Abstract
Significance: Maintenance of metabolic homeostasis is critical for cellular and organismal health. Proper regulation of mitochondrial functions represents a crucial element of overall metabolic homeostasis. Mitochondrial sirtuins (SIRT3, SIRT4, and SIRT5) play pivotal roles in promoting this homeostasis by regulating numerous aspects of mitochondrial metabolism in response to environmental stressors. Recent Advances: New work has illuminated multiple links between mitochondrial sirtuins and cancer. SIRT5 has been shown to regulate the recently described post-translational modifications succinyl-lysine, malonyl-lysine, and glutaryl-lysine. An understanding of these modifications is still in its infancy. Enumeration of SIRT3 and SIRT5 targets via advanced proteomic techniques promises to dramatically enhance insight into functions of these proteins. Critical Issues: In this review, we highlight the roles of mitochondrial sirtuins and their targets in cellular and organismal metabolic homeostasis. Furthermore, we discuss emerging roles for mitochondrial sirtuins in suppressing and/or promoting tumorigenesis, depending on the cellular and molecular context. Future Directions: Currently, hundreds of potential SIRT3 and SIRT5 molecular targets have been identified in proteomic experiments. Future studies will need to validate the major targets of these enzymes, and elucidate how acetylation and/or acylation modulate their functionality. A great deal of interest exists in targeting sirtuins pharmacologically; this endeavor will require development of sirtuin-specific modulators (activators and inhibitors) as potential treatments for cancer and metabolic disease. Antioxid. Redox Signal. 22, 1060–1077.
Introduction
Mitochondria are cytoplasmic organelles that play central roles in diverse intracellular processes such as energy production, metabolism, apoptosis, intracellular signaling, and pathogen responses (158). Mitochondria are responsible for generating the majority of cellular ATP through oxidative metabolism by the Krebs cycle, β-oxidation of fatty acids, and oxidative phosphorylation (OXPHOS). Consequently, mitochondria are the principal source of reactive oxygen species (ROS) within the cell (92, 163). Under normal physiological conditions, low levels of ROS can function as “redox messengers” in the regulation of specific signaling pathways (62), whereas excess ROS beyond the cell's detoxification capacity can damage cellular macromolecules and promote cell death via the intrinsic apoptotic pathway (28, 146). To neutralize the harmful effects of ROS, cells have evolved numerous antioxidant systems (123). To meet bioenergetic demands of the cell, mitochondrial number, configuration, and/or activity can change in response to a variety of physiological conditions (136). Mitochondrial defects, whether genetic or acquired, are associated with many common diseases, including diabetes and cancer (162, 169). Therefore, normal cellular function requires mechanisms to finely regulate mitochondrial physiology. In recent years, protein post-translational modifications (PTMs) such as ADP-ribosylation and lysine acetylation, succinylation, malonylation, and glutarylation on diverse mitochondrial proteins have emerged as a novel mechanism of mitochondrial regulation. These modifications are directly regulated by members of the sirtuin enzyme family (2, 34, 91, 121, 156).
Sirtuins are mammalian homologues of the yeast silent information regulator 2 (Sir2) protein (42). Sirtuins were initially described as class III histone deacetylases, functionally similar to other HDACs. The sirtuin-catalyzed deacetylation reaction consumes nicotinamide adenine dinucleotide (NAD+) as a co-substrate, and generates the sirtuin feedback inhibitor nicotinamide (NAM), 2′-O-acetyl-ADP-ribose, and a deacetylated substrate (66). However, certain sirtuins are now known to possess alternative catalytic functions—for example, deacylase, decrotonylase, desuccinylase, demalonylase, deglutarylase, and ADP-ribosyltransferase activities (Fig. 1)—that play crucial roles in the regulation of diverse cellular processes (2, 10, 32, 34, 36, 48, 65, 72, 121, 156).
FIG. 1.
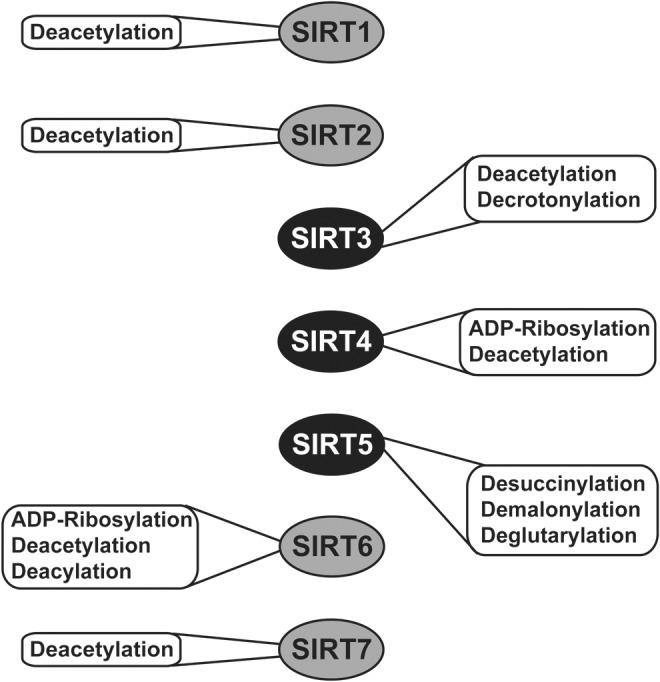
Catalytic activities of mammalian sirtuins. SIRT1, SIRT2, and SIRT7 function primarily as deacetylases, whereas other mammalian sirtuins catalyze alternative reactions, in addition to or instead of deacetylation. SIRT4 acts as both a deacetylase and an ADP-ribosyltransferase. SIRT5 catalyzes desuccinylation, demalonylation, and deglutarylation. SIRT6 catalyzes ADP-ribosylation and deacylation, in addition to deacetylation. Only activities shown to be biologically significant are depicted.
Owing to their NAD+ dependence, fluctuations in NAD+ levels modulate sirtuin catalytic activities. Calorie restriction (CR), a reduction in calorie intake without malnutrition, is an intervention that promotes extended healthy lifespan. CR promotes expression of nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzyme of NAD+ biosynthesis, and thus induces increased levels of intracellular NAD+ (98). Consistently, increased intracellular NAD+ occur during metabolic stresses such as CR or prolonged fasting (19, 20, 105), and are associated with sirtuin activation in a tissue-specific manner (63, 65). Moreover, NAMPT expression is linked to the circadian clock, thereby regulating NAD+ levels and sirtuin activity (120). Sirtuins are widely expressed in normal tissues (99) and play diverse biological roles such as regulating oxidative stress, DNA repair, genomic stability, cell survival, apoptosis, metabolism, aging, and longevity (47).
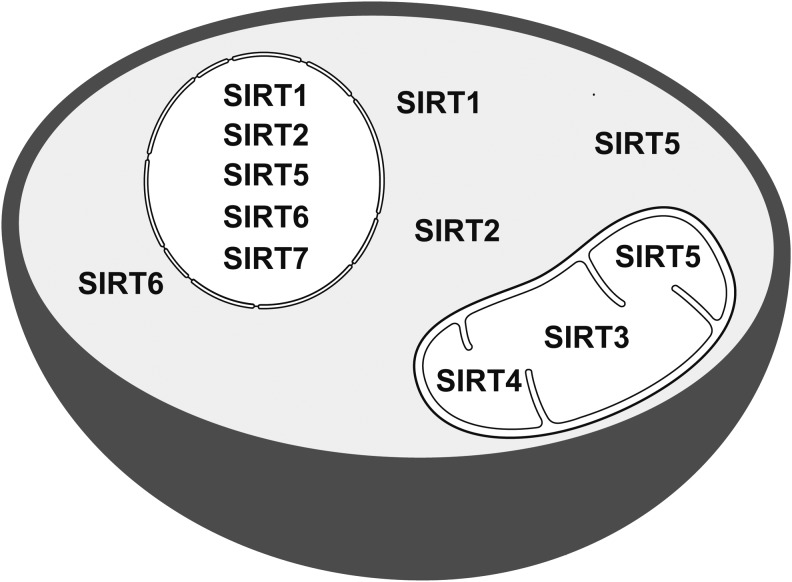
Seven sirtuin members (SIRT1–SIRT7) are encoded in mammalian genomes. These proteins possess a conserved NAD+-binding and catalytic domain, with distinct flanking N- and C-termini, and differ from one another with regard to catalytic activities (Fig. 1), subcellular localization (Fig. 2), protein targets, and biological functions (42, 47). SIRT6 and SIRT7 are found predominantly in the nucleus, whereas both SIRT1 and SIRT2 can be nuclear and cytosolic. SIRT3, SIRT4, and SIRT5 primarily reside in the mitochondrial matrix (25, 47). Here, we focus on SIRT3, SIRT4, and SIRT5: their catalytic activities, protein substrates, major target pathways, and roles in disease, particularly cancer.
FIG. 2.
Subcellular localization of mammalian sirtuins. SIRT7 is present in the nucleus, whereas SIRT1, SIRT2, and SIRT6 are both nuclear and cytosolic. SIRT3, SIRT4, and SIRT5 primarily reside in the mitochondrial matrix; SIRT5 is also found in the cytosol and the nucleus.
SIRT3: Activity, Expression, and Metabolic Regulation
SIRT3 activity
Among mitochondrial sirtuins, SIRT3 is, by far, the best characterized, and possesses robust deacetylase activity (93). SIRT3-deficient mice exhibit elevated mitochondrial protein acetylation (91). Sol et al. have shown that mouse embryonic fibroblasts (MEFs) derived from Sirt3 knockout (KO) mice display increased acetylation of more than 100 lysine sites, mostly on mitochondrial proteins (147). Using high-resolution mass spectrometry, two recent studies provided further support for the role of SIRT3 as a major regulator of the mitochondrial acetylome, particularly in response to CR or fasting (57, 128). Interestingly, SIRT3 deacetylase activity exhibits circadian rhythmicity, attributed to clock-driven oscillation in NAD+ levels in mouse liver (120). Mice with defects in the circadian clock displayed reduced SIRT3 activity, and increased acetylation of multiple mitochondrial enzymes, including well-known targets of SIRT3. NAD+ supplementation restored SIRT3 activity and increased oxygen consumption in mice with defects in the circadian clock (120).
Tan et al. described lysine crotonylation as novel histone PTM, which is specifically enriched at active gene promoters and potential enhancers in mammalian genome (155). A very recent study by Bao et al. reported that SIRT3 possesses decrotonylase activity, using a similar catalytic mechanism as for acetyl lysine hydrolysis (10, 36). siRNA-mediated SIRT3 knockdown (KD) causes accumulation of global histone crotonylation and enrichment of crotonylation on histone 3 lysine 4 (10). SIRT3 has been reported to interact with chromatin in U2OS cells, resulting in the repression of adjacent genes (68). Interestingly, SIRT3 KD leads to increased lysine crotonylation at five of seven genes analyzed in U2OS cells, along with a significant increase in mRNA levels of three candidate genes (10). These observations suggest that SIRT3 might repress the expression of target genes via modulation of histone lysine crotonylation. This implies the surprising conclusion that an active fraction of SIRT3 is present in the nucleus. Indeed, Bao et al. report that a substantial amount of full-length (unprocessed) SIRT3 is nuclear. However, unprocessed human SIRT3 has been reported to be catalytically inactive, at least with regard to deacetylation (140). It is conceivable that a fraction of catalytically active SIRT3 is present extra-mitochondrially. Alternatively, SIRT3 deficiency may modulate cellular metabolism via activity within mitochondria to promote higher intracellular crotonyl-CoA levels, in turn, leading to increased histone crotonylation. This could occur as a consequence of disordered mitochondrial butyrate or glutaryl metabolism (155). Clearly, more work is needed to fully elucidate the role of SIRT3 in modulating crotonyl-lysine levels, particularly in the extra-mitochondrial compartment.
SIRT3 expression may correlate with human longevity
SIRT3 is expressed abundantly in tissues with high oxidative capacity, such as liver, brain, kidney, skeletal muscle, and brown adipose tissue, and its expression is increased by CR, fasting, or exercise (58, 111, 139, 143). Interestingly, increased expression of SIRT3 may be linked with longevity in humans. The presence of SNP rs11555236 in linkage disequilibrium with the putative functional enhancer region of SIRT3, which is a part of variable number of tandem repeat (VNTR) element located within a SIRT3 intron (4, 12), has been reported to be associated with increased longevity in some human populations (4, 12, 131). SIRT3 expression was higher in peripheral blood mononuclear cells from individuals homozygous for this variant (4). Conversely, SIRT3 expression and activity decline in response to high-fat feeding (9, 59, 77, 111), in insulin resistance (174), and in human subjects with the metabolic syndrome (59). Mice with germline ablation of Sirt3 have no grossly apparent phenotype under nonstress conditions (91). Similarly, mice with muscle or liver specific Sirt3 gene deletion are metabolically similar to wild-type (WT) littermates when fed a normal or high-fat diet (HFD), or under conditions of fasting, exercise, or cold challenge (37). However, under stress conditions or with advancing age, SIRT3 plays a crucial role in attenuating the onset of multiple pathologies. In the next section, we will briefly describe the roles of SIRT3 in regulating mitochondrial metabolism and health (summarized in Fig. 3). For more details about SIRT3 biology, readers are referred elsewhere (93).
FIG. 3.
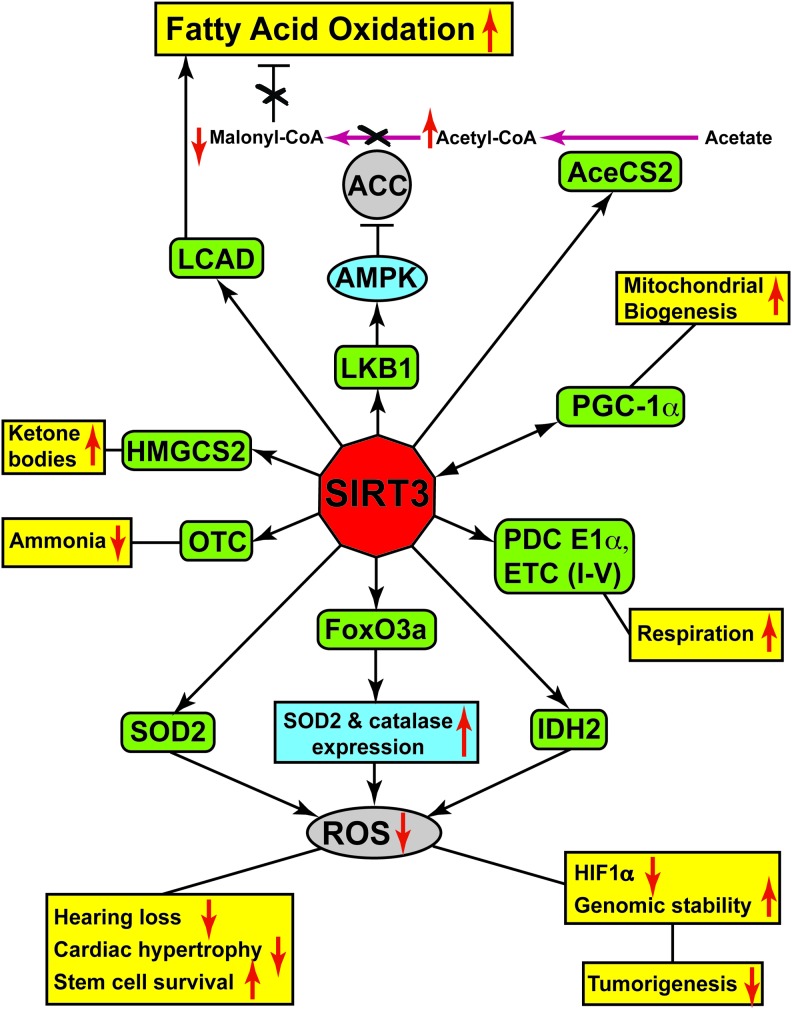
Schematic representation of SIRT3 targets and downstream functions. SIRT3 deacetylates and activates multiple targets (green rounded rectangles), which can either directly regulate key cellular and physiological processes (yellow) or alter the activity (blue ellipse) or expression levels (blue rectangle) of downstream factors. Upward and downward red arrows designate promotion or suppression of particular activity or expression. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
SIRT3 Regulates Multiple Metabolic Pathways
SIRT3 maintains ATP levels by regulating mitochondrial electron transport chain function
A major role of SIRT3 is to maintain cellular ATP levels by promoting mitochondrial electron transport chain (ETC) activity. Various studies have revealed that SIRT3 deacetylates multiple ETC proteins to enhance energy production and improve ETC efficiency, such as NDUFA9 (complex I), SDHA (complex II), and the ATP synthase subunit β (complex V) (1, 9, 27, 40, 77, 79, 125). Moreover, the activities of complexes III and IV are significantly reduced in Sirt3-null mice under high-fat feeding conditions (77). As a consequence, tissues from SIRT3-deficient mice show decreased mitochondrial respiration and reduced ATP levels (1).
Role of SIRT3 in acetate metabolism
Acetyl-CoA synthetase 2 (AceCS2), an enzyme catalyzing the conversion of acetate into acetyl-CoA, was the first SIRT3 substrate identified (53, 138). Under normal feeding conditions, AceCS2-deficient mice display no strong phenotypes; however, under fasting conditions, they show a 75% reduction in acetyl-CoA levels, along with a more than 5-fold increase in plasma acetate levels (134). Indeed, AceCS2 mRNA levels are significantly increased in brown adipose tissue from fasted WT mice (134) and similar to SIRT3, AceCS2 is abundantly expressed in tissues such as heart and skeletal muscle, and its expression is further induced under ketogenic conditions (43). SIRT3 deacetylates and activates AceCS2 (53, 138), indicating a role for SIRT3 in acetyl-CoA generation under fasting conditions in providing the fasting cells with an alternative source of energy.
SIRT3 promotes fatty acid oxidation
SIRT3 plays a pivotal role in regulating fatty acid β-oxidation by deacetylation and activation of long-chain-specific acyl-CoA dehydrogenase (LCAD) (58). Under fasting conditions, SIRT3-deficient mice show decreased fatty acid β-oxidation, resulting in elevated serum long-chain fatty acid levels (54, 58). Inefficient fatty acid oxidation may contribute to the reduced ATP levels of SIRT3-deficient mice under fasting conditions (1).
As previously mentioned, SIRT3 expression and activity decline in response to high-fat feeding (9, 59, 77, 111), and mice lacking SIRT3 show accelerated obesity, insulin resistance, hyperlipidemia, and hepatic steatosis when fed an HFD over a prolonged period (59). In this context, SIRT3-deficient mice fed a HFD expressed elevated levels of the lipogenic enzyme stearoyl-CoA desaturase 1 (SCD1) (59). Increased SCD1 levels are associated with obesity and type 2 diabetes (64). Another study demonstrated that SIRT3 suppresses hepatic lipid accumulation, via AMP-activated kinase (AMPK)-dependent phosphorylation and inhibition of acetyl-CoA carboxylase (ACC) (142). ACC is a biotin-dependent enzyme that plays a crucial role in fatty acid synthesis by catalyzing the production of malonyl-CoA by irreversible carboxylation of acetyl-CoA (133).
SIRT3 deacetylates PDC E1α to promote glucose oxidation
The pyruvate dehydrogenase complex (PDC) catalyzes oxidative decarboxylation of pyruvate into acetyl-CoA, which is subsequently used in Krebs cycle to generate ATP (110, 118). PDC plays a key role in glucose metabolism, linking glycolysis with the Krebs cycle. Roughly half of all caloric intake passes through PDC (117). Phosphorylation of the PDC E1α subunit by pyruvate dehydrogenase kinases (PDK1–4) inhibits PDC activity, whereas dephosphorylation by pyruvate dehydrogenase phosphatases (PDP1 and 2) restores its activity (110). Reduced PDC activity and increased glycolysis are common in cancer cells, as a part of an overall metabolic reprograming associated with malignancy (179). Fan et al. found that increased lysine acetylation of PDC E1α and PDP1 is common in diverse types of human cancer cells (35). Putative mitochondrial acetyltransferases are poorly characterized. Acetyl-CoA acetyltransferase 1 (ACAT1) is a mitochondrial enzyme involved in the final step of isoleucine catabolism, and it converts 2-methyl-acetoacetyl-CoA into propionyl-CoA and acetyl-CoA. Fan et al. found that recombinant ACAT1 directly acetylates PDC E1α and PDP1 in vitro (35). Further, treatment of PDC E1α and PDP1 with cell lysates from ACAT1 KD H1299 cells results in their decreased lysine acetylation and increased activity (35). Conversely, incubation of recombinant PDC E1α with cell lysates from SIRT3 KD H1299 leads to their increased acetylation and decreased activity (35). In EGF-treated cancer cells, phosphorylation of PDP1 results in dissociation of SIRT3 and recruitment of ACAT1, which acetylates PDC E1α and PDP1. This, in turn, induces dissociation of PDP1 from PDC E1α and recruitment of PDK1, leading to reduced PDC activity. (35). Indeed, Sirt3 deletion in skeletal muscle and myoblasts results in increased PDC E1α acetylation, increasing its phosphorylation and decreasing its activity (74). Decreased PDC activity promotes a switch of skeletal muscle metabolism from glucose oxidation toward lactate production and fatty acid utilization (74). Recently, a study by Ozden et al. confirmed the role of SIRT3 in directly deacetylating PDC E1α (110). A PDC E1α mutant mimicking the deacetylated state showed increased activity as compared with an acetylation mimic. Cells expressing an acetylation mimic showed increased proliferation, colony formation, and survival on treatment with ionizing radiation, all of which are characteristics of the transformed state (110).
SIRT3 facilitates ketone body production
SIRT3 also regulates the activity of 3-hydroxy-3-methylglutaryl CoA synthase 2 (HMGCS2) (144), the enzyme that performs the initial step in the conversion of acetyl-CoA into ketone bodies (acetoacetate and β-hydroxybutyrate) in the liver. Ketone bodies, in turn, serve as an important source of energy for tissues such as heart, skeletal muscle, and brain under fasting conditions. SIRT3-mediated HMGCS2 deacetylation promotes its activity and thus facilitates ketone body formation (144). Consequently, Sirt3 KO mice display reduced serum β-hydroxybutyrate levels during prolonged fasting (144).
SIRT3 regulates the urea cycle via ornithine transcarbamoylase activation
Using a high-throughput approach combining acetyl-peptide arrays with metabolomics analysis, Hallows et al. identified ornithine transcarbamoylase (OTC) as a SIRT3 substrate (54). OTC is an enzyme that catalyzes the second step of the urea cycle, the key process in the detoxification of ammonia generated by amino-acid catabolism. SIRT3 directly deacetylates OTC at lysine 88 and stimulates its activity, thus promoting urea cycle function (54). As a consequence, SIRT3-deficient mice display an inability to deacetylate and activate mitochondrial OTC, and show urea cycle functional deficiency in response to CR along with elevated urinary orotic acid levels (54), a well-known clinical marker of human OTC deficiency (38).
SIRT3 Promotes Diverse Aspects of Healthy Aging
SIRT3 prevents hearing loss during CR
Age-related hearing loss (AHL), characterized by a decline of auditory function with increasing age, is associated with attrition of spiral ganglion neurons and sensory hair cells in the cochlea of the inner ear (90, 148). Studies have shown that CR delays the onset of AHL and reduces cochlear pathology (141, 150) by reducing oxidative stress (150), which plays a major role in AHL (73, 149). Notably, SIRT3 is required for the beneficial effect of CR in preserving auditory function. Hearing function is normal in young SIRT3-deficient mice, but with age they lose cochlear cells, manifest hearing loss, even under CR conditions, and display hearing loss (151). SIRT3 maintains hearing by suppressing oxidative stress under CR, by deacetylation and activation of isocitrate dehydrogenase 2 (IDH2). Activation of IDH2, in turn, increases NADPH levels, used to regenerate reduced glutathione, a key component of cellular ROS defense (151, 176).
SIRT3 promotes hematopoietic stem cell survival by reducing oxidative stress
Brown et al. identified a role for SIRT3 in preserving the functions of hematopoietic stem cells (HSCs) during aging by lowering ROS levels (15). SIRT3 is dispensable for HSC function at a young age; however, SIRT3 deficiency results in a reduced HSC pool size in aged mice. Moreover, in a serial transplantation experiment, Sirt3 KO results in impaired HSC self-renewal and reconstitution (15). Indeed, HSCs from WT aged mice show reduced levels of SIRT3 expression and activity, contributing to an increase in ROS levels occurring in aged HSCs. SIRT3 overexpression suppresses ROS levels via deacetylation and activation of the mitochondrial antioxidant enzyme, manganese superoxide dismutase (SOD2), and increases the colony-forming ability of aged HSCs (15). Moreover, in competitive transplantation assay, SIRT3 overexpression results in increased functional reconstitution by HSCs (15). Thus, SIRT3 upregulation rejuvenates aged HSCs by partially rescuing their functional defects.
SIRT3 suppresses cardiac hypertrophy
SIRT3 has also emerged as a crucial factor in maintaining cardiac health (132). Under stress conditions, increased SIRT3 levels in cardiomyocytes are protective against genotoxic and oxidative stress-mediated cell death (154). Furthermore, SIRT3-deficient mice show cardiac hypertrophy basally, and this effect becomes more pronounced in response to hypertrophic stimuli (50, 153). Conversely, SIRT3-overexpressing mice are protected from hypertrophy induction (153). Mechanistically, the cardioprotective role of SIRT3 has been ascribed to multiple functions of this sirtuin. According to one proposed mechanism, SIRT3 reduces cellular ROS levels by deacetylating and activating forkhead box O3a (FoxO3a) (153), a transcription factor that promotes expression of antioxidant-encoding genes such as SOD2 and catalase (86, 157). Another study identified a role for SIRT3 in deacetylating cyclophilin D, a modulator of the mitochondrial permeability transition pore (mPTP), to suppress mPTP opening, thereby inhibiting induction of cell death in cardiomyocytes and potentially in other cell types (50, 145). In addition, SIRT3 catalyzes the deacetylation and activation of LKB1, a serine/threonine kinase that phosphorylates and activates AMPK. Activated AMPK blocks hypertrophy by a number of mechanisms, including promotion of catabolic pathways to upregulate ATP production (122).
SIRT3 regulates mitochondrial fusion
Mitochondrial quality control is crucial for overall cellular health. In this regard, mitochondria continuously undergo fusion and fission; a balance between these processes is necessary to maintain mitochondrial morphology and function (21). Optic atrophy protein 1 (OPA1), a dynamin-related GTPase, mediates the fusion of inner mitochondrial membranes (126). A recent study found that activity of OPA1 is regulated by SIRT3-mediated deacetylation (135). Stress conditions induce OPA1 hyperacetylation, leading to a reduction in its GTPase activity. SIRT3 binds to and deacetylates OPA1 to restore its activity. Consistently, OPA1 isolated from Sirt3 KO cells displayed reduced GTPase activity, and mitochondria isolated from Sirt3 KO hearts showed disorganized mitochondrial morphology (135), suggesting a role for SIRT3 in maintaining mitochondrial integrity via OPA1.
SIRT3 plays a role in mitochondrial biogenesis
Maintenance of mitochondrial function and integrity require the selective degradation of defective mitochondria and the generation of new mitochondria. While defective mitochondria are targeted for degradation by the lysosome for hydrolytic digestion by a process known as mitophagy, the process of mitochondrial biogenesis induces mitochondrial DNA (mtDNA) replication and synthesis of mitochondrial proteins, resulting in increased mitochondrial number and mass (14, 106). A major regulator of mitochondrial biogenesis is PGC-1α, which, by co-activating NRF-1 and NRF-2, induces the expression of nuclear encoded mitochondrial transcription factor A (TFAM) (14, 173). TFAM is an essential protein that binds to mtDNA, regulates mitochondrial transcription initiation, and participates in mitochondrial genome replication (17). A study by Kong et al. reported that PGC-1α promotes expression of the Sirt3 gene in muscle cells and hepatocytes, mediated by an estrogen-related receptor-binding element (ERRE) in the Sirt3 promoter (82). Interestingly, activation of Sirt3 gene expression via ERRE is required for PGC-1α-mediated mitochondrial biogenesis. Overexpression of PGC-1α results in increased mtDNA copy number and induces the transcription of cytochrome c oxidase subunits I, II, and VIIa. However, SIRT3 KD impairs PGC-1α-induced mitochondrial biogenesis and blocks PGC-1α-induced mitochondria-related gene expression (82). Another recent study by Dai et al. showed that treatment of rat cortical neurons with H2O2 causes oxidative stress-induced injury and significantly decreases mtDNA content (31). However, SIRT3 overexpression inhibits H2O2-induced neuronal damage and increases expression of PGC-1α, NRF-1, and TFAM, resulting in increased mtDNA content (31). These findings highlight roles of SIRT3 in regulating mitochondrial biogenesis.
SIRT3 regulates the mitochondrial unfolded protein response
Maintenance of protein homeostasis is essential for cell function and survival. Accumulation of misfolded and aggregated proteins in the mitochondria induces cellular proteotoxic stress and initiates the mitochondrial unfolded protein response (UPRmt) (75). The UPRmt activates expression of nuclear encoded protective genes to reduce proteotoxic stress and to re-establish mitochondrial homeostasis (56, 75). A recent study by Papa and Germain described a novel role for SIRT3 in the UPRmt to coordinate both the antioxidant defenses and mitophagy (112). Proteotoxic stress leads to increased levels of FoxO3a, SOD2, catalase, and the autophagy marker LC3B-II (112). Interestingly, proteotoxic stress also induces SIRT3 expression. siRNA-mediated SIRT3 inhibition expression significantly attenuates LC3B-II induction (112). Moreover, SIRT3 inhibition prevents upregulation of SOD2 and FoxO3a and results in a significant increase in mitochondrial O2− levels after proteotoxic stress. In addition, loss of SIRT3 leads to a decrease in the mitochondrial membrane potential, increases aggregation of mitochondrial proteins, and reduces the viability of cells undergoing proteotoxic stress (112). These observations indicate that SIRT3 acts as a major coordinator of UPRmt induced by mitochondrial proteotoxic stress.
Roles for SIRT3 in Cancer
SIRT3 functions as a tumor suppressor
The tumor suppressor role of SIRT3 was first identified with the observations that SIRT3-deficient cells are more easily transformed than WT controls, and SIRT3-deficient mice develop mammary tumors with a long latency (51, 79). SIRT3 expression is reduced in human breast cancer, colon carcinoma, osteosarcoma, and hepatocellular carcinoma (11, 39, 51, 79, 177, 178). Moreover, deletion of the SIRT3 locus is present in about 20% of all human cancers, and 40% of human breast and ovarian cancers, further supporting a tumor-suppressor role for this protein (39). SIRT3 KD in human cancer cells resulted in increased tumor size and reduced latency in xenografts, whereas SIRT3 overexpression decreased xenograft tumorigenicity, indicating that SIRT3 continues to retard tumor growth in the context of preformed cancer cells (11).
As previously noted, SIRT3 functions as a tumor suppressor, in part by suppressing the production of ROS via deacetylation and activation of antioxidant enzyme SOD2 (124, 159), IDH2 (151), and FoxO3a (153). Increased ROS levels promote nuclear and mitochondrial genome instability, and stabilize hypoxia-inducible factor (HIF) 1-alpha, a part of a protein complex that promotes a shift toward glycolysis, whose upregulation is associated with tumor development (11, 39, 79). SIRT3 also plays other functions that are relevant for tumor suppression. SIRT3 overexpression, by reducing ROS levels, decreases the expression of the transferrin receptor, TfR1, by inhibiting iron regulatory protein 1 (IRP1), thereby suppressing the proliferation of pancreatic cancer cells (70). IRP1, which serves as an ROS sensor (100), displays reduced binding to the iron response element (IRE) in SIRT3 overexpressing cells (70). IRE is found in the 5′ untranslated regions of mRNAs whose protein products are associated with iron metabolism. Furthermore, SIRT3 may also be involved in tumor suppression by modulating the activity of extra-mitochondrial factors. In this regard, Inuzuka et al. found that SIRT3 deacetylates the proto-oncoprotein S-phase kinase-associated protein 2 (Skp2) (67), which is overexpressed in multiple types of cancer and functions as an E3 ubiquitin ligase to target numerous tumor suppressors for proteasome-mediated degradation (41). SIRT3-mediated deacetylation leads to Skp2 nuclear import, thereby preventing its targeting of E-cadherin (67). Reduced E-cadherin expression occurs in many cancer types, and is a characteristic of epithelial-mesenchymal transition and cancer metastasis (160). Overall, these studies indicate that SIRT3 functions as a tumor suppressor by increasing mitochondrial respiration, repressing ROS production, promoting nuclear and mtDNA integrity, destabilizing HIF1-alpha, decreasing TfR1 expression, and promoting nuclear import of Skp2.
Potential role of SIRT3 in tumor promotion
As with other sirtuins (179), there are reports of tumor-promoting roles for SIRT3. SIRT3 is overexpressed in many oral squamous cell carcinomas (OSCCs) relative to normal oral mucosa, and SIRT3 depletion in these cells impairs their growth and proliferation, and sensitizes them to genotoxic therapy (5). However, another study reported that despite increased expression of SIRT3 in OSCC, its catalytic activity is significantly reduced (22). Further, it has been found that 23.8% of OSCC patients analyzed carried a germline point mutation in SIRT3, resulting in substitution of a valine residue with isoleucine at position 208 in the SIRT3 catalytic domain (22). Recombinant SIRT3 with this V208I mutation displayed reduced catalytic efficiency (22). Consistent with the notion that SIRT3 can play an oncogenic function, a recent study reported the presence of an extra copy of the SIRT3 locus in a family with Li-Fraumeni Syndrome (8), an inherited condition characterized by an increased risk of developing diverse cancer types. Consistently, Ashraf et al. reported that increased SIRT3 expression was associated with human lymph node-positive breast cancer (7). In context of these observations, ectopic expression of SIRT3 rescued p53-induced growth arrest in human bladder tumor-derived Ej-p53 cells (87). These opposing roles of SIRT3, as both a tumor suppressor and an oncogene, are context- and cell-type specific.
SIRT4-Regulated Processes: Targets and Physiological Implications
Although SIRT4 possesses a conserved sirtuin deacetylase domain (42), initial reports did not identify any deacetylase activity of this sirtuin (2, 52, 107). Recently, however, SIRT4 has been reported to possess specific deacetylase activity toward at least one particular substrate (see subsequent section on SIRT4 and fatty acid metabolism) (84). SIRT4 also exhibits strong ADP-ribosyltransferase activity (2, 52). In the next sections, we describe the effect of SIRT4-mediated regulation on various target substrates (Fig. 4) and their physiological functions.
FIG. 4.
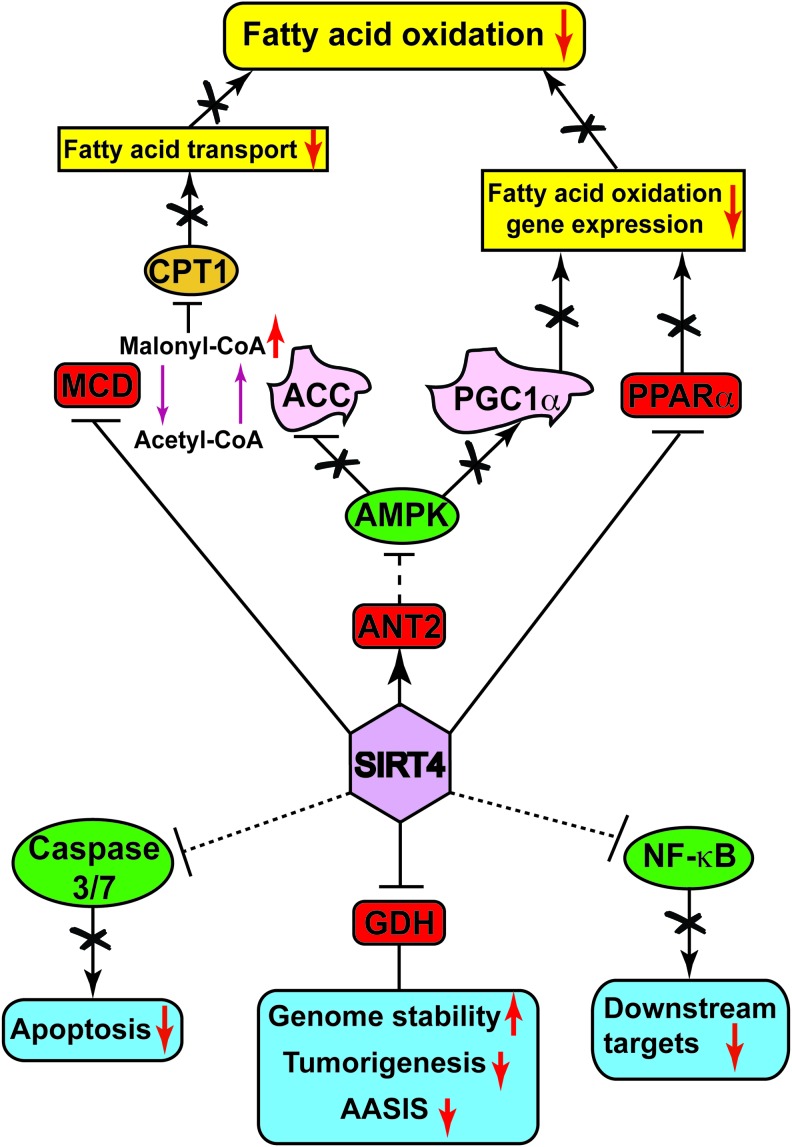
Overview of SIRT4 target substrates and cellular functions. SIRT4 directly (red rounded rectangles) or indirectly (green ellipses) modulates the activity of various target substrates, which either regulate aspects of fatty acid metabolism (yellow rectangles) or play crucial roles in other cellular processes (blue rounded rectangles). Upward and downward red arrows indicate the promotion or suppression of a particular activity, gene expression, or physiological activity. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
SIRT4 suppresses glutamate dehydrogenase activity
The first target of SIRT4 identified was glutamate dehydrogenase (GDH), a mitochondrial enzyme that catalyzes the second step in glutamine catabolism. Glutamine is initially hydrolyzed by glutamate synthase (GLS) to glutamate, which is then subsequently converted to a Krebs cycle intermediate, α-ketoglutarate, by the action of GDH (95). SIRT4 interacts with GDH in pancreatic β-cells and ADP-ribosylates GDH to repress its activity (52). Pancreatic islets isolated from Sirt4 KO mice show increased GDH activity compared with controls. The SIRT4-mediated decrease in GDH activity results in the repression of amino-acid-stimulated insulin secretion (AASIS) in pancreatic β-cells and, thus, Sirt4 KO mice show elevated circulating insulin levels (52). These observations suggest an inhibitory role of SIRT4 in glutamine metabolism by repressing the activity of GDH.
SIRT4 inhibits fatty acid metabolism
SIRT4 deacetylates and inhibits malonyl-CoA decarboxylase
Malonyl-CoA decarboxylase (MCD) is an enzyme that catalyzes the generation of acetyl-CoA from malonyl-CoA. Malonyl-CoA, in turn, allosterically inhibits the activity of carnitine palmitoyltransferase 1 (CPT1), the enzyme that catalyzes mitochondrial uptake of fatty acids for β-oxidation (133). Under nutrient-rich conditions, SIRT4 deacetylates MCD, thereby repressing its activity (84). SIRT4 overexpression in myocytes and adipocytes resulted in reduced MCD activity, whereas muscles and white adipose tissues from Sirt4 KO mice showed elevated MCD activity and a reduction in malonyl-CoA levels (84). As a consequence, Sirt4 KO mice showed increased fatty acid oxidation, associated with increased exercise capacity and resistance to diet-induced obesity (84). However, SIRT4-deficient mice did not show improved insulin sensitivity commensurate with their relative leanness.
SIRT4 inhibits β-oxidation gene expression by repressing PPARα activity
Peroxisome-activated receptor α (PPARα) is a ligand-activated transcription factor that promotes the expression of genes involved in fatty acid catabolism (78, 85). Nasrin et al. reported that SIRT4 KD in mouse primary hepatocytes and myotubes resulted in increased expression of genes encoding fatty acid metabolism enzymes, along with elevated SIRT1 expression (104). Consequently, a significant increase in fatty acid oxidation was observed upon SIRT4 KD in primary hepatocytes, which was found to be SIRT1 dependent, as SIRT1/SIRT4 double KD hepatocytes showed a blunted increase in fatty acid oxidation (104). Recently, SIRT4 overexpression has been shown to repress PPARα transcriptional activity, resulting in suppression of fatty acid catabolism gene expression, and, hence, reduced fatty acid oxidation (83). Conversely, PPARα target gene expression levels were remarkably elevated in Sirt4 KO livers (83). Sirt4 deletion resulted in increased NAD+ levels, leading to increased activity of SIRT1. Interestingly, SIRT1 has been shown to activate PPARα, and this activation was repressed by SIRT4 overexpression (83). Collectively, both studies identify SIRT4 as a suppressor of fatty acid oxidation by inhibiting SIRT1-mediated activation of PPARα.
SIRT4 negatively regulates AMPK activity to suppress fatty acid oxidation
AMPK plays a key role in promoting fatty acid oxidation, by phosphorylating and inhibiting ACC. As previously mentioned, ACC catalyzes the production of malonyl-CoA from acetyl-CoA. Thus, AMPK-driven ACC phosphorylation reduces malonyl-CoA levels, resulting in increased CPT1-mediated mitochondrial fatty acid uptake (3, 133, 172). A recent study showed that during fasting, increased SIRT4 levels inhibit AMPK activity and suppress fatty acid oxidation (61). AMPK also activates PGC-1α, a transcriptional co-activator of fatty acid oxidation genes (164). Consistent with these reports, livers from Sirt4 KO mice showed elevated levels of active AMPK, resulting in increased phosphorylated ACC and PGC1-α induction (61). ANT2 is a mitochondrial protein associated with the inner mitochondrial membrane that catalyzes the exchange of ATP generated in the mitochondria with cytosolic ADP (119). ANT2 KD in SIRT4-overexpressing cells rescues decreased AMPK activity, indicating that ANT2 plays a crucial role in SIRT4-dependent AMPK regulation (61). Altogether, SIRT4 suppresses fatty acid oxidation by modulating the activity of MCD, PPARα, and AMPK.
Other targets of SIRT4
Caspases are a family of cysteine proteases that play key roles in apoptosis. SIRT4 overexpression decreases the activities of caspases 3 and 7 under hypoxic conditions, and reduces induction of caspases 3 and 9 by hypoxia (89). Consequently, SIRT4 protects H9c2 cardiomyoblast cells against hypoxia-induced apoptosis (89). More recently, SIRT4 has been shown to inhibit the activity of NF-κB by inhibiting IkBα degradation (23). NF-κB is a transcription factor that plays a key role in inflammatory responses, and transcriptionally regulates the expression of surface adhesion molecules, such as VCAM-1 and E-selectin. Treatment of human pulmonary microvascular endothelial cells (HPMECs) with cigarette smoke extract (CSE) strongly induces expression of VCAM-1 and E-selectin (23). However, overexpression of SIRT4 in HPMECs inhibits the CSE-induced expression of these surface adhesion molecules and mononuclear cell adhesion (23). Tissues and cells from Sirt4 KO mice show reduced ATP levels, whereas overexpression of SIRT4 increases ATP content (61). The interaction of SIRT4 with ANT2 is essential for maintaining ATP homeostasis (61). However, there are no reports showing that SIRT4 biochemically modifies ANT2.
SIRT4 Acts as a Tumor Suppressor via Repression of Glutamine Metabolism
SIRT4 mRNA levels are reduced in several human cancers (13, 26, 45, 71, 170), and reduced SIRT4 mRNA levels correlate with inferior survival in patients with lung tumors (71). Recent studies confirmed that SIRT4 indeed acts as a tumor suppressor, by repressing glutamine metabolism and promoting genomic stability (30, 69, 71). Glutamine is a key amino acid required for diverse intracellular processes such as macromolecular synthesis, redox homeostasis, oxidative metabolism, and many others (95). Although most mammalian cells can synthesize glutamine, under conditions of rapid cell proliferation, such as cancer, a steady extracellular source of glutamine becomes essential. Glutamine serves as an anaplerotic substrate by replenishing the Krebs cycle via α-ketoglutarate, a product of glutamine catabolism. Consistently, many cancer cells are “glutamine addicted,” and require exogenous glutamine to support survival and proliferation (95). For example, cell cycle progression in HeLa cells is absolutely dependent on glutamine (29). Jeong et al. showed that genotoxic stress, which arrests cell cycle progression to allow DNA damage repair, induces SIRT4 expression, which, in turn, represses mitochondrial glutamine metabolism (71). Sirt4 KO MEFs show increased entry of glutamine-derived metabolites into the Krebs cycle, and are unable to repress cellular glutamine uptake in response to DNA damage (71). Moreover, these cells display an increased proliferation rate, a phenotype abrogated by inhibitors of glutamine metabolism, highlighting the glutamine-dependent proliferation of these cells (71). Consistently, HeLa cells, which use glutamine as a major energy source, show growth inhibition in response to SIRT4 overexpression (71). Moreover, SIRT4 deficiency is associated with larger tumor formation in a nude mice allograft model, and two independently derived strains of Sirt4 KO mice displayed increased incidence of spontaneous lung tumors (71).
The tumor-suppressor activity of SIRT4 was further evaluated in the context of Myc-driven human Burkitt lymphoma cells (69). c-Myc is a transcription factor that upregulates glutaminase expression, by reducing the expression of inhibitory microRNAs targeting this mRNA (95). Myc-driven cancers typically show marked glutamine dependence (44, 168). Overexpression of SIRT4 reduces glutamine utilization in Burkitt lymphoma cells, inhibits their proliferation, and sensitizes them to glucose depletion (69). Moreover, loss of SIRT4 in a mouse Burkitt lymphoma model increases lymphomagenesis and mortality. Malignant B cells derived from these mice display increased glutamine uptake and GDH activity (69).
Csibi et al. showed that a mechanistic target of rapamycin complex 1 (mTORC1) negatively regulates SIRT4 expression by promoting proteasome-mediated degradation of the SIRT4 transcriptional regulator CREB2 (30). Tuberous sclerosis 2 (TSC2) is a negative regulator of mTORC1; thus, Tsc2 KO MEFs show increased mTORC1 activation. Inhibition of mTORC1 activity by rapamycin results in increased SIRT4 expression and reduced GDH activity in Tsc2 KO cells (30). Consistently, SIRT4 overexpression inhibits transformation and proliferation of Tsc2 KO MEFs in vitro, and delays tumor development in xenograft models (30).
SIRT5 Regulates Newly Described PTMs
SIRT5 is phylogenetically most closely related to prokaryotic (so-called class III) sirtuins (42). SIRT5 is broadly expressed with the highest expression levels in brain, heart, liver, kidney, muscles, and testis (99, 102). SIRT5 is predominantly mitochondrial (33, 99, 102, 137); however, several reports have revealed the existence of functional extra-mitochondrial SIRT5 (46, 96, 116). In this regard, Park et al. reported that a significant amount of SIRT5 is present in the cytosol in mouse hepatocytes and human 293T cells, and that a number of cytosolic and nuclear proteins, in addition to many mitochondrial proteins, were hypersuccinylated in the absence of SIRT5 (116). To date, no strong phenotype or major metabolic abnormality has been described in Sirt5 KO mice (91, 101, 175). Thus, SIRT5 seems to be largely dispensable for gross metabolic homeostasis under basal, unstressed conditions, which is true of most sirtuins except SIRT1 and SIRT6. SIRT5 overexpression was reported to enhance ATP synthesis and oxygen consumption in HepG2 cells, whereas SIRT5 KD had no effect in this context (16). Conversely, SIRT5 KD human cells and mitochondria isolated from SIRT5-deficient mouse livers showed increased respiration in the presence of succinate and pyruvate, indicating that SIRT5 inhibits mitochondrial respiration under some conditions (116).
SIRT5 and Protein Deacylation
Based on homology to other sirtuins, SIRT5 was originally annotated as a deacetylase (42). However, recently SIRT5 has been shown to preferentially deacylate negatively charged modifications: malonylation, succinylation, and glutarylation (34, 116, 121, 127, 156). Du et al. showed that SIRT5 possesses minimal deacetylase activity compared with SIRT1 and SIRT3 (34); however, the catalytic efficiency of SIRT5 for demalonylation and desuccinylation was much higher than for deacetylation (34, 121). Protein lysates from SIRT5-deficient livers showed increased lysine malonylation and lysine succinylation, with little impact on lysine acetylation (121). Du et al. described the presence of an arginine residue (Arg105) and tyrosine residue (Tyr102) in the acyl-binding pocket of SIRT5, which are conserved in most class III sirtuins (34). The presence of arginine and tyrosine residues in the catalytic pocket of SIRT5 is likely responsible for their preference for negatively charged acyl groups.
Via mass spectrometry approaches, two independent studies have identified multiple SIRT5 succinylated targets in mouse liver mitochondria (127), and globally in MEFs and liver tissues (116). Among the quantifiable sites identified by Park et al., more than 90% showed hypersuccinylation in Sirt5 KO cells, strongly suggesting that SIRT5 is a major regulator of lysine succinylation in mammals (116). Similarly, Rardin et al. reported that SIRT5 deficiency resulted in hypersuccinylation of 32% sites in 56% of mitochondrial proteins overall (127). The preference of SIRT5 for negatively charged acyl groups was further corroborated by another recent study (156). Tan et al. identified and validated lysine glutarylation as an evolutionary conserved post-translational modification; similar to lysine succinylation, this modification was also regulated by SIRT5 (156). Proteomic screening of liver extracts from Sirt5 KO mice revealed hyperglutarylation of 683 lysine sites on 191 proteins; more than three quarters of these proteins were found to be mitochondrial (156). These studies emphasize the major cellular role of SIRT5 in removing negatively charged lysine modifications, primarily within the mitochondrial matrix.
Although many metabolic enzymes have been identified as malonylated or succinylated, the significance of these PTMs is still unclear. It has been proposed that protein acylation results from the nonenzymatic lysine modification (167), due to accumulation of intrinsically reactive carbon metabolites, which can negatively impact protein function and, hence, disrupt cellular homeostasis (166). Therefore, by removing these lysine PTMs, sirtuins may contribute to maintaining the quality of the proteome, especially in mitochondria.
Metabolic Targets of SIRT5
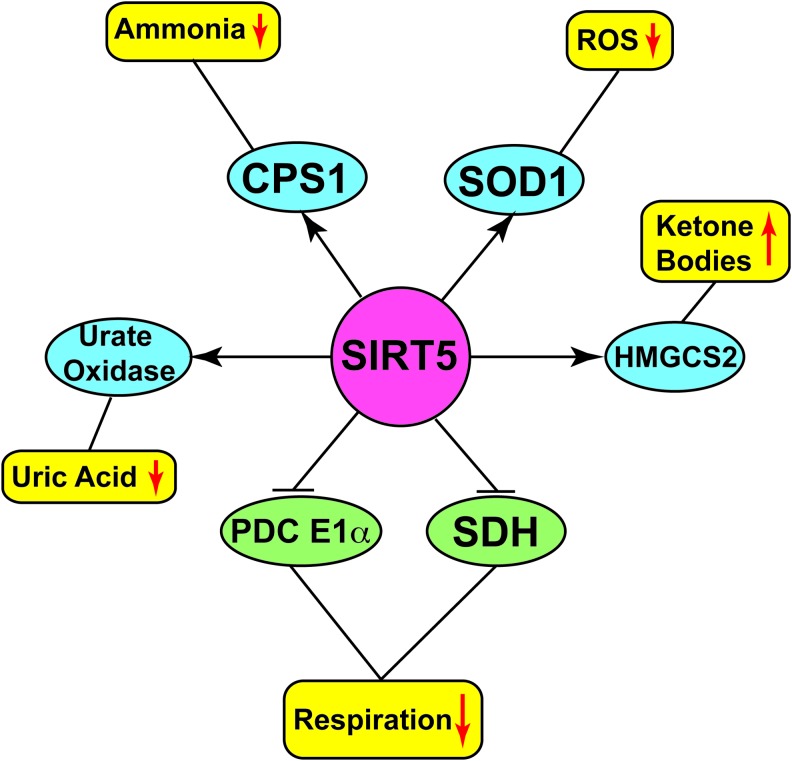
Similar to other sirtuins, SIRT5 targets a number of protein substrates (Fig. 5) implicated in diverse metabolic pathways. Cytochrome c, a mitochondrial protein with central roles in oxidative metabolism and apoptosis, was the first reported target of SIRT5 (137). However, no in vivo evidence indicates a role for SIRT5 in regulating functions of this protein. Carbamoyl phosphate synthetase 1 (CPS1) is the enzyme catalyzing the initial step of the urea cycle for ammonia detoxification and disposal (55, 97). Deacetylation (102), desuccinylation (34), and deglutarylation (156) of CPS1 by SIRT5 result in its increased enzymatic activity. Sirt5-null mice display lower CPS1 activity (34, 102) and have reduced capacity to detoxify ammonia. During conditions of high amino-acid catabolism (fasting, CR, or a high protein diet), SIRT5-deficient mice showed elevated blood ammonia levels (102). These findings were further complemented by another study, which revealed increased expression of Sirt5 mRNA in the livers of WT mice during fasting, and, in addition, increased CPS1 activity in the livers of SIRT5-overexpressing transgenic mice (108). Analysis of mitochondrial proteins in the livers of SIRT5-overexpressing transgenic mice identified urate oxidase as another target of SIRT5 (103). Urate oxidase catalyzes the conversion of urate to allantoin, the last step of purine catabolism in most mammals (but not humans) (6). In the livers of SIRT5-overexpressing transgenic mice, urate oxidase showed decreased acetylation and increased activity (103).
FIG. 5.
Major targets regulated by SIRT5. SIRT5 deacetylates, desuccinylates, demalonylates, and/or deglutarylates multiple metabolic enzymes to activate (blue ellipses) or inhibit (green ellipses), either increasing (yellow rounded rectangle with upward red arrow) or decreasing (yellow rounded rectangles with downward red arrow) the levels/activity of particular compound/cellular activity. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
In the context of ROS management, it was recently shown that SIRT5 binds to, desuccinylates, and activates Cu/Zn superoxide dismutase (SOD1). SOD1 is a key cellular antioxidant enzyme, and SOD1-mediated ROS detoxification is significantly increased when SOD1 is co-overexpressed with SIRT5 (88). A number of observations potentially implicate SOD1 in tumorigenesis (114). For example, increased SOD1 expression was found in a panel of breast cancer cell lines (113); overexpression of SOD1 promotes growth of lung cancer cells (152); and inhibition of SOD1 induces cell death in the lung carcinoma cell line A549 (49). Mutation of the SOD1 succinylation site inhibited the growth of lung cancer cells (88), suggesting a role for SIRT5-mediated SOD1 desuccinylation and activation in promoting tumorigenesis.
In their global analysis of lysine succinylation, Park et al. reported widespread succinylation in diverse mitochondrial metabolic enzymes (116). Among a large number of putative SIRT5 targets, they further analyzed the role of SIRT5 in regulating PDC E1α and succinate dehydrogenase (SDH), an enzyme that catalyzes the oxidation of succinate to fumarate. SIRT5 robustly desuccinylated PDC E1α in vitro and repressed its activity. Consequently, SIRT5 KD resulted in elevated PDC E1α activity, along with a substantial increase in SDH activity and elevated cellular respiration (116). Rardin et al. showed hypersuccinylation of HMGCS2 in the absence of SIRT5 (127). HMGCS2 is the rate-limiting enzyme of ketone body synthesis, and hypersuccinylation decreases its activity. Consequently, there is a mild defect in ketone body formation during fasting in SIRT5-deficient animals (127). Loss of SIRT5 also leads to hypersuccinylation of proteins involved in fatty acid β-oxidation; liver and skeletal muscle from Sirt5 KO mice show modest accumulation of medium- and long-chain acylcarnitines (127).
Overall, SIRT5 target a number of protein substrates involved in glucose oxidation, ketone body formation, fatty acid oxidation, ammonia detoxification, and ROS management. One way to rationalize the functions of SIRT5 may be that this protein suppresses glucose oxidation while facilitating use of alternative energy sources, such as fatty acids, ketone bodies, and amino acids. These conditions occur during fasting and CR. However, no role for SIRT5 in CR has been directly identified as yet.
SIRT5 and Cancer
Until recently, no reports assessed roles for SIRT5 in any type of malignancy. A recent analysis of human high-grade serous ovarian carcinomas found that the region encompassing the SIRT5 locus was amplified in 30% of these tumors (18). Altered activities of SIRT5 targets, PDC and SDH, are implicated in neoplasia and cancer cell metabolic reprogramming. As previously noted, PDC activity is frequently suppressed in tumor cells, in part via reversible phosphorylation (60, 76, 80, 81, 109, 115, 129, 130). It is possible that SIRT5 contributes to metabolic reprogramming in cancer cells, potentially by participating in PDC inhibition. Very recently, Lu et al. showed that SIRT5 is overexpressed in advanced non-small cell lung cancer (NSCLC), and that SIRT5 KD repressed the growth rate of NSCLC cell lines (94). SIRT5 KD in these cells increased their susceptibility to genotoxic drugs (94). NRF2 is a transcription factor that regulates development and drug resistance of human NSCLC (24). NSCLCs show NRF2 upregulation, and SIRT5 KD resulted in reduced NRF2 levels, and expression of downstream targets of NRF2 (94). These observations highlighted the potential role of SIRT5 in promoting lung cancer growth and drug resistance, by promoting the expression of NRF2 and its downstream targets (94). Another study proposed that SIRT5-mediated SOD1 desuccinylation and activation might be relevant in lung tumor cell growth (88). SOD1 KD lung tumor cells expressing succinylation-resistant SOD1 mutant displayed reduced proliferation (88), potentially indicating that regulation of SOD1 via succinylation is critical for lung tumor cell growth. Clearly, further studies are warranted to explore the possible involvement of SIRT5 in tumorigenesis and reprograming of cancer cell metabolism.
An Interplay Between Mitochondrial Sirtuins Contributes to Metabolic Homeostasis
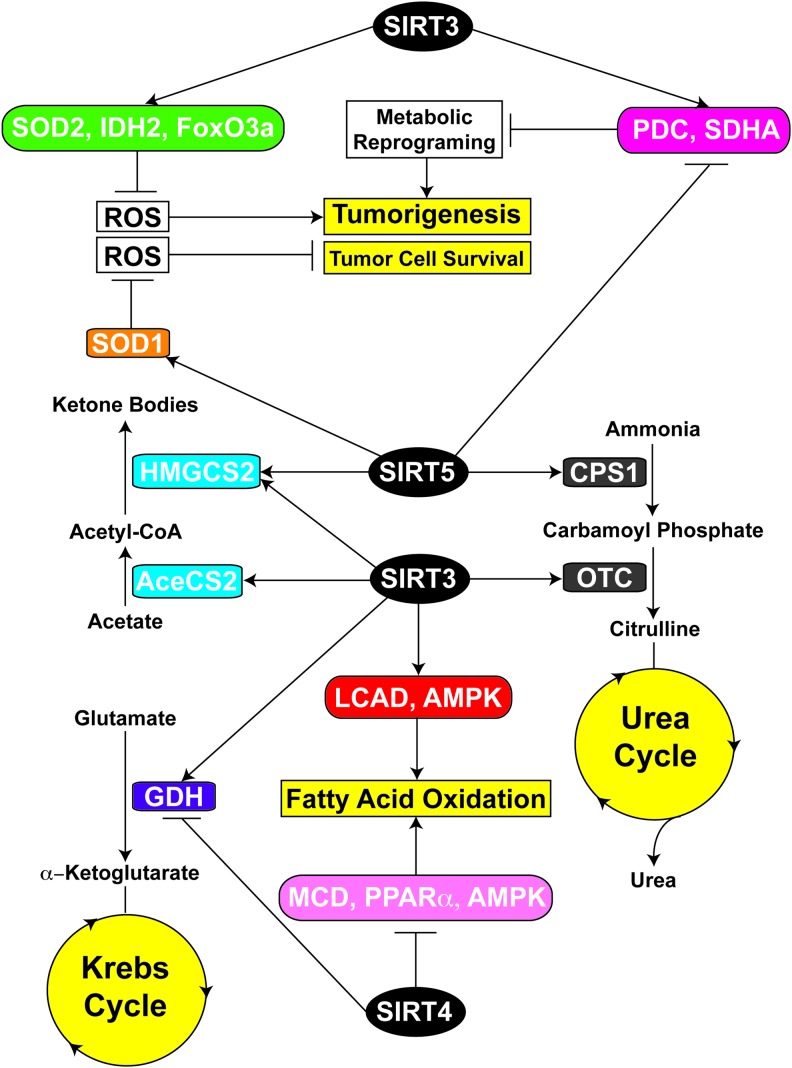
Maintenance of metabolic homeostasis is crucial for proper cellular function, and it is achieved by tightly regulated interactions among different metabolic pathways under diverse physiological conditions. Mitochondrial sirtuins are critical regulators of multiple metabolic processes, and they influence numerous aspects of metabolic homeostasis in a carefully orchestrated manner (Fig. 6). For instance, under chow feeding conditions, SIRT4 represses AASIS by inhibiting the activity of GDH (52), which catalyzes the conversion of glutamate into the Krebs cycle intermediate α-ketoglutarate (95), thereby promoting cellular glucose metabolism. However, during CR, SIRT4 activity is suppressed, and thus GDH is released from SIRT4-mediated inhibition (52, 165). Moreover, it has been reported that acetylation of GDH also reduces its activity, whereas SIRT3-mediated deacetylation activates it (91, 137). Therefore, it is possible that during CR, increased SIRT3 activity induces hepatic glucose production from amino acids by activating GDH-mediated conversion of glutamate into α-ketoglutarate (91, 137), which fuels the Krebs cycle. Another example where coordination between SIRT3 and SIRT4 has been observed is the regulation of FAO. As noted, during fasting, SIRT3 promotes FAO through LCAD and AMPK (58, 142). Conversely, SIRT4 suppresses FAO by negatively regulating the activities of MCD, PPARα, and AMPK (83, 84). Further, SIRT4 KD increases the expression of SIRT1, which activates PPARα and increases FAO gene expression (83, 104). Collectively, these results suggest that under conditions of glucose deprivation, crosstalk between SIRT3 and SIRT4 may stimulate utilization of alternative energy sources such as amino acids and fatty acids.
FIG. 6.
Overview of interplay between mitochondrial sirtuins. Enzymes involved in ketogenesis (blue boxes) and urea cycle (gray boxes) are activated by both SIRT3 and SIRT5. While SIRT5 inhibits the enzymatic activities of glucose metabolism (magenta box), SIRT3 activates them to suppress metabolic reprogramming and tumorigenesis. SIRT3 also suppresses tumorigenesis by decreasing ROS levels through activation of antioxidant machinery (green box). SIRT5 activates SOD1 (orange box) to maintain ROS below toxic levels to support tumor cell survival. SIRT3 also shares common target/pathways with SIRT4. SIRT3 promotes FAO by activating LCAD and AMPK (red box), whereas SIRT4 represses FAO through inhibition of MCD, PPARα, and AMPK (pink box). Similarly, SIRT3 activates GDH (violet box), which is inhibited by SIRT4. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Recent studies illustrate potential crosstalk between SIRT3 and SIRT5, both of which activate the enzymes involved in ketogenesis and the urea cycle. Ketone bodies, an important source of energy for extrahepatic tissues under fasting conditions, are produced in mitochondria from acetyl-CoA. SIRT3-mediated activation of AceCS2 results in enhanced acetyl-CoA production (53, 138). Acetyl-CoA is then converted into 3-hydroxy-3-methylglutaryl-CoA by HMGCS2, which is finally converted into ketone bodies. HMGCS2 is activated by both SIRT3-mediated deacetylation (144) and SIRT5-mediated desuccinylation (127). In addition, under the conditions of fasting, amino acids are catabolized as a carbon source for gluconeogenesis. Toxic ammonia is generated in this process, requiring conversion into urea for proper disposal via the urea cycle. During CR, SIRT3 and SIRT5 play pivotal roles in the urea cycle by activating OTC (54) and CPS1 (34, 102, 156), respectively. CPS1 catalyzes the initial rate-limiting step in the urea cycle and converts ammonia into carbamoyl phosphate (171). OTC is the second enzyme involved in the mitochondrial urea cycle, and catalyzes the conversion of carbamoyl phosphate into citrulline (171). Although modulation of enzymatic activities in ketogenesis and urea cycle by SIRT3 and SIRT5 highlights their important roles in the adaptive response to fasting, mechanistic understanding of this coordination is incomplete.
We have already discussed roles of SIRT3 and SIRT5 in ROS regulation, generated as a by-product of OXPHOS, and also in the modulation of activities of enzymes involved in glucose metabolism. While ROS are produced as a product of normal cellular functioning, increased ROS levels often result in oxidative stress and adversely affect genomic stability, promoting tumorigenesis (161). Moreover, increased ROS levels stabilize HIF1α, which promotes a shift toward glycolysis, providing advantages to rapidly dividing tumor cells (11, 39, 79). As described earlier, SIRT3 enhances the ability of mitochondria to detoxify ROS by activating IDH2 (151) and SOD2 (124, 159) and potentially by increasing the expression of antioxidants through an interaction with FoxO3a (153). Therefore, by reducing the ROS levels, SIRT3 functions as a tumor suppressor by maintaining genomic stability and destabilizing HIF1α. As previously noted, elevated ROS promote carcinogenesis; however, excessive ROS levels beyond a toxic threshold may overwhelm cellular antioxidant capacity and trigger cell cycle arrest and apoptosis (161). Therefore, reduced SIRT3 expression in cancer cells is likely to impose increased demand on the antioxidant machinery to protect cells from the deleterious effects of elevated ROS levels. Studies from several groups indicate an important role of SOD1 in tumor initiation and progression (114). SOD1 is overexpressed in many types of cancer cells (113, 152), and its activity may be essential for limiting ROS levels to a level that is consistent with robust cellular proliferation. In this regard, a recent study by Lin et al. found that succinylation of SOD1 leads to decreases in its activity (88). SIRT5 binds to, desuccinylates, and activates SOD1. Expression of SIRT5 potentiates SOD1-mediated ROS scavenging (88). These results highlight a potential role of SIRT5 in the defense mechanisms of cancer cells against ROS-induced apoptosis, by promoting SOD1 activity. In this context, Lu et al. showed that SIRT5 is overexpressed in NSCLC, and SIRT5 KD results in the repression of NSCLC cells growth (94). In addition, SIRT5 represses the activities of PDC and SDH (116), both of which have a critical role in cancer cell metabolic reprogramming, and PDC activity is suppressed in many types of tumor cells. This represents an example of mutual antagonism between SIRT3 and SIRT5, as SIRT3 increases the activities of PDC (35) and SDHA (27, 40).
Concluding Remarks
Recent findings in sirtuin biology have highlighted the importance of mitochondrial sirtuins in regulating multiple metabolic pathways. Owing to their important roles in metabolic regulation, mitochondrial sirtuins may represent attractive candidates for development of therapeutic interventions against cancer and other diseases. However, many protein targets are common among mitochondrial sirtuins, and the degree to which these proteins functionally interact with each other has yet to be addressed. Additional work is also required to better understand potential redundancy in their functions, and the ways they respond to different environmental stimuli. Finally, the identification of novel lysine acyl modifications regulated by mitochondrial sirtuins and the diverse array of their putative targets suggest that our understanding of these important regulators is truly still in its infancy.
Abbreviations Used
- AASIS
amino acid stimulated insulin secretion
- ACAT1
acetyl-CoA acetyltransferase 1
- ACC
acetyl-CoA carboxylase
- AceCS2
acetyl-CoA synthetase 2
- AHL
age-related hearing loss
- AMPK
AMP-activated kinase
- ANT2
ATP/ADP translocase 2
- CPS1
carbamoyl phosphate synthetase 1
- CPT1
carnitine palmitoyltransferase 1
- CR
calorie restriction
- CREB2
cAMP response element binding protein 2
- CSE
cigarette smoke extract
- ERRE
estrogen-related receptor-binding element
- ETC
electron transport chain
- FoxO3a
forkhead box protein O3a
- GDH
glutamate dehydrogenase
- GLS
glutamate synthase
- HFD
high-fat diet
- HIF1α
hypoxia-inducible factor 1-alpha
- HMGCS2
3-hydroxy-3-methylglutaryl CoA synthase 2
- HPMECs
human pulmonary microvascular endothelial cells
- HSCs
hematopoietic stem cells
- IDH2
isocitrate dehydrogenase 2
- IRE
iron response element
- IRP1
iron regulatory protein 1
- KD
knockdown
- KO
knockout
- LCAD
long-chain-specific acyl-CoA dehydrogenase
- MCD
malonyl-CoA decarboxylase
- MEFs
mouse embryonic fibroblasts
- mPTP
mitochondrial permeability transition pore
- mtDNA
mitochondrial DNA
- mTORC1
mechanistic target of rapamycin complex 1
- NAD+
nicotinamide adenine dinucleotide
- NAM
nicotinamide
- NAMPT
nicotinamide phosphoribosyltransferase
- NDUFA9
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 9
- NSCLC
non-small cell lung cancer
- OPA1
optic atrophy protein 1
- OSCC
oral squamous cell carcinoma
- OTC
ornithine transcarbamoylase
- PDC
pyruvate dehydrogenase complex
- PDK
pyruvate dehydrogenase kinase
- PDP1
pyruvate dehydrogenase phosphatase 1
- PGC-1α
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- PPARα
peroxisome-activated receptor α
- PTM
post-translational modification
- ROS
reactive oxygen species
- SCD1
stearoyl-CoA desaturase 1
- SDH
succinate dehydrogenase
- SDHA
succinate dehydrogenase subunit A
- Skp2
S-phase kinase-associated protein 2
- SOD1
Cu/Zn superoxide dismutase
- SOD2
Mn superoxide dismutase
- TFAM
mitochondrial transcription factor A
- TSC2
tuberous sclerosis 2
- UPRmt
mitochondrial unfolded protein response
- WT
wild type
Acknowledgments
The authors thank William Giblin and Dr. Jeongsoon Park for their critical comments on this article and helpful discussion, and apologize to investigators whose work was not cited due to space limitations. Work in our laboratory is supported by the National Institutes of Health (R01GM101171 and R21CA177925 to D.L.), the Glenn Foundation for Medical Research, and the Discovery Fund of the University of Michigan Comprehensive Cancer Center.
References
- 1.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, and Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A 105: 14447–14452, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, and Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem 282: 33583–33592, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Alam N. and Saggerson ED. Malonyl-CoA and the regulation of fatty acid oxidation in soleus muscle. Biochem J 334 (Pt 1): 233–241, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albani D, Ateri E, Mazzuco S, Ghilardi A, Rodilossi S, Biella G, Ongaro F, Antuono P, Boldrini P, Di Giorgi E, Frigato A, Durante E, Caberlotto L, Zanardo A, Siculi M, Gallucci M, and Forloni G. Modulation of human longevity by SIRT3 single nucleotide polymorphisms in the prospective study “Treviso Longeva (TRELONG)”. Age (Dordr) 36: 469–478, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D'Silva NJ, and Kapila YL. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer 117: 1670–1678, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alvarez-Lario B. and Macarron-Vicente J. Uric acid and evolution. Rheumatology (Oxford) 49: 2010–2015, 2010 [DOI] [PubMed] [Google Scholar]
- 7.Ashraf N, Zino S, Macintyre A, Kingsmore D, Payne AP, George WD, and Shiels PG. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer 95: 1056–1061, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aury-Landas J, Bougeard G, Castel H, Hernandez-Vargas H, Drouet A, Latouche JB, Schouft MT, Ferec C, Leroux D, Lasset C, Coupier I, Caron O, Herceg Z, Frebourg T, and Flaman JM. Germline copy number variation of genes involved in chromatin remodelling in families suggestive of Li-Fraumeni syndrome with brain tumours. Eur J Hum Genet 21: 1369–1376, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao J, Scott I, Lu Z, Pang L, Dimond CC, Gius D, and Sack MN. SIRT3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radic Biol Med 49: 1230–1237, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao X, Wang Y, Li X, Li XM, Liu Z, Yang T, Wong CF, Zhang J, Hao Q, and Li XD. Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. Elife 3, 2014; DOI: 10.7554/eLife.02999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell EL, Emerling BM, Ricoult SJ, and Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene 30: 2986–2996, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, Greco V, Maggiolini M, Feraco E, Mari V, Franceschi C, Passarino G, and De Benedictis G. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics 85: 258–263, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, Devries S, Koppie T, Pejavar S, Carroll P, and Waldman FM. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res 11: 4044–4055, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Brenmoehl J. and Hoeflich A. Dual control of mitochondrial biogenesis by sirtuin 1 and sirtuin 3. Mitochondrion 13: 755–761, 2013 [DOI] [PubMed] [Google Scholar]
- 15.Brown K, Xie S, Qiu X, Mohrin M, Shin J, Liu Y, Zhang D, Scadden DT, and Chen D. SIRT3 reverses aging-associated degeneration. Cell Rep 3: 319–327, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buler M, Aatsinki SM, Izzi V, Uusimaa J, and Hakkola J. SIRT5 is under the control of PGC-1alpha and AMPK and is involved in regulation of mitochondrial energy metabolism. FASEB J 28: 3225–3237, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Campbell CT, Kolesar JE, and Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta 1819: 921–929, 2012 [DOI] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, and Auwerx J. AMPK regulates energy expenditure by modulating NAD(+) metabolism and SIRT1 activity. Nature 458: 1056–1060, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, and Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab 11: 213–219, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan DC. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol 22: 79–99, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Chen IC, Chiang WF, Liu SY, Chen PF, and Chiang HC. Role of SIRT3 in the regulation of redox balance during oral carcinogenesis. Mol Cancer 12: 68, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Wang H, Luo G, and Dai X. SIRT4 inhibits cigarette smoke extracts-induced mononuclear cell adhesion to human pulmonary microvascular endothelial cells via regulating NF-kappaB activity. Toxicol Lett 226: 320–327, 2014 [DOI] [PubMed] [Google Scholar]
- 24.Chen Z, Ye X, Tang N, Shen S, Li Z, Niu X, Lu S, and Xu L. The histone acetylranseferase hMOF acetylates Nrf2 and regulates anti-drug responses in human non-small cell lung cancer. Br J Pharmacol 171: 3196–3211, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi JE. and Mostoslavsky R. Sirtuins, metabolism, and DNA repair. Curr Opin Genet Dev 26C: 24–32, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi YL, Tsukasaki K, O'Neill MC, Yamada Y, Onimaru Y, Matsumoto K, Ohashi J, Yamashita Y, Tsutsumi S, Kaneda R, Takada S, Aburatani H, Kamihira S, Nakamura T, Tomonaga M, and Mano H. A genomic analysis of adult T-cell leukemia. Oncogene 26: 1245–1255, 2007 [DOI] [PubMed] [Google Scholar]
- 27.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, and Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry 49: 304–311, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Circu ML. and Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic Biol Med 48: 749–762, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colombo SL, Palacios-Callender M, Frakich N, Carcamo S, Kovacs I, Tudzarova S, and Moncada S. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc Natl Acad Sci U S A 108: 21069–21074, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, Henske EP, Haigis MC, Cantley LC, Stephanopoulos G, Yu J, and Blenis J. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 153: 840–854, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai SH, Chen T, Wang YH, Zhu J, Luo P, Rao W, Yang YF, Fei Z, and Jiang XF. Sirt3 protects cortical neurons against oxidative stress via regulating mitochondrial Ca2+ and mitochondrial biogenesis. Int J Mol Sci 15: 14591–14609, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominy JE, Jr., Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP, Ruan HB, Feldman J, Pierce K, Mostoslavsky R, Denu JM, Clish CB, Yang X, Shulman GI, Gygi SP, and Puigserver P. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell 48: 900–913, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dryden SC, Nahhas FA, Nowak JE, Goustin AS, and Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol 23: 3173–3185, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, and Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334: 806–809, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, Gu TL, Aguiar M, Lonning S, Chen H, Mohammadi M, Britton LM, Garcia BA, Aleckovic M, Kang Y, Kaluz S, Devi N, Van Meir EG, Hitosugi T, Seo JH, Lonial S, Gaddh M, Arellano M, Khoury HJ, Khuri FR, Boggon TJ, Kang S, and Chen J. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol Cell 53: 534–548, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feldman JL, Dittenhafer-Reed KE, and Denu JM. Sirtuin catalysis and regulation. J Biol Chem 287: 42419–42427, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandez-Marcos PJ, Jeninga EH, Canto C, Harach T, de Boer VC, Andreux P, Moullan N, Pirinen E, Yamamoto H, Houten SM, Schoonjans K, and Auwerx J. Muscle or liver-specific Sirt3 deficiency induces hyperacetylation of mitochondrial proteins without affecting global metabolic homeostasis. Sci Rep 2: 425, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finkelstein JE, Hauser ER, Leonard CO, and Brusilow SW. Late-onset ornithine transcarbamylase deficiency in male patients. J Pediatr 117: 897–902, 1990 [DOI] [PubMed] [Google Scholar]
- 39.Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, and Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell 19: 416–428, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finley LW, Haas W, Desquiret-Dumas V, Wallace DC, Procaccio V, Gygi SP, and Haigis MC. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS One 6: e23295, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frescas D. and Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer 8: 438–449, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun 273: 793–798, 2000 [DOI] [PubMed] [Google Scholar]
- 43.Fujino T, Kondo J, Ishikawa M, Morikawa K, and Yamamoto TT. Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J Biol Chem 276: 11420–11426, 2001 [DOI] [PubMed] [Google Scholar]
- 44.Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, and Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, and Petersen I. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A 98: 13784–13789, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geng YQ, Li TT, Liu XY, Li ZH, and Fu YC. SIRT1 and SIRT5 activity expression and behavioral responses to calorie restriction. J Cell Biochem 112: 3755–3761, 2011 [DOI] [PubMed] [Google Scholar]
- 47.Giblin W, Skinner ME, and Lombard DB. Sirtuins: guardians of mammalian healthspan. Trends Genet 30: 271–286, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gil R, Barth S, Kanfi Y, and Cohen HY. SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic Acids Res 41: 8537–8545, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Glasauer A, Sena LA, Diebold LP, Mazar AP, and Chandel NS. Targeting SOD1 reduces experimental non-small-cell lung cancer. J Clin Invest 124: 117–128, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CM, Rosenzweig A, and Sinclair DA. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY) 2: 914–923, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haigis MC, Deng CX, Finley LW, Kim HS, and Gius D. SIRT3 is a mitochondrial tumor suppressor: a scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res 72: 2468–2472, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, and Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell 126: 941–954, 2006 [DOI] [PubMed] [Google Scholar]
- 53.Hallows WC, Lee S, and Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A 103: 10230–10235, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, Zhao S, Guan KL, and Denu JM. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell 41: 139–149, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haussinger D. Nitrogen metabolism in liver: structural and functional organization and physiological relevance. Biochem J 267: 281–290, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haynes CM. and Ron D. The mitochondrial UPR—protecting organelle protein homeostasis. J Cell Sci 123: 3849–3855, 2010 [DOI] [PubMed] [Google Scholar]
- 57.Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, and Coon JJ. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell 49: 186–199, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr., Alt FW, Kahn CR, and Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464: 121–125, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV, Jr., Kahn CR, and Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell 44: 177–190, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, Ge Q, Gu TL, Polakiewicz RD, Roesel JL, Chen GZ, Boggon TJ, Lonial S, Fu H, Khuri FR, Kang S, and Chen J. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell 44: 864–877, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ho L, Titus AS, Banerjee KK, George S, Lin W, Deota S, Saha AK, Nakamura K, Gut P, Verdin E, and Kolthur-Seetharam U. SIRT4 regulates ATP homeostasis and mediates a retrograde signaling via AMPK. Aging (Albany NY) 5: 835–849, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holmstrom KM. and Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol 15: 411–421, 2014 [DOI] [PubMed] [Google Scholar]
- 63.Houtkooper RH, Canto C, Wanders RJ, and Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev 31: 194–223, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hulver MW, Berggren JR, Carper MJ, Miyazaki M, Ntambi JM, Hoffman EP, Thyfault JP, Stevens R, Dohm GL, Houmard JA, and Muoio DM. Elevated stearoyl-CoA desaturase-1 expression in skeletal muscle contributes to abnormal fatty acid partitioning in obese humans. Cell Metab 2: 251–261, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Imai S, Armstrong CM, Kaeberlein M, and Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800, 2000 [DOI] [PubMed] [Google Scholar]
- 66.Imai S. and Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci 31: 212–220, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Inuzuka H, Gao D, Finley LW, Yang W, Wan L, Fukushima H, Chin YR, Zhai B, Shaik S, Lau AW, Wang Z, Gygi SP, Nakayama K, Teruya-Feldstein J, Toker A, Haigis MC, Pandolfi PP, and Wei W. Acetylation-dependent regulation of Skp2 function. Cell 150: 179–193, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iwahara T, Bonasio R, Narendra V, and Reinberg D. SIRT3 functions in the nucleus in the control of stress-related gene expression. Mol Cell Biol 32: 5022–5034, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jeong SM, Lee A, Lee J, and Haigis MC. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J Biol Chem 289: 4135–4144, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeong SM, Lee J, Finley LW, Schmidt PJ, Fleming MD, and Haigis MC. SIRT3 regulates cellular iron metabolism and cancer growth by repressing iron regulatory protein 1. Oncogene 2014. [Epub ahead of print]; DOI: 10.1038/onc.2014.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jeong SM, Xiao C, Finley LWS, Lahusen T, Souza AL, Pierce K, Li Y-H, Wang X, Laurent G, German NJ, Xu X, Li C, Wang R-H, Lee J, Csibi A, Cerione R, Blenis J, Clish CB, Kimmelman A, Deng C-X, and Haigis MC. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell 23: 450–463, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, and Lin H. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496: 110–113, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang H, Talaska AE, Schacht J, and Sha SH. Oxidative imbalance in the aging inner ear. Neurobiol Aging 28: 1605–1612, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jing E, O'Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S, Bain JR, Lee KY, Verdin EM, Newgard CB, Gibson BW, and Kahn CR. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes 62: 3404–3417, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jovaisaite V, Mouchiroud L, and Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J Exp Biol 217: 137–143, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaal EM, Cascante M, Shlomi T, Gottlieb E, and Peeper DS. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498: 109–112, 2013 [DOI] [PubMed] [Google Scholar]
- 77.Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Van Hove JL, Watson PA, Birdsey N, Bao J, Gius D, Sack MN, Jing E, Kahn CR, Friedman JE, and Jonscher KR. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J 433: 505–514, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, and Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest 103: 1489–1498, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, and Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17: 41–52, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim JW, Gao P, Liu YC, Semenza GL, and Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol 27: 7381–7393, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim JW, Tchernyshyov I, Semenza GL, and Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3: 177–185, 2006 [DOI] [PubMed] [Google Scholar]
- 82.Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, and Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One 5: e11707, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Laurent G, de Boer VC, Finley LW, Sweeney M, Lu H, Schug TT, Cen Y, Jeong SM, Li X, Sauve AA, and Haigis MC. SIRT4 represses peroxisome proliferator-activated receptor alpha activity to suppress hepatic fat oxidation. Mol Cell Biol 33: 4552–4561, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, Lovitch SB, Sharpe AH, Kurland IJ, Steegborn C, Gygi SP, Muoio DM, Ruderman NB, and Haigis MC. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell 50: 686–698, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leone TC, Weinheimer CJ, and Kelly DP. A critical role for the peroxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci U S A 96: 7473–7478, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li M, Chiu JF, Mossman BT, and Fukagawa NK. Down-regulation of manganese-superoxide dismutase through phosphorylation of FOXO3a by Akt in explanted vascular smooth muscle cells from old rats. J Biol Chem 281: 40429–40439, 2006 [DOI] [PubMed] [Google Scholar]
- 87.Li S, Banck M, Mujtaba S, Zhou MM, Sugrue MM, and Walsh MJ. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS One 5: e10486, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin ZF, Xu HB, Wang JY, Lin Q, Ruan Z, Liu FB, Jin W, Huang HH, and Chen X. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem Biophys Res Commun 441: 191–195, 2013 [DOI] [PubMed] [Google Scholar]
- 89.Liu B, Che W, Xue J, Zheng C, Tang K, Zhang J, Wen J, and Xu Y. SIRT4 prevents hypoxia-induced apoptosis in H9c2 cardiomyoblast cells. Cell Physiol Biochem 32: 655–662, 2013 [DOI] [PubMed] [Google Scholar]
- 90.Liu XZ. and Yan D. Ageing and hearing loss. J Pathol 211: 188–197, 2007 [DOI] [PubMed] [Google Scholar]
- 91.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr., Weissman S, Verdin E, and Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27: 8807–8814, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lombard DB, Tishkoff DX, and Bao J. Mitochondrial sirtuins in the regulation of mitochondrial activity and metabolic adaptation. Handb Exp Pharmacol 206: 163–188, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lombard DB. and Zwaans BM. SIRT3: as simple as it seems? Gerontology 60: 56–64, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu W, Zuo Y, Feng Y, and Zhang M. SIRT5 facilitates cancer cell growth and drug resistance in non-small cell lung cancer. Tumour Biol 35: 10699–10705, 2014 [DOI] [PubMed] [Google Scholar]
- 95.Lukey MJ, Wilson KF, and Cerione RA. Therapeutic strategies impacting cancer cell glutamine metabolism. Future Med Chem 5: 1685–1700, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matsushita N, Yonashiro R, Ogata Y, Sugiura A, Nagashima S, Fukuda T, Inatome R, and Yanagi S. Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes Cells 16: 190–202, 2011 [DOI] [PubMed] [Google Scholar]
- 97.Meijer AJ, Lamers WH, and Chamuleau RA. Nitrogen metabolism and ornithine cycle function. Physiol Rev 70: 701–748, 1990 [DOI] [PubMed] [Google Scholar]
- 98.Meynet O. and Ricci JE. Caloric restriction and cancer: molecular mechanisms and clinical implications. Trends Mol Med 20: 419–427, 2014 [DOI] [PubMed] [Google Scholar]
- 99.Michishita E, Park JY, Burneskis JM, Barrett JC, and Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16: 4623–4635, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mueller S. Iron regulatory protein 1 as a sensor of reactive oxygen species. Biofactors 24: 171–181, 2005 [DOI] [PubMed] [Google Scholar]
- 101.Nakagawa T. and Guarente L. Urea cycle regulation by mitochondrial sirtuin, SIRT5. Aging (Albany NY) 1: 578–581, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nakagawa T, Lomb DJ, Haigis MC, and Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137: 560–570, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nakamura Y, Ogura M, Ogura K, Tanaka D, and Inagaki N. SIRT5 deacetylates and activates urate oxidase in liver mitochondria of mice. FEBS Lett 586: 4076–4081, 2012 [DOI] [PubMed] [Google Scholar]
- 104.Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, Ren X, Wu Z, Streeper RS, and Bordone L. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem 285: 31995–32002, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, and Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science 310: 314–317, 2005 [DOI] [PubMed] [Google Scholar]
- 106.Nisoli E. and Valerio A. Healthspan and longevity in mammals: a family game for cellular organelles? Curr Pharm Des 20: 5663–5670, 2014 [DOI] [PubMed] [Google Scholar]
- 107.North BJ, Marshall BL, Borra MT, Denu JM, and Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 11: 437–444, 2003 [DOI] [PubMed] [Google Scholar]
- 108.Ogura M, Nakamura Y, Tanaka D, Zhuang X, Fujita Y, Obara A, Hamasaki A, Hosokawa M, and Inagaki N. Overexpression of SIRT5 confirms its involvement in deacetylation and activation of carbamoyl phosphate synthetase 1. Biochem Biophys Res Commun 393: 73–78, 2010 [DOI] [PubMed] [Google Scholar]
- 109.Oudijk L, Gaal J, Korpershoek E, van Nederveen FH, Kelly L, Schiavon G, Verweij J, Mathijssen RH, den Bakker MA, Oldenburg RA, van Loon RL, O'Sullivan MJ, de Krijger RR, and Dinjens WN. SDHA mutations in adult and pediatric wild-type gastrointestinal stromal tumors. Mod Pathol 26: 456–463, 2013 [DOI] [PubMed] [Google Scholar]
- 110.Ozden O, Park SH, Wagner BA, Song HY, Zhu Y, Vassilopoulos A, Jung B, Buettner GR, and Gius D. Sirt3 deacetylates and increases pyruvate dehydrogenase activity in cancer cells. Free Radic Biol Med 76: 163–172, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Ward JL, 3rd, Goodyear LJ, and Tong Q. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging (Albany NY) 1: 771–783, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Papa L. and Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol 34: 699–710, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Papa L, Hahn M, Marsh EL, Evans BS, and Germain D. SOD2 to SOD1 switch in breast cancer. J Biol Chem 289: 5412–5416, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Papa L, Manfredi G, and Germain D. SOD1, an unexpected novel target for cancer therapy. Genes Cancer 5: 15–21, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Papandreou I, Cairns RA, Fontana L, Lim AL, and Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3: 187–197, 2006 [DOI] [PubMed] [Google Scholar]
- 116.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, Lombard DB, and Zhao Y. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell 50: 919–930, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Patel MS. and Korotchkina LG. Regulation of the pyruvate dehydrogenase complex. Biochem Soc Trans 34: 217–222, 2006 [DOI] [PubMed] [Google Scholar]
- 118.Patel MS, Nemeria NS, Furey W, and Jordan F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J Biol Chem 289: 16615–16623, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pebay-Peyroula E. and Brandolin G. Nucleotide exchange in mitochondria: insight at a molecular level. Curr Opin Struct Biol 14: 420–425, 2004 [DOI] [PubMed] [Google Scholar]
- 120.Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, Levine DC, Bacsik DJ, Gius D, Newgard CB, Goetzman E, Chandel NS, Denu JM, Mrksich M, and Bass J. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342: 1243417, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, and Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics 10: M111. 012658, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pillai VB, Sundaresan NR, Kim G, Gupta M, Rajamohan SB, Pillai JB, Samant S, Ravindra PV, Isbatan A, and Gupta MP. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol Chem 285: 3133–3144, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pirinen E, Lo Sasso G, and Auwerx J. Mitochondrial sirtuins and metabolic homeostasis. Best Pract Res Clin Endocrinol Metab 26: 759–770, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Qiu X, Brown K, Hirschey MD, Verdin E, and Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab 12: 662–667, 2010 [DOI] [PubMed] [Google Scholar]
- 125.Rahman M, Nirala NK, Singh A, Zhu LJ, Taguchi K, Bamba T, Fukusaki E, Shaw LM, Lambright DG, Acharya JK, and Acharya UR. Drosophila Sirt2/mammalian SIRT3 deacetylates ATP synthase beta and regulates complex V activity. J Cell Biol 206: 289–305, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ranieri M, Brajkovic S, Riboldi G, Ronchi D, Rizzo F, Bresolin N, Corti S, and Comi GP. Mitochondrial fusion proteins and human diseases. Neurol Res Int 2013: 293893, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, Uppala R, Fitch M, Riiff T, Zhu L, Zhou J, Mulhern D, Stevens RD, Ilkayeva OR, Newgard CB, Jacobson MP, Hellerstein M, Goetzman ES, Gibson BW, and Verdin E. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab 18: 920–933, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, and Gibson BW. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci U S A 110: 6601–6606, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ricketts C, Woodward ER, Killick P, Morris MR, Astuti D, Latif F, and Maher ER. Germline SDHB mutations and familial renal cell carcinoma. J Natl Cancer Inst 100: 1260–1262, 2008 [DOI] [PubMed] [Google Scholar]
- 130.Ricketts CJ, Forman JR, Rattenberry E, Bradshaw N, Lalloo F, Izatt L, Cole TR, Armstrong R, Kumar VK, Morrison PJ, Atkinson AB, Douglas F, Ball SG, Cook J, Srirangalingam U, Killick P, Kirby G, Aylwin S, Woodward ER, Evans DG, Hodgson SV, Murday V, Chew SL, Connell JM, Blundell TL, Macdonald F, and Maher ER. Tumor risks and genotype-phenotype-proteotype analysis in 358 patients with germline mutations in SDHB and SDHD. Hum Mutat 31: 41–51, 2010 [DOI] [PubMed] [Google Scholar]
- 131.Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, and De Benedictis G. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol 38: 1065–1070, 2003 [DOI] [PubMed] [Google Scholar]
- 132.Sack MN. Emerging characterization of the role of SIRT3-mediated mitochondrial protein deacetylation in the heart. Am J Physiol Heart Circ Physiol 301: H2191–H2197, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu Rev Nutr 28: 253–272, 2008 [DOI] [PubMed] [Google Scholar]
- 134.Sakakibara I, Fujino T, Ishii M, Tanaka T, Shimosawa T, Miura S, Zhang W, Tokutake Y, Yamamoto J, Awano M, Iwasaki S, Motoike T, Okamura M, Inagaki T, Kita K, Ezaki O, Naito M, Kuwaki T, Chohnan S, Yamamoto TT, Hammer RE, Kodama T, Yanagisawa M, and Sakai J. Fasting-induced hypothermia and reduced energy production in mice lacking acetyl-CoA synthetase 2. Cell Metab 9: 191–202, 2009 [DOI] [PubMed] [Google Scholar]
- 135.Samant SA, Zhang HJ, Hong Z, Pillai VB, Sundaresan NR, Wolfgeher D, Archer SL, Chan DC, and Gupta MP. SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Mol Cell Biol 34: 807–819, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schiff M, Benit P, Coulibaly A, Loublier S, El-Khoury R, and Rustin P. Mitochondrial response to controlled nutrition in health and disease. Nutr Rev 69: 65–75, 2011 [DOI] [PubMed] [Google Scholar]
- 137.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, and Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol 382: 790–801, 2008 [DOI] [PubMed] [Google Scholar]
- 138.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, and Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A 103: 10224–10229, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, Giallourakis C, Comb MJ, Alt FW, and Lombard DB. Calorie restriction alters mitochondrial protein acetylation. Aging Cell 8: 604–606, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schwer B, North BJ, Frye RA, Ott M, and Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol 158: 647–657, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Seidman MD. Effects of dietary restriction and antioxidants on presbyacusis. Laryngoscope 110: 727–738, 2000 [DOI] [PubMed] [Google Scholar]
- 142.Shi T, Fan GQ, and Xiao SD. SIRT3 reduces lipid accumulation via AMPK activation in human hepatic cells. J Dig Dis 11: 55–62, 2010 [DOI] [PubMed] [Google Scholar]
- 143.Shi T, Wang F, Stieren E, and Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem 280: 13560–13567, 2005 [DOI] [PubMed] [Google Scholar]
- 144.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, and Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab 12: 654–661, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Shulga N, Wilson-Smith R, and Pastorino JG. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci 123: 894–902, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 146.Sohal RS. and Weindruch R. Oxidative stress, caloric restriction, and aging. Science 273: 59–63, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sol EM, Wagner SA, Weinert BT, Kumar A, Kim HS, Deng CX, and Choudhary C. Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS One 7: e50545, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Someya S. and Prolla TA. Mitochondrial oxidative damage and apoptosis in age-related hearing loss. Mech Ageing Dev 131: 480–486, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Someya S, Xu J, Kondo K, Ding D, Salvi RJ, Yamasoba T, Rabinovitch PS, Weindruch R, Leeuwenburgh C, Tanokura M, and Prolla TA. Age-related hearing loss in C57BL/6J mice is mediated by Bak-dependent mitochondrial apoptosis. Proc Natl Acad Sci U S A 106: 19432–19437, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Someya S, Yamasoba T, Weindruch R, Prolla TA, and Tanokura M. Caloric restriction suppresses apoptotic cell death in the mammalian cochlea and leads to prevention of presbycusis. Neurobiol Aging 28: 1613–1622, 2007 [DOI] [PubMed] [Google Scholar]
- 151.Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, and Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143: 802–812, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Somwar R, Erdjument-Bromage H, Larsson E, Shum D, Lockwood WW, Yang G, Sander C, Ouerfelli O, Tempst PJ, Djaballah H, and Varmus HE. Superoxide dismutase 1 (SOD1) is a target for a small molecule identified in a screen for inhibitors of the growth of lung adenocarcinoma cell lines. Proc Natl Acad Sci U S A 108: 16375–16380, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, and Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest 119: 2758–2771, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, and Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol 28: 6384–6401, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, and Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146: 1016–1028, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, and Zhao Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab 19: 605–617, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tan WQ, Wang K, Lv DY, and Li PF. Foxo3a inhibits cardiomyocyte hypertrophy through transactivating catalase. J Biol Chem 283: 29730–29739, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tang X, Luo YX, Chen HZ, and Liu DP. Mitochondria, endothelial cell function, and vascular diseases. Front Physiol 5: 175, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, and Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell 40: 893–904, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tiwari N, Gheldof A, Tatari M, and Christofori G. EMT as the ultimate survival mechanism of cancer cells. Semin Cancer Biol 22: 194–207, 2012 [DOI] [PubMed] [Google Scholar]
- 161.Trachootham D, Alexandre J, and Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 8: 579–591, 2009 [DOI] [PubMed] [Google Scholar]
- 162.Turner N. and Heilbronn LK. Is mitochondrial dysfunction a cause of insulin resistance? Trends Endocrinol Metab 19: 324–330, 2008 [DOI] [PubMed] [Google Scholar]
- 163.Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol 552: 335–344, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vega RB, Huss JM, and Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol 20: 1868–1876, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Verdin E, Hirschey MD, Finley LW, and Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci 35: 669–675, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wagner GR. and Hirschey MD. Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell 54: 5–16, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wagner GR. and Payne RM. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem 288: 29036–29045, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wahlstrom T. and Arsenian Henriksson M. Impact of MYC in regulation of tumor cell metabolism. Biochim Biophys Acta 2014. [Epub ahead of print]; DOI: 10.1016/j.bbagrm.2014.07.004 [DOI] [PubMed] [Google Scholar]
- 169.Wallace DC. Mitochondria and cancer. Nat Rev Cancer 12: 685–698, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan DW, Tang HM, and Peng ZH. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol 29: 77–83, 2012 [DOI] [PubMed] [Google Scholar]
- 171.Watford M. The urea cycle: a two-compartment system. Essays Biochem 26: 49–58, 1991 [PubMed] [Google Scholar]
- 172.Winder WW, Wilson HA, Hardie DG, Rasmussen BB, Hutber CA, Call GB, Clayton RD, Conley LM, Yoon S, and Zhou B. Phosphorylation of rat muscle acetyl-CoA carboxylase by AMP-activated protein kinase and protein kinase A. J Appl Physiol (1985) 82: 219–225, 1997 [DOI] [PubMed] [Google Scholar]
- 173.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, and Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98: 115–124, 1999 [DOI] [PubMed] [Google Scholar]
- 174.Yechoor VK, Patti ME, Ueki K, Laustsen PG, Saccone R, Rauniyar R, and Kahn CR. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc Natl Acad Sci U S A 101: 16525–16530, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yu J, Sadhukhan S, Noriega LG, Moullan N, He B, Weiss RS, Lin H, Schoonjans K, and Auwerx J. Metabolic characterization of a Sirt5 deficient mouse model. Sci Rep 3: 2806, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yu W, Dittenhafer-Reed KE, and Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem 287: 14078–14086, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang CZ, Liu L, Cai M, Pan Y, Fu J, Cao Y, and Yun J. Low SIRT3 expression correlates with poor differentiation and unfavorable prognosis in primary hepatocellular carcinoma. PLoS One 7: e51703, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang YY. and Zhou LM. Sirt3 inhibits hepatocellular carcinoma cell growth through reducing Mdm2-mediated p53 degradation. Biochem Biophys Res Commun 423: 26–31, 2012 [DOI] [PubMed] [Google Scholar]
- 179.Zwaans BM. and Lombard DB. Interplay between sirtuins, MYC and hypoxia-inducible factor in cancer-associated metabolic reprogramming. Dis Model Mech 7: 1023–1032, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]