Abstract
A long-standing quest in the autophagy field is to define the membrane origin of the autophagosome. We have established a cell-free assay based on LC3 lipidation that recapitulates multiple regulatory hallmarks of early autophagosome biogenesis. Using a systematic membrane fractionation approach, we have identified the ER-Golgi intermediate compartment (ERGIC) as the most efficient membrane substrate for LC3 lipidation. Further studies indicate that the ERGIC plays an essential role to trigger LC3 lipidation and autophagosome biogenesis by recruiting the key early autophagic factor ATG14.
Keywords: autophagy, autophagosome, ER-Golgi intermediate compartment, LC3, lipidation
Autophagosome biogenesis requires a substantial membrane investment, likely originating from one or more organelles within the cell. The source of this membrane has been under intense investigation. Over the years, many sources of membrane for the initiation and propagation of the phagophore, the precursor to the autophagosome, have been investigated including endoplasmic reticulum (ER), mitochondria, Golgi, plasma membrane (PM) and vesicles containing ATG9. In spite of this, the organelle that triggers autophagosome biogenesis has remained elusive. Membrane precursors may reside in an organelle lacking a distinctive feature that could be identified by visual techniques. Therefore, a more functional biogenesis assay may be useful in defining the membrane of origin.
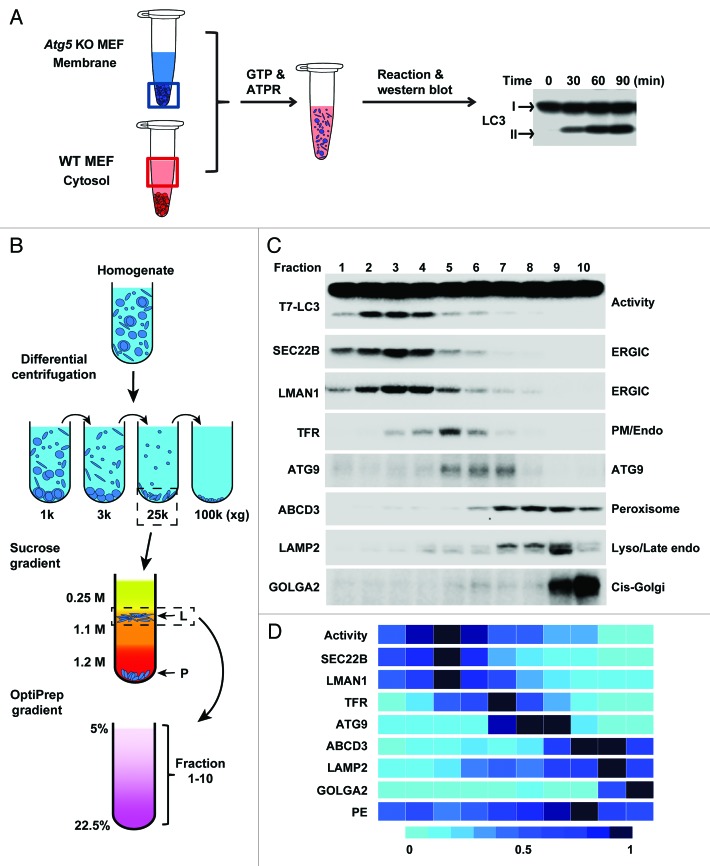
A key step in autophagosome biogenesis is the covalent linkage of a cytosolic protein, LC3-I, to phosphatidylethanolamine (PE) by a ubiquitin-like conjugation system. This process starts at the time of phagophore nucleation when the ATG12–ATG5 protein conjugate, the E3 enzyme for LC3 lipidation, docks on a certain membrane and proceeds until the completion of a mature autophagosome. Therefore, lipidation of LC3 provides a specific landmark to biochemically monitor autophagy. In the current study, we developed a functional approach based on LC3 lipidation to assay autophagosome biogenesis in a cell-free reaction. Atg5 knockout (KO) mouse embryonic fibroblasts (MEFs) are deficient in lipidation and in phagophore formation. By incubating pure recombinant LC3 with cytosol from wild-type (WT) cells and membrane from Atg5 KO MEFs, in the presence of GTP and an ATP-regeneration system, we observed de novo generation of lipidated LC3 (Fig. 1A). The in vitro LC3 lipidation is governed by the pathways regulating cellular autophagy in that: 1) it is stimulated by cytosol and membrane from starved, rapamycin- or Torin 1-treated cells, and is attenuated in the absence of ULK1, suggesting the involvement of the MTORC1-ULK1 pathway; 2) phosphatidylinositol-3 kinase (PtdIns3K) inhibitors, wortmannin and 3-methyladenine, and a phosphatidylinositol-3 phosphate (PtdIns3P) blocker peptide containing the FYVE domain, prevent in vitro LC3 lipidation, implying a requirement for the PtdIns3K pathway; 3) LC3 lipidation is totally abolished in the absence of ATG5, ATG3, or ATG7 demonstrating a dependence on the ubiquitin-like conjugation system. Therefore, the cell-free reaction recapitulates an early step of autophagosome formation.
Figure 1. In vitro LC3 lipidation and the 3-step membrane fractionation. (A) In vitro reconstitution of LC3 lipidation. Atg5 knockout (KO) MEF membrane was combined with wild-type (WT) cell cytosol plus GTP and an ATP regeneration system (ATPR). The generation of lipidated LC3 (LC3-II) in each time point was analyzed by immunoblot. (B) Fractionation scheme to look for the most active membrane for LC3 lipidation. Total membrane of Atg5 KO MEFs was subjected to differential centrifugations followed by the lipidation activity test. The 25,000 × g (25 k) pellet membrane, which has the highest activity, was layered on a sucrose gradient to separate into to L (light) and P (pellet) fractions followed by the lipidation activity test. The L fraction, which contains the majority of the activity, was further resolved on an OptiPrep gradient after which 10 fractions from the top were collected and the lipidation activity in each fraction was examined. (C and D) Activity of the 10 fractions derived from the last step with respect to the distribution of the indicated markers (C and D) and PE (D). TFR, transferrin receptor; Lyso, lysosome; endo, endosome; PM, plasma membrane. This figure was modified from Figures 1, 4, and 7 of Ge et al., eLife 2013;2:e00947.
To test the contribution of each individual organelle to autophagosome biogenesis, we designed a three-step membrane purification approach to separate cellular membranes which were assayed for their ability to support lipidation in the in vitro reaction (Fig. 1B). Membranes enriched in any of the previously characterized autophagosome origins including ER, Golgi, PM, mitochondria, ATG9 vesicles, or the mitochondrial-associated endoplasmic reticulum membranes (MAM) are depleted in LC3 lipidation activity. Instead, a membrane fraction with ER-Golgi intermediate compartment (ERGIC) marker proteins displays the highest activity for LC3 lipidation (Fig. 1C). Immuno-depletion and -isolation approaches confirmed that ERGIC can directly induce LC3 lipidation. Moreover, lipidation of the ERGIC-enriched fraction is stimulated by cytosol prepared from starved cells and inhibited by PtdIns3K inhibitors, indicating ERGIC is an autophagically-regulated membrane template for LC3 lipidation. Although PE is the substrate for LC3 lipidation, the ERGIC fraction is neither enriched in PE (Fig. 1D) nor any autophagic factors directly contributing to LC3 lipidation, implying that the ERGIC may possess a novel lipidation-inducing property, possibly an integral membrane protein receptor.
Drugs such as H89 and clofibrate that block anterograde transport and deplete ERGIC in cultured cells abolish the potency of the membranes that induced LC3 lipidation in vitro. Removal of the drugs from cell culture restores ERGIC and the cell-free lipidation activity with equivalent kinetics. In cells, H89 and clofibrate block the formation of starvation-induced LC3 puncta. The same is also observed in cells transfected with SAR1 mutants (H79G or T39N) that disperse ERGIC by blocking COPII vesicle budding. Moreover, H89 and clofibrate also diminish ATG16L1 puncta (a phagophore marker), suggesting ERGIC is required for phagophore formation.
Several studies have shown that the membrane recruitment of ATG14 (an autophagy-specific PtdIns3K marker), activates the autophagy-specific PtdIns3K complex to generate PtdIns3P. In starved cells, we found that ATG14 and ZFYVE1/DFCP1 (a PtdIns3P-binding omegasome marker) colocalize with markers of the ERGIC membrane. Autophagic puncta marked by ATG14 and ZFYVE1 are not formed in cells treated with H89 or expressing a SAR1 mutant (H79G or T39N). Using the cell-free reaction, we found that ATG14 and ZFYVE1 associate with a buoyant membrane that is depleted from H89-treated cells. Therefore, the ERGIC acts as a membrane platform for ATG14 recruitment and subsequent PtdIns3P generation. We suggest that this platform matures into an autophagosome by the recruitment of additional membrane from other sources such as the Golgi membrane, the endosome, the plasma membrane, and the mitochondrion.
The ERGIC is a recycling compartment characterized by a tubulovesicular structure that is located between the ER and cis-Golgi compartments and is subject to a constant flux of membrane traffic. A subset of the ERGIC may become diverted to specialized events such as the formation of a phagophore membrane in starved cells. ATG14 and subsequent markers of autophagy may be recruited to the ERGIC through interaction with a specific receptor protein, lipid, or a highly-curved area in the membrane.
Subdomains of the ER in contact with mitochondria provide a lipid acquisition environment for autophagosome formation. Considering the location of the ERGIC adjacent to the ER, a burst initiation of autophagosome biogenesis could quickly mobilize the ER for further membrane acquisition and expansion. During this process, membranes from multiple sources may contribute to the expansion of a phagophore through fusion events dependent on SNARE proteins. Details of this maturation process including the source of additional membrane may be amenable to a cell-free reconstitution approach as we have described for the early events.
In summary, we have established a cell-free assay to reconstitute an early stage of autophagosome biogenesis and defined the ERGIC as a primary membrane determinant to trigger LC3 lipidation. This approach should permit the isolation of functional forms of the cytosolic and membrane proteins and lipids that build a functional phagophore membrane.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Qing Zhong, Nicholas Ktistakis, Noboru Mizushima, Masaaki Komatsu, David King, Kundu Mondira, Eisaku Hokazono, Jennifer Lippincott-Schwartz, Bob Lesch, David Melville, Min Zhang, Ann Fisher, Xiaozhu Zhang, Jeremy Thorner, Ta-Yuan Chang, David Sabatini, Roberto Zoncu, and Xuejun Jiang for reagents, technical assistance, and advice on the study. LG was supported as a fellow of the Human Frontier Science Program and now as a fellow of the Jane Coffin Childs Fund. RS is an investigator of the HHMI and a senior fellow of the UC Berkeley Miller Institute.