Abstract
The torI gene has been identified by using a genetic multicopy approach as a negative regulator of the torCAD operon that encodes the trimethylamine N-oxide reductase respiratory system in Escherichia coli. The negative effect was due to a previously unidentified small ORF (66 aa) of phage origin that we called torI for Tor inhibition. Overexpression of torI led to an 8-fold decrease of the torCAD operon transcription. This operon is positively regulated, in the presence of trimethylamine N-oxide, by a four-step phosphorelay involving the TorS sensor and the TorR response regulator. Epistatic experiments showed that TorI acts downstream of TorS and needs the presence of TorR. In vitro experiments showed that it is neither a TorR phosphatase nor a histidine kinase inhibitor and that it binds to the effector domain of TorR. Unexpectedly, TorI did not impede TorR DNA binding, and we propose that it may prevent RNA polymerase recruitment to the torC promoter. This study thus reveals a previously uncharacterized class of response regulator inhibitors.
In bacteria, two-component signal-transduction systems constitute the main device for signal detection, allowing adaptative responses to changes in the environmental conditions. Such a system comprises a ligand responsive histidine kinase and a response regulator, usually a transcription factor, and molecular communication between them arises through phosphotransfer reactions (or His–Asp phosphorelay) (1, 2). The sensor kinase autophosphorylates on a histidine residue and then transfers the phosphoryl group to a conserved aspartate in the receiver domain of the response regulator. Two types of scenarios can be found: a phosphotransfer in (i) two steps involving a classical sensor kinase or (ii) four steps when the kinase contains two to three phosphorylation sites (hybrid or unorthodox kinases, respectively) (2). In the four-step type, an additional domain called HPt (histidine phosphotranferase) is required for the phosphoryl transfer to the response regulator (2, 3). The sensor kinases often play the role of aspartate phosphatases for their cognate response regulators, allowing a rapid return to the inactivated state and both classical and complex sensor kinases have been described with such activity. The ratio between the kinase and phosphatase activities of the sensor protein thus determines the phosphorylation level of the response regulator (2). In the four-step phosphorelays, additional phosphatase proteins can be found to regulate signal transduction at intermediate checkpoints. This is the case of the aspartate phosphatase Spo0E family of proteins (4) and some of the Rap proteins (5) both in Bacillus subtilis, and the only known histidine phosphatase SixA in Escherichia coli (6). Another strategy to avoid signal transduction through phosphorelays is to block the autophosphorylation step. A few histidine kinase inhibitors have been described so far and include KipI and Sda in B. subtilis as well as FixT in Sinorhizobium meliloti (7–9). All of these strategies are used to modulate the signal-transduction pathways, allowing an integrated response.
The TorR protein in E. coli is a well characterized transcriptional activator of the OmpR response regulator family (10, 11). In response to the presence of trimethylamine N-oxide (TMAO) in the environment, the sensor kinase TorS autophosphorylates and transfers a phosphoryl group to TorR through a four-step phosphorelay leading to the expression of the torCAD operon (12). This operon encodes the structural proteins involved in TMAO anaerobic respiration, which comprises the c-type cytochrome TorC, the reductase TorA, and the TorA-specific chaperone TorD (13–16). TorR binding to the tor boxes, which are direct repeats located in the torR–torC intergenic sequence, has two consequences: (i) binding to the high-affinity binding site (boxes 1 and 2) that overlaps with torR -10 promoter sequence provokes a negative autoregulation loop, and (ii) binding to the four tor boxes allows RNA polymerase recruitment and torCAD operon expression (10, 17).
The complex phosphorelay occurring between TorS and TorR led us to suspect the presence of intermediate checkpoints in the TMAO signal-transduction pathway. Thus, to identify negative regulators of this pathway, we used a multicopy plasmid library that was screened for negative effect on the torCAD operon expression. This approach did not lead to the isolation of any phosphatase or histidine kinase inhibitor of the TorS/TorR system, but it permitted the identification of the apoform of TorC as a negative regulator (18). Indeed, immature TorC blocks the TMAO signal-transduction pathway by binding to the sensor domain of TorS (19). Using the same approach, we characterized a new response regulator inhibitor, which we called TorI (for Tor inhibition). We showed that TorI interferes with transcription activation of the torC promoter by binding to the effector domain of TorR without affecting its DNA-binding ability. A model is proposed in which binding of TorI to TorR could prevent RNA polymerase recruitment to the torC promoter.
Materials and Methods
Bacterial Strains, Plasmids, Media, and Growth Conditions. Bacterial strains and plasmids are listed in Table 1. For plasmid library screening, MacConkey medium containing lactose (2%) and TMAO (10 mM) was used (18). For β-galactosidase activity determination, strains were grown overnight anaerobically in Luria broth medium at 37°C, and activities were measured as described (20). Values represent the average of at least three independent determinations with a variation of no more than 15% from the mean. When necessary, 50 μg/ml ampicillin, 10 mM TMAO, 0.04% arabinose, or 1 mM isopropyl β-d-thiogalactoside (IPTG) were added.
Table 1. Strains and plasmids used in this study.
| Name | Characteristics | Source |
|---|---|---|
| Strains | ||
| LCB620 | MC4100 torA-lacZ | 13 |
| LCB726 | LCB620 torS726 | 24 |
| LCB506 | MC4100 pcnB | 17 |
| Plasmids | ||
| pUW15 | pUC18 containing region 2,474,268 to 2,477,125 of E. coli chromosome | This work |
| pUW15ΔSphI | pUW15 3′ end insert deletion at SphI | This work |
| pUW15ΔSacI | pUW15 5′ end insert deletion at SacI | This work |
| pUW15ΔEcoRI | pUW15 5′ end insert deletion at EcoRI | This work |
| pJFi | pJF119EH containing torI coding sequence | This work |
| pBtorI | pBAD33 containing torI coding sequence | This work |
| pETsI | pET22(b) containing torI coding sequence | This work |
| pBtorR | pBAD33 containing torR coding sequence | This work |
| pET-RN | pET22(b) containing 122 N-terminal codons of torR | This work |
| pET-RC | pET22(b) containing an ATG codon followed by 108 C-terminal codons of torR | This work |
| pPtor16 | pGE593 containing the torC promoter (—86 to +276 relative to torC transcription start site) fused to lacZ | 10 |
| pPtor46 | pPtor16 with a modified —35 box (GTGCCG→TTGACA) | This work |
| pPR1 | pGE593 containing the torR promoter (—124 to +15 relative to torR transcription start site) fused to lacZ | 17 |
Plasmid Construction. pUW15 plasmid deletions were constructed by hydrolysis of the original plasmid with EcoRI, SalI, or SacI followed by purification of the largest DNA fragments and self-ligation of these products using T4 DNA ligase (New England Biolabs, Beverly, MA). To construct plasmids pJFi, pETsI, and pBtorI, torI coding sequence was PCR-amplified by using MC4100 chromosomal DNA as a template and appropriate restriction-site-containing primers, and then cloned into pJF119EH, pET22(b), and pBAD33 vectors, respectively. Plasmid pBtorR was constructed by cloning a PCR-amplified torR gene, by using chromosomal DNA and appropriate primers, into pBAD33 vector. To create plasmids pET-RN and pET-RC, the corresponding regions of torR (122 N-terminal codons and 108 C-terminal codons, respectively) were PCR-amplified by using MC4100 chromosomal DNA as a template and cloned into pET22(b) vector, creating a His6 tag at the C termini of TorRN and TorRC. Promoter fusions were constructed by amplification of MC4100 chromosomal DNA by using appropriate primers and a proofreading DNA polymerase to produce blunt-ended PCR products and cloned directly into the SmaI site of pGE593 as described elsewhere (10, 17). The same strategy was used for pPtor46 except that one primer contained the mutated -35 region (Table 1). Sequence accuracy of the cloned inserts was checked by sequencing. All primer sequences are available upon request to the authors.
Protein Purifications. TorR was overproduced and purified as described (11). TorRN and TorRC were produced from BL21(DE3) harboring plasmids pET-RN or pET-RC, respectively. Cells were grown in Luria broth medium containing ampicillin until the OD600 reached 0.6 unit. IPTG (1 mM) was then added, and the cells were grown for an additional 2 h at 37°C. For TorRN, French-pressed extract was equilibrated with 40 mM Tris buffer (pH 7.4) and loaded onto a HiTrap chelating Ni2+ column (Amersham Pharmacia). The protein was eluted with a step gradient of imidazole, and TorRN was eluted with 250 mM imidazole; the buffer was then exchanged for 40 mM Tris buffer (pH 7.4)/0.4 M KCl by using a Microcon 3000 (Millipore). For TorRC, French-pressed extract was equilibrated with 40 mM Tris buffer (pH 7.4) and loaded onto a HiTrap heparin column (Amersham Pharmacia). The protein was eluted with a step gradient of KCl and was found in the fraction containing 0.5 M KCl. TorI protein was produced from BL21(DE3) harboring plasmid pETsI. Cells were grown in Luria broth medium until the OD600 reached 0.8 unit, and IPTG (1 mM) was added for 2 h at 37°C. French-pressed extract was equilibrated with 40 mM Tris buffer (pH 7.4) and loaded onto a HiTrap SP column (Amersham Pharmacia). The protein was eluted with a step gradient of KCl and was found in the fraction containing 0.3 M KCl.
Chemical Cross-Linking Studies. Experiments were carried out using bismaleimidohexane (BMH) as a cross-linker agent (Pierce). Proteins (10–80 μM) were incubated for 30 min at room temperature in 1× PBS buffer containing 300 mM KCl and 1 mM BMH. Interactions were analyzed by 12% Tris-Tricine SDS/PAGE followed by Coomassie blue staining. Matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry was performed on bands cut from the gel at the Proteomic Platform (Institut de Biologie Structurale et Microbiologie, Centre National de la Recherche Scientifique, Marseille, France).
Phosphorylation Assay. Phosphorylation assay was carried out as described (12) except that 5 μM purified TorI was added before ATP when indicated. To check the stability of the phosphorylated form of TorR in the presence of TorI, TorR-P was first purified by passage through a HiTrap heparin column and eluted with a step gradient of KCl from 0.1 to 1.0 M. TorR-P was found in the fraction containing 0.8 M KCl. TorR-P was then incubated for 2 h at room temperature in the presence or in the absence of 5 μM purified TorI. TorR-P was then analyzed by Tris-Tricine SDS/PAGE, followed by direct detection on a PhosphorImager screen (Molecular Dynamics).
DNase I Footprinting. About 1 nM 32P-end-labeled DNA encompassing positions -218 up to +56 relative to the torC transcription start site was used in 50 μl of binding mix [10 mM Tris· HCl, pH 7.5/50 mM NaCl/2.5 mM MgCl2/0.5 mM DTT/4% glycerol/30 ng of poly(dI-dC)·poly(dI-dC) per μl]. Different amounts of proteins (purified TorR or TorI and RNA polymerase from Sigma) were then added as indicated, and the footprint assay was carried out as described (10).
Results
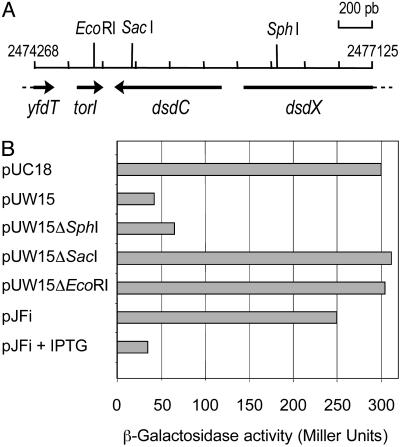
Isolation of a New Gene (torI) That Decreased torCAD Operon Transcription. To identify negative regulators of the torCAD operon, we transformed strain LCB620 with a multicopy plasmid library (21) and isolated white clones, indicating a decreased torA–lacZ expression, on MacConkey lactose plates containing TMAO. After sequencing of the plasmids, the inserts provoking the most drastic effect were found to cover only two regions of the E. coli chromosome. One class contained the torC gene (18), and the second class carried a region located at 2,475 kb on the chromosome (Fig. 1A). The plasmid containing the shortest insert (pUW15) was further analyzed and proved to contain a single annotated gene: dsdC. As shown in Fig. 1B, deletion of the 3′ end of the insert (pUW15ΔSphI) did not modify significantly the negative effect observed with the intact plasmid (pUW15). Surprisingly, deletions on the other side (5′) of the insert (pUW15ΔSacI and pUW15ΔEcoRI) led in both cases to the loss of the negative effect on torA–lacZ expression, indicating that dsdC was not involved in the negative control of the tor promoter. Analysis of the DNA sequence upstream of dsdC revealed the presence of a previously undetected ORF of 201 bp with a putative promoter and a SD sequence correctly positioned (data not shown). This predicted gene was then cloned into an expression vector and transformed into strain LCB620. Upon induction with IPTG, the expression of the torA–lacZ fusion showed a dramatic decrease (Fig. 1B, pJFi + IPTG), demonstrating the role of this gene in the negative regulation of the tor operon. It was consequently named torI for Tor inhibition.
Fig. 1.
Identification of the torI gene. (A) Genetic map of the insert carried by plasmid pUW15. Positions on the E. coli chromosome are indicated at both extremities. The restriction enzyme sites used for plasmid deletion are indicated. (B) Effect of torI overexpression on the torCAD promoter activity. Expression of the torA–lacZ fusion (strain LCB620) in the presence of the various plasmids was measured in cells grown anaerobically in Luria broth medium containing TMAO.
Characteristics of the TorI Protein. The torI gene was predicted to encode a 66-aa protein containing a high proportion of basic residues leading to a predicted pI of 9.52 (Fig. 2). We looked for TorI homologues by using the tblastn algorithm at National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST/) on finished and unfinished bacterial genomes and found two categories of predicted proteins. The first one contained proteins that show 100% identity with TorI and are the products of gene hkaC in the coliphage HK620 genome and gene 18 in the genome of the Shigella flexneri phage Sf6 (22). So far, no biological function has been assigned to these predicted proteins. By comparing the three genes, only a few differences at the nucleotide level were observed (data not shown), indicating a transversal acquisition of these genes or a recent evolution from a common ancestor. In the second category, several proteins with more than 25% identity to TorI were found (Fig. 2). None of these proteins has a known function, although some of them are predicted to contain a DNA-binding motif by homology with a putative transcriptional regulator in phage P4. Analysis of the DNA sequences surrounding the genes for these proteins, revealed that they are located near a phage integrase encoding gene (BAB36936 of E. coli O157:H7, AAF93670 of Vibrio cholerae), in a pathogenicity island (AJ236887 of Yersinia pseudotuberculosis), or in a characterized prophage region (AAG57758 protein of E. coli O157:H7). torI itself belongs to the defective prophage KplE1 (or CPS-53) genome sequence (23). Thus, TorI seems to belong to a phage-related protein family.
Fig. 2.
TorI homologue alignment. TorI homologues retrieved by tblastn search and showing more than 25% identity with TorI by using a fasta local alignment are shown. Predicted proteins with 100% identity (HkaC in HK620 and the product of gene 18 in phage Sf6) were not shown. Conserved residues are indicated according to the percentage of representation as follows: +, >60%; *, >80%; actual residue letter, 100%. The proteins used for the alignment are as follows: TorI, protein of E. coli K-12; GenBank accession no. ZP_00132755 of Haemophilus somnus; accession nos. AAG55333, AAG57758, and BAB36936 of E. coli O157:H7; accession no. CAC89727 of Yersinia pestis; accession nos. AAF93670 and AAF94934 of V. cholerae; accession no. BAC93026 of Vibrio vulnificus; accession no. AAF84594 of Xylella fastidiosa; accession no. AJ236887 of Y. pseudotuberculosis.
TorI Acts Downstream of the TorS Kinase and Its Effect Is TorR-Dependent. The torCAD operon is strictly regulated by the presence of TMAO, and this regulation pathway involves the two-component system TorS/TorR (12). To determine whether TorI is acting at the level of the TorS/TorR phosphorelay or independently of it, we checked the effect of TorI overproduction in two different genetic backgrounds (LCB726 and LCB620/pBtorR). The LCB726 strain carries the torS726 allele, which confers a TMAO-independent expression of the tor operon (24), and the same phenotype is obtained by arabinose induction of the torR gene in strain LCB620/pBtorR. In both cases, overexpression of torI upon induction by IPTG proved to be dominant over either the torS726 constitutive mutation or the overproduction of TorR (Table 2). Indeed, the ratios observed in those genetic contexts are similar to the one measured in LCB620 in the presence of TMAO. Thus, TorI seems to act downstream of the TorS/TorR phosphorelay. We then asked if the effect of TorI could be observed in the absence of TorR. For this purpose, we used a plasmid born promoter (pPtor46) with a consensus -35 box leading to the constitutive expression of the tor promoter independently of the presence of TorR or TMAO (data not shown). Into a strain carrying pPtor46, the torI gene was introduced on a compatible plasmid (pBtorI) under the control of an arabinose-inducible promoter. As shown in Table 3, the expression of the pPtor46 promoter remained constant whether TorI was produced (+ara) or not (-ara), whereas the wild-type promoter in pPtor16 showed a significantly decreased activity (4-fold) under the same conditions. This result strongly suggests that TorR is necessary for TorI to down-regulate the expression of the tor operon.
Table 2. Effect of TorI on torA–lacZ fusion expression.
| β-Galactosidase activity, Miller units
|
|||
|---|---|---|---|
| pJFi* in strains: | —IPTG | +IPTG | Ratio |
| LCB620† | 250 | 35 | 7.1 |
| LCB726 | 655 | 94 | 7.0 |
| LCB620/pBtorR‡ | 248 | 28 | 8.9 |
Expression of torI is under the control of an IPTG-inducible promoter (ptac)
Expression of the torA—lacZ fusion was measured in the presence of TMAO
torR expression was induced with arabinose
Table 3. Effect of TorI overexpression on various plasmid-born promoters.
| β-Galactosidase activity, Miller units
|
|||
|---|---|---|---|
| pBtorI* in strains: | —ara | +ara | Ratio |
| LCB506/pPtor16† | 424 | 104 | 4.1 |
| LCB506/pPtor46 | 150 | 162 | 0.9 |
| LCB506/pPR1‡ | 20 | 18 | 1.1 |
Expression of torI is under the control of an arabinose-inducible promoter (pBAD)
Expression of the torC—lacZ fusion was measured in the presence of TMAO
torR—lacZ fusion
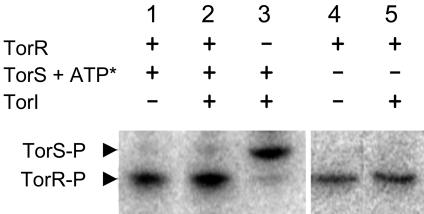
TorI Is Neither a Phosphatase Nor an Anti-Kinase of the TorS/TorR Two-Component System. To confirm these in vivo results and to further characterize the role of TorI in vitro, purification of TorI near to homogeneity was achieved in one step by using a cation exchange chromatography column (Materials and Methods). As previously described (12), TorS was able to promote the transphosphorylation of TorR in the presence of [γ-32P]ATP (Fig. 3, lane 1). The addition of TorI before ATP in the in vitro assay did not affect the phosphorylation level of either TorS or TorR (Fig. 3, lanes 2 and 3), indicating that TorI was not a histidine kinase inhibitor of the TorS/TorR two-component system. Moreover, when the phosphorylated form of TorR was purified from TorS and ATP, we observed that the aspartate-phosphate of TorR was as stable in the presence of TorI as in its absence (Fig. 3, compare lanes 4 and 5), suggesting that TorI had no aspartate-phosphate phosphatase activity either. Taken together, these results showed that TorI did not affect the TorS/TorR phosphorelay.
Fig. 3.
In vitro effect of TorI on the TorS/TorR phosphorelay. Phosphorylation of TorS (2 μM of purified TorS726) and TorR (2 μM) proteins preincubated with or without TorI (5 μM) was started by the addition of [γ-32P]ATP (ATP*) to the reaction mixture and incubated at 37°C for 30 min before SDS/PAGE analysis (lanes 1–3). The stability of TorR-P was checked by incubation at room temperature of purified TorR-P in the presence or absence of TorI (5 μM) for 2 h before SDS/PAGE analysis (lanes 4 and 5). The positions of TorS-P and TorR-P are indicated by arrows.
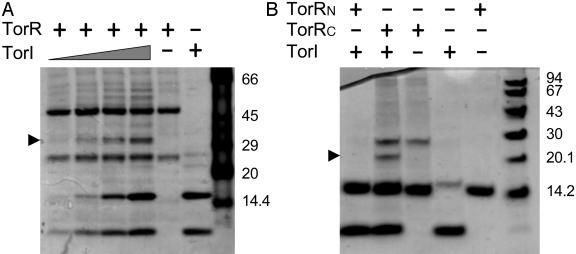
TorI Binds to the Effector (C-Terminal) Domain of TorR. Because TorI had no effect on the phosphorylation pathway but required TorR for the down-regulation of the torCAD promoter, we hypothesized that TorI could bind directly to the TorR response regulator. Purified TorI and TorR were submitted to in vitro crosslinking by using BMH as a cross-linker. Addition of BMH to each protein independently led to the detection of two main bands, suggesting that both TorI and TorR are found as monomers and dimers in solution (Fig. 4A). Addition of increasing amounts of TorI (10–80 μM) to TorR (20 μM) led to the detection of a main additional band, the intensity of which increased as the TorI/TorR ratio increased (Fig. 4A). The apparent molecular mass of the complex (close to 35 kDa) is compatible with the presence of one monomer of TorR (26.1 kDa) plus one monomer of TorI (7.7 kDa). To further characterize this complex, we submitted it to MALDI-TOF analysis, and both TorI and TorR proved to be present in the analyzed band (data not shown). Because TorR is made of two distinct domains, a receiver domain that contains the conserved phosphorylable aspartate and an effector domain, we then ask to which domain TorI was binding. The TorR protein was split into two domains of 122 and 108 residues, respectively. Each domain of TorR was produced with a His6 tag, purified, and then incubated with TorI in the presence of BMH. As shown in Fig. 4B, no additional band was detected when the receiver domain of TorR was used (TorRN). In contrast, a supplementary band, with an apparent molecular mass of ≈20 kDa, appeared upon incubation of TorI with the C-terminal domain of TorR (TorRC), and the size of this complex was compatible with the addition of one monomer of TorRC (13.3 kDa) and one monomer of TorI (7.7 kDa). MALDI-TOF analysis of this complex confirmed that it contained both TorRC and TorI (data not shown). These results showed that TorI was able to bind efficiently to the effector domain of TorR, and because TorRC is not the phospho-accepting domain, this conclusion is in agreement with the fact that TorI does not impede the phosphorylation of TorR. Moreover, phosphorylation of TorR with acetylphosphate did not modify the interaction pattern of TorI and TorR (data not shown).
Fig. 4.
In vitro crosslinking of TorR and TorI proteins. Purified proteins were incubated with BMH at room temperature for 30 min before SDS/PAGE analysis and Coomassie blue staining. (A) Full-length TorR (20 μM) was incubated with increasing amounts of TorI (10, 20 40, and 80 μM). Control lanes contain either 20 μM TorR or 80 μM TorI incubated with BMH. (B) TorI (40 μM) was incubated with a 20 μM concentration of either TorRN or TorRC as indicated. Control lanes contain each protein incubated with BMH only. Arrows point to the different complexes.
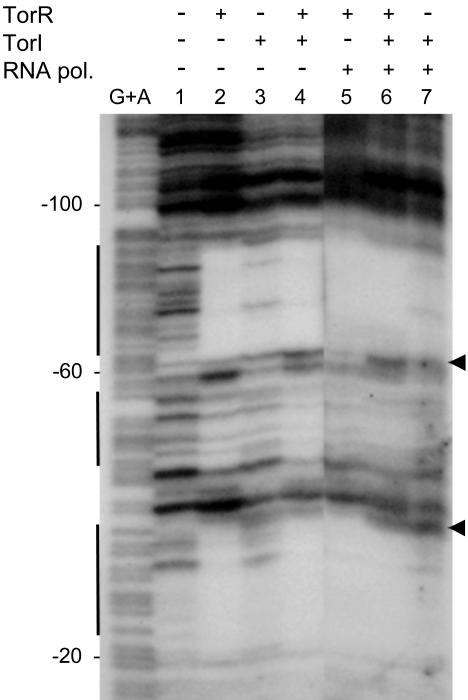
TorI Does Not Prevent TorR Binding to Its DNA Targets. The TorR effector domain has distinct roles in DNA binding and recruitment of the RNA polymerase to the promoter. To decipher which of these functions could be affected by TorI binding, we performed the DNaseI footprinting assay shown in Fig. 5. For this purpose, a DNA fragment, corresponding to positions -218 through +56 relative to torC transcription start site, was 32P-labeled and used as the probe. As previously shown (10), TorR alone protected three regions encompassing the tor boxes and spanning positions -87 through -64, -55 through -43, and -33 through -22 (lane 2). This experiment was carried out with 5 μM TorR, and addition of a 4-fold excess of TorI (20 μM) did not modify TorR protection of the tor boxes (lane 4), indicating that TorI did not affect the DNA-binding capacities of TorR in vitro. In the presence of TorI alone (lane 3), the intensity of the footprint obviously decreased all along the track, suggesting an unspecific binding activity of TorI to the tor promoter that is probably due to its basic properties. This unspecific binding was confirmed by the absence of a specific bandshift by using in vitro DNA gel retardation (data not shown).
Fig. 5.
Effect of TorI on TorR footprint on the torC promoter. A 272-bp labeled DNA fragment encompassing the torC promoter region was digested with DNaseI after incubation with (lanes 5–7) or without (lanes 1–4) RNA polymerase (0.6 μM) and: (lane 1) no TorR and TorI proteins; (lanes 2 and 5) TorR (5 μM); (lanes 3 and 7) TorI (20 μM); or (lanes 4 and 6) both TorR (5 μM) and TorI (20 μM). G+A sequencing ladder is indicated. Numbering is relative to torC transcription start site (+1). Vertical bars indicate the TorR protected regions. The arrows point to the extended protection in the presence of RNA polymerase.
To confirm in vivo that TorI did not prevent TorR from binding to DNA, we studied the effect of TorI overproduction on the torR promoter activity. Indeed, torR is negatively autoregulated through TorR binding to its high-affinity binding site (17). Consequently, if TorI interferes with TorR binding, torR promoter derepression would be expected. In fact, expression of the torR promoter on plasmid pPR1 remained insensitive to the overproduction of TorI, whereas in the same conditions, the torC promoter (pPtor16) was clearly down-regulated (Table 3). All together these results showed that TorI binding to TorR did not prevent TorR from binding to the torR and torC promoters. To check whether TorI had any influence on TorR–RNA polymerase interaction at the torC promoter, we performed additional footprinting experiments wherein the RNA polymerase was added (Fig. 5, lanes 5–7). Addition of the RNA polymerase to TorR in the reaction provoked an extended print on the target DNA pointed by arrows (lane 5). Upon addition of TorI to the reaction, the print on DNA was similar to that observed in the absence of RNA polymerase (compare lanes 4 and 6), suggesting that TorI binding to TorR might interfere with RNA polymerase recruitment.
Discussion
In this study, we describe the identification of a new response regulator inhibitor that interferes with TorR activity by binding to its effector domain (Fig. 4). Interestingly, TorI does not affect TorR DNA binding (Fig. 5 and Table 3) and has no impact either on the signal detection or on the phosphorelay mechanism (Table 2 and Fig. 3).
Among the proteins that interfere with response regulator activity is the Rap family of proteins that have been described as aspartate-phosphatases in B. subtilis (5). Recently, two members of this family (RapC and RapG) that are not aspartate-phosphatases have been described. Indeed, in contrast to the other Rap proteins, RapC and RapG prevent the ComA and DegU response regulators, respectively, from interacting with their DNA targets (25, 26). The mode of action of TorI is novel because its interaction with the effector domain of TorR has no effect on its DNA-binding activity. Moreover, in vitro experiments suggest that the TorR–RNA polymerase extended print on the target DNA was affected in the presence of TorI (Fig. 5). Thus we propose that binding of TorI to the effector domain of TorR might impede interactions with the RNA polymerase, preventing its recruitment to the torC promoter. The inhibitory affect of TorI is reminiscent of that of the Spx protein in B. subtilis. Although Spx is not a response regulator inhibitor, it interferes with response-regulator-stimulated transcription by interacting with the RNA polymerase α-CTD (27). Thus both TorI and Spx interfere with response-regulator-transcription activation either by binding to the response regulator itself (TorI) or by binding to the RNA polymerase (Spx).
In vivo experiments confirmed that only the transcription-activation function of TorR is targeted by TorI because there is no effect on torR-transcription repression (Table 3). Accordingly, we propose that TorI binds to the effector domain of TorR in a region involved in interacting with RNA polymerase. TorR belongs to the OmpR response-regulator family, in which the C-terminal domain includes a winged helix–turn–helix DNA-binding structure (28–30). The second helix of this motif is thought to interact with the major groove of the DNA, whereas the wings allow close contact with the minor grooves. In the 3D structure of OmpR and PhoB, a protruding loop located between the two helices proved to be part of the RNA-polymerase-binding region (28, 31, 32). Considering the high degree of similarity in the structure of the OmpR family of proteins, the corresponding loop in TorR is a candidate for TorI binding, and ongoing studies focus on the characterization of TorI/TorR interactions.
torI was identified as a negative regulator of the tor operon when present on a multicopy plasmid (Fig. 1), and we found that torI encodes a protein that proved to be a potent inhibitor of torC transcription. The torI gene was not previously annotated in the E. coli genome sequence, probably because of its small size (33). In fact, torI is not part of the actual E. coli chromosome sequence because it is located between the two border sequences of the defective prophage KplE1 (23). Moreover, the closest TorI homologues belong to phage genome sequences (hkaC in HK620 and gene18 in Sf6) (22). Thus, TorI seems to be part of a protein family of phage origin. In this paper, we demonstrated that TorI had a regulatory function involving protein–protein interaction in E. coli. It would be of interest to determine whether this is also the case for the other members of this family and if this regulatory function is a general strategy developed by phages during infection. Interestingly, most of the TorI homologues are found in bacteria capable of TMAO respiration, including E. coli O157:H7 and Vibrio species (34), or in bacteria that contain a close homologue of the TorA reductase such as Yersinia species and Haemophilus somnus. In addition, all of these bacteria possess response regulators highly similar to TorR, and an attractive hypothesis is that the TorI homologues also interact with response regulators of the TorR family in these organisms. Another possibility is that proteins of the TorI family inhibit more than one response regulator and are thus able to alter some aspects of the bacterial metabolism to profit the phage cycle.
Acknowledgments
We thank all of the members of our laboratory for useful discussions and D. Dubnau, C. Iobbi-Nivol, and C. Jourlin-Castelli for helpful comments on the manuscript. We are grateful to C. Bordi for contributing to plasmid construction, and to C. Bohn and P. Bouloc for the gift of the E. coli plasmid library. This work was supported by the Centre National de la Recherche Scientifique and the Aix-Marseille II University.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TMAO, trimethylamine N-oxide; IPTG, isopropyl β-d-thiogalactoside; BMH, bismaleimidohexane.
References
- 1.Stock, A. M., Robinson, V. L. & Goudreau, P. N. (2000) Annu. Rev. Biochem. 69, 183-215. [DOI] [PubMed] [Google Scholar]
- 2.Inouye, M. & Dutta, R. (2003) Histidine Kinases in Signal Transduction (Academic, San Diego).
- 3.Ikegami, T., Okada, T., Ohki, I., Hirayama, J., Mizuno, T. & Shirakawa, M. (2001) Biochemistry 40, 375-386. [DOI] [PubMed] [Google Scholar]
- 4.Perego, M. (2001) Mol. Microbiol. 42, 133-143. [DOI] [PubMed] [Google Scholar]
- 5.Perego, M., Glaser, P. & Hoch, J. A. (1996) Mol. Microbiol. 19, 1151-1157. [DOI] [PubMed] [Google Scholar]
- 6.Ogino, T., Matsubara, M., Kato, N., Nakamura, Y. & Mizuno, T. (1998) Mol. Microbiol. 27, 573-585. [DOI] [PubMed] [Google Scholar]
- 7.Wang, L., Grau, R., Perego, M. & Hoch, J. A. (1997) Genes Dev. 11, 2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkholder, W. F., Kurtser, I. & Grossman, A. D. (2001) Cell 104, 269-279. [DOI] [PubMed] [Google Scholar]
- 9.Garnerone, A. M., Cabanes, D., Foussard, M., Boistard, P. & Batut, J. (1999) J. Biol. Chem. 274, 32500-32506. [DOI] [PubMed] [Google Scholar]
- 10.Simon, G., Jourlin, C., Ansaldi, M., Pascal, M. C., Chippaux, M. & Méjean, V. (1995) Mol. Microbiol. 17, 971-980. [DOI] [PubMed] [Google Scholar]
- 11.Simon, G., Méjean, V., Jourlin, C., Chippaux, M. & Pascal, M. C. (1994) J. Bacteriol. 176, 5601-5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jourlin, C., Ansaldi, M. & Méjean, V. (1997) J. Mol. Biol. 267, 770-777. [DOI] [PubMed] [Google Scholar]
- 13.Méjean, V., Iobbi-Nivol, C., Lepelletier, M., Giordano, G., Chippaux, M. & Pascal, M. C. (1994) Mol. Microbiol. 11, 1169-1179. [DOI] [PubMed] [Google Scholar]
- 14.Gon, S., Giudici-Orticoni, M. T., Méjean, V. & Iobbi-Nivol, C. (2001) J. Biol. Chem. 276, 11545-11551. [DOI] [PubMed] [Google Scholar]
- 15.Pommier, J., Méjean, V., Giordano, G. & Iobbi-Nivol, C. (1998) J. Biol. Chem. 273, 16615-16620. [DOI] [PubMed] [Google Scholar]
- 16.Ilbert, M., Méjean, V., Giudici-Orticoni, M. T., Samama, J. P. & Iobbi-Nivol, C. (2003) J. Biol. Chem. 278, 28787-28792. [DOI] [PubMed] [Google Scholar]
- 17.Ansaldi, M., Simon, G., Lepelletier, M. & Méjean, V. (2000) J. Bacteriol. 182, 961-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ansaldi, M., Bordi, C., Lepelletier, M. & Méjean, V. (1999) Mol. Microbiol. 33, 284-295. [DOI] [PubMed] [Google Scholar]
- 19.Gon, S., Jourlin-Castelli, C., Théraulaz, L. & Méjean, V. (2001) Proc. Natl. Acad. Sci. USA 98, 11615-11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, J. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Cold Spring Harbor, NY).
- 21.Bohn, C. & Bouloc, P. (1998) J. Bacteriol. 180, 6072-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark, A. J., Inwood, W., Cloutier, T. & Dhillon, T. S. (2001) J. Mol. Biol. 311, 657-679. [DOI] [PubMed] [Google Scholar]
- 23.Rudd, K. E. (1999) Res. Microbiol. 150, 653-664. [DOI] [PubMed] [Google Scholar]
- 24.Jourlin, C., Bengrine, A., Chippaux, M. & Méjean, V. (1996) Mol. Microbiol. 20, 1297-1306. [DOI] [PubMed] [Google Scholar]
- 25.Core, L. & Perego, M. (2003) Mol. Microbiol. 49, 1509-1522. [DOI] [PubMed] [Google Scholar]
- 26.Ogura, M., Shimane, K., Asai, K., Ogasawara, N. & Tanaka, T. (2003) Mol. Microbiol. 49, 1685-1697. [DOI] [PubMed] [Google Scholar]
- 27.Nakano, S., Nakano, M. M., Zhang, Y., Leelakriangsak, M. & Zuber, P. (2003) Proc. Natl. Acad. Sci. USA 100, 4233-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondo, H., Nakagawa, A., Nishihira, J., Nishimura, Y., Mizuno, T. & Tanaka, I. (1997) Nat. Struct. Biol. 4, 28-31. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Hackert, E. & Stock, A. M. (1997) J. Mol. Biol. 269, 301-312. [DOI] [PubMed] [Google Scholar]
- 30.Kenney, L. J. (2002) Curr. Opin. Microbiol. 5, 135-141. [DOI] [PubMed] [Google Scholar]
- 31.Okamura, H., Hanaoka, S., Nagadoi, A., Makino, K. & Nishimura, Y. (2000) J. Mol. Biol. 295, 1225-1236. [DOI] [PubMed] [Google Scholar]
- 32.Makino, K., Amemura, M., Kawamoto, T., Kimura, S., Shinagawa, H., Nakata, A. & Suzuki, M. (1996) J. Mol. Biol. 259, 15-26. [DOI] [PubMed] [Google Scholar]
- 33.Blattner, F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453-1474. [DOI] [PubMed] [Google Scholar]
- 34.Proctor, L. M. & Gunsalus, R. P. (2000) Environ. Microbiol. 2, 399-406. [DOI] [PubMed] [Google Scholar]