Abstract
Much recent and ongoing research is focused on understanding the mechanisms and regulation of autophagy, a cellular self-degradation pathway with many links to human health. Although many assays exist to measure the total magnitude of autophagy, electron microscopy remains the tool of choice for the determination of the size and the number of autophagosomes formed in a given mutant or under given induction conditions. Here we present a detailed protocol for measuring autophagic bodies in the yeast Saccharomyces cerevisiae by electron microscopy. Furthermore, we present an improved mathematical method for estimating body size and a new method for estimating body number. Finally, we include a discussion of the merits and limitations of these methods and an example of their application to autophagic bodies formed in the ume6∆ strain.
Keywords: autophagosome, autophagy, electron microscopy, stereology, yeast, vacuole
Introduction
Macroautophagy (hereafter called autophagy) is a conserved pathway by which eukaryotic cells transport cytoplasmic components to the lysosome/vacuole for degradation. Starvation-induced autophagy begins with the formation of a double-membrane compartment, the phagophore, that expands into a vesicle that in baker’s yeast, Saccharomyces cerevisiae, is ~200 to 500 nm in diameter, 1 termed an autophagosome, which nonselectively envelops bulk cytoplasm. The outer membrane of the autophagosome fuses with the vacuole, delivering the inner vesicle, subsequently termed an autophagic body, into its lumen where it and its contents are degraded by resident proteases and lipases.
Autophagy is a protective response against a variety of cellular stresses, and as such plays a role in many important diseases. 2 Therefore, there is great interest in understanding both the basic mechanisms and the regulatory control of autophagy. Yeast has emerged as one of the primary model systems for the study of autophagy, largely due to its genetic tractability; most known autophagy-related (ATG) genes were first discovered in yeast, and it remains a major tool for the continued study of autophagic regulation. 3
One key aspect of the regulatory process is the magnitude of autophagic flux—that is, how much autophagy is actually taking place. Autophagic flux is regulated by many factors, which can potentially act either by controlling autophagosome size, autophagosome number, or both. Although a variety of assays can determine overall autophagic flux, electron microscopy is the best way to determine whether a change in flux is due to a change in the size or the number of the autophagosomes formed.
The mechanisms controlling autophagosome size are largely unknown, although recent theoretical work suggests that it could depend on the rate of membrane addition to the expanding phagophore as compared with the rate of its spontaneous closure into a double-membrane vesicle. 4 One factor that controls autophagosome size is Atg8, whose expression levels correlate directly with the size of autophagosomes formed. 5 Atg17 is also necessary for the formation of normal-sized autophagosomes, possibly due to its stimulation of Atg1 kinase activity. 6 , 7 Understanding the regulatory mechanisms that control autophagosome size, and which pathways feed into size control vs. number control, will be a key step in advancing our overall knowledge of autophagic regulation.
It is relatively difficult to measure the size of autophagosomes directly, both because of the low abundance of completed autophagosomes that have not yet fused with the vacuole as well as the challenge of preserving and identifying membrane structures in the cytoplasm. Instead, it is customary to use an indirect method: measuring the size of autophagic bodies inside the vacuole. Normally, autophagic bodies are quickly degraded; however, when vacuolar degradation is blocked such as by deletion of the gene encoding a key activating protease, Pep4, autophagic bodies accumulate in the vacuole. These bodies can be easily visualized by electron microscopy (EM) after a simple chemical fixation, and their size and abundance provides a measure of the size and number of autophagosomes that were formed. 5 Although this measure is somewhat indirect, and there is the possibility that the chemical fixation may affect the apparent size of the body, it nonetheless should give a reasonable estimate of autophagosome size, and is particularly useful for comparing the relative sizes of autophagosomes formed in wild-type and mutant strains when processed in parallel.
The structures that can be seen in an electron micrograph do not represent the entire autophagic body, but instead are a random section through a larger structure. Therefore, the average observed radius of the bodies in the micrograph does not correspond to the actual average radius of original autophagic bodies. Instead, sectioning introduces a number of biases, such as: 1
1) The section most likely was not taken through the equator of the original body, but instead nearer to one of the poles. This will lead to an underestimation of the size of the original body.
2) Larger bodies are more likely to be sampled by a random section than are smaller bodies. This will lead to an overestimation of the average size of the original bodies.
3) There is a minimum size for a body section that can be accurately identified in a micrograph; smaller body sections will be missed, leading to an overestimation of the average size of the original bodies.
Mathematical methods to correct for these biases and estimate the size of the original bodies have been previously described. 1 , 8 Here we present improved versions of these methods that are more accurate and easier to use, and extend them to also provide an estimate of the number of original bodies. We also present a detailed protocol, which includes instructions for 1) performing EM, 2) analyzing the images, and 3) using the mathematical methods (with templates provided as Microsoft Excel files) to estimate autophagic body size and number. The discussion section includes a comparison of the improved methods with those described previously, along with recommendations regarding sample size. Finally, we give an example of how to use these methods by reanalyzing the larger autophagic bodies formed during nitrogen starvation in yeast lacking the transcriptional repressor Ume6. 9
1. Materials
1.1 Fixation and embedding of yeast samples for EM
1.1.1 General equipment
1. Yeast strain with PEP4 (and optionally VPS4) deleted (see Note 1).
2. Shaking incubator set at 30 °C, 250 RPM for growing yeast cultures.
3. Spectrophotometer for measuring absorbance at 600 nm (A600).
4. Low-speed centrifuge with a minimum 50-ml capacity for harvesting cultures.
5. Microcentrifuge for pelleting cells during fixation and embedding.
6. Millipore “Milli-Q” or equivalent water filtration system (0.22-µm pore size), or 0.22-µm syringe filters (Fisher Scientific, 05-713-386), for preparing particulate-free purified water.
7. Nutating mixer (Fisher Scientific, 22-363-152) or rotator for gentle agitation of 1.5 ml tubes. Nutation is preferred to end-over-end rotation because it is gentler.
8. EM facility with staff to section samples and help with imaging.
1.1.2 Culture media
1. YPD (growth medium): 1% yeast extract (ForMedium, YEM04), 2% peptone (ForMedium, PEP04), 2% glucose.
2. SD-N (nitrogen starvation medium): 0.17% yeast nitrogen base without amino acids and without ammonium sulfate (ForMedium, CYN0501), 2% glucose.
1.1.3 Fixation
1.5% KMnO4 in water, APHA grade (Lab Chem, LC199401).
2. 1.5 ml microcentrifuge tubes.
1.1.4 Dehydration and embedding
1. An unopened bottle of acetone (Fisher Scientific, A18-4), ACS-grade or EM-grade (see Note 2).
2. Low Viscosity Spurr Kit (Ted Pella, 18300-4221) or its equivalent, containing nonenyl succinic anhydride (Ted Pella, 18301), ERL 4221 epoxide resin (Ted Pella, 18306-4221), diglycidyl ether of poly (propylene glycol) 736 (Ted Pella, 18310), and dimethylaminoethanol (Ted Pella, 18315).
3. Thin-walled 0.65 ml PCR tubes (Denville Scientific, C18065).
1.2 Image analysis
1. Adobe Photoshop (Adobe Systems) or GIMP (http://www.gimp.org/downloads/).
2. ImageJ (http://rsbweb.nih.gov/ij/download.html).
1.3 Estimation of original autophagic body size and number
1.3.1 Size estimation
1. Microsoft Excel.
2. Excel workbook “Size_Estimation.xlsx” (File SX1).
1.3.2 Number estimation
1. Excel workbook “Number_Estimation.xlsx” (File SX2).
2. Methods
2.1 Fixation and embedding of yeast samples for EM
2.1.1 Culture
1. Inoculate 5 ml of YPD medium with a single colony of the pep4Δ vps4Δ genotype from a freshly streaked YPD plate (see Note 3). Grow this starter culture 6–8 h, at 30 °C, with shaking (250 rpm), without allowing the culture to reach saturation (see Note 4).
2. Use this culture to inoculate a 30 ml YPD culture to a starting A600 = 0.0001. Grow at 30 °C, with shaking.
3. When culture has reached A600 = 1.0 (approximately 16 h; See Note 5), transfer the cells to a 50-ml conical tube and harvest by centrifugation for 5 min, at 1600 × g.
4. Resuspend the cells in 30 ml sterile water to wash; harvest again by centrifugation.
5. Resuspend the cells in 30 ml SD-N for starvation, and grow at 30 °C with shaking for 1–4 h, as desired (See Note 6).
6. Harvest the cells by centrifugation for 5 min, at 1600 × g.
7. Resuspend the cells by pipetting up and down in 30 ml purified water prewarmed to 30 °C (See Note 7).
8. Repeat steps 6 and 7 for an additional wash step, then harvest again by centrifugation.
2.1.2 Fixation
1. Prepare a 1.5% KMnO4 solution by diluting the 5% stock to a final concentration of 1.5% v/v in purified water and passing the solution through a syringe filter.
2. Resuspend the cells by pipetting up and down in 1 ml of freshly prepared 1.5% KMnO4 solution (see Note 8). Transfer the cell suspension to a 1.5 ml centrifuge tube.
3. Add additional 1.5% KMnO4 solution until the tube is entirely filled and air is excluded as much as possible.
4. Incubate for 30 min at 4 °C with gentle agitation (e.g., nutation).
5. Harvest the cells by centrifugation at 1500 × g for 3 min. Remove the supernatant fraction by decanting and discard it.
6. Repeat steps 2 and 3 with a new 1.5 ml 1.5% KMnO4 solution.
7. Incubate overnight at 4 °C with gentle agitation.
2.1.3 Dehydration and embedding
1. Harvest the cells by centrifugation for 3 min at 1500 × g and remove the supernatant fraction by decanting.
2. Resuspend the pellet in 1 ml purified water by pipetting up and down. Collect the cells by centrifugation for 3 min at 1500 × g and remove the supernatant fraction by suction.
3. Repeat step 2, 4 additional times to wash.
4. Resuspend the cells by pipetting up and down in 1 ml 10% v/v acetone/water, incubate 20 min at room temperature with gentle nutation, then harvest as in step 2.
5. Repeat step 4 with 1 ml 30% v/v acetone/water, then 50%, 70%, 90%, and 95% v/v acetone/water.
6. Resuspend the cells by pipetting up and down in 1 ml 100% dry acetone (see Note 2), incubate 20 min at room temperature with gentle nutation, then harvest as in step 2.
7. Repeat step 6, 2 additional times to remove all traces of water.
8. Make fresh Spurr resin by weighing out the following components into a 50-ml conical tube: 8.2 g ERL 4221, 1.9 g diglycidyl ether of poly (propylene glycol) 736, 11.8 g nonenyl succinic anhydride, and 0.2 g dimethylaminoethanol. 10 Mix with a disposable pipette and protect from light; store at room temperature for up to 2 d. (This recipe makes enough resin to process ~6 samples). Spurr resin is very toxic! See Note 9 for safe handling and disposal instructions.
9. Resuspend the cells by pipetting up and down in 1 ml 20% v/v Spurr resin/acetone. Incubate at least 1 h at room temperature with gentle agitation. Harvest by centrifugation at 2400 × g for 3 min, and remove resin by pipette to a disposal container.
10. Repeat step 9, using 50% v/v Spurr resin/acetone.
11. Resuspend the cells by pipetting up and down in 1 ml 100% Spurr resin. Incubate at least 8 h at room temperature with gentle agitation. Harvest by centrifugation at 2400 × g for 5 min, and remove resin by pipette to a disposal container.
12. Repeat step 11 with a new aliquot of 100% Spurr resin.
13. Resuspend the cells in ~0.5 ml of 100% Spurr resin, and transfer to a 0.65-ml thin-walled PCR tube. Be careful to avoid introducing bubbles into the resin during the resuspension or transfer, and ensure that the resin entirely fills the bottom of the tube. Add additional resin until the tube is full.
14. Pellet cells by centrifugation at 2400 × g for 5 min, but do not remove the resin.
15. Incubate the tube for 12–16 h at 70 °C in a PCR machine with a heated lid or a drying oven to polymerize the resin.
16. Use a razor blade to cut away the 0.65 ml PCR tube and release the polymerized block.
2.1.4 Sectioning, staining, and imaging
1. The above procedure should result in a conical-shaped block with the cells concentrated in a pellet at the pointed end. A trained electron microscopist should be able to trim, face, and section this block by standard procedures without much difficulty. We recommend 70-nm sections.
2. Sections should be stained for 5 min each with 1% aqueous uranyl acetate followed by Reynolds lead citrate using standard procedures. This step can also be performed by any trained electron microscopist.
3. Any standard biological electron microscope can be used for imaging the cells. In order to get a representative sample, the microscopist should choose a region to image, and take images of every cell with a visible vacuole within that region (see Note 10). Do not include cells where no vacuole is visible, but do include cells with no autophagic bodies. Make sure that all of the images are taken at the same magnification (we recommend ~10000×), and that a scale bar is included on the image or you have some other way of knowing the exact magnification.
4. If you also want to estimate the number of autophagic bodies, you will need an estimate of the mean size of the vacuole. For this, it is helpful to also take some images at a lower magnification (we recommend ~1000×).
2.2 Image analysis
2.2.1 Outlining autophagic bodies
See also Video SV1 (Tutorial 1: Outlining Autophagic Bodies).
1. Keep a backup copy of the original images in a separate folder/disk. Open an image file in Photoshop. (This entire procedure can also be performed in GIMP, although the software interface is slightly different.)
2. Create a new layer (ctrl+shift+n).
3. Using the brush tool (see Note 11), outline each autophagic body. Autophagic bodies are defined as structures inside the vacuole with a density similar to that of the cytoplasm and a dark membrane boundary (see Note 12). Care must be taken to outline each body completely, without any gaps. Also, the outlines must not be touching each other, but instead separated by empty space. Since the autophagic bodies themselves are usually touching, the layer on which the outlines are being drawn should be shifted after each outline is completed to make a clear space for the next body to be outlined (Fig. 1).
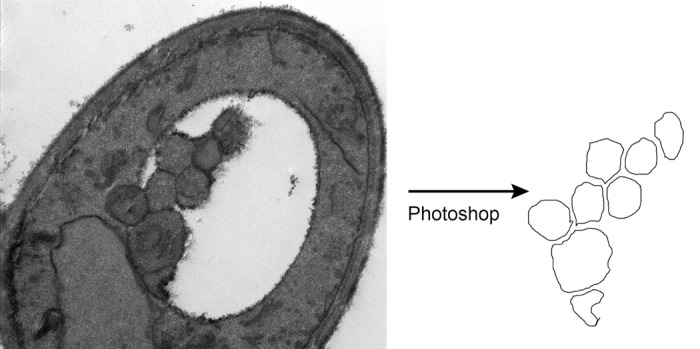
Figure 1. Example of autophagic body outlines derived from an EM image.
4. Hide the background layer, so that only the outlines are showing.
5. Save the file as a grayscale TIFF image.
2.2.2 Measuring the outlines
See also Video SV2 (Tutorial 2: Measuring Outlines in ImageJ).
1. Open an outlined image in ImageJ.
2. In the Analyze menu, go to “Set Scale” and enter how many pixels correspond to how many nanometers in your EM images (see Note 13). Check the “global” box so that this will apply to all of your images (see Note 14).
3. In the Analyze menu, go to “Set Measurements” and make sure “area” is checked (and other options are unchecked).
4. In the Image menu, go to adjust > Threshold to bring up the threshold palette. Click “auto” on this palette to automatically adjust the threshold of the image.
5. In the Analyze menu, select “Analyze Particles.” Check “Display Results” and “Clear Results” and “Include Holes.” Press “Analyze” (see Note 15).
6. Check the displayed results to make sure that there are the same number of results as there are outlines, and that the relative values make sense (e.g., an image with 4 large outlines and 2 small ones should give 4 large results and 2 small results; if not, see Note 16).
7. Copy and paste the results into Microsoft Excel.
8. Open a new outlined image in ImageJ (The “Open Next” command is useful for this). Repeat steps 4–7 to measure these bodies and copy the results into Excel.
9. Repeat step 8 for all images.
2.2.3 Outlining vacuoles
1. The procedure for outlining and measuring vacuoles is essentially the same as for outlining and measuring autophagic bodies, but is more easily done on a lower magnification image (~1000×). Be sure to outline the entire vacuole in each cell section, including not only any clear space but also any space that is filled with autophagic bodies. Remember to also include vacuole sections that are entirely filled with autophagic bodies. Any cell with no visible vacuole can of course be ignored. Very small vacuoles, such as those sometimes seen ringing the nucleus, can also be ignored, as they very rarely contain autophagic bodies (see Note 10).
2.3 Estimation of original autophagic body size
2.3.1 Size estimation
See also Video SV3 (Tutorial 3: Size Estimation).
1. Copy the list of autophagic body cross sectional areas generated in step 8 of 1.1.2 (maximum 2000 values) into cell J4 (“Area”) of the worksheet “Results” of the workbook “Size_Estimation.xlsx.”
2. Adjust the empirical parameters (Section Thickness, Recognition Threshold, and Calculation Range) in the upper left of the worksheet as necessary (see Note 17).
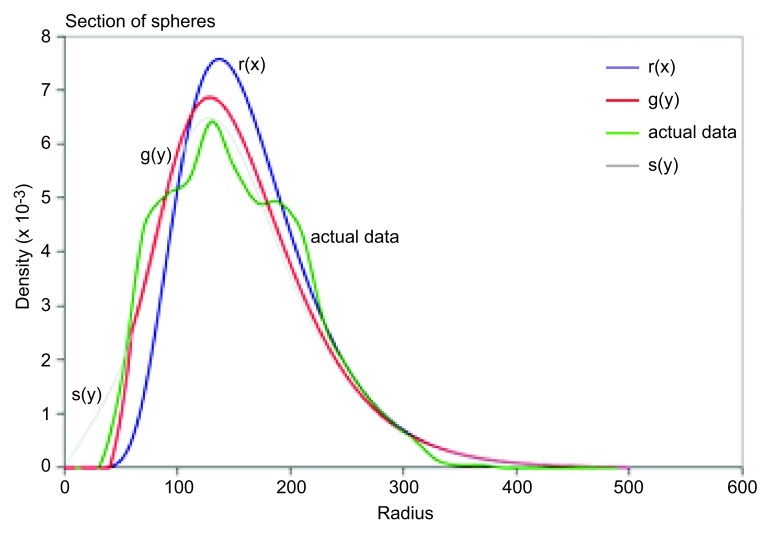
3. Your data are displayed on the worksheet “Density Plot.” Also displayed is a log-normal distribution r(x) defined by mu and sigma (cells A15 and B15), along with the calculated observed distribution of bodies s(y) based on this r(x), and the adjusted observed distribution g(y) based on s(y). As a first pass, adjust the values of mu and sigma in cells A15 and B15 until the graph of g(y) approximately matches your actual data (Fig. 2).
Figure 2. Example of manually fitting the graph of g(y) to the actual data.
4. Cells F31 and G31 display the mean and standard deviation of g(y). By systematically varying mu and sigma (cells A15 and B15), you can calculate the values of mu and sigma that best fit your data using a linear regression of the mean and standard deviation of your data against the means and standard deviations of g(y). To do this, choose 3 values of mu (generally differing by 0.1) and 3 values of sigma (generally differing by 0.05) based on the estimate you generated manually in step 3. You may need to adjust the Calculation Range, depending on the values of mu and sigma you choose (see Note 17).
5. Enter the first combination of mu and sigma that you chose into cells A15 and B15, copy the results in cells B31-G31 and paste them as values into cells B43-G43. Enter the second combination of mu and sigma into A15 and B15, and copy cells B31-G31 into cells B44-G44, and so on until the results from all nine combinations have been copied. The worksheet will then run the linear regression and display the best-fit mu and sigma for your data in cells B40 and C40.
6. Enter the best-fit values of mu and sigma from B40 and C40 into A15 and B15 in order to calculate the mean and standard deviation of the radii and the mean volume of the original autophagic bodies (cells D16-G16). Also record the best-fit mu and sigma and the sample number (cell O7) for later use. (File SX3 contains an example of a completed analysis.)
2.3.2 Number estimation
See also Video SV4 (Tutorial 4: Number Estimation).
1. First, estimate the mean radius of the vacuole. This can be done using the “Size_Estimation” workbook, following exactly the same procedure as for estimating the mean size of the bodies (section 2.3.1, but using the data from section 2.2.3—see File SX4 for an example). Be sure to adjust the Recognition Threshold and Calculation Range as necessary (see Note 17). Copy the estimated mean radius (cell D16, step 6 of 2.3.1) for use in step 3 below.
2. Open the workbook “Number_Estimation” and adjust the empirical parameters. The Calculation Range and the Autophagic Body Recognition Thresholds should be the same as were used in step 2 of 2.3.1, while the Vacuole Recognition Threshold should be the same as the lower (“ignore below”) recognition threshold used when estimating the mean vacuole radius.
3. For mu and sigma, use the best-fit values of mu and sigma generated for the autophagic bodies in step 5 of 2.3.1. For the vacuole radius, use the mean vacuole radius estimated in step 1 above.
4. Calculate the number of observed autophagic bodies per vacuole section from your data by dividing the total number of autophagic body outlines (“sample number” from step 6 of 2.3.1) by the total number of vacuoles imaged, including empty vacuoles. Enter this number into cell C23.
5. The estimated total number of autophagic bodies per vacuole will be calculated and displayed in cell C27. File SX5 contains an example of a completed analysis.
3. Notes
1. The PEP4 deletion is crucial, as otherwise the autophagic bodies will be broken down and not accumulate in the vacuole. The VPS4 deletion is not crucial, but is recommended, as this will block delivery of small vesicles to the vacuole via the multivesicular body pathway. The presence of these small vesicles makes it somewhat more difficult to distinguish the outlines of the autophagic bodies.
2. It is important that the acetone used in these final dehydration steps and the first embedding steps be free from moisture. We use a fresh, unopened bottle of regular ACS grade acetone, which gives satisfactory results. EM grade, extra-dry acetone is also available from various EM-supply vendors, and may be preferable if difficulties are encountered with the quality of the embedding.
3. Chemical fixation may induce changes in the apparent size of the autophagic bodies. In addition, both the size and number of autophagic bodies formed as well as the size and shape of the vacuole is dependent on the exact growth conditions. Therefore, samples from all strains to be compared should be grown and processed in parallel.
4. It is crucial that the cells be growing under optimal conditions prior to fixation. It is recommended to restreak a fresh plate, grow it at 30 °C, and use this fresh plate to inoculate the starter culture directly without ever having placed the plate at 4 °C. If using a slightly older plate (1–3 wk old, stored at 4 °C), the starter culture should be grown for at least 24 h, with dilution into fresh YPD as necessary to maintain it continually in log phase (A600 ≤ 1.0), before using it to inoculate the 30-ml culture.
5. These numbers are for SEY6210 pep4Δ vps4Δ yeast, with a doubling time of ~1.5 h. They may need to be adjusted for other strains with different doubling times. Either the starting dilution or the growth time may be adjusted as desired; the only critical point is that the culture is maintained continually in log phase. This protocol is for 30 units (30 ml of culture at A600 = 1) of yeast; as few as 10 units (e.g., 30 ml of culture at A600 = 0.33) also gives satisfactory results. More units simply provide more material, making sectioning easier.
6. One h of nitrogen starvation is enough to see some accumulation of autophagic bodies, although 2 or 4 h of starvation are more typical time points. Both the number and the size of the autophagic bodies will increase with longer starvation times. If desired, other autophagy-inducing conditions can be used, such as glucose starvation or rapamycin treatment, although in this case the optimal time of treatment may be different.
7. From this point forward, it is essential that the purified water used be free of particulate contaminants. It should either be collected directly from the spout of the Milli-Q system or else passed through a 0.22-µm syringe filter prior to use.
8. From this point forward the cells should be treated gently. The cells are particularly fragile during the dehydration procedure. Resuspension should always be by gentle pipetting up and down, with care taken to avoid introducing bubbles, and never by vortex.
9. Spurr resin, like most EM resins, is toxic in its unpolymerized form. Care should be taken when handling both the individual components and the mixed resin to avoid both skin contact and inhalation. Gloves should be worn at all times, and it is advisable to wear a face mask, or work in a fume hood. Waste resin should first be allowed to completely polymerize (> 1 d at 70 °C, or > 1 wk at room temperature), after which it can be disposed of in the trash. Tubes or gloves contaminated with resin should be placed in a sealed plastic bag and likewise given time to polymerize before disposal. Waste resin/acetone mixtures should be placed in an open container in a fume hood to allow the acetone to evaporate and the resin to polymerize before disposal. Check with your institutional hazardous waste office (or EM facility) to see if they recommend any modification to these instructions.
10. It might be more convenient to ignore vacuole sections below a certain size. This should not affect the results as long as the same size threshold is used both when taking images for the body outlining and when outlining the vacuoles. This minimum threshold would then be used as the recognition limit when estimating vacuole size and body number.
11. We recommend a 6-point black brush. Alternately, using a colored brush makes it easier to see the outlines while painting. This requires converting the image from grayscale to RGB before outlining, and then reverting back to grayscale afterwards (ImageJ needs a grayscale image of the outlines for the next steps). Also note that this manual outlining is very sensitive to variations in technique. Therefore, we recommend that if comparing 2 different samples, the researcher should alternate between outlining images from one sample and images from the other, and be cautious about comparing outlines made at different times. Moreover, we would not ever recommend directly comparing outlines generated by 2 different researchers.
12. Due to imperfect preservation of the EM samples, some autophagic bodies will be easier to distinguish than others. Common problems include “incomplete” bodies (where the membrane and perhaps part of the contents appears to have been erased from one side), “hollow” bodies, which have a distinct membrane but no contents, and conglomerates of bodies where it can be difficult to distinguish the exact boundaries between one body and the next. The most accurate measurement of the size of the autophagic bodies would result from measuring only clearly defined autophagic bodies free of these problems. However, using these strict criteria will give an undercounting of the number of autophagic bodies per vacuole. Therefore, if the user wishes to estimate both the size and the number of autophagic bodies, it is better to relax these criteria somewhat and make a best guess as to the outlines of each autophagic body.
13. One way to determine the scale is to open the original image in Photoshop and carefully select just the scale bar, copy it to a new canvas that is set to measure in pixels, and read how many pixels long is that copied scale bar.
14. The next time an image is opened, ImageJ will display an error message warning you that the global calibration conflicts with the calibration of the image. Uncheck “disable global calibration,” and instead check “disable these messages.”
15. The goal here is to measure the area of each outlined autophagic body, including every bit within the boundary. As ImageJ is being constantly updated, some of the options might change to different names with a different version. Nevertheless, it should be fairly straightforward to find the right options by experimenting with the checkboxes. Also, it is possible to record a short “macro” in ImageJ to automatically perform these steps, speeding up this part of the analysis.
16. If there are fewer measurements than there are outlines, this is likely because some of the outlines were too close together and were counted as a single outline by ImageJ. Reopen the outlined image in Photoshop and adjust or redraw the outlines as necessary so that they are clearly separated. If there are more measurements than there are outlines, these are probably the result of stray dots or lines made while outlining. These stray images should give very small values, which can be ignored. If there are the right number of outlines, but some of the values are unexpectedly low (e.g., a large outline giving the same value as a very small one), this probably indicates a gap in the outline. Open the outlined image in Photoshop and fill in any gaps.
17. For Section Thickness, fill in the actual thickness of the ultrathin sections used for the EM. The Recognition Threshold generates g(y) from s(y) by throwing out all of the values below the “ignore below” value and adding a linear bias against the values between the “Ignore Below” and “Full Recognition Above” values. You should decide on the best values to enter here by looking at your actual data (you can use Excel to sort it from smallest to largest and find the threshold below which few or no values fall). The Calculation Range is the range of autophagic body sizes over which the numerical integration is done. The optimal range depends on the values of mu and sigma being tested. If the calculation range too is set too small, this can cause a dramatic underestimate of g(y) for large values of mu and sigma. If the range is set too large, this will result in undersampling and gradually degrade the accuracy of the calculation, particularly for smaller values of mu and sigma. Generally, a calculation range of 3 times the mean radius of the body cross sections is a good place to start. If the calculation range is set too large or too small, a warning message will appear in cell H5.
4. Discussion
4.1 An improved method for estimating the size of autophagic bodies
This work began with the adaptation of a previously published simulation method for estimating the size of autophagic bodies. 1 The previous method took into account the 2 major biases that affect the observed size of the cross sections: namely, that the section may have not been taken through the equator of the original body, and that larger bodies are more likely to be sampled than smaller bodies. In addition, that method attempted to account for a minor bias expected to result from the fact that an electron micrograph section has a given thickness, typically ~70 nm, and that autophagic bodies tend to cluster together, and therefore may partially obscure each other in the projected image, causing the area of their cross sections to appear slightly smaller than they actually are.
A major limitation of the previous method is that the simulation takes hours to run, even on relatively modern hardware. Closer examination revealed that the steps to cluster the vesicles and to resolve the obscuring effect are the most computationally intensive. When the code was modified to skip these steps and directly take the projection of the spherical segments/caps as the readout, the results only differed approximately 1–2%, suggesting that the obscuring effect is largely negligible for our purposes. In addition, in the previous simulation code the slice was always taken from the center of the vesicle cluster. Altering the code to take a random slice led to a ~6–8% reduction in estimated body size. These improvements are included in the attached R code (File S6).
Furthermore, by ignoring the small obscuring effect, the relationship between random sections and the original vesicle population can be described algebraically. 11 This in turn allowed us to solve the problem by a faster numerical method in Microsoft Excel. 8 Our extensive testing demonstrated that the results given by this direct calculation method were nearly identical to those given by the updated simulation method (Fig. S1A). Based on its speed and ease of use, we think that the calculation method is more suitable for daily use by most cell biologists, and thus the protocol given here makes use of this method. The simulation method is better reserved for method validation in case the algebraic equations need to be modified for different experimental settings.
4.1.1 Mathematical background for size estimation
The direct calculation method relies on the same assumptions as the simulation, namely that the bodies are spherical 12 and that the probability density function of the body radii, r(), is a log normal distribution dependent on mu (µ) and sigma (σ):
If the bodies are randomly positioned in space, resulting in size-biased sampling when taking a random slice of the space, and the thickness in the sample slice is 2T, then the density function of the radii of the projected body sections s() is a function of r(): 8 , 11
Note that in the special case where T = 0, only the latter part remains:
And in the basic case where all bodies are of the same radius R, we have the equivalent j(), which s() is built upon:
For a given piece of body cross section, whether it can be reliably recognized and collected as data by a human being is affected by its size. In practice, very small ones are generally not collected. Assuming that below a lower recognition threshold, L, all are ignored, above an upper recognition threshold, U, all are collected, and in between, the possibility of being collected is proportional to the size, then the collection possibility function c() is:
Combining s() and c(), the density function of the radii of body sections a researcher can expect to get, g(), is:
For a given r(), s() and g() can be calculated using numerical integration. Now a linear regression can be used to fit μ and σ of r() based on the mean and standard deviation of the experimentally observed body cross sections.
4.2 A method for estimating the number of autophagic bodies
In order to correlate EM measurements with autophagic flux, it is necessary to know not only the actual size of the autophagic bodies, but also their number. Any estimate of number is complicated by the fact that the observed number of body cross sections depends not only on the actual number of bodies, but also on the size of the bodies, as larger bodies are more likely to be captured in a random section. Moreover, the proportion of bodies captured in a given slice through the vacuole also depends on the size of the vacuole. Therefore, a method was also needed that would take these factors into account to provide an estimate of the number of autophagic bodies.
In order to use the direct calculation method to estimate body number, we need to introduce one more factor, as there is the implicit assumption that we are talking about the number within a certain unit of space. In cases where the cell is fairly large, this unit of space is often a random cube within the cell. For yeast cells, the vacuole is generally a more convenient choice.
First, we consider the cube case. This case applies when a researcher is randomly casting a square on a random slice of certain tissue, then counting the number of body cross sections within the square. For simplicity, we assume that both the bodies and the thickness of the slice are relatively small vs. the size of the square/cube. This assumption saves us from dealing with the complicated sampling strategy when objects are near the border of the square/cube. Assuming the half height of the unit cube is H, and the total number of bodies per unit cube is N, then the expectation of the number of observed bodies in such a random square is:
Alternatively:
In the unrealistic case where human recognition of body sections is 100%, i.e., c() = 1, then we have the much simpler equation:
When T = 0 (which is another way of saying T < < E(r)), the equation becomes:
During autophagy-inducing conditions, most yeast cells contain a single, roughly spherical vacuole. 13 If we assume that all vacuoles are spheres with radius of R, then the situation is not much different (with H = R, as the shape of sampled area does not really matter).
For convenience, generally only vacuolar sections above a certain size, R1, are considered. In this case, instead of a complete sphere, only a spherical segment is sampled. Assuming the half height of the spherical segment is H1, its volume is V1, and the number of bodies within this segment is N1, then the expected number of observed bodies n1 can be expressed as:
N1 can be obtained from: , where V is the volume of a complete vacuole. Thus:
Alternatively:
In order to verify the accuracy of this direct calculation method, we compared it to the results given by the simulation method. We adapted the simulation method to place the bodies in a spherical vacuole of a defined size and recorded the number of simulated cross sections captured by each random slice. The original simulation method arranged the bodies into a clump in the center of the vacuole prior to taking the slice. However, we found that omitting this section of the code and treating each body independently had no significant effect on the mean number of observed cross sections (Fig. S1B), but decreased the computation time > 1000 fold, so we used the method without the clumping routine (File S7) for further comparisons.
Varying the number of bodies used for each simulation allowed us to calculate n1/N by linear regression, and compare it to the n1/N estimate derived from the direct calculation method. We found the results given by each method to be very similar (Fig. S1C). The direct calculation method is easier to use, and therefore we make use of it in this protocol.
4.3 Recommended sample sizes
The accuracy of the estimate of the size of the autophagic bodies is of course dependent on the number of body cross sections measured. We looked at the effect of sample size using a large sample of 1600 body cross sections. Random subsets of this larger sample were taken to simulate smaller sample sizes, and these subsamples were used to estimate the original body size. These results were compared with those obtained using the entire 1600 data points (Fig. 3A). While a sample size of 100–200 body cross sections generally gives a reasonable estimate, suitable for a first pass or if comparing 2 very different samples, a sample size of > 800 cross sections is recommended for situations where a more accurate estimate is needed.
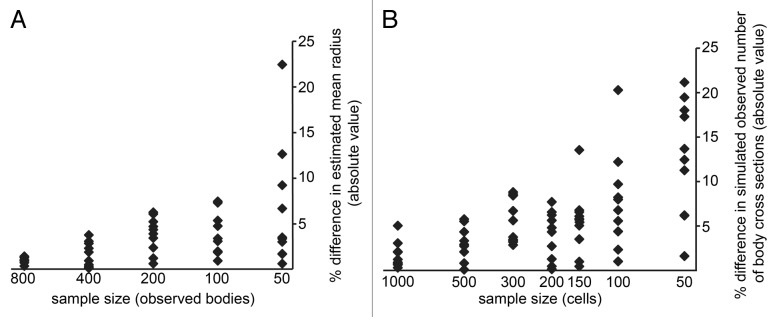
Figure 3. Effect of sample size on the results obtained by these methods. (A) Variation in estimated mean radius due to sample size. For each sample size, 10 random subsamples of that size were taken from a given sample of 1600 measurements and used to estimate the mean radius. The percent difference between this estimate and the reference estimate (obtained using the entire 1600 measurements) is plotted. (B) Variation in estimated body number due to sample size. A simulation method including a clumping routine (File S6; mu = 5, sigma = 0.4, balls = 80, recognition threshold = 30) was used to generate a set of simulated cross sections for a varying number of simulated cells. The percent difference between the number of cross sections per cell predicted by each run of the simulation and the reference number (the average of the 1000-cell simulations) is plotted.
When estimating the number of autophagic bodies, it is the number of vacuole cross sections, not body cross sections, that determines the sample size. Since under most conditions there are multiple body cross sections per vacuole section, this means that more cells must be imaged in order to accurately estimate the number of bodies than to accurately estimate their size. To judge the effect of sample size on estimating autophagic body number, we used the simulation code (including the clumping routine, which did not change the mean number of body sections per vacuole section but did change the standard deviation) to generate simulated samples of sizes ranging from 100 to 1000 cells. The results of each simulation run were compared with the average of all 7 of the 1000 cell simulation runs (Fig. 3B). Based on this, we recommend using a sample size of at least 200 cells to estimate body number.
4.4 Reanalysis of autophagosome size and number in ume6∆ cells
The deletion of the transcriptional repressor Ume6 leads to higher levels of Atg8 and an increase in autophagic flux as measured by the Pho8∆60 assay (Fig. 4A). 9 Analysis of EM images of autophagic bodies (Fig. 4B) using the previously published simulation method showed that larger autophagosomes were formed in ume6∆ cells than in the isogenic wild type. 9 We noticed at the time that ume6∆ cell sections also contained somewhat more autophagic body cross sections, but we could not determine whether there were actually more autophagic bodies in the ume6∆ cells or whether this was just a sampling bias introduced by the increase in autophagic body size.
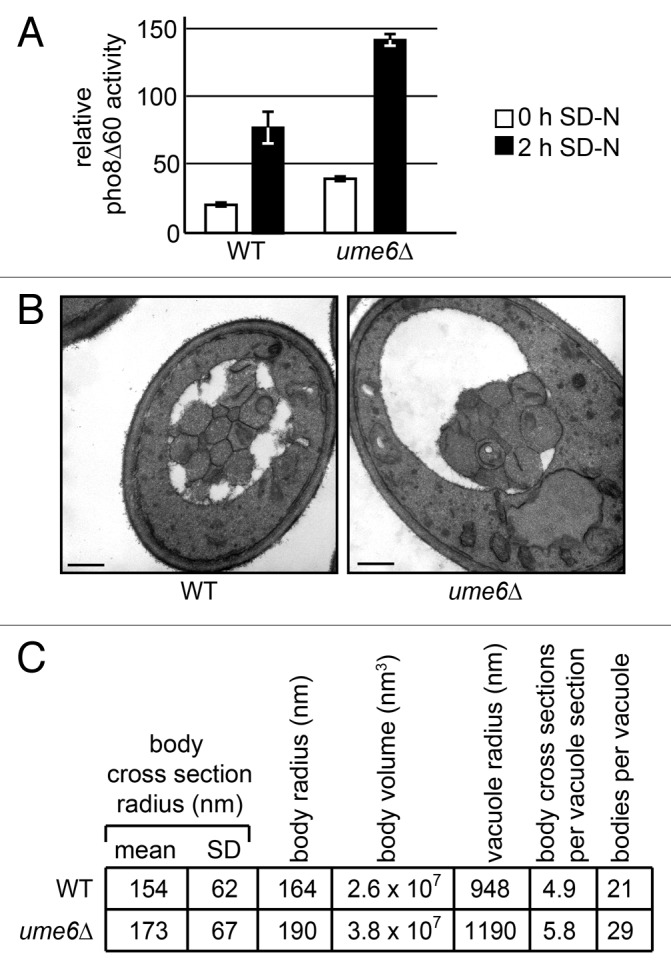
Figure 4. Analysis of autophagosome size and number in ume6∆ cells. (A) Pho8∆60 assay of autophagic flux during nitrogen starvation in SEY6210 pho13∆ pho8∆60 (WT), and ume6∆ cells. (B) Autophagic bodies in SEY6210 pep4∆ vps4∆ (WT) and ume6∆ cells after 2 h of nitrogen starvation. Scale bars: 500 nm. (C) Results from the analysis of the autophagic bodies following the protocol described in this paper. N > 200 cells, and > 1000 bodies. SD, standard deviation. (A and B) were adapted from reference 9 (copyright National Academy of Sciences [USA]).
Therefore, we applied the new methods described here for estimating both size and number to the wild-type and ume6∆ samples. We again saw that autophagic bodies in the ume6∆ strain were larger than those in the wild type (~45% larger by volume). In addition, we now estimate that there are ~35% more autophagic bodies formed in ume6∆ than in wild-type cells after 2 h of nitrogen starvation (Fig. 4C).
The reader should remember that these methods rest on certain simplifying assumptions (e.g., the treatment of the bodies and vacuoles as spherical) and thus these results are only an estimate. However, the 1.9-fold total increase in autophagic flux calculated from these results is very similar to the 1.8-fold increase in Pho8∆60 activity seen at the 2 h time point (Fig. 4A). This suggests that, at least in this case, these estimates are reasonably accurate.
Wild-type cells were estimated to contain approximately 21 bodies per vacuole after 2 h of nitrogen starvation. Baba et al. 12 performed serial sectioning of 6 slp1∆ cells, in which the fusion of autophagosomes with the vacuole is blocked, and reported an average of 20 bodies per cell after 3 h of nitrogen starvation. Although this is less than would be expected from our estimation, this could be a difference in strain background or the result of the small sample size used in that case. Observation of the rate of autophagosome formation by fluorescence microscopy has suggested that ~9 autophagosomes are formed per hour during nitrogen starvation at room temperature. 5 This would suggest that ~18 autophagosomes would be formed after 2 h of starvation, although we would expect the rate to be somewhat faster at 30 °C. Overall, the methods described here give an estimate of autophagic body number that is within the same general range as has been previously published.
We think these estimates should be sufficient for most purposes, and these methods are fast and easy to use. Therefore, we expect them to be useful in the continued investigation of autophagy and its regulation.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Mark J Backues for helpful suggestions in relation to the simulation of autophagic bodies within the vacuole. This work was supported by NIH grants GM053396 (DJK), and 1-F32-GM-101748-01 (SKB), the Natural Science Foundation of China (31222034, 31171285; ZX), the National Key Basic Research Program of China (2011CB910100; ZX), and the Ministry of Education of China (NCET-12-0283; ZX).
Glossary
Abbreviations:
- A600
absorbance at 600 nm
- ATG
autophagy-related
- EM
electron microscopy
References
- 1. Xie Z, Nair U, Geng J, Szefler MB, Rothman ED, Klionsky DJ. . Indirect estimation of the area density of Atg8 on the phagophore. Autophagy 2009; 5:217 - 20; http://dx.doi.org/ 10.4161/auto.5.2.7201; PMID: 19088501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Levine B, Kroemer G. . Autophagy in the pathogenesis of disease. Cell 2008; 132:27 - 42; http://dx.doi.org/ 10.1016/j.cell.2007.12.018; PMID: 18191218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Klionsky DJ. . Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 2007; 8:931 - 7; http://dx.doi.org/ 10.1038/nrm2245; PMID: 17712358 [DOI] [PubMed] [Google Scholar]
- 4. Knorr RL, Dimova R, Lipowsky R. . Curvature of double-membrane organelles generated by changes in membrane size and composition. PLoS One 2012; 7:e32753; http://dx.doi.org/ 10.1371/journal.pone.0032753; PMID: 22427874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xie Z, Nair U, Klionsky DJ. . Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 2008; 19:3290 - 8; http://dx.doi.org/ 10.1091/mbc.E07-12-1292; PMID: 18508918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. . Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell 2005; 16:2544 - 53; http://dx.doi.org/ 10.1091/mbc.E04-08-0669; PMID: 15743910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang C-W, Klionsky DJ. . Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell 2005; 16:3438 - 53; http://dx.doi.org/ 10.1091/mbc.E04-10-0894; PMID: 15901835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Z. The function and trafficking of Atg8 during autophagosome formation. 2008. PhD dissertation, University of Michigan. [Google Scholar]
- 9. Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, et al. . Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci U S A 2012; 109:11206 - 10; http://dx.doi.org/ 10.1073/pnas.1200313109; PMID: 22733735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ellis E. . Corrected formulation for Spurr low viscosity embedding medium using the replacement epoxide ERL 4221. Microsc Microanal 2006; 12:288 - 9; http://dx.doi.org/ 10.1017/S1431927606062660 [DOI] [Google Scholar]
- 11. Feuerverger A, Menzinger M, Atwood HL, Cooper RL. . Statistical methods for assessing the dimensions of synaptic vesicles in nerve terminals. J Neurosci Methods 2000; 103:181 - 90; http://dx.doi.org/ 10.1016/S0165-0270(00)00315-0; PMID: 11084211 [DOI] [PubMed] [Google Scholar]
- 12. Baba M, Takeshige K, Baba N, Ohsumi Y. . Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J Cell Biol 1994; 124:903 - 13; http://dx.doi.org/ 10.1083/jcb.124.6.903; PMID: 8132712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cardenas ME, Heitman J. . FKBP12-rapamycin target TOR2 is a vacuolar protein with an associated phosphatidylinositol-4 kinase activity. EMBO J 1995; 14:5892 - 907; PMID: 8846782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.