel-shaped structure in (A) and endocytic CHRN-containing puncta at the upper right corner of the NMJ. GFP-MAP1LC3A signals in (B), (C) overlay of CHRN (red) and MAP1LC3A (green). (D–F) Blow-up of the upper right corner of NMJ as depicted in (A–C). (G–J) Confocal slices showing exemplary colocalization patterns for CHRN, GFP-MAP1LC3A, and TRIM63-mCherry. Mostly, MAP1LC3A capped CHRN- and TRIM63-double-positive puncta at two ends, but also more complex patterns were observed (J). Similar marker distributions were observed upon at least 2 different transfections.
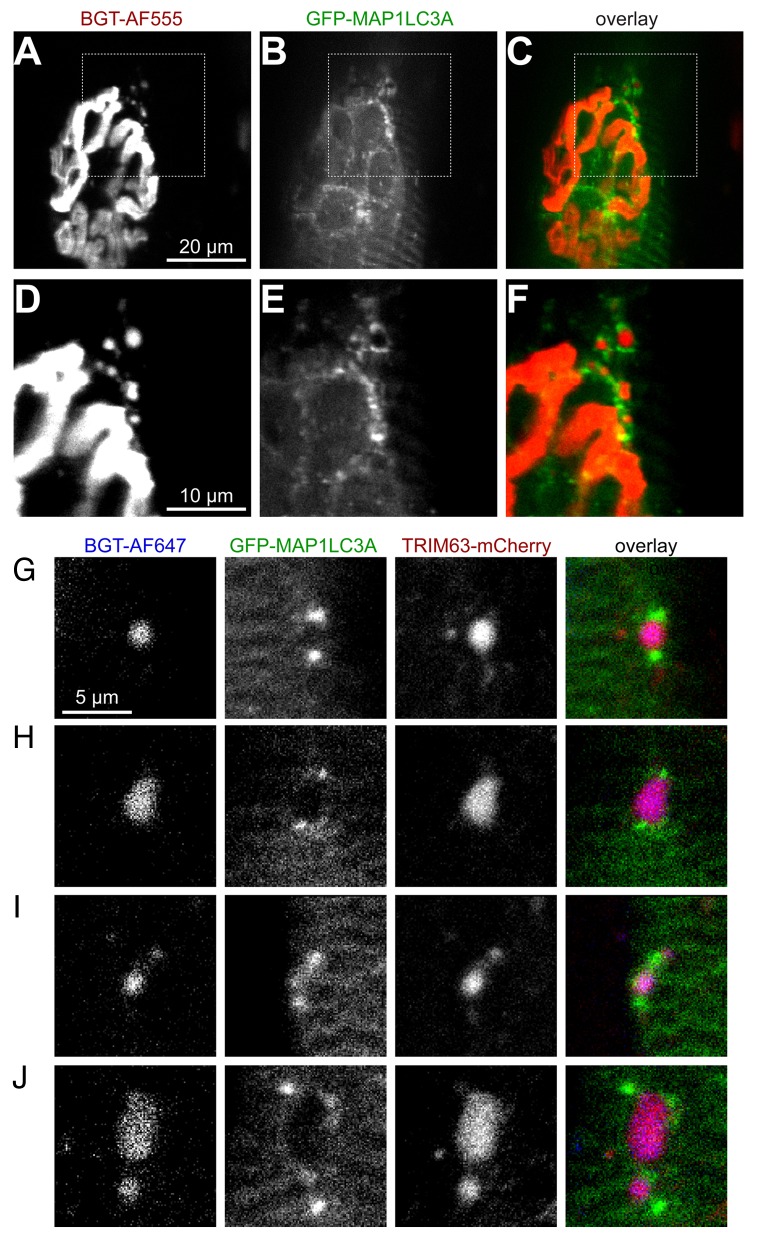
Internalized CHRN are accompanied by MAP1LC3A
Despite the extraordinary physiological importance of CHRN, surprisingly little is known about many of its lifecycle aspects, including the regulation of its degradation. Our previous extensive work on the mechanisms underlying CHRN recycling 25 , 34 , 35 and CHRN degradation identified an interaction between CHRN and the E3 ligase TRIM63. 28 Furthermore, the previously reported direct interaction between TRIM63 and SQSTM1 29 prompted us to speculate that TRIM63 may induce selective autophagy as a principal route for CHRN removal. To address this issue, we first transfected the autophagic marker GFP-MAP1LC3A into live mouse muscle. In this construct, GFP was fused N-terminally to MAP1LC3A to avoid separation of the fluorescent tag from the autophagic marker protein in the course of the conjugation process that leads to the recruitment of MAP1LC3A to the phagophore. 36 Few days later, CHRN were labeled with BGT-AF555 and then imaged in vivo. This revealed an intimate relationship between endocytic carriers bearing CHRN and MAP1LC3A. Indeed, MAP1LC3A was not only found in a typical striated pattern corresponding to the sarcomeric organization of skeletal muscle, but it was also enriched in the region of the NMJ (Fig. 4A to C). Notably, many BGT-positive, i.e., internalized CHRN-containing puncta were surrounded by 1, 2, or many point- or crescent-shaped MAP1LC3A signals (Fig. 4D to F), as it is to be expected during the process of phagophore formation. To understand if TRIM63 is involved in these processes, we then cotransfected GFP-MAP1LC3A and TRIM63-mCherry in muscle and imaged the NMJ in the presence of BGT-AlexaFluor 647. This clearly demonstrated the codistribution of CHRN and TRIM63 signals, which in most cases were again accompanied by MAP1LC3A-positive puncta (Fig. 4G to J). In some cases, MAP1LC3A almost completely surrounded the CHRN-TRIM63 complex (Fig. 5A–A', see insert).
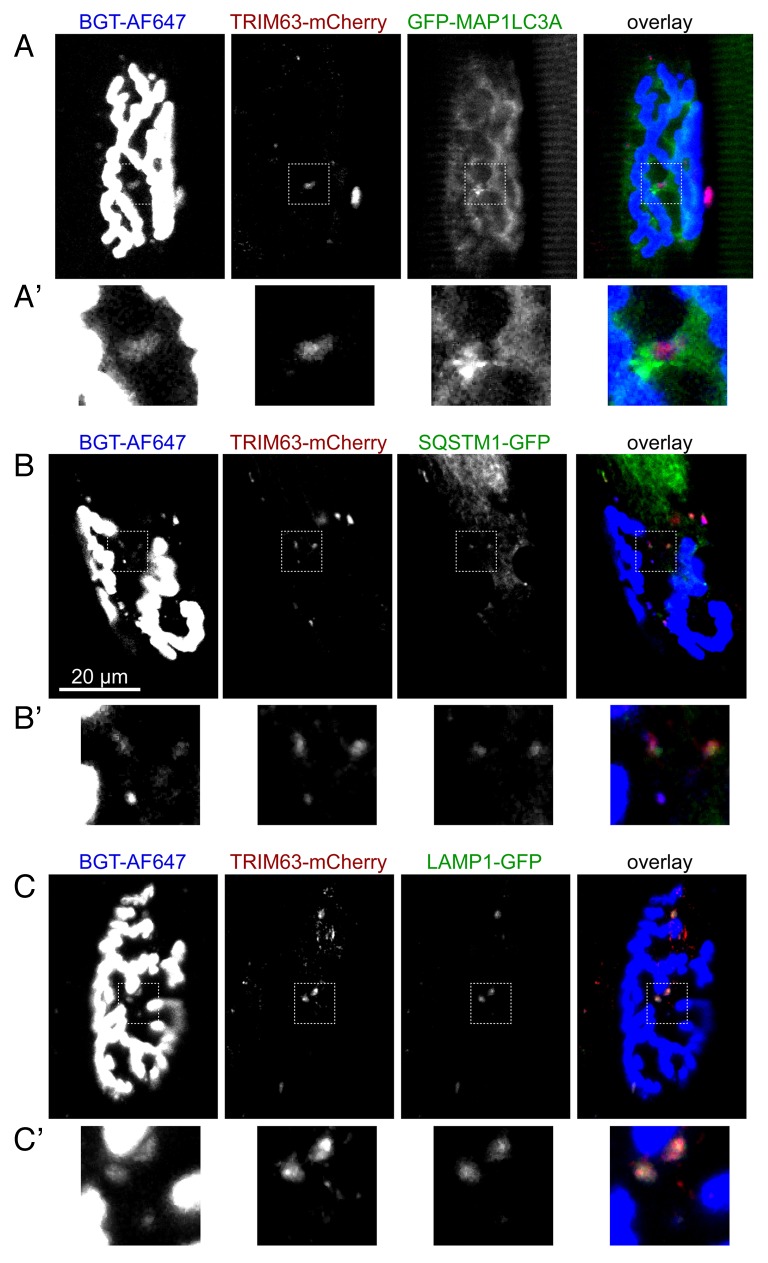
Figure 5. Endo/lysosomal carriers containing CHRN colocalize with a panel of autophagic and lysosomal markers. Mouse tibialis anterior muscles were cotransfected with TRIM63-mCherry and either GFP-MAP1LC3A (A), SQSTM1-GFP (B), or LAMP1-GFP (C). After 7 d, CHRNs were marked with BGT-AF647 and 24 h later, muscles were imaged using in vivo confocal microscopy. All panels show single optical slices with CHRN signals (left column), TRIM63-mCherry signals (middle column) and GFP-constructs (right column) followed by an overlay. Similar marker distributions were observed upon at least 2 different transfections.
CHRN colocalizes in vivo with selective autophagy markers
To fulfill the requirements of selective autophagy the CHRN-TRIM63 complex is expected to interact with selective autophagy receptors and, upon further progression to the autolysosome, with lysosomal markers. To investigate these points, we cotransfected SQSTM1-GFP or LAMP1-GFP with TRIM63-mCherry and visualized muscles in vivo in the presence of BGT-AF647. In both cases, colocalization of all 3 markers was observed in immediate vicinity of the NMJ (Fig. 5B and C), further consolidating the assumption that CHRN is targeted to autophagic decay. In the case of SQSTM1-GFP one elongated structure showed 3 different domains in which either CHRN, TRIM63, or SQSTM1 were most prominent (see upper left of 3 TRIM63-positive puncta in Fig. 5B') with TRIM63 appearing to act as a linker between the flanking domains enriched in CHRN or SQSTM1.
SH3GLB1 colocalizes with TRIM63 and targets to autophagosomes containing CHRN
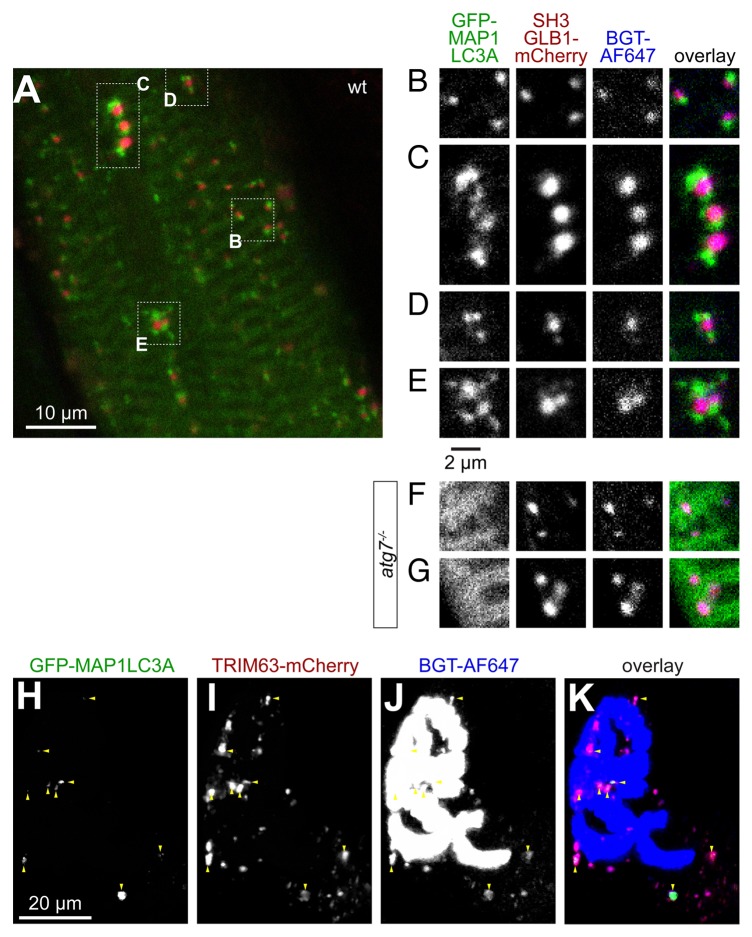
Recently, another player in autophagy, SH3GLB1/Bif-1/endophilin B1, 37 was found to directly interact with TRIM63 and to be present in endocytic CHRN vesicles. 28 To address whether SH3GLB1 might be involved in autophagy of CHRN in skeletal muscle, we cotransfected muscles with GFP-MAP1LC3A and SH3GLB1-mCherry and performed in vivo imaging after BGT-AlexaFluor 647 labeling of CHRN. As expected, SH3GLB1-mCherry nicely colocalized with endocytic CHRN vesicles and was capped by GFP-MAP1LC3A (Fig. 6A to E). To address whether the capping of CHRN-SH3GLB1 double-positive puncta by MAP1LC3A is linked to its lipidation we tested the distribution patterns of CHRN, SH3GLB1-mCherry, and GFP-MAP1LC3A upon transfection of atg7−/− mice. 38 In contrast to WT muscle, the typical capping of CHRN-SH3GLB1 double-positive puncta by GFP-MAP1LC3A was here almost completely abolished and GFP-MAP1LC3A was mostly localized in sarcomeric striation patterns (Fig. 6F and G) or larger aggregates (not shown). To prove codistribution of TRIM63 and SH3GLB1 we then performed in vivo imaging in the presence of TRIM63-GFP, SH3GLB1-mCherry, and BGT-AlexaFluor 647. This revealed that most TRIM63-positive puncta were also positive for CHRN and SH3GLB1 (Fig. 6F–I).
Figure 6. SH3GLB1 perfectly matches endocytic CHRN carriers and is capped by MAP1LC3A in an ATG7-dependent manner. Tibialis anterior muscles of WT (A–E and H–K) or atg7−/− mice (F and G) were cotransfected with either SH3GLB1-mCherry and GFP-MAP1LC3A (A–G) or with SH3GLB1-mCherry and TRIM63-GFP (H–K). After 7 d, CHRNs were marked with BGT-AF647 and 24 h later, muscles were imaged using in vivo confocal microscopy. (A) Overview single optical slice showing the general codistribution of MAP1LC3A (green) and SH3GLB1 (red) in WT muscle. (B–E) Details from (A) showing exemplary colocalization patterns. While SH3GLB1 and CHRN colocalize perfectly and almost quantitatively, MAP1LC3A accompanies SH3GLB1- and CHRN-positive puncta either on 1 side (B), on 2 ends (C), or with more complex arrangements (D and E). (F and G) In atg7−/− muscle MAP1LC3A is mostly distributed on sarcomeric striations and does not accompany SH3GLB1 and CHRN double-positive vesicles. (H–K) Maximum z-projection of a double-transfected fiber at the level of the NMJ. Fluorescence signals as indicated on top right angle of panels. In the overlay (I), TRIM63 signals are shown in green, SH3GLB1 in red, CHRN in blue. Triple-positive signals are indicated with red arrowheads.
Proliferation of autophagic CHRN-containing structures upon atrophy induction is dependent on TRIM63
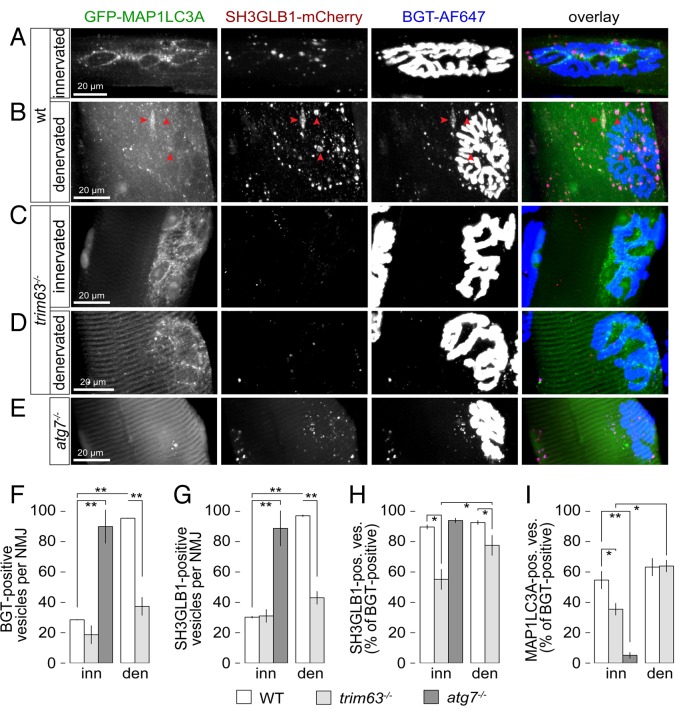
Next, we tested the hypothesis of an involvement of TRIM63 in the autophagic removal of CHRN by using trim63−/− mice. Thus, WT or trim63−/− mice were cotransfected with GFP-MAP1LC3A and SH3GLB1-mCherry. To discriminate between basal and atrophic states, hindlimb muscle atrophy was induced by sciatic denervation and 5 d later in vivo microscopy in the presence of the CHRN marker, BGT-AF647, was performed. First, quantitative analysis of the diameters of all visualized fibers showed significantly smaller numbers in denervated vs. innervated muscles of WT but not trim63−/− mice (Table 1). Second, in WT muscles, the amounts of endocytic CHRN-positive and SH3GLB1-positive puncta per NMJ region strongly increased upon denervation (Fig. 7A and B; Fig. 7F and G). In denervated WT muscles CHRN and SH3GLB1 puncta often aggregated in sac-like structures that were surrounded by a multitude of MAP1LC3A puncta (see red arrowheads in Fig. 7B), reminiscent of multivesicular bodies or amphisomes. 39 Quantitative analysis revealed a triplication of CHRN and SH3GLB1 punctate structures in WT muscles upon denervation as compared with the innervated control state (Fig. 7F and G). Colocalization of CHRN and SH3GLB1 was around 90% in innervated and denervated WT muscles (Fig. 7H) demonstrating a very strong link between both partners. About 60% of CHRN-positive puncta were accompanied by enhanced MAP1LC3A signals and this value did not vary between innervated and denervated muscles (Fig. 7I). Upon starvation, colocalization of CHRN-positive puncta with SH3GLB1 and MAP1LC3A was similar, although the increase in CHRN-positive vesicles was less pronounced as compared with denervated muscle (not shown). Notably, in trim63−/− muscles the denervation-induced massive increase in endocytic CHRN- and SH3GLB1-containing vesicles was completely absent (Fig. 7C and D; Fig. 7F and G). Conversely, the relative proportion of MAP1LC3A-capped structures was only mildly or not at all reduced (Fig. 7I). Colocalization between CHRN and SH3GLB1 was consistently reduced but not zeroed (Fig. 7H). In summary, these data demonstrate an important role of TRIM63 in the atrophy-induced but not the basal autophagy of CHRN. To further substantiate the hypothesis of an involvement of autophagy in CHRN removal we also worked with autophagy-deficient atg7−/− mice. Notably, the amounts of CHRN or SH3GLB1 positive carriers in these innervated muscles were comparable to the values in denervated WT muscles (Fig. 7F and G). While the absence of ATG7 had no impact on the cotargeting of CHRN and SH3GLB1 (Fig. 7H), the link between MAP1LC3A and CHRN puncta was grossly reduced (Fig. 7I). Together, these findings further corroborate an important role of autophagic processing in CHRN turnover.
Table 1. Effect of denervation on fiber diameters in tibialis anterior muscles.
| Fiber diameter (µm)a | SEM | n | |
|---|---|---|---|
| WT, innervated | 55.3 | 5.85 | 3 |
| WT, denervated | 33.3b | 5.44 | 3 |
| trim63 −/−, innervated | 54.5 | 8.80 | 3 |
| trim63 −/−, denervated | 53.1 | 3.48 | 3 |
a Maximal fiber diameters were obtained from confocal microscopy pictures of WT and trim63−/− mice that were denervated for 5 d and then imaged in vivo. b P < 0.05 for the difference between WT innervated vs. denervated, according to Welch test.
Figure 7. Endocytic CHRN carriers proliferate upon denervation in a TRIM63-dependent manner. Mouse tibialis anterior muscles of WT, trim63−/−, or atg7−/− mice were cotransfected with SH3GLB1-mCherry and GFP-MAP1LC3A. Five days before imaging, WT and trim63−/− muscles were unilaterally denervated. Twenty-four hours before in vivo confocal microscopy imaging CHRNs were stained with BGT-AF647. (A–E) Maximum z-projections of individual SH3GLB1 and MAP1LC3A double-positive fibers at the level of their NMJs from innervated (A, C, and E) or denervated muscles (B and D) of either WT (A and B), trim63−/− (C and D), or atg7−/− mice. Red arrowheads in (B) depict sac-like triple-positive structures, which resemble multivesicular bodies or amphisomes. (F–I) Quantitative analyses as indicated. All graphs depict mean ± SEM (WT and trim63−/−: n = 3 muscles, between 616 and 1415 structures were analyzed per condition. atg7−/−: n = 4 muscles, 2800 structures were analyzed). *P < 0.05; **P < 0.01 according to the Welch test.
Biochemical, immunofluorescence, and imaging evidence strengthen the view of a functional link between CHRN, TRIM63, and selective autophagy
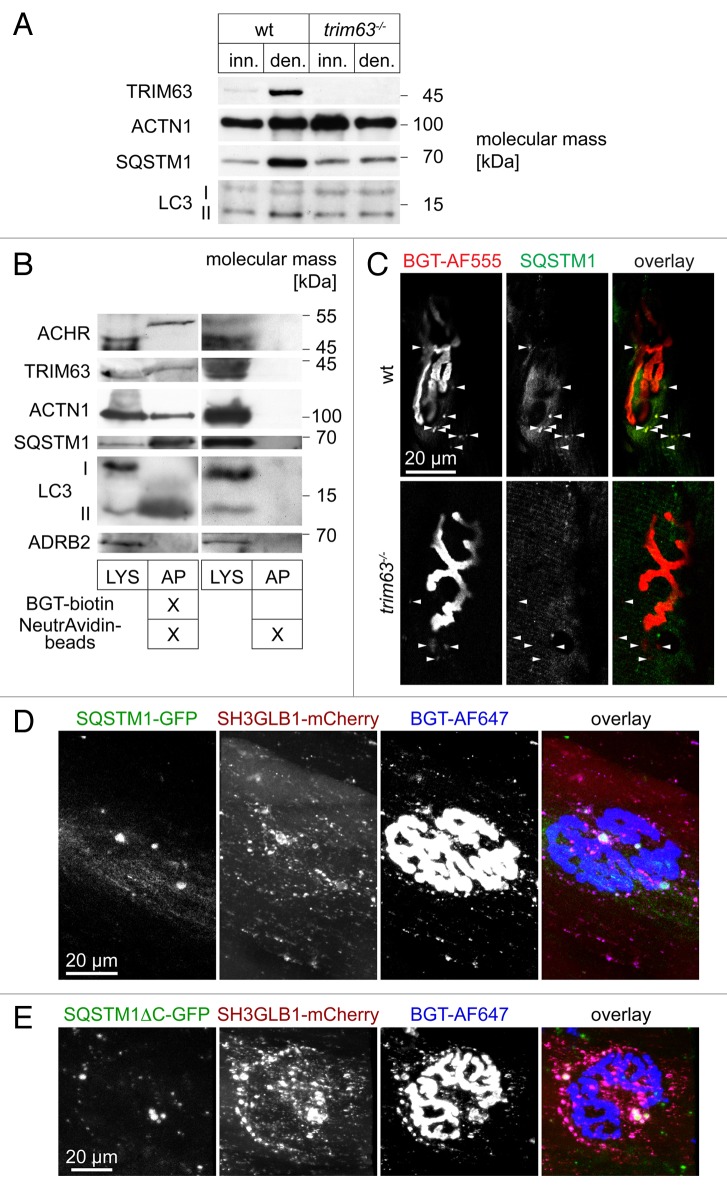
All previously shown data strongly suggest a central function of TRIM63 in autophagic removal of CHRN under atrophic conditions. To consolidate this assumption a set of additional ascertainments were performed. First, western blot analysis of innervated and denervated muscles showed that SQSTM1 and LC3-II are upregulated upon denervation in a TRIM63-dependent manner (Fig. 8A). Second, we performed affinity precipitation of endogenous surface-labeled CHRN using in vivo injection of BGT coupled to biotin. This revealed cosedimentation of TRIM63, ACTN1, SQSTM1 and LC3-II, but not of ADRB2 (negative control) (Fig. 8B). Third, immunostaining of denervated muscle slices yielded colocalization between endogenous SQSTM1 and CHRN-positive puncta in the presence but not in the absence of TRIM63 (Fig. 8C). Fourth, overexpression of heterologous SQSTM1-GFP in innervated WT muscles induced a massive increase in the total amount of SH3GLB1- and CHRN-positive carriers (Fig. 8D). Quantitative analysis detected 121.6 ± 11.2 (mean ± SEM; n = 3 muscles; 2674 structures analyzed) BGT-positive vesicles per NMJ, i.e., about 4 times more than under comparable conditions lacking SQSTM1-GFP (see Fig. 7F). Notably, vesicle number was again doubled to a value of 224.8 ± 20.1 (mean ± SEM; n = 4 muscles; 5973 structures analyzed) BGT-positive vesicles per NMJ in the presence of the mutant SQSTMΔC-GFP. This lacks both the ubiquitin-associated domain and LC3 interacting region and targets to the autophagosome formation site in a process involving self-oligomerization but not necessarily interaction with MAP1LC3A. 40
Figure 8. SQSTM1 is upregulated in a TRIM63-dependent manner, it colocalizes with endocytic CHRN and regulates CHRN carrier formation. (A) Representative western blot signals against TRIM63, ACTN1 (loading control), SQSTM1 and MAP1LC3A from lysates of WT or trim63−/− gastrocnemius muscles, that were either innervated (inn.) or denervated for 5 d (den.). (B) WT mouse gastrocnemius muscles were injected with BGT-biotin (left 2 lanes) or with saline (right 2 lanes). Five hours later, mice were sacrificed, and muscles harvested and lysed. Subsequently, BGT-biotin labeled CHRN were sedimented with NeutrAvidin-coupled beads. Depicted are representative western blot immunosignals using antibodies against CHRN, SQSTM1, MAP1LC3A, TRIM63, ACTN1 (positive control), and ADRB2 (negative control). LYS, lysate; AP, biotin-neutravidin affinity precipitate. (C) Denervated EDL muscles from WT and trim63−/− mice were sectioned and stained with BGT-AF555 against CHRN and with an antibody against SQSTM1. Shown are single confocal sections depicting parts of individual NMJs surrounded by some punctate CHRN-positive structures (arrowheads). While in the WT most of these puncta also exhibit SQSTM1 immunofluorescence signals, this is not the case in muscle lacking TRIM63. In the overlay pictures, BGT and anti-SQSTM1 signals appear in red and green, respectively. (D and E) WT tibialis anterior muscles were cotransfected with SH3GLB-mCherry and either SQSTM1-GFP (C) or SQSTM1ΔC-GFP (D). After 7 d, BGT-AF647 was injected and 24 h later, muscles were imaged in vivo. Shown are en face maximum z-projections. In the overlay pictures, GFP-, mCherry- and BGT-signals appear in green, red, and blue, respectively.
Discussion
Skeletal muscle atrophy is a process that can be triggered by a variety of factors including disuse, fasting, denervation, and aging, as well as by several diseases like cardiac and renal failure, diabetes, AIDS, or cancer. Skeletal muscle constitutes about 40% to 60% of our body mass and is also a major metabolic organ system. Therefore, chronic muscle atrophy is increasingly perceived as a major obstacle during the re-convalescence in an increasing and diverse group of patients. 41 , 42 While the most upstream triggers might vary between different atrophy induction processes, the basal protein network mediating protein breakdown that is consistently induced upon several forms of atrophic stimuli was previously identified and its constituents were called “atrogenes.” 31 A principal member of the atrogene family is the E3 ubiquitin ligase TRIM63, 30 and it was proposed to be a major driving force for the ubiquitin-proteasomal degradation of primarily sarcomeric muscle proteins. 19 However, this function is increasingly challenged by different observations. Exemplary is that the expression profile of TRIM63 does not in all cases match to observed muscle protein breakdown. 43 Furthermore, a recent report shows that proteasomal activity upon muscle denervation is actually increased and not decreased in the absence of TRIM63. 44
We have recently shown that endogenous TRIM63 is highly enriched at the nerve-muscle synapse, the neuromuscular junction, and heterologous expression of TRIM63-GFP is detected by in vivo microscopy in puncta colocalizing with internalized CHRN, the major postsynaptic ion channel of the NMJ. 28 Furthermore, since CHRN half-life upon denervation is significantly stabilized in trim63−/− mice and TRIM63 coprecipitates with CHRN, a crucial role of TRIM63 for CHRN turnover was claimed. 28 Despite the enormous physiological relevance of CHRN, deplorably little is known about the mechanisms driving its turnover. In particular, the pathway leading to the degradation of this pentameric ion channel remains largely elusive. Evidence for lysosomal degradation of CHRN comes from studies using iodinated protein flux 45 , 46 and from the effects of lysosomal inhibitors on CHRN levels in C2C12 muscle cells. 18 , 47 Given the recent discussions about the physiological roles of TRIM63 in muscle atrophy and the findings of its possible function in the turnover of the CHRN, we focused on this process in more detail.
First, we observed a direct impact of full starvation but not of amino acid depletion on the stability of CHRN (Figs. 2 and 3). Notably, TRIM63 protein was clearly more abundant upon total fasting but not upon amino acid deprivation (Fig. 3C). While amino acid deprivation is thought to primarily block protein synthesis through the mechanistic target of rapamycin complex 1 (MTORC1) via amino acid-sensing mechanisms, 33 full starvation negatively acts on the class I phosphoinositide 3-kinase-AKT/PKB pathway (thus blocking MTORC1) on the one hand, and acts to induce FOXO3 as a master switch in skeletal muscle atrophy on the other hand. 48 FOXO3, in turn, activates the atrogene program and, consequently, TRIM63 expression. 48 Notably, as revealed by in vivo imaging of mouse muscle the full starvation protocol significantly increased the amount of endocytic carriers containing CHRN per NMJ (Fig. 4), which were then also found to be largely positive for TRIM63 and a set of autophagic/lysosomal markers, including MAP1LC3A, SQSTM1, and LAMP1 (Figs. 5–7). While all other markers in this list typically showed point-to-point colocalization with CHRN signals, MAP1LC3A was rarely directly on top of the CHRN puncta. Rather, it was mostly accompanying the CHRN-positive puncta where it displayed punctual or crescent shapes. This is in perfect agreement with a function of MAP1LC3A in forming phagophores that are caught in the process of engulfment. Notably, such a localization pattern was almost entirely dependent on the presence of ATG7 (Figs. 6 and 7), suggesting that only cleaved and lipidated LC3 (LC3-II) interacts with CHRN containing carriers. One of the most frequent features was the “double cap” where MAP1LC3A signals were found at 2 ends of a CHRN-positive structure (see e.g., Fig. 4G and H). In summary, these findings demonstrate that CHRN is targeted into autophagic compartments.
Recently, SH3GLB1 was described to be intimately involved in the autophagic process. It was suggested to regulate ATG9 trafficking and to mediate fission of autophagic donor membranes to trigger starvation-induced autophagy. 37 , 49 , 50 Consistent with the in vitro interaction of SH3GLB1 and TRIM63 as suggested by yeast two-hybrid and pull-down experiments, we found SH3GLB1-GFP in close proximity to the NMJ in CHRN-containing puncta. 28 Most SH3GLB1- and CHRN-positive puncta were exactly overlaying with TRIM63 (Fig. 6) and accompanied by MAP1LC3A (Fig. 6). Notably, under control conditions nearly all SH3GLB1-positive puncta were also positive for CHRN, but only about 60% of these structures were surrounded by MAP1LC3A (Fig. 7). This suggests that in muscle SH3GLB1 is more likely codistributing with the autophagic cargo than with the autophagic donor membranes, although we cannot completely exclude that also the latter occurs. Strikingly, denervation led to a massive increase of SH3GLB1 and CHRN double-positive puncta (Fig. 7), which was almost completely blocked in trim63−/− mice (Fig. 7), showing that TRIM63 plays a crucial role in atrophy-induced endocytic retrieval of CHRN and its subsequent autophagic processing. This also provides an explanation for the stabilization of CHRN lifetime upon denervation in a trim63−/− background. 28 It is noteworthy that the rate of colocalization between SH3GLB1 and CHRN as well as between MAP1LC3A and CHRN was significantly decreased in the trim63−/− under control conditions. This is in agreement with earlier data that connected SH3GLB1 to endocytic processing rather than autophagy assistance, 51 - 53 and also suggests that TRIM63 controls a pool of autophagic carriers under unchallenged conditions.
Mechanistically, the data provided in this study suggest that CHRN is targeted to the LC3-II-positive autophagosome in a process of selective autophagy that is regulated and mediated by TRIM63, SH3GLB1, and SQSTM1. Yet, a couple of questions are still open. First, selective autophagy entails target ubiquitination. But what is the target? There are different possibilities, including CHRN, TRIM63 or other components. While TRIM63 is well known to autoubiquitinate itself, CHRN ubiquitination has been investigated so far only in cell cultures and this revealed smears of ubiquitinated protein. 26 , 27 Our own attempts used exclusively whole muscle extracts and both, precipitation of ubiquitinated proteins or of BGT-biotin surface-labeled CHRN followed by western blot against CHRN or polyubiquitinated protein, respectively (Fig. S1). This showed discrete bands, in particular one band at about 55-kDa molecular mass. It is unlikely that this band is unspecific, since it was found with 2 different methods and controls (e.g., test for unspecific binding to resin and for unspecific secondary antibody binding) were all negative. Thus, one can ask whether this represents mono- or oligoubiquitinated CHRN that could serve as a target for selective autophagy. Given that this band was also found in trim63−/− preparations (not shown) this option may seem rather improbable. However, it needs to be stressed that CHRN turnover appears to be regulated only upon atrophy by TRIM63, while basal turnover might be controlled by another E3 ligase. Unfortunately, our precipitation experiments were then not quantitative enough to demonstrate a TRIM63-dependent difference of the CHRN ubiquitination pattern between basal and atrophic conditions (not shown). Another point to be further investigated concerns the functions of SQSTM1 and SH3GLB1. The multiple conjectures described here between these 2 proteins and TRIM63 and CHRN very strongly suggest a direct role of both in the autophagic turnover of CHRN. Based on our data we speculate that SH3GLB1 is important in initial endocytic CHRN vesicle formation and then escorts the receptor trough multiple further steps. SQSTM1 also displayed a strong CHRN vesicle-inducing propensity, which was levered in the presence of the SQSTM1 mutant lacking the ubiquitin-associated domain and LC3 interacting region (Fig. 8). This suggests that SQSTM1 is involved in CHRN vesicle formation and might serve in progressing these carriers to the autophagosome. The increase in vesicle number in the presence of the SQSTM1ΔC mutant suggests that this progression is not completed and thus previously formed carriers accumulate.
In summary, using live mouse imaging, radiolabeling approaches, and biochemistry we here deliver evidence that CHRN degradation is dependent on the metabolic state of the muscle, and that this is likely mediated by selective autophagy, both under normal as well as under atrophic conditions. While TRIM63 is essential for this process upon atrophy induction, basal autophagy of CHRN seems to be only partially controlled by TRIM63 and might involve additional E3 ligases. Future studies should address their identity and further clarify the mechanistic interplay between TRIM63, SQSTM1, and SH3GLB1and functional regulation network that controls these interactions.
Materials and Methods
Animals
Experiments used a total of 105 adult male C57BL/10 J, trim63−/−, 29 and atg7−/− 38 mice with a typical weight between 30 and 36 g. Animals were maintained in the local animal facilities. Use and care of animals were as approved by German authorities and were according to national law (TierSchG7). Anesthesia was administered using either inhalation of Isoflurane (cp-pharma, AP/DRUGS/220/96) or intraperitoneal (i.p.) injection of Xylavet® 20 mg/ml (cp-pharma) and Zoletil® 100 (Laboratoires Virbac). For total starvation, litter was exchanged and food removed, water was supplied ad libitum. Since the effect of fasting on the reduction of mouse tibialis anterior muscle was described previously (loss of approximately 15% to 20% of cross-sectional area), 54 and we needed to maintain the animals after the fasting period for further analyses, muscle loss could not be demonstrated by standard cross-sectional area or muscle weight determinations, but was only indirectly addressed by weighing the animals before, during, and after the fasting period. Amino acid-deprivation experiments used ad libitum access to special protein-free chow (SSniff, E15200-14) or the corresponding control food (SSniff, E15000-40). Transfections and sciatic denervations were as previously described. 34 , 55 Loss of mouse skeletal muscle mass upon denervation was previously measured in other studies and accounted for about 30% (tibialis anterior muscle) and 56% (quadriceps muscle) after 6 19 and 14 d of denervation, 28 respectively. Although muscle weights were not taken in the present study, we determined fiber diameters for all measured fibers from in vivo confocal images. Therefore, the fiber width was determined at the biggest width in the image stacks showing GFP-MAP1LC3A signals. Furthermore, successful and persistent denervation was checked at the end of each experiment by careful visual inspection of the initially dissected pelvic region.
Biochemical analyses
For Figure 2, WT mice were, either, fed, starved for 48 h, or supplied with protein-free chow for 5 to 7 d. Subsequently, mice were killed, complete gastrocnemius muscles prepared, shock frozen in liquid nitrogen, and stored at −80 °C. Then, tissues were levigated under liquid nitrogen and homogenized in extraction buffer (8 M urea, 2 M thiourea, 3% SDS, 50 mM Tris/Cl pH 6.8, 0.015% bromophenol, 75 mM DTT). After incubation at 60 °C for 20 min, extracts were centrifuged for 10 min at 18,000 × g. Supernatant fractions were aliquoted and frozen at −80 °C until use. For western blots, samples were run on 4–12% BisTris-Gels (Invitrogen, NP0322BOX) and blotted onto nitrocellulose membranes. Detection of TRIM63 was performed with chicken anti-TRIM63 (Myomedix, MuRF1-3), and anti-chicken-alkaline phosphatase (Jackson, 303-055-003). For affinity coprecipitation (Fig. 8B; Fig. S1) gastrocnemius and tibialis anterior muscles were injected with biotinylated BGT (Invitrogen, B1196) and then surgically extracted 6 h later. Tissue lysis, affinity precipitation with NeutrAvidin agarose (Thermo Scientific, 29202), and western blot (Fig. 8B; Fig. S1) were all performed as described previously. 25 , 35 Antibody combinations used for detection of various proteins were as follows: chicken anti-TRIM63 (Myomedix, MuRF1-3) and anti-chicken-horseradish peroxidase (HRP) (Jackson, 303-035-003), guinea pig anti-SQSTM1 (Progen, GP62-C) and anti-guinea pig-HRP (Thermo Scientific, PA1-28597), rabbit anti-MAP1LC3A (Abgent, AP1802a) or rabbit anti-ADRB2 (Santa Cruz, Sc-569) and anti-rabbit-HRP (Daco, P0448), mouse anti-ACTN1 (Sigma Aldrich, A7811) or mouse anti-CHRN (BD Bioscience, Affinity precipitation, 610989) and anti-mouse-HRP (Daco, P0447). For pulldown of polyubiquitinated proteins, TUBEs kit purchased from LifeSensors (UM102) was used as per the supplier’s protocol. Western blot analysis was performed as for Figure S1. CHRN protein bands were detected using rat anti-CHRN antibody (Covance, MAb210) and anti-rat-HRP (Thermo Scientific, 31470). Polyubiquitin was probed using mouse anti-polyubiquitin antibody (MBL, MBL-D058-3) and anti-mouse-HRP antibody (DACO, P0447).
Radioiodine assay for measuring CHRN half-life and data analysis
The radioiodine approach was performed as previously described. 23 In brief, 0.7 pmol of 125I-BGT with high specific activity from Perkin Elmer (NEX126H050UC) was injected intramuscularly to the tibialis anterior muscle at day 0 and subsequent 125I-emission measurements from entire hind legs were performed under mild isoflurane anesthesia using liquid nitrogen-cooled GX-3018 (Canberra GmbH) and electrically cooled GEM-FX5825P4-S (AMETEK GmbH, ORTEC Division) Germanium semiconductor detectors.
In vivo visualization and measurement of CHRN turnover rate
CHRN turnover was measured as described previously. 25 , 34 , 35 In brief, infrared fluorescent BGT-AF647 (25 pmol, Life Technologies, B35450) and red fluorescent BGT-AF555 (25 pmol, Life Technologies, B35451) were sequentially injected at a temporal distance of 10 d, to label the old and new receptor pool respectively. After the second injection the superficial 200 µm of tibialis anterior muscles were examined in vivo with an upright Leica SP2 (Leica Microsystems) confocal microscope equipped with a 63×/1.2 NA water immersion objective. Automated analysis of CHRN turnover was done as previously described. 25 Briefly, 3D stacks at 512 × 512 pixel resolution were taken of BGT-AexaFluor647 (“old receptors”) and of BGT-AF555 signals (“new receptors”) using a 63× objective and confocal in vivo imaging. The 3D stacks were automatically segmented using a custom-made algorithm, and pixel signal intensity values for AF555 and AF647 were extracted. Then, the fraction of pixels per NMJ, having AF555 signal intensity higher than that of AF647 was calculated.
Immunofluorescence stainings
Denervated WT and trim63−/− EDL muscles were harvested and immediately fixed in freshly prepared 4% paraformaldehyde in phosphate-buffered saline (PBS, 137 mM, NaCl, 2.7 mM KCl, 10 mM Na2HPO4 × 2 H2O, 2 mM KH2PO4, pH 7.4). Muscles were embedded in 2% agarose gel and 50-μm thick sections were prepared using a vibratome (Leica VT1000S). Sections were then permeabilized using 0.5% Triton X-100 (ROTH, 3051.2) in PBS and blocked with 2% bovine serum albumin (GENTAUR, PAA-K41-001) in PBS. Primary antibody against SQSTM1 (Progen, GP62-C) was incubated over night at 4 °C, followed by thorough washing and incubation with anti-guinea pig-AF488 secondary antibody and BGT-AF555 (both LifeTechnologies, A-11073 and B-35451, respectively) for 8 h. Finally, sections were washed in 2% bovine serum albumin in PBS and then imbedded in MOWIOL (Calbiochem, 475904) on glass slides for confocal imaging.
Plasmids, in vivo transfection, and endocytic CHRN pool labeling
Constructs encoding heterologous fusion proteins EGFP-MAP1LC3A and LAMP1-EGFP, SH3GLB1-mCherry, TRIM63-mCherry, SQSTM1-EGFP (Addgene, 38277 40 ), and SQSTM1ΔC-EGFP (Addgene, 38282 40 ) were transfected into the tibialis anterior muscle as described. 34 , 55 Eight days later, the transfected muscles were exposed and superficial parts of the muscles were visualized under the microscope, but 24 h prior to microscopy, the transfected muscles were injected either with BGT-AF647 (for EGFP and mCherry-fusion protein-cotransfected muscles) or BGT-AF555 (for EGFP-fusion protein-transfected muscles) to label the surface exposed receptors, some of which are subsequently endocytosed.
Data processing and statistics
Images were electronically processed using ImageJ software (NIH). For quantitative analysis of vesicle numbers and colocalizations, the following procedure was used. First, images were cropped to just leave the NMJ-spanning width of a fiber complete. Then, the channel containing SH3GLB1-mCherry signals was median filtered (kernel 1 × 1) and thresholded to just exclude the background. Next, all objects larger than 10 pixels were segmented and imported into the ROI manager, visual inspection excluded unspecific signals or signals from neighboring fibers. Subsequently, the channel containing CHRN signals was screened for additional CHRN-positive structures and these ROIs were back-screened in the SH3GLB1 channel. This procedure yielded total amounts of CHRN and SH3GLB1 positive structures. Finally, all segments were screened in all channels for local maxima to yield the quantity of structures being positive for CHRN, SH3GLB1 and/or MAP1LC3A. In the case of MAP1LC3A, those structures were counted as positive which clearly displayed dot-, crescent- or circular-shaped local maxima fitting to the segment. Image composition used Adobe Photoshop and Adobe Illustrator (both Adobe Systems Software), and ImageJ. Graphic representation of data was achieved by Microsoft Excel:mac2008 and subsequent incorporation into Adobe Illustrator composites. Statistical analysis employed SigmaPlot v.12 (Systat Software Inc.) for calculation of regressions of radioiodine experiments. Significance was tested with the Student t test or Welch test, where applicable. Therefore, Kolmogorov-Smirnow-test for normal distribution and F-test for homo/heteroscedasticity were performed.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
RR and SL are supported by DFG (grant RU923-7/1, LA668-15/1) and by the EU-FP7 (SarcoSi).
Glossary
Abbreviations:
- AF
AlexaFluor®
- CHRN
cholinergic receptor, nicotinic
- ATG7
autophagy-related 7
- BGT
α-bungarotoxin
- GFP
green fluorescent protein
- LAMP1
lysosomal-associated membrane protein 1
- MAP1LC3A
microtubule-associated protein 1 light chain 3A
- NMJ
neuromuscular junction
- PBS
phosphate-buffered saline
- SEM
standard error of the mean
- SH3GLB1
SH3-domain GRB2-like endophilin B1
- SQSTM1
sequestosome 1
- SQSTM1 ΔC
sequestosome 1 lacking ubiquitin-associated domain and LC3 interacting region
- TRIM63
tripartite motif containing 63
- E3
ubiquitin protein ligase
- WT
wild type
References
- 1. Shaid S, Brandts CH, Serve H, Dikic I. . Ubiquitination and selective autophagy. Cell Death Differ 2013; 20:21 - 30; http://dx.doi.org/ 10.1038/cdd.2012.72; PMID: 22722335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. . p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171:603 - 14; http://dx.doi.org/ 10.1083/jcb.200507002; PMID: 16286508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. . A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009; 33:505 - 16; http://dx.doi.org/ 10.1016/j.molcel.2009.01.020; PMID: 19250911 [DOI] [PubMed] [Google Scholar]
- 4. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. . p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131 - 45; http://dx.doi.org/ 10.1074/jbc.M702824200; PMID: 17580304 [DOI] [PubMed] [Google Scholar]
- 5. Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K. . Alteration of autophagosomal proteins (LC3, GABARAP and GATE-16) in Lewy body disease. Neurobiol Dis 2011; 43:690 - 7; http://dx.doi.org/ 10.1016/j.nbd.2011.05.022; PMID: 21684337 [DOI] [PubMed] [Google Scholar]
- 6. Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, et al. . Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 2010; 11:45 - 51; http://dx.doi.org/ 10.1038/embor.2009.256; PMID: 20010802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein Á, Bloor S, Rutherford TJ, Freund SM, Komander D, Randow F. . An essential role for the ATG8 ortholog LC3C in antibacterial autophagy. Autophagy 2013; 9:784 - 6; http://dx.doi.org/ 10.4161/auto.23698; PMID: 23434839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shehata M, Matsumura H, Okubo-Suzuki R, Ohkawa N, Inokuchi K. . Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J Neurosci 2012; 32:10413 - 22; http://dx.doi.org/ 10.1523/JNEUROSCI.4533-11.2012; PMID: 22836274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karlin A. . Emerging structure of the nicotinic acetylcholine receptors. Nat Rev Neurosci 2002; 3:102 - 14; http://dx.doi.org/ 10.1038/nrn731; PMID: 11836518 [DOI] [PubMed] [Google Scholar]
- 10. Fambrough DM. . Control of acetylcholine receptors in skeletal muscle. Physiol Rev 1979; 59:165 - 227; PMID: 375254 [DOI] [PubMed] [Google Scholar]
- 11. Green WN, Millar NS. . Ion-channel assembly. Trends Neurosci 1995; 18:280 - 7; PMID: 7571003 [PubMed] [Google Scholar]
- 12. Millar NS, Harkness PC. . Assembly and trafficking of nicotinic acetylcholine receptors (Review). Mol Membr Biol 2008; 25:279 - 92; http://dx.doi.org/ 10.1080/09687680802035675; PMID: 18446614 [DOI] [PubMed] [Google Scholar]
- 13. Engel AG, Lindstrom JM, Lambert EH, Lennon VA. . Ultrastructural localization of the acetylcholine receptor in myasthenia gravis and in its experimental autoimmune model. Neurology 1977; 27:307 - 15; http://dx.doi.org/ 10.1212/WNL.27.4.307; PMID: 557772 [DOI] [PubMed] [Google Scholar]
- 14. Fumagalli G, Engel AG, Lindstrom J. . Ultrastructural aspects of acetylcholine receptor turnover at the normal end-plate and in autoimmune myasthenia gravis. J Neuropathol Exp Neurol 1982; 41:567 - 79; http://dx.doi.org/ 10.1097/00005072-198211000-00001; PMID: 6982313 [DOI] [PubMed] [Google Scholar]
- 15. Akaaboune M, Culican SM, Turney SG, Lichtman JW. . Rapid and reversible effects of activity on acetylcholine receptor density at the neuromuscular junction in vivo. Science 1999; 286:503 - 7; http://dx.doi.org/ 10.1126/science.286.5439.503; PMID: 10521340 [DOI] [PubMed] [Google Scholar]
- 16. Bruneau E, Sutter D, Hume RI, Akaaboune M. . Identification of nicotinic acetylcholine receptor recycling and its role in maintaining receptor density at the neuromuscular junction in vivo. J Neurosci 2005; 25:9949 - 59; http://dx.doi.org/ 10.1523/JNEUROSCI.3169-05.2005; PMID: 16251443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bruneau EG, Esteban JA, Akaaboune M. . Receptor-associated proteins and synaptic plasticity. FASEB J 2009; 23:679 - 88; http://dx.doi.org/ 10.1096/fj.08-107946; PMID: 18978155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Libby P, Bursztajn S, Goldberg AL. . Degradation of the acetylcholine receptor in cultured muscle cells: selective inhibitors and the fate of undegraded receptors. Cell 1980; 19:481 - 91; http://dx.doi.org/ 10.1016/0092-8674(80)90523-1; PMID: 7357615 [DOI] [PubMed] [Google Scholar]
- 19. Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, et al. . Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 2001; 294:1704 - 8; http://dx.doi.org/ 10.1126/science.1065874; PMID: 11679633 [DOI] [PubMed] [Google Scholar]
- 20. Yampolsky P, Pacifici PG, Witzemann V. . Differential muscle-driven synaptic remodeling in the neuromuscular junction after denervation. Eur J Neurosci 2010; 31:646 - 58; http://dx.doi.org/ 10.1111/j.1460-9568.2010.07096.x; PMID: 20148944 [DOI] [PubMed] [Google Scholar]
- 21. Levitt TA, Loring RH, Salpeter MM. . Neuronal control of acetylcholine receptor turnover rate at a vertebrate neuromuscular junction. Science 1980; 210:550 - 1; http://dx.doi.org/ 10.1126/science.7423205; PMID: 7423205 [DOI] [PubMed] [Google Scholar]
- 22. Stanley EF, Drachman DB. . Denervation accelerates the degradation of junctional acetylcholine receptors. Exp Neurol 1981; 73:390 - 6; http://dx.doi.org/ 10.1016/0014-4886(81)90274-0; PMID: 7262244 [DOI] [PubMed] [Google Scholar]
- 23. Strack S, Petersen Y, Wagner A, Röder IV, Albrizio M, Reischl M, Wacker IU, Wilhelm C, Rudolf R. . A novel labeling approach identifies three stability levels of acetylcholine receptors in the mouse neuromuscular junction in vivo. PLoS One 2011; 6:e20524; http://dx.doi.org/ 10.1371/journal.pone.0020524; PMID: 21655100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nelson PG, Lanuza MA, Jia M, Li MX, Tomas J. . Phosphorylation reactions in activity-dependent synapse modification at the neuromuscular junction during development. J Neurocytol 2003; 32:803 - 16; http://dx.doi.org/ 10.1023/B:NEUR.0000020625.70284.a6; PMID: 15034269 [DOI] [PubMed] [Google Scholar]
- 25. Röder IV, Choi KR, Reischl M, Petersen Y, Diefenbacher ME, Zaccolo M, Pozzan T, Rudolf R. . Myosin Va cooperates with PKA RIalpha to mediate maintenance of the endplate in vivo. Proc Natl Acad Sci U S A 2010; 107:2031 - 6; http://dx.doi.org/ 10.1073/pnas.0914087107; PMID: 20133847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Christianson JC, Green WN. . Regulation of nicotinic receptor expression by the ubiquitin-proteasome system. EMBO J 2004; 23:4156 - 65; http://dx.doi.org/ 10.1038/sj.emboj.7600436; PMID: 15483627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mouslim C, Aittaleb M, Hume RI, Akaaboune M. . A role for the calmodulin kinase II-related anchoring protein (αkap) in maintaining the stability of nicotinic acetylcholine receptors. J Neurosci 2012; 32:5177 - 85; http://dx.doi.org/ 10.1523/JNEUROSCI.6477-11.2012; PMID: 22496563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rudolf R, Bogomolovas J, Strack S, Choi KR, Khan MM, Wagner A, Brohm K, Hanashima A, Gasch A, Labeit D, et al. . Regulation of nicotinic acetylcholine receptor turnover by MuRF1 connects muscle activity to endo/lysosomal and atrophy pathways. Age (Dordr) 2013; 35:1663 - 74; http://dx.doi.org/ 10.1007/s11357-012-9468-9; PMID: 22956146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Witt CC, Witt SH, Lerche S, Labeit D, Back W, Labeit S. . Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO J 2008; 27:350 - 60; http://dx.doi.org/ 10.1038/sj.emboj.7601952; PMID: 18157088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, Bang ML, Trombitas K, Granzier H, Gregorio CC, et al. . Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J Mol Biol 2001; 306:717 - 26; http://dx.doi.org/ 10.1006/jmbi.2001.4448; PMID: 11243782 [DOI] [PubMed] [Google Scholar]
- 31. Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL. . Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J 2004; 18:39 - 51; http://dx.doi.org/ 10.1096/fj.03-0610com; PMID: 14718385 [DOI] [PubMed] [Google Scholar]
- 32. Sandri M. . Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 2008; 23:160 - 70; http://dx.doi.org/ 10.1152/physiol.00041.2007; PMID: 18556469 [DOI] [PubMed] [Google Scholar]
- 33. Hietakangas V, Cohen SM. . Regulation of tissue growth through nutrient sensing. Annu Rev Genet 2009; 43:389 - 410; http://dx.doi.org/ 10.1146/annurev-genet-102108-134815; PMID: 19694515 [DOI] [PubMed] [Google Scholar]
- 34. Röder IV, Petersen Y, Choi KR, Witzemann V, Hammer JA 3rd, Rudolf R. . Role of Myosin Va in the plasticity of the vertebrate neuromuscular junction in vivo. PLoS One 2008; 3:e3871; http://dx.doi.org/ 10.1371/journal.pone.0003871; PMID: 19057648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Choi KR, Berrera M, Reischl M, Strack S, Albrizio M, Röder IV, Wagner A, Petersen Y, Hafner M, Zaccolo M, et al. . Rapsyn mediates subsynaptic anchoring of PKA type I and stabilisation of acetylcholine receptor in vivo. J Cell Sci 2012; 125:714 - 23; http://dx.doi.org/ 10.1242/jcs.092361; PMID: 22331361 [DOI] [PubMed] [Google Scholar]
- 36. Klionsky DJ. . For the last time, it is GFP-Atg8, not Atg8-GFP (and the same goes for LC3). Autophagy 2011; 7:1093 - 4; http://dx.doi.org/ 10.4161/auto.7.10.15492; PMID: 21993240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mulé JJ, et al. . Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 2007; 9:1142 - 51; http://dx.doi.org/ 10.1038/ncb1634; PMID: 17891140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. . Autophagy is required to maintain muscle mass. Cell Metab 2009; 10:507 - 15; http://dx.doi.org/ 10.1016/j.cmet.2009.10.008; PMID: 19945408 [DOI] [PubMed] [Google Scholar]
- 39. Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerød L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. . Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol 2007; 179:485 - 500; http://dx.doi.org/ 10.1083/jcb.200702115; PMID: 17984323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Itakura E, Mizushima N. . p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol 2011; 192:17 - 27; http://dx.doi.org/ 10.1083/jcb.201009067; PMID: 21220506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Forbes SC, Little JP, Candow DG. . Exercise and nutritional interventions for improving aging muscle health. Endocrine 2012; 42:29 - 38; http://dx.doi.org/ 10.1007/s12020-012-9676-1; PMID: 22527891 [DOI] [PubMed] [Google Scholar]
- 42. Adegoke OA, Abdullahi A, Tavajohi-Fini P. . mTORC1 and the regulation of skeletal muscle anabolism and mass. Appl Physiol Nutr Metab 2012; 37:395 - 406; http://dx.doi.org/ 10.1139/h2012-009; PMID: 22509811 [DOI] [PubMed] [Google Scholar]
- 43. Foletta VC, White LJ, Larsen AE, Léger B, Russell AP. . The role and regulation of MAFbx/atrogin-1 and MuRF1 in skeletal muscle atrophy. Pflugers Arch 2011; 461:325 - 35; http://dx.doi.org/ 10.1007/s00424-010-0919-9; PMID: 21221630 [DOI] [PubMed] [Google Scholar]
- 44. Gomes AV, Waddell DS, Siu R, Stein M, Dewey S, Furlow JD, Bodine SC. . Upregulation of proteasome activity in muscle RING finger 1-null mice following denervation. FASEB J 2012; 26:2986 - 99; http://dx.doi.org/ 10.1096/fj.12-204495; PMID: 22508689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hubbard AL, Cohn ZA. . Externally disposed plasma membrane proteins. II. Metabolic fate of iodinated polypeptides of mouse L cells. J Cell Biol 1975; 64:461 - 79; http://dx.doi.org/ 10.1083/jcb.64.2.461; PMID: 163834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hubbard AL, Cohn ZA. . Externally disposed plasma membrane proteins. I. Enzymatic iodination of mouse L cells. J Cell Biol 1975; 64:438 - 60; http://dx.doi.org/ 10.1083/jcb.64.2.438; PMID: 163833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Valkova C, Albrizio M, Röder IV, Schwake M, Betto R, Rudolf R, Kaether C. . Sorting receptor Rer1 controls surface expression of muscle acetylcholine receptors by ER retention of unassembled alpha-subunits. Proc Natl Acad Sci U S A 2011; 108:621 - 5; http://dx.doi.org/ 10.1073/pnas.1001624108; PMID: 21187406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. . Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004; 117:399 - 412; http://dx.doi.org/ 10.1016/S0092-8674(04)00400-3; PMID: 15109499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takahashi Y, Meyerkord CL, Wang HG. . Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ 2009; 16:947 - 55; http://dx.doi.org/ 10.1038/cdd.2009.19; PMID: 19265852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Takahashi Y, Meyerkord CL, Hori T, Runkle K, Fox TE, Kester M, Loughran TP, Wang HG. . Bif-1 regulates Atg9 trafficking by mediating the fission of Golgi membranes during autophagy. Autophagy 2011; 7:61 - 73; http://dx.doi.org/ 10.4161/auto.7.1.14015; PMID: 21068542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cheung ZH, Ip NY. . Endophilin B1: Guarding the gate to destruction. Commun Integr Biol 2009; 2:130 - 2; PMID: 19704909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang C, Li A, Gao S, Zhang X, Xiao H. . The TIP30 protein complex, arachidonic acid and coenzyme A are required for vesicle membrane fusion. PLoS One 2011; 6:e21233; http://dx.doi.org/ 10.1371/journal.pone.0021233; PMID: 21731680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang C, Li A, Zhang X, Xiao H. . A novel TIP30 protein complex regulates EGF receptor signaling and endocytic degradation. J Biol Chem 2011; 286:9373 - 81; http://dx.doi.org/ 10.1074/jbc.M110.207720; PMID: 21252234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, Milan G, Masiero E, Del Piccolo P, Foretz M, et al. . Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J 2010; 29:1774 - 85; http://dx.doi.org/ 10.1038/emboj.2010.60; PMID: 20400940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Donà M, Sandri M, Rossini K, Dell’Aica I, Podhorska-Okolow M, Carraro U. . Functional in vivo gene transfer into the myofibers of adult skeletal muscle. Biochem Biophys Res Commun 2003; 312:1132 - 8; http://dx.doi.org/ 10.1016/j.bbrc.2003.11.032; PMID: 14651990 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.