Abstract
The Kaposi's sarcoma-associated herpesvirus (KSHV, also called human herpesvirus 8) has been linked to KS and primary effusion lymphoma (PEL) in immunocompromised individuals. We report that PEL cell lines have constitutive active alternative NF-κB pathway and demonstrate high-level expression of NF-κB2/p100 precursor and its processed subunit p52. To elucidate the mechanism of activation of the alternative NF-κB pathway in PEL cells, we have investigated the role of KSHV-encoded viral Fas-associated death domain-like IL-1β-converting enzyme inhibitory protein (vFLIP) K13. We demonstrate that stable expression of K13, but not other FLIPs, in a variety of cell lines constitutively up-regulates p100/NF-κB2 expression and leads to its processing into the p52 subunit. K13-induced up-regulation and processing of p100 critically depends on the IκB kinase (IKK)α/IKK1 subunit of the IKK complex, whereas IKKβ/IKK2, receptor-interacting protein, and NF-κB-inducing kinase are dispensable for this process. Silencing of endogenous K13 expression by siRNA inhibits p100 processing and cellular proliferation. Our results demonstrate for the first time, to our knowledge, that KSHV vFLIP K13 is required for the growth and proliferation of PEL cells and alternative NF-κB pathway plays a key role in this process. Therapeutic agents targeting the alternative NF-κB pathway may have a role in the treatment of KSHV-associated lymphomas.
Kaposi's sarcoma associated herpes virus (KSHV), also known as human herpes virus 8, is a γ-2 herpes virus that is the most frequent cause of malignancy among AIDS patients (1). In addition to KS, KSHV has been associated with multicentric Castleman's disease and primary effusion lymphoma (PEL). One hallmark of KSHV is its extensive molecular piracy of key cell regulatory genes. The KSHV genome is known to encode for homologs of several cytokines, chemokines, and their receptors, as well as proteins involved in the regulation of cell cycle and apoptosis (1). Although these viral proteins resemble their cellular counterparts in sequence, they have evolved to perform functions critical to the life cycle of the virus and have acquired functional attributes not possessed by their cellular homologs (1). One such KSHV protein with unique functional properties is encoded by the ORF K13 (also called orf71).
The K13 protein contains two homologous copies of a death effector domain that is also present in the prodomain of caspase 8 [also known as Fas-associated death domain IL-1β-converting enzyme (also known as FLICE)]. Proteins with two death effector domains have been discovered in other viruses as well and are collectively referred to as viral FLICE inhibitory proteins (vFLIPs) (2). We and others recently demonstrated that vFLIP K13 has acquired the unique ability to activate the classical NF-κB pathway by virtue of its ability to physically associate with and activate a 700-kDa IκB kinase (IKK) signalosome complex consisting of IKK1/IKKα, IKK2/IKKβ, and NEMO/IKKγ (3–5). The activated IKK complex phosphorylates IκBα, leading to its ubiquitination and subsequent degradation via the proteasome pathway. Degradation of IκBα releases NF-κB of its inhibitory influence, thereby freeing it to migrate to the nucleus and turn on the expression of its target genes.
NF-κB is composed of homo- and heterodimers of five subunits, including c-Rel, NF-κB1 (p50), NF-κB2 (p52), p65 (RelA), and RelB (6). NF-κB1 and NF-κB2 are produced as p105 and p100 precursors, respectively, that contain the DNA-binding and dimerization Rel-homology domains in their N-terminal halves and inhibitory IκB-like domains in their C-terminal halves (7). Recently, an alternative (or noncanonical) pathway of NF-κB activation that involves proteasome-mediated processing of p100/NF-κB2 into p52 subunit, has been described (7). The alternative NF-κB pathway is essential for development and organization of lymphoid tissues and is activated by a number of receptors involved in lymphocyte development and survival, such as BAFF-R, CD40, and LTβR (8–10).
In this report, we present evidence that the alternative NF-κB pathway is constitutively active in PEL-derived cells and is essential for their growth and proliferation. Moreover, we demonstrate that KSHV vFLIP is a key mediator of alternative NF-κB activation in PEL cells and activates this pathway by means of an NF-κB-inducing kinase (NIK)- and IKK2-independent and an IKK1-dependent process that involves up-regulation, phosphorylation, and ubiquitination of p100/NF-κB2.
Materials and Methods
Plasmids and Cell Lines. Plasmids encoding K13-Flag, NIK, IKK1, and its kinase-inactive mutant (IKK1-KM) have been described (3). Expression constructs encoding wild-type and mutant p100 have been described and were obtained from S.-C. Sun (Pennsylvania State University College of Medicine, Hershey, PA) (11). An N-terminal hemagglutinin (HA)-tagged p100 construct was made in the pCDNA3 vector. Human non-small-cell lung cancer cell line NCI-H460 (H460) was provided by J. Minna of our institution. PEL cell lines (BC-1, BC-3, and BCBL-1), CEM (T cell leukemia), BJAB (B-cell lymphoma), and HeLa (human ovarian carcinoma) were obtained from the American Type Culture Collection. H460, BC-1, BC-3, BCBL-1, CEM, BJAB, and Jurkat cells were cultured in RPMI medium 1640 supplemented with 10% FCS. Wild-type and NIK-/- murine embryonic fibroblast (MEF) have been described (12) and were obtained from R. Schreiber (Washington University School of Medicine, St. Louis). Wild type, IKK1-/-, IKK2-/-, and NEMO-/- MEF cells were obtained from I. Verma (The Salk Institute for Biological Sciences, La Jolla, CA) and R. Gaynor (Eli Lilly and Company, Indianapolis). MEF cells were cultured in DMEM with 10% FBS. Wild-type and receptor-interacting protein (RIP)-deficient Jurkat cells were obtained from B. Seed (Massachusetts General Hospital, Boston) and were cultured in RPMI medium 1640 supplemented with 10% FCS (13). Retrovirus constructs containing C-terminal Flag epitope-tagged KSHV vFLIP K13, cellular FLIP (cFLIP)L [Mach-related inducer of toxicity (MRIT)-α1], cFLIPs (MRIT-β1), MC159, and MC160 were generated in murine stem cell virus neo-based retroviral vector and used for infection as described (4).
Western Blot. Western blot analysis was performed essentially as described (4). Primary antibodies used in these experiments were: p52 (mouse monoclonal, Upstate Biotechnology, Lake Placid, NY); p52 (rabbit polyclonal, Santa Cruz Biotechnology); p-IκBα (Cell Signaling Technology, Beverly, MA); total IκBα (Santa Cruz Biotechnology); actin (goat polyclonal, Santa Cruz Biotechnology); tubulin (mouse monoclonal, Sigma); IKK1 (Santa Cruz Biotechnology); IKK2 (Santa Cruz Biotechnology); anti-GFP (Santa Cruz Biotechnology); mouse monoclonal M2 Flag, and rabbit polyclonal Flag (Sigma). A rabbit polyclonal antibody against full-length K13 protein was generated in our laboratory. FLAG and control mouse IgG beads were obtained from Sigma and used in coimmunoprecipitation studies as described (4).
Cell Viability Assay. Cell viability was measured by using (3–4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt as described (14). Percent cell survival was calculated based on the reading of uninfected cells as 100%. Cell viability was also measured by using the Cell titer-Glo luminescent cell viability reagent (Promega).
RNA Interference. Short interfering (si)RNA hairpins against K13, p100, and a control siRNA were expressed under human U6 promoter and were generated by using a described lentiviral vector (15) modified to encode a GFP-blasticidin fusion protein. The sequences of the sense strand targeted by the siRNA hairpins are as follows. K13 (5′-CGTGTTCATACCTCAACCCACAC-3′); p100 (5′-CTCCTCCATTGTGGAACCCAAGGAGC-3′); and control (5′-CAGCATGAAGATCTCAATGAAGTAGCC-3′). Recombinant lentiviruses were generated and used to infect BC-3 cells according to the instructions of the manufacturer (Invitrogen). At 48–72 h after infection, cells were examined under a fluorescent microscope or by flow cytometry to determine the degree of viral infection based on the expression of GFP. Approximately 70–90% of the cells were found to be GFP-positive after a single round of infection in different experiments.
Results
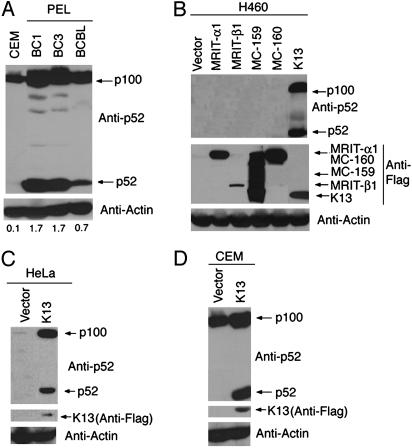
Constitutive Up-Regulation and Processing of p100 in PEL Cell Lines. The canonical NF-κB pathway is constitutively active in PEL-derived cell lines (4, 16). To investigate the status of the alternative NF-κB pathway in PEL cell lines, we examined the expression of p100/NF-κB2, and its processed subunit, p52, in three KSHV-infected PEL cell lines: BC-1, BC-3, and BCBL-1, respectively. As shown in Fig. 1A, we observed high-level expression of p100 precursor in all three PEL-derived cell lines as compared with CEM cell line, a non-PEL lymphoma cell line that is not infected with KSHV. More importantly, in addition to up-regulation of p100/NF-κB2 precursor, all three PEL cell lines demonstrated significant expression of the p52 subunit, indicating that a p100 precursor is being processed into p52 subunit by the activity of the alternative NF-κB pathway. Collectively, the above results indicated that the alternative NF-κB pathway is constitutively active in PEL cell lines.
Fig. 1.
Status of alternating NF-κB pathway in vFLIP K13-expressing cells. (A) Alternative NF-κB pathway is constitutively active in PEL cell lines. Western blots demonstrating constitutive p100 up-regulation and processing in three PEL cell lines (BC-1, BC-3, and BCBL-1) as compared with CEM cells. The actin blot shows equal protein loading. The numbers below the blots represent the intensity of the p52 band relative to the actin band. (B) Expression of KSHV vFLIP K13, but not of other FLIPs, leads to p100 up-regulation and its processing into the p52 subunit. Cell lysates from H460 cells expressing an empty vector or the indicated Flag epitope-tagged FLIPs were analyzed by immunoblotting with an antibody against p52. The blot was reprobed with Flag and actin antibodies to demonstrate the expression of various FLIPs and equal loading of the lanes, respectively. (C and D) Retroviral-mediated expression of vFLIP K13 leads to p100 up-regulation and its processing into p52 subunit in HeLa and CEM cells, as measured by Western blotting by using a p52 antibody.
Activation of Alternative NF-κB Pathway by vFLIP K13. We have reported that HHV 8-encoded vFLIP K13 is unique among the vFLIPs in possessing the ability to activate the canonical NF-κB pathway (3). Because vFLIP K13 is one of the few KSHV-encoded proteins to be expressed in latently infected PEL cells, it is a prime candidate for causing the constitutive activation of the alternative NF-κB pathway as well. We decided to test this hypothesis by ectopically expressing vFLIP in several hematopoietic and solid-tumor-derived cell lines by retroviral-mediated gene transfer. As shown in Fig. 1B, expression of vFLIP K13 in the H460 (non-small-cell lung cancer) cell line led to upregulation of p100 expression and generation of a p52 subunit. Interestingly, these effects were restricted to vFLIP K13 and were not observed with other FLIPs, such as cFLIPL (MRIT-α1), cFLIPS (MRIT-β), vFLIP MC159, and vFLIP MC160 (Fig. 1B). Furthermore, activation of the alternative NF-κB pathway by vFLIP K13 was not limited to the H460 cells because it led to up-regulation of p100 expression and generation of its active p52 subunits in multiple additional solid tumor and hematopoietic cell lines, such as HeLa, CEM, Jurkat, and 293 cells (Fig. 1 C and D and data not shown).
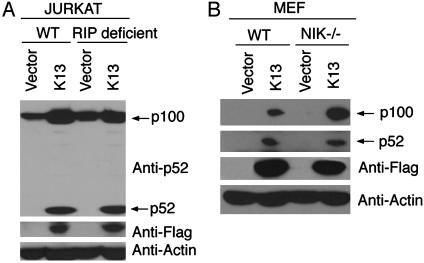
Role of RIP in vFLIP K13-Induced Alternative NF-κB Activation. RIP, a serine-threonine protein kinase, has been shown to play an essential role in NF-κB activation through tumor necrosis factor receptor 1 (13, 17). We have demonstrated that RIP and vFLIP K13 physically interact with each other (3, 4). To examine the involvement of RIP in a K13-induced alternative NF-κB pathway, we took advantage of mass populations of wild-type and RIP-deficient Jurkat cells expressing an empty vector or Flag-tagged K13 (18). Consistent with the above results, expression of K13 led to up-regulation of p100 and its processing into p52 fragment in the wild-type Jurkat cells. However, we found near equivalent up-regulation of p100 expression and its processing into p52 in the RIP-deficient Jurkat cells as well (Fig. 2A), thereby arguing against a major role of RIP in K13-induced activation of the alternative NF-κB pathway.
Fig. 2.
RIP and NIK are not required for K13-induced p100 up-regulation or processing. (A) Wild-type (WT) and RIP-deficient Jurkat cells were transduced with an empty retroviral vector or a vector expressing Flag epitope-tagged K13. Expression of p100/p52 (Top), Flag-K13 (Middle), and actin (Bottom) was determined by Western blotting with the indicated antibodies. (B) Wild-type (WT) and NIK knockout (NIK-/-) MEF cells were transduced with an empty retroviral vector or a vector-expressing Flag epitope-tagged K13. Expression of p100/p52, Flag-K13, and actin was determined by Western blotting with the indicated antibodies.
Role of NIK in vFLIP K13-Induced Alternative NF-κB Activation. NIK is a serine-threonine kinase that plays a key role in lymphotoxin β-receptor (12, 19–21)-, CD40 (10)-, BAFF-R (8)-, and LMP-1-induced p100/NF-κB2 processing (22–24). NIK is also known to coimmunoprecipitate with vFLIP K13 on transient transfection in 293T cells (3). However, we found that K13 expression leads to p100 up-regulation and near equivalent generation of the p52 fragment in the wild-type and NIK-deficient MEF cells (Fig. 2B). Thus, similar to the situation with human T cell leukemia virus-1 Tax (11), vFLIP K13 can induce p52 processing independent of NIK.
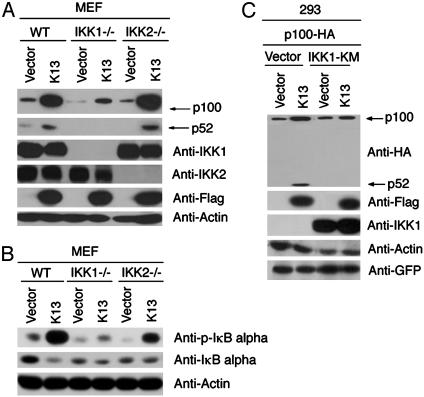
Role of IKK1 and IKK2 in vFLIP K13-Induced Alternative NF-κB Activation. Recent studies (9, 25, 26) suggest an essential role of IKK1 in NIK-induced processing of p100/NF-κB2 subunit and induction of a subset of NF-κB target genes. Because K13 is known to interact with the IKK complex, we investigated whether IKK1 and IKK2 are required for K13-induced activation of the alternative NF-κB pathway. We have previously generated stable clones of wild-type, IKK1-, and IKK2-deficient MEF cells expressing an empty vector or vFLIP K13 (18). As shown in Fig. 3A, expression of K13 in the wild-type and IKK2-deficient MEF cells led to a significant increase in the expression of p100 and the generation of its p52 subunit. However, only a small increase in p100 expression was detected in K13-expressing IKK1-deficient cells and this increase was not accompanied by the generation of p52 fragment. The above results point to a key role of IKK1 in K13-induced up-regulation of p100 level and indicate that IKK2 is not required for this process.
Fig. 3.
vFLIP K13-mediated p100 up-regulation and processing depends on IKK1/IKKα but is independent of IKK2/IKKβ. (A) Cellular extracts from MEFs (WT, IKK1-/-, and IKK2-/-) stably expressing empty vector or Flag-K13 were subjected to Western blot analysis with the indicated antibodies. K13-mediated p100 up-regulation and processing is markedly reduced in IKK1-/- cells, whereas loss of IKK2 has no significant effect. (B) Role of IKK1 and IKK2 in K13-induced canonical NF-κB activation as measured by phosphorylation of IκBα. Cellular extracts from MEF cells expressing an empty vector or K13 were immunoblotted with the indicated antibodies. (C) Kinase activity of IKK1 is required for p100 processing. The indicated constructs were transfected into 293T cells. The amount of dominant-negative IKK1 (IKK1-KM) construct (1.5 μg) was three times the amount of K13-Flag (0.5 μg) and p100-HA (0.5 μg) constructs, respectively, and the total amount of transfected DNA was kept constant by adding an empty vector. Approximately 36 h after transfection, cell lysates were prepared and immunoblotted with the indicated antibodies. The IKK1-KM mutant effectively blocks K13-mediated p100 processing.
The up-regulation of p100 expression is believed to be mediated by the canonical NF-κB pathway (9). The inability of K13 to significantly up-regulate the expression of p100 in IKK1-deficient cells suggested that IKK1 plays a major role in K13-induced activation of the canonical NF-κB pathway as well. Consistent with this hypothesis, we observed that expression of K13 led to significant phosphorylation of IκBα, which was associated with a decrease in the steady-state level of total IκBα in the wild-type MEF cells (Fig. 3B). Whereas significant phosphorylation of IκBα upon K13 expression was observed in the IKK2-deficient cells, K13 expression led to only a minor increase in the phosphorylation of IκBα in the IKK1-deficient MEFs (Fig. 3B). The above results are consistent with our recently published results (18) demonstrating that IKK1 plays a major role in K13-induced canonical NF-κB activation.
Next, we tested whether K13 could induce the processing of p100 into p52 subunit independent of its effect on p100 upregulation and whether this function required the kinase activity of IKK1. To test this hypothesis, we transiently transfected 293T cells with plasmids encoding NH2-terminal HA-tagged p100 along with an empty vector or Flag-tagged K13 and in the absence or presence of a kinase-inactive mutant of IKK1 (IKK1-KM). We observed significant processing of the exogenously expressed p100 into p52 subunit in cells cotransfected with a K13-encoding plasmid but not in those transfected with an empty vector (Fig. 3C, lanes 1 and 2). Importantly, processing of p100 into p52 fragment was completely abolished in cells transfected with the kinase-inactive mutant of IKK1 (Fig. 3C, lane 4). Taken together, the above results demonstrate that vFLIP K13 can induce the processing of p100 into p52 independent of its effect on p100 up-regulation and that the kinase activity of IKK1 is essential for this process.
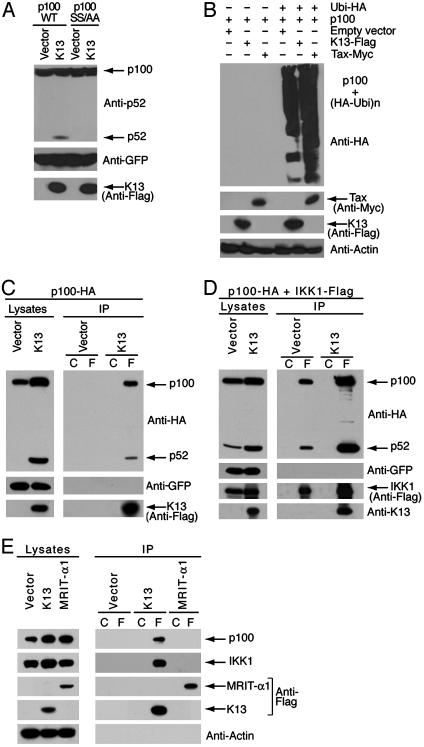
vFLIP K13-Mediated p52 Production Involves p100 Phosphorylation and Ubiquitination. NIK, IKK1, and Tax have been shown to phosphorylate p100 at serines 866 and 870, which serves as a trigger for its ubiquitination and subsequent processing into the p52 subunit (25). To determine whether the same residues are also involved in the ubiquitination and processing of p100 by K13, we transiently transfected 293 cells with either wild-type p100 or its mutant harboring mutations at the above residues (p100SS866/870AA) along with plasmids expressing an empty vector or vFLIP-K13. Remarkably, mutations at the serine 866 and 870 sites abolished processing of p100 by K13 (Fig. 4A). In an independent experiment, we observed that coexpression of K13 with HA-ubiquitin and wild-type p100 led to the appearance of a smear of high molecular weight adducts of p100, consistent with its polyubiquitinated forms. The human T cell leukemia virus oncoprotein Tax was used as a positive control in this experiment and induced p100 ubiquitination (Fig. 4B), as reported (11). Taken together, the above results suggest that K13-induced phosphorylation of p100 at serine 866 and 870 residues triggers its polyubiquitination, which is known to be a critical step in the inducible processing of p100.
Fig. 4.
vFLIP K13 directly interacts with p100 and induces p100 ubiquitination. (A) p100 phosphorylation is required for p100 processing into its p52 subunit. Wild-type p100 (p100-WT) and its mutant construct (p100-SS/AA; where Ser residues at sites 866 and 870 are mutated to Ala; SS866/870AA) were used to transfect 293 cells along with an empty vector or Flag-tagged K13. A GFP-encoding plasmid was cotransfected to monitor transfection efficiency. After 48 h, cells lysates were prepared and immunoblotted with the indicated antibodies. vFLIP K13 induces the processing of p100-WT but not of the p100SS/AA mutant. (B) vFLIP K13-induced p100 processing is associated with its ubiquitination. A quantity of 293 cells were cotransfected, as indicated with 500 ng of vector encoding Flag-p100 together with an empty vector or 500 ng of plasmid encoding HA-ubiquitin (Ubi-HA) and 500 ng of Flag-tagged vFLIP-K13 or Myc tagged-Tax. After 48 h, cells lysates were prepared and immunoblotted with the indicated antibodies. (C) KSHV vFLIP K13 directly interacts with p100. Whole-cell extracts from 293 cells transfected with HA-tagged p100 along with either an empty vector or Flag-K13 were immunoprecipitated by using mouse IgG control beads (lane C) or Flag beads (lane F). The interaction between K13 and p100 in the immunoprecipitates was detected by using rabbit polyclonal HA antibody. Western blot with a GFP antibody confirms equal transfection efficiency and antibody against the Flag-tag confirms the expression of Flag-tagged vFLIP K13. (D) vFLIP K13 enhances the interaction between p100 and IKK1 and stimulates p100 processing. Whole-cell extracts from 293 cells transfected with Flag-tagged IKK1 and HA-tagged p100 along with either an empty vector or K13 were immunoprecipitated by using mouse IgG control beads (lane C) or Flag beads (lane F). The interaction between IKK1 and p100 in the immunoprecipitates was detected by using rabbit polyclonal HA antibody. Western blot with a GFP antibody confirms equal transfection efficiency and with a rabbit polyclonal antibody against the K13 confirms the expression of vFLIP K13. (E) vFLIP K13 specifically interacts with endogenous p100 and IKK1. BC-3 cells were transduced with an empty retroviral vector or vectors expressing Flag epitope-tagged vFLIP K13 or Flag-tagged cFLIPL/MRIT-α1. Lysates from stably transduced cells were immunoprecipitated by using mouse IgG control beads (lane C) or Flag beads (lane F), and the presence of coimmunoprecipitated p100 and IKK1 was analyzed by Western blotting with the indicated antibodies.
vFLIP K13 Physically Interacts with p100 and Stimulates Its Processing. We investigated whether processing of p100 by K13 involves physical interaction between the two proteins. To address this question, we cotransfected 293 cells with Flag-tagged K13 along with HA-tagged p100. vFLIP K13 was immunoprecipitated from the cellular lysates by using Flag or control antibody beads and coimmunoprecipitated p100 and p52 were detected by immunoblotting with a HA antibody. As shown in Fig. 4C, we readily detected an interaction between K13 and full-length p100 and its processed p52 subunit.
Because K13 is known to physically interact with the IKK complex (4, 5), the above results raised the possibility that it might stimulate p100 processing by targeting it to the IKK complex, as was recently shown for Tax (11). Therefore, we asked whether expression of vFLIP K13 would augment the interaction between IKK1 and p100. For this purpose, we cotransfected 293 cells with K13 along with Flag-tagged IKK1 and HA-tagged p100. IKK1 was immunoprecipitated from the cellular lysates by using Flag or control antibody beads and coimmunoprecipitated p100 and p52 were detected by immunoblotting with an HA antibody. As shown in Fig. 4D, coexpression of K13 led to a significant increase in the interaction between IKK1 and p100 as compared with control vector-transfected cells, which was also associated with increased processing of p100 into p52.
vFLIP K13 Interacts with Endogenous p100 and IKK1. We sought to determine whether vFLIP-K13 can interact with endogenous p100 and IKK1 in PEL cells. To overcome the problem of lack of K13 antibodies suitable for immunoprecipitation, we engineered BC-3 cells to express either Flag-tagged vFLIP K13 or Flag-tagged cFLIPL/MRIT-α1 (control) by using retroviral gene transfer and immunoprecipitated the tagged proteins by using Flag or control antibody beads. As shown in Fig. 4E, vFLIP K13 interacted specifically with endogenous p100 and endogenous IKK1, whereas no interaction was seen between cFLIPL/MRIT-α1 and endogenous p100 or IKK1.
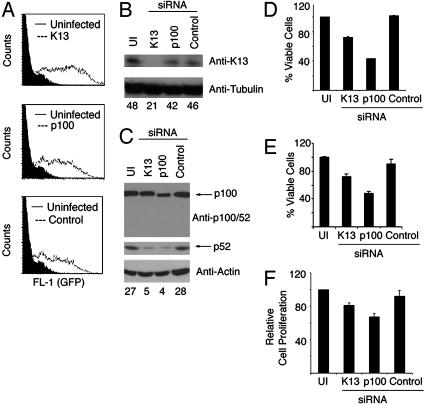
vFLIP K13 Is a Key Mediator of p100 Processing in KSHV-Infected PEL Cells. Next, we sought to determine whether K13 is responsible for the activation of the alternative NF-κB pathway observed in PEL cells and its role in the growth and proliferation of these cells. To address this question, we generated lentiviral vectors encoding siRNA hairpins targeted against K13 and p100, respectively. The lentiviral constructs also expressed a GFP-blasticidin fusion protein to help with monitoring the infection efficiency. We chose BC-3 cells for our studies because, unlike BC-1 cells, they are infected only with KSHV and are not coinfected with Epstein–Barr virus. In multiple experiments, we observed that ≈70–90% of BC-3 cells became GFP-positive after a single round of infection with the siRNA-encoding lentiviruses (Fig. 5A). Western blot analysis on cellular lysates prepared 72 h after infection demonstrated significant down-regulation of endogenous K13 expression in cells infected with the lentiviral vector encoding siRNA against this protein (Fig. 5B). However, there was no decrease in K13 expression in the cells infected with lentiviruses encoding p100 or control siRNAs, thereby arguing against the possibility of a generalized suppression of protein expression by siRNA-induced IFN response pathway (Fig. 5B). High-level expression of GFP in the lentiviral infected cells and a lack of effect on tubulin expression (Fig. 5B) further suggested that K13 siRNA leads to sequence-specific down-regulation of K13 expression.
Fig. 5.
Effects of siRNA-mediated K13 down-regulation in a BC-3 PEL cell line. (A) Flow cytometry analysis demonstrating high-efficiency infection of BC-3 PEL cells with siRNA-encoding lentiviral vectors as determined by GFP expression. Analysis was performed 72 h after infection with the indicated lentiviral constructs. (B and C) BC-3 cells were left uninfected (UI) or infected with lentiviral vectors encoding a control siRNA or siRNAs directed against K13 and p100. Approximately 72 h after infection, cell lysates were prepared and used in Western blot with the indicated antibodies. Equal loading of protein was demonstrated by Western blotting with antibodies against actin or tubulin. The numbers below the blots represent the intensity of the K13 and p52 bands relative to tubulin and actin bands, respectively. (D and E) Effect of vFLIP-K13 and p100 gene silencing on BC-3 cell number. BC-3 cells were either left uninfected or infected with the indicated siRNA-encoding lentiviral constructs. At 72 h after infection, viable cell numbers were measured by using (3–4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (D), and Cell titer-Glo luminescence reagent (E). The data represents mean ± SD of a representative of three independent experiments with similar results. (F) Effect of vFLIP-K13 and p100 gene silencing on the proliferation of BC-3 cells. Cells were infected with the indicated siRNA-encoding lentiviral vectors and were grown for 24 h in the presence of BrdUrd reagent. ELISA was performed to detect the amount of BrdUrd-labeled DNA as suggested by the manufacturer (Exalpha Biologicals). The data represent mean ± SD of a representative of three independent experiments with similar results.
We studied the effect of down-regulation of K13 expression on p100 expression and processing in BC-3 cells. Cells expressing K13 siRNA demonstrated a marked decrease in p100 processing into a p52 subunit (Fig. 5C). However, we did not observe a significant decrease in p100 expression with K13 siRNA. This finding could reflect the presence of alternative mechanisms to maintain p100 expression in the PEL cells. As expected, a lentivirus construct expressing p100 siRNA led to a decrease in the expression of both p100 and p52 (Fig. 5C). During the course of these experiments, we observed that infection with lentiviral constructs encoding siRNAs against K13 and p100 led to a significant decrease in cell number as compared with infection with a control siRNA-encoding construct or uninfected cells (Fig. 5 D and E). In repeated experiments, we failed to observe any significant increase in apoptosis in K13 and p100 siRNA-expressing cells (data not shown). Therefore, we sought to determine whether vFLIP and p100 silencing could effect cellular proliferation. An ELISA-based BrdUrd cell proliferation assay showed a significant reduction in cell proliferation in cells expressing siRNAs against vFLIP and p100, respectively (Fig. 5F). Collectively, our results demonstrate that vFLIP K13-induced p100 processing contributes to the growth and proliferation of KSHV-infected cells.
Discussion
Based on their sequence homology to the death effector domain present in Fas-associated death domain-like IL-1β-converting enzyme and its homologs, vFLIPs were originally believed to primarily protect virally infected cells from death receptor-induced apoptosis (2, 27). However, we and other researchers (3–5) subsequently demonstrated that the main biological function of K13 is to interact with and activate the IKK complex, which leads to phosphorylation and degradation of IκBα and activation of the classical p65/p50 NF-κB pathway. In the present study, we demonstrate that KSHV vFLIP can also up-regulate p100/NF-κB2 expression and its processing into the active p52 subunit. Thus, similar to human T cell leukemia virus-1-transforming protein Tax (11) and Epstein–Barr virus-transforming protein LMP1 (22, 24), KSHV vFLIP activates both canonical and alternative NF-κB pathways. Simultaneous activation of both NF-κB pathways probably contributes to viral persistence in the infected lymphocytes.
Genetic studies have established that alternative NF-κB activation through the tumor necrosis factor family receptors, and LMP1 involves a signaling pathway that depends on NIK and IKK1 (8, 10, 12, 19–22, 24). In the present study, we demonstrate that alternative NF-κB activation through K13 also involves IKK1 but is independent of NIK. In this respect, K13 resembles the human T cell leukemia virus oncoprotein Tax, which has been also shown to induce p100 processing independent of NIK (11). The similarity in the mechanism of alternative NF-κB activation by K13 and Tax may be explained by our results demonstrating that K13 resembles Tax in physically interacting with both IKK1 and p100.
We have observed that K13 expression leads to significant up-regulation of p100 expression, a phenomenon previously observed upon Tax and LMP1 expression as well and attributed to the activation of the canonical NF-κB pathway (11, 22). Interestingly, unlike LMP-1 (22), up-regulation of p100 by K13 primarily depends on IKK1 and is independent of IKK2. Furthermore, K13-induced phosphorylation and degradation of IκBα is markedly reduced in IKK1-deficient cells (Fig. 3B) and is associated with marked reduction in NF-κB DNA-binding and transcriptional activities (18). Taken together, the above results suggest that IKK1 is primarily responsible for the activation of both the canonical and alternate NF-κB pathways through K13, whereas IKK2 might contribute to the maximal activation of the K13-induced canonical NF-κB pathway (18). However, it is important to point out that K13 might up-regulate the expression of p100 independent of canonical NF-κB activation, for example, by interacting with and stabilizing this protein. Finally, our results with exogenously expressed p100 demonstrate that K13 can induce p100 processing independent of its effect on p100 up-regulation (Figs. 3C and 4A).
We recently demonstrated that the K13 protein could transform rodent fibroblasts in both in vitro and in vivo assays (28). Although the exact role of the canonical vs. alternative NF-κB pathway in the transforming activity of K13 is not clear at present, it is likely that the alternative pathway plays a major role in this process. This conclusion is supported by recent studies (25, 29) in which deregulated p52 expression has been linked to abnormal lymphocyte proliferation and oncogenic transformation. Along the same lines, in the present study, we demonstrate that siRNA-mediated down-regulation of K13 expression blocks the proliferation of a PEL cell line that was associated with the down-regulation of p52 processing. Whereas inhibition of the canonical NF-κB pathway and the effect of K13 on other growth promoting pathways might have contributed to the observed antiproliferative effect, our results with siRNA against p100 suggest that inhibition of the alternative pathway plays a major role in this process. Inhibitors of the alternative NF-κB pathway may have a role in the treatment of KSHV-associated lymphomas.
Acknowledgments
We thank Drs. Richard Gaynor and Inder Verma for providing the wild-type and IKK-deficient mouse embryonic fibroblast cell lines; Dr. Brian Seed for providing RIP-deficient Jurkat cells; Dr. Robert D. Schreiber for providing wild-type and NIK-deficient mouse embryonic fibroblast cell lines; Dr. Shao-Cong Sun for providing p100 expression plasmids; Catherine Howard for critical reading of the manuscript; and Alejandra Herrera for preparation of figures. This work was supported by National Institutes of Health Grant CA85177.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: KHSV, Kaposi's sarcoma-associated herpesvirus; HA, hemagglutinin; MEF, murine embryonic fibroblast; siRNA, short interfering RNA; vFLIP, viral Fas-associated death domain-like IL-1β-converting enzyme inhibitory protein; cFLIP, cellular FLIP; IKK, IκB kinase; RIP, receptor-interacting protein; NIK, NF-κB-inducing kinase; MRIT, Mach-related inducer of toxicity.
References
- 1.Moore, P. S. & Chang, Y. (2003) Annu. Rev. Microbiol. 57, 609-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thome, M., Schneider, P., Hofmann, K., Fickenscher, H., Meinl, E., Neipel, F., Mattmann, C., Burns, K., Bodmer, J. L., Schroter, M., et al. (1997) Nature 386, 517-521. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhary, P. M., Jasmin, A., Eby, M. T. & Hood, L. (1999) Oncogene 18, 5738-5746. [DOI] [PubMed] [Google Scholar]
- 4.Liu, L., Eby, M. T., Rathore, N., Sinha, S. K., Kumar, A. & Chaudhary, P. M. (2002) J. Biol. Chem. 277, 13745-13751. [DOI] [PubMed] [Google Scholar]
- 5.Field, N., Low, W., Daniels, M., Howell, S., Daviet, L., Boshoff, C. & Collins, M. (2003) J. Cell Sci. 116, 3721-3728. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh, S., May, M. J. & Kopp, E. B. (1998) Annu. Rev. Immunol. 16, 225-260. [DOI] [PubMed] [Google Scholar]
- 7.Pomerantz, J. L. & Baltimore, D. (2002) Mol. Cell 10, 693-695. [DOI] [PubMed] [Google Scholar]
- 8.Claudio, E., Brown, K., Park, S., Wang, H. & Siebenlist, U. (2002) Nat. Immunol. 3, 958-965. [DOI] [PubMed] [Google Scholar]
- 9.Dejardin, E., Droin, N. M., Delhase, M., Haas, E., Cao, Y., Makris, C., Li, Z. W., Karin, M., Ware, C. F. & Green, D. R. (2002) Immunity 17, 525-535. [DOI] [PubMed] [Google Scholar]
- 10.Coope, H. J., Atkinson, P. G., Huhse, B., Belich, M., Janzen, J., Holman, M. J., Klaus, G. G., Johnston, L. H. & Ley, S. C. (2002) EMBO J. 21, 5375-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao, G., Cvijic, M. E., Fong, A., Harhaj, E. W., Uhlik, M. T., Waterfield, M. & Sun, S. C. (2001) EMBO J. 20, 6805-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin, L., Wu, L., Wesche, H., Arthur, C. D., White, J. M., Goeddel, D. V. & Schreiber, R. D. (2001) Science 291, 2162-2165. [DOI] [PubMed] [Google Scholar]
- 13.Ting, A. T., Pimentel-Muinos, F. X. & Seed, B. (1996) EMBO J. 15, 6189-6196. [PMC free article] [PubMed] [Google Scholar]
- 14.Sun, Q., Matta, H. & Chaudhary, P. M. (2003) Blood 101, 1956-1961. [DOI] [PubMed] [Google Scholar]
- 15.Matta, H., Hozayev, B., Tomar, R., Chugh, P. & Chaudhary, P. M. (2003) Cancer Biol. Ther. 2, 206-210. [DOI] [PubMed] [Google Scholar]
- 16.Keller, S. A., Schattner, E. J. & Cesarman, E. (2000) Blood 96, 2537-2542. [PubMed] [Google Scholar]
- 17.Kelliher, M. A., Grimm, S., Ishida, Y., Kuo, F., Stanger, B. Z. & Leder, P. (1998) Immunity 8, 297-303. [DOI] [PubMed] [Google Scholar]
- 18.Matta, H., Sun, Q., Moses, G. & Chaudhary, P. M. (2003) J. Biol. Chem. 278, 52406-52411. [DOI] [PubMed] [Google Scholar]
- 19.Mordmuller, B., Krappmann, D., Esen, M., Wegener, E. & Scheidereit, C. (2003) EMBO Rep. 4, 82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller, J. R. & Siebenlist, U. (2003) J. Biol. Chem. 278, 12006-12012. [DOI] [PubMed] [Google Scholar]
- 21.Yilmaz, Z. B., Weih, D. S., Sivakumar, V. & Weih, F. (2003) EMBO J. 22, 121-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atkinson, P. G., Coope, H. J., Rowe, M. & Ley, S. C. (2003) J. Biol. Chem. 278, 51134-51142. [DOI] [PubMed] [Google Scholar]
- 23.Eliopoulos, A. G., Caamano, J. H., Flavell, J., Reynolds, G. M., Murray, P. G., Poyet, J. L. & Young, L. S. (2003) Oncogene 22, 7557-7569. [DOI] [PubMed] [Google Scholar]
- 24.Luftig, M., Yasui, T., Soni, V., Kang, M. S., Jacobson, N., Cahir-McFarland, E., Seed, B. & Kieff, E. (2004) Proc. Natl. Acad. Sci. USA 101, 141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao, G., Harhaj, E. W. & Sun, S. C. (2001) Mol. Cell 7, 401-409. [DOI] [PubMed] [Google Scholar]
- 26.Senftleben, U., Cao, Y., Xiao, G., Greten, F. R., Krahn, G., Bonizzi, G., Chen, Y., Hu, Y., Fong, A., Sun, S. C. & Karin, M. (2001) Science 293, 1495-1499. [DOI] [PubMed] [Google Scholar]
- 27.Bertin, J., Armstrong, R. C., Ottilie, S., Martin, D. A., Wang, Y., Banks, S., Wang, G. H., Senkevich, T. G., Alnemri, E. S., Moss, B., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 1172-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun, Q., Zachariah, S. & Chaudhary, P. M. (2003) J. Biol. Chem. 278, 52437-52445. [DOI] [PubMed] [Google Scholar]
- 29.Ishikawa, H., Carrasco, D., Claudio, E., Ryseck, R. P. & Bravo, R. (1997) J. Exp. Med. 186, 999-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]