Abstract
Purpose
To compare the expression profile of developmentally important genes between hand-made cloned buffalo embryos produced from reprogramming of donor cell with oocyte extracts and selection of recipient cytoplast through brilliant cresyl blue staining and in vitro fertilized (IVF) embryos.
Methods
Hand-made cloned embryos were produced using oocyte extracts treated donor cells and brilliant cresyl blue (BCB) stained recipient cytoplasts. IVF embryos were produced by culturing 15–20 COCs in BO capacitated sperms from frozen thawed buffalo semen and the mRNA expression patterns of genes implicated in metabolism (GLUT1), pluripotency (OCT4), DNA methylation (DNMT1), pro- apoptosis (BAX) and anti-apoptosis (BCL2) were evaluated at 8- to16- cell stage embryos.
Results
A significantly (P < 0.05) higher number of 8- to16- cell and blastocyst stages (73.9 %, 32.8 %, respectively) were reported in hand-made cloning (HMC) as compared to in vitro fertilization (49.2 %, 24.2 %, respectively). The amount of RNA recovered from 8- to 16- cell embryos of HMC and in vitro fertilization did not appear to be influenced by the method of embryo generation (3.76 ± 0.61 and 3.82 ± 0.62 ng/μl for HMC and in vitro fertilization embryos, respectively). There were no differences in the expression of the mRNA transcripts of genes (GLUT1, OCT4, DNMT1, BAX and BCL2) were analysed by real-time PCR between hand-made cloned and IVF embryos.
Conclusions
Pre-treatment of donor cells with oocyte extracts and selection of developmentally competent oocytes through BCB staining for recipient cytoplast preparations may enhance expression of developmentally important genes GLUT1, OCT4, DNMT1, BAX, and BCL2 in hand-made cloned embryos at levels similar to IVF counterparts. These results also support the notion that if developmental differences observed in HMC and in vitro fertilization produced foetuses and neonates are the results of aberrant gene expression during the pre-implantation stage, those differences in expression are subtle or appear after the maternal to zygotic transition stage of development.
Keywords: Buffalo, Oocyte extracts, Brilliant cresyl blue, Hand-made cloning, In vitro fertilization
Introduction
Somatic cell nuclear transfer (SCNT) through zona-free approach or hand-made cloning (HMC) has emerged as a more efficient as well as economical technique in comparison to the traditional micromanipulator- based approaches [1], and has been successfully used to produce cloned offsprings’ in several livestock species including cattle [2], pigs [3] and buffalo [4]. Although HMC has been performed in buffalo in certain laboratories, reconstruction of embryos remains one of the most difficult and demanding part of SCNT procedures in this species [5]. The efficiency of obtaining live offspring from cloned buffalo embryos is still very low [5]. Although some information is available on HMC in buffalo regarding the comparison between in vitro culture conditions and examining the effect of source of donor nucleus [6] and much more information needs to be generated to enable large scale application of HMC technology to this species.
Reprogramming of a differentiated cell nucleus by SCNT is an inefficient process, following nuclear transfer, the donor nucleus often fails to express early embryonic genes and establish a normal embryonic pattern of chromatin modifications [7–9]. Relative studies have provided evidence that this low efficiency was due to incomplete reprogramming of donor nuclei [10–12]. Recent reports suggested that Xenopus laevis egg extracts had the ability to reprogram mammalian somatic cells to express stem cell gene OCT4 and the cells could be partially reprogrammed to an embryonic state [13]. The remodelled cells acquired the capacity of dedifferentiation through cell culture [14–16]. The embryos reconstructed from cumulus cell nuclei treated with oocytes cytoplasmic lysates resulted in high quality blastocyst at a rate about three times greater than control in mouse model [17]. Extracts of porcine oocytes system activated pluripotent marker gene, especially NANOG, and induced partial differentiation of fibroblast cell culture [18]. Extract of bovine oocytes induced epigenetic reprogramming of yak fibroblasts, had made them suitable donors for yak inter-species SCNT [19]. However, the gene expression profiles of developmentally important genes in cloned embryos derived from extract treated donor cells have not been studied.
The follicular oocytes used in SCNT are commonly recovered from ovaries of slaughtered bovine of unknown age, breed, health status and reproductive performance [20]. The immature oocytes are routinely selected based on compaction of cumulus corona investment and the homogeneity of the ooplasm, this may reduce the yield of transferable embryos, as some oocytes with apparently normal morphology are in the early stage of degeneration [21]. Therefore, finding a non-invasive and non-perturbing method for selection of oocytes prior to culture has become of prime importance. It is generally believed that glucose-6-phospahte dehydrogenase (G6PDH) protein is active in the growing oocytes, but has decreased activity in oocytes that have finished their growth phase [22]. Brilliant cresyl blue (BCB) test based on the capability of G6PDH to convert the BCB stain from blue to colorless, thus oocytes that have finished their growth phase show a decreased G6PDH activity and exhibit a cytoplasm with a blue coloration (BCB+) while growing oocytes are expected to high level of active G6PDH which results colorless cytoplasm (BCB−) [23]. It was reported that BCB+ oocytes yielded significantly higher blastocyst development rather than BCB− and control oocytes in pig [24], goat [25], bovine [26, 27] and sheep [28]. However, whether BCB+ oocytes enhance the viability of HMC buffalo embryos in terms of developmentally important genes expression is unknown.
In mammalian development, two major reprogramming events occur, both involving epigenetic modification. The first occurs during germ cell development and second during pre-implantation development [29]. The latter reprogramming event which is critical for zygote development is initiated around 8- to 16- cell stage in bovine and is termed maternal to zygotic transition (MZT) [30, 31]. During MZT, the embryo undergoes several important physiological events including modification of chromatin structure [32] and de novo methylation of DNA [33]. One challenge in SCNT pre-implantation embryos is to not only initiate new gene expression at MZT, but also to turn off the expression of somatic donor cell gene at the same time [34].
Gene expression analysis of pre-implantation embryonic development provides important information to understand the changes an organism undergoes in the transition from gametes to a pluripotent embryo and into the earliest stages of cell differentiation [35]. Gene expression varies according to the respective in vitro embryo production system and nuclear transfer (NT) protocol [36–38]. Studies of the effects of SCNT on gene transcripts in SCNT-derived embryos showed that various modifications of the NT protocol (source of donor cells, passage number of donor cells, cell cycle stage of donor cells, activation protocol) have distinct effects on embryonic gene expression patterns [39, 40]. The delayed onset or absence of embryonic transcription of genes in NT embryos indicates an incomplete reprogramming of the donor nuclei with which the embryos were reconstructed, in addition, the identification of genes whose expressions profiles are frequently abnormal in cloned embryos will provide markers for the diagnosis of cloned embryo viability prior to embryo transfer and, therefore, potentially negate the time and money-consuming transfer of non-viable embryos to recipient animals [41]. Although the morphologic appearance of SCNT embryos does not differ from that of in vitro fertilized (IVF) embryos, the potential to develop to term dramatically differs between SCNT and IVF embryos [42, 43].
Markers that will be useful for predicting the potential of NT embryos to develop into young ones are needed [44]. So far, no studies have been conducted at the developmentally important gene expression levels in buffalo HMC embryos at MZT period. Here, mainly focus is on the differences in developmentally important gene expression patterns in pre-implantation zona-free SCNT derived buffalo embryos at MZT period using IVF embryos as control. Because this is the stage at which the embryonic genome becomes transcriptionally active, failures in reprogramming of the donor nucleus may become readily apparent and such studies also evaluate the “normality” of in vitro produced or manipulated embryos [34]. The products of these genes play important roles during pre-implantation and post-implantation development and, hence, have the potential to be used as genetic markers for embryo viability [45, 46]. They are thought to be involved in numerous biological processes including metabolism (GLUT1), pluripotency (OCT4), DNA methylation (DNMT1), pro-apoptosis (BAX) and anti-apoptosis (BCL2).
IVF embryos are used as control in this study, because in vivo water buffalo embryos are difficult to obtain. Although IVF embryos have less developmental competence than those produced in vivo, they do not suffer the extreme embryonic ⁄ foetal loss that is commonly seen in SCNT studies [45]. Additionally in bovine SCNT studies, IVF embryos are commonly compared with SCNT embryos which are more readily obtained [10, 34, 41, 46, 47]. The successful reprogramming of a somatic cell by factors present in a cytoplast would result in an embryo with the same profile of gene transcription as that seen in embryos produced by in vitro fertilization procedures [41]. Therefore, comparing IVF embryos with those produced by SCNT is a meaningful and well accepted alternative.
Keeping in view the aforesaid realities, the present study is conducted with the objective to investigate the in vitro developmental competence of hand-made cloned buffalo embryos derived from pre-treatment of donor fibroblast with oocytes extract and selection of developmentally competent oocytes through BCB staining for recipient cytoplast preparations in terms of difference in gene expression profile of developmentally important genes during MZT period using IVF embryos as control. The results of these studies may help in evaluating the viability of manipulated hand-made cloned buffalo embryos.
Materials and methods
Chemicals
The different ready to use liquid media used in the present study for the culture of oocytes/embryos and somatic cells, which included tissue culture medium-199 (TCM-199), Dulbecco’s modified Eagle’s medium (DMEM) were purchased from Sigma Chemical Co., St. Louis, MO, USA, unless otherwise indicated. Various supplements, which included bovine serum albumin (BSA), antibiotics (gentamicin, penicillin and streptomycin), L- glutamine, sodium pyruvate, vitamins, non essential amino acids, fatty acid-free BSA, heparin, caffeine and other chemicals used for in vitro embryo production and for culture of cells in various experiments were also purchased from Sigma Chemical Co., St. Louis, MO, USA, unless otherwise indicated. Most of the chemicals used in the present study were of embryo or cell culture tested grade. Fetal bovine serum (FBS) was from Hyclone (Thermo Scientific, Wilmington, DE, USA), FBS used was from the same batch throughout the study. The reagents for molecular biology work were of biotechnology grade and purchased from Invitrogen (USA) and Fermentas (USA) unless otherwise indicated.
Collection of oocytes
In brief, ovaries from reproductive organs of adult, apparently healthy female buffaloes collected from abattoir within 30 min of slaughter were washed three times with warm isotonic saline (35 °C –37 °C) containing 400 IU/ml penicillin and 500 μg/ml streptomycin and transported to the laboratory within 4–6 h and aspiration of cumulus oocyte complexes (COCs) were performed as described by Chauhan et al. [48] with some modifications. Oocytes from follicles (2 to 8 mm) were aspirated with 18 gauge needle attached to 10 ml syringe (Sigma, Cat. No. Z248029) loaded with aspiration medium (TCM-199 containing 0.3 % BSA, 0.1 mg/ml glutamine and 50 μg/ml gentamicin). The oocytes were washed four to six times with the washing medium, which consisted of TCM-199 with 10 % FBS, 0.09 mg/ml sodium pyruvate, 0.1 mg/ml L- glutamine and 50 μg/ml gentamicin. COCs having a compact and unexpanded cumulus mass with equal to or greater than three layers of cumulus cells and homogenous granular ooplasm were selected.
Brilliant cresyl blue staining
Brilliant cresyl blue staining of selected COCs performed as described by Jiamin et al. [21] with some modifications. Briefly to carry out the BCB test, immediately after oocytes were selected, they were washed three times in Dulbecco’s PBS, modified by the addition of 0.4 % BSA (mDPBS). The oocytes were exposed to 26 μM BCB diluted in mDPBS for 90 min at 38.5 °C in a humidified air atmosphere. The oocytes were then transferred to mDPBS, washed twice, and examined under a stereo zoom microscope. They were distributed into two groups according to their cytoplasm coloration; those with or without blue coloration of the cytoplasm were designated as BCB+ and BCB−, respectively. The concentration of 26 μM had earlier been found to be effective for oocytes of pig [24], goat [25] and bovine [26] as it was supportive of a high rate of in vitro maturation of selected oocytes without apparent loss of viability. Oocytes of the control group were selected on the basis of COCs having equalled to or more than 3 layers of cumulus cells, and with homogenous, evenly granular ooplasam without exposure to BCB.
In vitro maturation (IVM)
COCs from BCB+, BCB− and control groups were subjected to maturation in IVM media consisted of TCM-199 + sodium pyruvate (0.80 mM) + L-glutamine (2 mM) + 10 % FBS + 5 % follicular fluid (FF) + PMSG (20 IU/ml) + hCG (10 IU/ml) + gentamicin (50 μg/ml). The pH of the media was adjusted to 7.4 and filtered through 0.22 μm membrane filter (Pall life Sciences, Ann Arbor, USA) immediately before use. The COCs were washed several times with IVM medium and group of 15–20 COCs were placed independently in 100 μl droplets of IVM medium covered with sterilized mineral oil in 35 mm petri dishes and cultured for 21 h under 5 % CO2 at 38.5 °C.
In vitro embryo production through in vitro fertilization
In vitro maturation
COCs having a compact and unexpanded cumulus mass cells and homogenous granular ooplasam were subjected to IVM for 24 h in previously mentioned IVM media.
Sperm capacitation
Spermatozoa were capacitated using Bracket and Oliphant medium (BO) [49]. Semen was obtained from the semen lab of Central Institute for Research on Buffaloes (Hisar, India). In all experiments, frozen semen from the same bull was used. Frozen semen from buffalo bulls stored in 0.25 ml straws were thawed in water bath at 37 °C for 1 min. Spermatozoa were washed twice at 2,500 rpm for 5 min using semen washing solution of BO medium supplemented with 10 μg/ml heparin, 137 μg/ml sodium pyruvate and 1.942 mg/ml caffeine sodium benzoate. The pellet was resuspended in around 0.5 ml of the washing BO medium and the sperm number was counted using haemocytometer and number of spermatozoa were adjusted to be 2 × 106/ml.
In vitro fertilization procedure
For in vitro fertilization, matured oocytes were washed twice in oocytes washing solution of BO medium and transferred to 50 μl droplets of capacitation and fertilization BO medium (Washing medium supplemented with 10 mg/ml of fatty acid free BSA). The spermatozoa in 50 μl of the capacitation and fertilization BO medium was then added to the droplet containing oocytes, covered with sterile mineral oil and placed in CO2 incubator at 38.5 °C for 18 h for IVF.
In vitro culture (IVC)
At the end of sperm - oocyte 18 h co-incubation, prior to transfer to the IVC droplets, presumed zygotes were washed four times in 400 μl of Research Vitro Cleave medium (K-RVCL- 50, Cook®, Australia) supplemented with 1 % fatty acid-free (FAF) BSA and cultured in this medium in a humidified CO2 incubator at 38.5 °C; embryo production rate was examined under inverted microscope (Nikon Inc., Tokyo, Japan) to record the number of cleaved embryos at 48 h post-insemination (h.p.i), percentage of embryos with 8- to 16- cells at 94–96 h.p.i and blastocyst formation at 168–192 h.p.i.
Establishment of fibroblast cell culture
Primary ear fibroblast culture of Murrah buffalo calf established and prepared for HMC as reported earlier by Shah et al. [6] with some modifications. Briefly ear skin samples from new born Murrah buffalo calf were obtained aseptically in sterile phosphate buffered saline (PBS) with 1 % antibiotic-antimycotic solution and transferred to laboratory within 10 min. Tissue samples were washed six times with Dulbecco’s phosphate buffered saline (DPBS) and chopped into small pieces (0.5 mm) and the pieces of tissue were transferred to 25 cm2 culture flasks and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 2 mM L-glutamine, 15 % FBS, 1 % non essential amino acids, 1 % vitamins and 50 μg/ml gentamicin in a CO2 incubator (5 % CO2 in air) at 37 °C. Cells were passaged upon reaching 60–70 % confluence by partial trypsinization. Cells forming a confluent monolayer (passage 2–5) were allowed to grow further for 3 days to achieve over-confluence.
Oocytes extract preparation
Buffalo oocytes extract preparation was carried out as described by Xiong et al. [19] with some modifications. Briefly, COCs with expanded cumulus mass were transferred into a 1.5 ml microcentrifuge tube containing 500 μl hyaluronidase (0.5 mg/ml) in T2 (everywhere T denotes HEPES modified TCM-199 supplemented with 2.0 mM L-glutamine, 0.2 mM sodium pyruvate, 50 μg/ml gentamicin and the following number denotes 2 % FBS) and incubated for 1 min at 38.5 °C, followed by vortexing (2–3 min). Entirely denuded oocytes with uniformly granular cytoplasm were selected and incubated in pronase (2.0 mg/ml in T10) for 3–5 min at 38.5 °C. Oocytes with totally digested zona pellucida were washed twice with extraction buffer (50 mM KCl, 5 mM MgCl2, 5 mM ethylene glycol tetraacetic acid (EGTA), 2 mM β-mercaptoethanol, 0.1 mM PMSF, protease inhibitor cocktail, and 50 mM HEPES, pH 7.6) containing an energy regeneration system (ERS: 1 mM ATP, 10 mM phosphocreatine, 25 μg/ml creatine kinase, pH 7.4). Nearly 400 zona- free oocytes were transferred into 10 μl of ERS in a 0.2 ml eppendorf tube and centrifuged at 20,879 ×g for 20 min at 4 °C. The supernatant was used as extract and stored at −80 °C. The procedure was repeated to prepare sufficient extract.
Permeabilization of cell membranes, extract treatment and donor cell preparations
Donor cells pre-treatment with buffalo oocyte extracts as described by Shuang et al. [50] with some modifications. Briefly, donor cells suspensions were washed twice in DPBS (Ca2+, Mg2+ free) by spun down at 1,000 rpm for 5 min and supernatant discarded carefully. Cells suspended in 500 μl DPBS (Ca2+, Mg2+ free) containing 1.5 U/ml streptolycin-O at 38.5 °C for 30 min. The aliquots of permeable cells (about 1 × 106 cells/aliquot) were resuspended in 10 μl oocyte extracts and incubated at 38.5 °C for 30 min. The extract treated cells were cultured in DMEM supplemented with 2 mM L-glutamine, 15 % FBS, 1 % non-essential amino acids, 1 % vitamins and 50 μg/ml gentamicin in a CO2 incubator (5 % CO2 in air) at 37 °C. Morphological features, together with shape and size of cells, their tendency to form attachment to the culture flask were recorded at an interval 24 h and medium was replaced every 3 days. Viability of the cells was monitored by standard protocol of exclusion of tryplan blue dye, and the cells were counted with a haemocytometer. The cells were cultured up to 1 week. Immediately before use, dispersed to single cells by treatment for 5 min by 0.25 % trypsin-EDTA, subsequently washed by centrifugation and resuspended in T20 media for use as nucleus donor cells.
Preparation of recipient cytoplast and hand-made cloning
The recipient cytoplast preparations from BCB selected/control in vitro matured oocytes and the procedures for HMC as described previously by Shah et al. [51].
Embryo culture
The activated embryos were cultured using the same culture protocol as described for the IVF embryos and the number of cleaved embryos, 8- to-16- cell and blastocyst stages at 48 h post activation (h.p.a), 94–96 h.p.a and 168–192 h.p.a, respectively were recorded.
Embryo collection and RT-PCR Analysis
Morphologically normal 8- to16- cell stage hand-made cloned embryos from BCB+ oocytes with extracts treated cells (ETCs) and IVF embryos collected at 94–96 h.p.a and 94–96 h.p.i, respectively were separately treated using a cell to cDNA kit (Ambion Inc, The RNA Company, Austin, TX) according to the manufacturers’ protocol immediately after taking photographs. 10–12 embryos at 8-to 16- cell stage were analysed in each group. The embryos were washed with 200 μl ice cold PBS after which 50 μl of chilled cell lysis buffer was added and the mixture was incubated at 75 °C for 10 min in a thermal cycler. Genomic DNA was degraded by incubating the cell lysates in DNase-I at 37 °C for 30 min and the remaining activity of DNase-I was inactivated by heating at 75 °C for 5 min. For cDNA synthesis, 10 μl of the cell lysates (RNA), 4 μl dNTP mix (2.5 mM each dNTP), 2 μl random decamer taken in 200 μl PCR tube. The reaction mixture was mixed and incubated at 70 °C for 1 min to denature RNA for easier binding of primer in a thermal cycler, immediately cool the tubes on ice and add remaining reverse transcriptase reagents; 2 μl 10X RT buffer, 1 μl MMLV reverse transcriptase and 1 μl RNase inhibitor. The reaction mixture was again mixed and incubated in a thermal cycler at 42 °C for 60 min and 95 °C for 10 min to inactivate the reverse transcriptase. The synthesized cDNA was stored at −80 °C until used for amplification step.
PCR reaction was carried out in a 50 μl final volume containing 45 μl platinum PCR supermix (Invitrogen), and 5 μl of primer (200 nM each, Sigma) and template DNA solution, nuclease free water instead of template DNA was taken as negative control. GAPDH was used as the reference gene and the primers sequences were used for GLUT1, OCT4, DNMT1, BAX, BCL2 and GAPDH are mentioned in Table 1. The PCR conditions were same except the annealing temperature (Table 1), as 94 °C for 2 min (Initial denaturation), denaturation at 94 °C for 30 s, elongation at 72 °C for 1 min (35 cycles). The amplified DNA fragments were resolved on 2 % agarose gel containing 0.5 μg⁄ml ethidium bromide against a 50-bp ladder and visualized under gel documentation system (Alpha Imager, Alpha Innotech,San Leandro, CA, USA).
Table 1.
Detail of primers used for expression of developmentally important genes in hand-made cloned and IVF embryos
| Sr No | Gene | Primer sequence | Annealing temp (°C) | Size (bp) | Source | Accession No |
|---|---|---|---|---|---|---|
| 1. | GAPDH | CCTGCCAAGTATGATGAGA (F) | 53 | 131 | Bubalus bubalis | GU324291 |
| GAAGGTAGAAGAGTGAGTGT (R) | ||||||
| 2. | GLUT1 | CCGTCTCTTCCTATCCAA (F) | 56 | 194 | Bubalus bubalis | HQ434959 |
| GGTCTTCTTGAATAGTGAGTT (R) | ||||||
| 3. | OCT4 | GACAAGGAGAAGCTGGAG (F) | 54 | 75 | Bubalus bubalis | JF898834.1 |
| GCAAATTGTTCAAGGTCTTTC (R) | ||||||
| 4. | DNMT1 | ATAAGTAAGATAGTGGTTGAGTTC (F) | 55 | 100 | Bos taurus | NM_182651 |
| TTG AGC ATA CAA GGA GGA A (R) | ||||||
| 5. | BAX | ATCCACCAAGAAGCTGAG (F) | 52 | 82 | Bubalus bubalis | HE661581 |
| CTGCGATCATCCTCTGTA (R) | ||||||
| 6. | BCL2 | GGC CCCTGTTTGATTTCT (F) | 56 | 97 | Bos taurus | U92434 |
| CTTATGGCCCAGATAGGC (R) |
Real-time PCR for relative quantification
Real-time PCR (Applied Biosystems 7500 Real-Time PCR system) was performed using SYBR green qPCR supermix (Invitrogen SYBR Green qPCR supermix: Carlsbad, CA, USA) as a double – standard DNA specific fluorescent dye in 25 μl reaction to assess the gene expression of GLUT1, OCT4, DNMT1, BAX, and BCL2 relative to housekeeping gene GAPDH. All gene of interest were analysed in triplicate with different samples. The amplification was carried out in 25 μl volume reaction mixture containing 12.5 μl of SYBR Green qPCR mastermix, 1 μl of primer (10 pM each forward and reverse primer), 2 μl of cDNA template and 9.5 μl of nuclease free water. Samples not exposed to reverse transcriptase (RT) were used as negative controls. For PCR, sample were activated at 95 °C for 10 min, followed by 40 cycles of denaturing at 95 °C for 45 s, then annealing at the specific primer annealing temperature (Table 1) for 30 s and extension at 72 °C for 30 s. The comparative CT method was used for relative quantification of target gene expression levels. Quantification was normalized to the internal control GAPDH gene. Within long-linear phase region of the amplification curve, each cycle doubled the amplified product. The ΔCT value was determined by subtracting the GAPDH CT value for each sample from the target gen CT value. Calculation of ΔΔCT value involved using the highest sample method ΔCT as an arbitrary constant to subtract from all other ΔCT samples values. Fold changes in relative mRNA expression of the target genes were determined using the formula 2- ΔΔCT.
Statistical analysis
The data were analyzed using SPSS (SPSS Inc. IL, USA). All the values are presented as mean ± SEM unless indicated otherwise. Differences among means were analyzed by one way ANOVA after arcsine transformation of the percentage data. The differences were considered significant at P < 0.05.
Results
In vitro development of hand- made cloned and IVF embryos
Developmental rates of hand- made cloned and IVF buffalo embryos are summarized in Table 2. A total number of 506 demi-cytoplasts were used to reconstruct 126 hand-made cloned embryos with ETCs; control oocytes (n = 50), BCB+ oocytes (n = 54) and BCB− oocytes (n = 22). A total number of 140 presumptive zygotes were incubated for in vitro fertilization experiment. In terms of the proportion of cleaved embryos recorded after NT, significant (P < 0.05) differences were found among the BCB+ oocytes and BCB− oocytes with ETCs (79.6 %, 54.2 %, respectively), whereas no significant (P > 0.05) differences in cleavage rate among NT embryos derived from control oocytes with ETCs, the BCB+ oocytes with ETCs (71.6 %,79.6 %, respectively); BCB+ oocytes with ETCs and IVF embryos (79.6 %, 82.1 %, respectively). Whereas a significant (P < 0.05) difference were found between IVF embryos and NT embryos derived from control oocytes with ETCs and BCB− oocytes with ETCs (82.1 %, 71.6 %, 54.2 %, respectively). NT embryos derived from BCB+ oocytes with ETCs produced significantly (P < 0.05) higher numbers of embryos at 8–16 cell stage (73.9 %) than NT embryos derived from BCB−oocytes with ETCs (33.3 %) and IVF embryos (49.2 %), no significant (P > 0.05) differences, however were observed between the BCB+ oocytes with ETCs (73.9 %) and control oocytes with ETCs (64.3 %). The percentage of NT blastocysts derived from the BCB+ oocytes with ETCs was significantly (P < 0.05) higher (32.8 %) as compared to the control oocytes with ETCs (22.2 %) and the BCB− oocytes with ETCs (6.3 %). There was no significant (P > 0.05) difference for blastocyst rate between NT embryos derived from BCB+ oocytes with ETCs (32.8 %) and IVF embryos (24.2 %). In addition, the rate of blastocyst development in NT embryos derived from the control oocytes with ETCs and IVF embryos were significantly (P < 0.05) higher than that for NT embryos derived BCB− oocytes with ETCs (22.2 %, 24.2 %, 6.3 %, respectively). Representative photographs of IVF and hand-made cloned buffalo embryos at different stages shown in Fig. 1.
Table 2.
Developmental rates of buffalo hand- made cloned and IVF embryos
| Group | Embryos cultured | No. Cleaved embryos (%) | No. 8–16 cell embryo (%) | No. Blastocysts (%) |
|---|---|---|---|---|
| Control oocytes+ETCs | 50 | 36 (71.6 ± 4.2)a | 32 (64.3 ± 8.4)a | 10 (22.2 ± 8.5)a |
| BCB+oocytes+ETCs | 54 | 43 (79.6 ± 7.1)ab | 40 (73.9 ± 6.3)a | 17 (32.8 ± 3.8)b |
| BCB− oocytes+ETCs | 22 | 12 (54.2 ± 4.2)b | 07 (33.3 ± 5.9)b | 02 (6.3 ± 6.3)c |
| IVF | 140 | 115 (82.1 ± 9.2)ab | 67 (49.2 ± 9.2)c | 33 (24.2 ± 4.7)ab |
Figures quoted as percent mean ± SEM
Cleavage, 8–16 cell stage and blastocyst values having different superscripts in the same column differ significantly (P < 0.05)
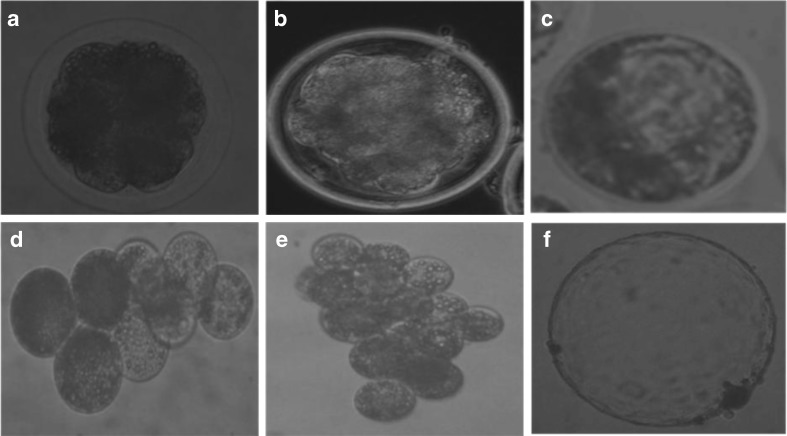
Fig. 1.
Development in vitro of buffalo IVF (a–c) and hand-made cloned (d–f) embryos (200×). a and d. 8–cell stage embryo. b and e. 16– cell stage embryo, c and f blastocyst
Gene expression profile of hand-made cloned and IVF embryos
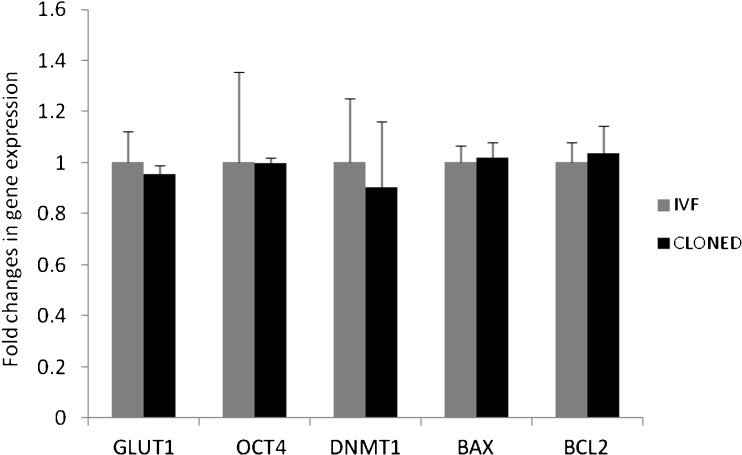
The gene expression differences we were interested primarily at 8- to 16-cell stage between hand-made cloned from BCB+ oocytes with ETCs and IVF embryos. The amount of RNA recovered from 8- to16- cell embryos of both groups did not appear to be influenced by the method of embryo generation (3.76 ± 0.61 and 3.82 ± 0.62 ng/μl for hand-made cloned, n = 10–12 and IVF embryos, n = 10–12). RT-PCR analysis revealed amplicons of GLUT1, OCT4, DNMT1, BAX, BCL2 and GAPDH with an expected length of 194 bp, 75 bp, 100 bp, 82 bp, 97 bp and 133 bp, respectively, were detected in buffalo hand-made cloned and IVF embryos at 8- to16- cell stages (Fig. 2). Relative expression levels of mRNA transcripts of developmentally important gens revealed there were no significant (P > 0.05) differences in the expression of the mRNA transcripts of genes (GLUT1, OCT4, DNMT1, BAX and BCL2) were analysed by real-time PCR between hand-made cloned and IVF, 8- to16- cell embryos (Fig. 3).
Fig. 2.
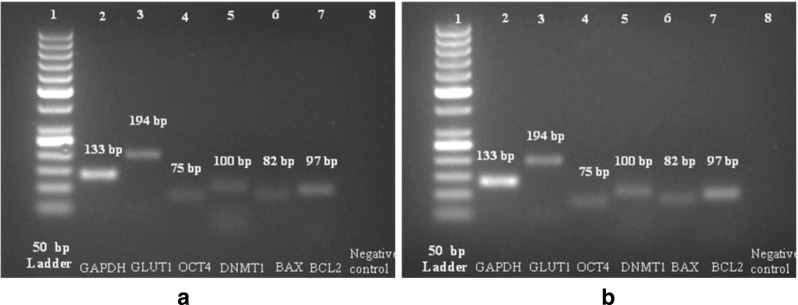
RT- PCR analysis of developmentally important genes in IVF (a) and handmade cloned (b) embryos at 8-to16- cell stages. (a & b). Lane 1–50 bp Ladder, Lane 2-GAPDH, Lane 3-GLUT1, Lane 4- OCT4, Lane 5-DNMT1, Lane 6-BAX, Lane7-BCL2 and Lane 8- Negative control
Fig. 3.
Relative expression profile of developmentally important genes in IVF and hand-made cloned embryos at 8-to16- cell stages
Discussion
The differences in developmentally important gene expression patterns in pre-implantation 8- to 16- cell embryos between hand-made cloned from oocyte extracts pre-treated donor cells and selection of recipient cytoplasts through BCB staining and IVF were studied in order to evaluate their expression during MZT in both group of embryos. Such studies evaluate the viability of manipulated hand-made cloned embryos. The genes under study were selected because they participate in key cellular processes during pre-implantation development and have been reported to become transcriptionally active during MZT [45, 46]. The present experiment evaluated the genes encoding metabolism (GLUT1), pluripotency (OCT4), DNA methylation (DNMT1), pro -apoptosis (BAX) and anti-apoptosis (BCL2).
It has been shown that embryo metabolism may be up regulated by in vitro environment. This shift in metabolism has been suggested to be associated with a reduction in viability [52]. Glycolysis and lactate production increased in embryos during in vitro culture [52]. Study in bovine indicates that there were significant differences in the expression of GLUT1 between SCNT and IVF embryos [46]. The reduction of OCT4 expression in bovine cloned blastocysts has been observed [53, 54]. Variation in methylation level of OCT4 was also reported for cloned bovine embryos [55]. In mice, it is generally accepted that OCT4 is down regulated in cloned pre-implantation embryos [7]. The DNA methylation has been studied in cloned embryos from numerous species, with the exception of the pig; all species have exhibited global DNA hypermethylation [11, 56]. DNA methylation has been shown to suppress gene expression, which in turn leads to the developmental failure of the cloned embryos, it is essential to identify DNA methylation-related gene expression profiles; especially for the DNA methyl transferases (DNMTs) related genes, which may provide insight between the DNA methylation state and the efficiency of SCNT embryos [57]. The occurrence of apoptosis in pre-implantation embryos has been considered one of the most important parameter for evaluation of embryo health [58, 59]. Apoptosis was first observed in bovine NT embryos at the 4-cell stage, but for in vitro fertilization embryos at 6- to 8-cell stages [60]. Expression of BCL2 and BAX can be correlated with apoptosis, BCL2 inhibits apoptosis by regulating the release of cytochrome-c and other proteins from mitochondria [61] and BAX is pro- apoptotic and accelerates S-phase progression [62]. Study in porcine embryos has shown that there was significantly lower BCL2 mRNA expression in SCNT derived as compared to IVF embryos and correspondingly, higher apoptotic incidence [57]. In several studies, the mRNA expression levels of BAX are used to evaluate embryos quality developed in vitro [46, 57, 63].
We thought that gene expression analysis at the 8- to16- cell stages for two different types of embryos might offer better insight into the reprogramming process and gene expression patterns of pre-implantation NT embryos. It is possible that gene expression analysis at 8- to 16- cell stages in the present study might correspond to gene expression analysis at MZT between hand-made cloned and IVF embryos. Camargo et al. [34] used 8- cell stage embryos collected at 70–74 h.p.a for gene expression analysis of bovine NT embryos at MZT. Therefore in the present study, as an attempt to examine the developmentally important gene expression pattern differences at MZT between two groups of embryos, we collected 8- to16- cell hand-made cloned and IVF embryos. To the best of our knowledge, there are no reports comparing developmentally important gene expression between hand-made cloned embryos derived from BCB stained oocytes with ETCs and IVF embryos.
In the present study a significantly (P < 0.05) higher 8- to16- cell embryos and blastocyst development rate was observed in hand-made cloned embryos reconstructed from BCB+ oocytes with ETCs (73.9 %, 32.8 %, respectively) as compared to IVF embryos (49.2 %, 24.2 %, respectively). The possible reason for our results may be oocyte extracts have changed chromatin structure of donor nuclei so that recipient cytoplasm was easier to reprogram them or the ability of the BCB stain to be able to differentially select the developmentally competent oocyte. Xenopus oocyte extract have employed to induce somatic cell reprogramming and several reprogramming factors have been identified, e.g., ISWI, FRGY2a/b, BRG1, NPM, and DJ-1 [64]. The use of BCB staining based on the existence of active G6PDH in immature oocytes has recognized to be proficient tool to screen developmentally competent or incompetent oocytes for various species including bovine [26, 27]. Furthermore, when yalk fibroblast were used for interspecies NT after bovine oocyte extract treatment, 8- cell and blastocyst formation rates significantly exceeded [19]. Additionally, in our experiments, the combination of developmentally competent oocytes with ETCs may be further reason for better 8- to16- cell embryos and blastocyst yield in buffalo HMC as compared to in vitro fertilization.
The results from published reports show that the embryonic genome becomes transcriptionally active for GLUT1, OCT4, DNMT1, BAX and BCL2 at the MZT stage [45, 46]. Similar expression profiles for all developmentally important genes at 8- to 16- cell embryos in HMC and in vitro fertilization were observed in our study. Result indicates that the hand-made cloned embryos expressing the same level of transcription activity for these genes as their IVF counterparts at 8- to16- cell stages. This suggests that a similar reprogramming of these genes occurred at 8- to16- cell stages between hand-made cloned and IVF embryos. This may be due to improvement of epigenetic reprogramming of donor nuclei in hand-made cloned embryos by factors in the oocyte extracts. The pre-treatment of donor cells with oocyte extracts and selection of developmentally competent oocytes through BCB staining for recipient cytoplast preparations enhanced the expression of developmentally important genes GLUT1, OCT4, DNMT1, BAX, and BCL2 in hand-made cloned embryos at levels similar to IVF counterparts which indicates hand-made cloned embryos reconstructed using oocytes extract treated donor cells and selection of recipient cytoplasts through BCB staining are still so fragile. Additionally, the fact that hand-made cloned and IVF embryos were cultured in vitro under the same conditions. Our finding are also consistent with the statement that the level of expression of the genes evaluated at MZT stage may be influenced more by in vitro environment than by the methods used to produce the embryos.
Amarnath et al. [46] reported a significant difference in the expression of GLUT1 between NT and in vitro fertilization derived blastocysts. Studies in mouse model suggest that with in vivo embryos, GLUT1 decreased 50 % in in vitro blastocysts [65]. These reports clearly suggest that viable embryos have higher GLUT1 expression. In our study, the relative expression of GLUT1 in between two types of embryos at MZT stage support the notion that hand-made cloned embryos reconstructed with oocytes extract treated donor cells and selection of recipient cytoplasts through brilliant cresyl blue staining are identical to IVF embryos in their ability to develop to term. Decreased glucose transporter expression triggers BAX- dependent apoptosis in murine blastocysts [66]; hence it is possible that, the identical level of GLUT1 expression observed in the present study is usually related to similar levels of apoptotic incidence in both the embryos at MZT. The onset of OCT4 embryonic gene expression has been suggested to occur at the 16- cell stage [67]. Interestingly, although some studies did not find expression of OCT4 at the 8- cell, or even at the 16- cell stage [67, 68], we were able to detect it in all embryos at 8- to16-cell stage. In the present study, we used real time-PCR, which may have enabled us to measure small amount of this transcript even in a fraction of (1/16) of the cDNA obtained from 8- to 16-cell embryos. This suggests that embryonic expression of OCT4 may start before the 16-cell stage.
It has been reported that the in vitro culture of embryos has great influence not only on gene expression, but also on postnatal growth and behaviour [69]. Bovine embryos produced in vitro have shown different gene expression when compared with their in vivo counterparts [70]. The embryo culture conditions seem to induced these differences in gene expression [71]. The changes in gene expression in bovine embryos produced by NT are associated more with in vitro culture conditions than with NT, was reported by Park et al. [72]. Other studies by semi-quantitative or competitive reverse transcription-PCR (RT-PCR) have reported differences in gene expression between NT and IVF embryos [34, 73]. This divergence among experiments may be due to different protocols used to produce and culture embryos, different methods to assess gene expression profile.
In assisted reproductive technology, blastocyst morphology is quite often used as measures for selection of viable embryos produced in vitro for embryo transfer. Embryo transfer of the selected NT embryos allows for average of 4.7 % of the transferred embryos to develop to term in buffalo [74]. Evaluation of gene expression profiles in our study suggest that no differences in the gene expression profiles among the morphologically dissimilar embryos at MZT may contribute to the less variations observed in their potential to develop to blastocyst. Obviously, many other genes are involved in the pre-implantation and successful development of a mammalian embryos and additional research will increase our understanding of both nuclear reprogramming events following somatic cell NT and embryonic development in this species. In addition, the identification of genes whose expression profiles are frequently abnormal in cloned embryos will provide markers for diagnosis of cloned embryos viability prior to embryo transfer and therefore, potentially negate the time and money-consuming transfer of non-viable embryos to recipient animals. Moreover, most of the study related to the evaluation of differences between NT and IVF embryos reported in previous work have been carried out considering the morula and blastocyst stage, were difference in gene expression profile may be more obvious compare to MZT stage [34, 46, 75], so future study can also be consider to find out any differences in the relative abundance of transcripts of these genes between these two groups of embryos at morula or blastocyst stage.
The results concluded that, HMC derived embryos from pre-treatment of donor cells with oocyte extracts and selection of developmentally competent recipient cytoplasts through BCB staining revealed expression of developmentally important genes GLUT1, OCT4, DNMT1, BAX, and BCL2 at levels similar to IVF counterparts at 8- to 16- cell stages. These results also support the notion that if developmental differences observed in HMC and in vitro fertilization produced foetuses and neonates are the results of aberrant gene expression during the pre-implantation stage, those differences in expression are subtle or appear after the MZT stage of development.
Acknowledgments
The authors would like to thank Dr. Inderjeet Singh, Director, Central Institute for Research on Buffaloes, for providing the necessary facilities for carrying out this work. We are also thankful to Dr. Jerome A. for reviewing the manuscript. Funding support from Indian Council of Agriculture and Research (ICAR), New Delhi is greatly acknowledged.
Footnotes
Capsule Gene expression profiles of developmentally important genes in hand-made cloned buffalo embryos using IVF embryos as control might helpful to evaluate the viability of manipulated hand-made cloned embryos.
References
- 1.Vajta G. Handmade cloning: the future way of nuclear transfer. Trends Biotechnol. 2007;25:250–3. doi: 10.1016/j.tibtech.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Tecirlioglu RT, Cooney MA, Lewis IM. Comparison of two approaches to nuclear transfer in the bovine: hand-made cloning with modifications and the conventional nuclear transfer technique. Reprod Fertil Dev. 2005;17:573–85. doi: 10.1071/RD04122. [DOI] [PubMed] [Google Scholar]
- 3.Du Y, Kragh PM, Zhang Y. Piglets born from hand-made cloning, an innovative cloning method without micromanipulation. Theriogenology. 2007;68:1104–10. doi: 10.1016/j.theriogenology.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Saha A, Panda SK, Chauhan MS, Manik RS, Palta P, Singla SK. Birth of cloned calves from vitrified-warmed zona-free buffalo (Bubalus bubalis) embryos produced by hand-made cloning. Reprod Fertil Dev. 2012;25:860–5. doi: 10.1071/RD12061. [DOI] [PubMed] [Google Scholar]
- 5.Selokar NL, Shah RA, Saha AP, Muzaffar M, Saini M, Chauhan MS, et al. Effect of post-fusion holding time, orientation and position of somatic cell-cytoplasts during electrofusion on the development of hand-made cloned embryos in buffalo (Bubalus bubalis) Theriogenology. 2012;78:930–6. doi: 10.1016/j.theriogenology.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Shah RA, George A, Singh MK, Kumar D, Anand T, Chauhan MS, et al. Pregnancies established from hand-made cloned balstocyst recontstuectd using skin fibroblast in buffalo (Bubalus bubalis) Theriogenology. 2009;71:1215–9. doi: 10.1016/j.theriogenology.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Bortvin A, Eggan K, Skaletsky H, Akutsu H, Berry DL, Yanagimachi R, et al. Incomplete reactivation of OCT4-related genes in mouse embryos cloned from somatic nuclei. Development. 2003;130:1673–80. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- 8.Santos F, Dean W. Epigenetic reprogramming during early development in mammals. Reproduction. 2004;127:643–51. doi: 10.1530/rep.1.00221. [DOI] [PubMed] [Google Scholar]
- 9.Blelloch R, Wang Z, Meissner A, Pollard S, Smith A, Jaenisch R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells. 2006;24:2007–13. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourc’his D, Le Bourhis D, Patin D, Niveleau A, Comizzoli P, Renard JP, et al. Delayed and incomplete reprogramming of chromosome methylation patterns in bovine cloned embryos. Curr Biol. 2001;11:1542–6. doi: 10.1016/S0960-9822(01)00480-8. [DOI] [PubMed] [Google Scholar]
- 11.Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, WolF E, et al. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci U S A. 2001;98:13734–8. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos F, Zakhartchenko V, Stojkovic M. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr Biol. 2003;3:1116–21. doi: 10.1016/S0960-9822(03)00419-6. [DOI] [PubMed] [Google Scholar]
- 13.Byrne JA, Simonsson S, Western PS, Gurdon JB. Nuclei of adult mammalian somatic cells are directly reprogrammed to OCT4 stem cell gene expression by amphibian oocytes. Curr Biol. 2003;13:1206–13. doi: 10.1016/S0960-9822(03)00462-7. [DOI] [PubMed] [Google Scholar]
- 14.Hansis C, Barreto G, Maltry N, Niehrs C. Nuclear reprogramming of human somatic cells by Xenopus egg extract requires BRG1. Curr Biol. 2004;14:1475–80. doi: 10.1016/j.cub.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 15.Alberio R, Johnson AD, Stick R, Campbell KH. Differential nuclear remodeling of mammalian somatic cells by Xenopus laevis oocyte and egg cytoplasm. Exp Cell Res. 2005;307:131–41. doi: 10.1016/j.yexcr.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 16.Miyamoto K, Nagao Y, Minami N, Yamada M, Ohsumi K, Imai H. Nuclear reprogramming of porcine fibroblast cells by Xenopus egg extracts. Reprod Fertil Dev. 2006;18:110. doi: 10.1071/RDv18n2Ab3. [DOI] [Google Scholar]
- 17.Bui HT, Wakayama S, Kishigami S, Kim JH, Thuan NV, Wakayama T. The cytoplasm of mouse germinal stage oocytes can enhance somatic cell nuclear reprogramming. Development. 2008;135:3935–45. doi: 10.1242/dev.023747. [DOI] [PubMed] [Google Scholar]
- 18.Miyamoto K, Tsukiyama T, Yang Y, Li N, Minami N, Yamada M, et al. Cell-Free extracts from mammalian oocytes partially induce nuclear reprogramming in somatic cells. Biol Reprod. 2009;80:935–43. doi: 10.1095/biolreprod.108.073676. [DOI] [PubMed] [Google Scholar]
- 19.Xiong XR, Li J, Fu M, Gao C, Wang Y, Zhong JC. Oocyte extract improves epigenetic reprogramming of yak fibroblast cells and cloned embryo development. Theriogenology. 2013;79:462–9. doi: 10.1016/j.theriogenology.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 20.Bhojwani S, Alm H, Torner H, Kanitz W, Poehland R. Selection of developmentally competent oocytes through brilliant cresyl blue stain enhances blastocyst development rate after bovine nuclear transfer. Theriogenology. 2007;67:341–54. doi: 10.1016/j.theriogenology.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Jianmin SU, Yongsheng W, Ruizhe L, Hui P, Song H, Qian L, et al. Oocytes selected uing BCB staining enhance nuclear reprogramming and the in vivo development of SCNT embryos in cattle. Plos One. 2012;7:e36181. doi: 10.1371/journal.pone.0036181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian WN, Braunstein LD, Pang J, Stublmeier KM, Xi QC, Tian X. Importance of glucose -6- phosphate dehydrogenase activity for cell growth. J Biol Chem. 1998;273:10609–17. doi: 10.1074/jbc.273.17.10609. [DOI] [PubMed] [Google Scholar]
- 23.Ericsson SA, Boic ML, Funahashi H, Day BN. Assessment of porcine oocytes using brilliant cresyl blue. Theriogenology. 1993;39:214. doi: 10.1016/0093-691X(93)90069-H. [DOI] [Google Scholar]
- 24.Roca J, Martinez E, Vazquez JM, Lucas X. Selection of immature pig oocytes for homologous in vitro penetration assays with the brilliant cresyl blue test. Reprod Fertil Dev. 1998;10:479–85. doi: 10.1071/RD98060. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Gonzalez E, Lopez-Bejar M, Velilla E, Paramio MT. Selection of prepubertal goat oocytes using the brilliant cresyl blue test. Theriogenology. 2002;57:1397–409. doi: 10.1016/S0093-691X(02)00645-3. [DOI] [PubMed] [Google Scholar]
- 26.Alm H, Torner H, Loehrke B, Viergutz T, Ghoneim IM, Kanitz W. Bovine blastocyst development rate in vitro is influenced by selection of oocytes by brilliant cresyl blue staining before IVM as indicator for glucose-6-phosphate dehydrogenase activity. Theriogenology. 2005;63:2194–205. doi: 10.1016/j.theriogenology.2004.09.050. [DOI] [PubMed] [Google Scholar]
- 27.Torner H, Ghanem N, Ambros C, Holker M, Tomek W. Molecular and sub cellular characterisation of oocytes screened for their developmental competence based on glucose-6-phosphate dehydrogenase activity. Reproduction. 2008;135:197–212. doi: 10.1530/REP-07-0348. [DOI] [PubMed] [Google Scholar]
- 28.Catala MG, Izquierdo D, Uzbekova S, Morato R, Roura M. Brilliant cresyl blue stain selects largest oocytes with highest mitochondrial activity, maturation promoting factor activity and embryo developmental competence in prepubertal sheep. Reproduction. 2011;142:517–27. doi: 10.1530/REP-10-0528. [DOI] [PubMed] [Google Scholar]
- 29.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 30.Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev. 1990;26:90–100. doi: 10.1002/mrd.1080260113. [DOI] [PubMed] [Google Scholar]
- 31.Schultz RM, Davis W, Jr, Strein P, Svoboda P. Reprogramming of gene expression during pre-implanation development. J Exp Zool. 1999;285:276–82. doi: 10.1002/(SICI)1097-010X(19991015)285:3<276::AID-JEZ11>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 32.Kanka J. Gene expression and chromatin structure in the pre-implantation embryo. Theriogenology. 2003;59:3–19. doi: 10.1016/S0093-691X(02)01267-0. [DOI] [PubMed] [Google Scholar]
- 33.Niemann H, Wrenzycki C. Alterations of expression of developmentally important genes in pre-implantation bovine embryos by in vitro culture conditions: implications for subsequent development. Theriogenology. 2000;53:21–34. doi: 10.1016/S0093-691X(99)00237-X. [DOI] [PubMed] [Google Scholar]
- 34.Camargo LS, Powell AM, do Vale Filho VR, Wall RJ. Comparision of gene expression in individual pre-implantaion bovine embryos produced by in vitro fertilization or somatic cell nuclear transfer. Reprod Fertil Dev. 2005;17:487–96. doi: 10.1071/RD04128. [DOI] [PubMed] [Google Scholar]
- 35.Steuerwald N, Cohen J, Herrera RJ. Analysis of gene expression in single oocytes and embryos by real-time rapid cycle fluorescence monitored RT-PCR. Mol Hum Reprod. 1999;5:1034–9. doi: 10.1093/molehr/5.11.1034. [DOI] [PubMed] [Google Scholar]
- 36.Wrenzycki C, Herrmann D, Lucas-Hahn L, Lemme E, Korsawe K, Niemann H. Gene expression patterns in in vitro produced and somatic nuclear transfer derived pre-implantation bovine embryos: relationship to the large offspring syndrome. Anim Reprod Sci. 2004;82–83:593–603. doi: 10.1016/j.anireprosci.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Niemann H, Wrenzycki C, Lucas-Hahn A, Brambrink T, Kues WA, Carnwath JW. Gene expression patterns in bovine in vitro produced and nuclear transfer derived embryos and their implications for early development. Cloning Stem Cells. 2002;4:29–38. doi: 10.1089/153623002753632020. [DOI] [PubMed] [Google Scholar]
- 38.Lonergan P, Rizos D, Gutierrez-Adan A, Fair T, Boland MP. Oocyte and embryo quality: effect of origin, culture conditions and gene expression patterns. Reprod Dome Anim. 2003;38:259–67. doi: 10.1046/j.1439-0531.2003.00437.x. [DOI] [PubMed] [Google Scholar]
- 39.Wrenzycki C, Wells D, Herrmann D, Miller A, Oliver J, Tervit R. Nuclear transfer protocol affects messenger RNA expression patterns in cloned bovine blastocysts. Biol Reprod. 2001;65:309–17. doi: 10.1095/biolreprod65.1.309. [DOI] [PubMed] [Google Scholar]
- 40.Wrenzycki C, Nieman H. Epigenetic reprogramming in early embryonic development: effects of in vitro production and somatic nuclear transfer. Reprod Biomed Online. 2003;7:649–56. doi: 10.1016/S1472-6483(10)62087-1. [DOI] [PubMed] [Google Scholar]
- 41.Daniels R, Hall V, Trounson AO. Analysis of gene transcription in bovine nuclear transfer embryos reconstructed with granulosa cell nuclei. Bio Rep. 2000;63:1034–40. doi: 10.1095/biolreprod63.4.1034. [DOI] [PubMed] [Google Scholar]
- 42.Li XP, Kato Y, Tsunoda Y. Comparative studies on the mRNA expression of development-related genes in an individual mouse blastocyst with different developmental potential. Cloning Stem Cells. 2006;8:214–24. doi: 10.1089/clo.2006.8.214. [DOI] [PubMed] [Google Scholar]
- 43.Kato Y, Li XP, Amarnath D, Uuhizawa K, Hashizume K, Tokunaga T, et al. Comparative gene expression analysis of bovine nuclear-transferred embryos with different developmental potential by cDNA microarray and real-time PCR to determine genes that might reflect calf normality. Cloning Stem Cells. 2007;9:495–511. doi: 10.1089/clo.2007.0014. [DOI] [PubMed] [Google Scholar]
- 44.Kato Y, Tsunoda Y. Role of the donor nuclei in cloning efficiency: can the ooplasm reprogram any nucleus. Int J Dev Biol. 2010;54:1623–9. doi: 10.1387/ijdb.103203yk. [DOI] [PubMed] [Google Scholar]
- 45.Li XP, Amarnath D, Kato Y, Tsunoda Y. Analysis of development- related gene expression in cloned bovine blastocysts with different developmental potential. Cloning Stem Cells. 2006;8:41–50. doi: 10.1089/clo.2006.8.41. [DOI] [PubMed] [Google Scholar]
- 46.Amarnath D, Li X, Kato Y, Tsunoda Y. Gene expression in individual bovine somatic cell cloned embryos at the 8- cell and blastocyst stages of pre-implantation development. J Reprod Dev. 2007;53:1247–63. doi: 10.1262/jrd.19096. [DOI] [PubMed] [Google Scholar]
- 47.Pandey A, Gupta SC, Singh N, Rana JS, Gupta N. Efficiency of SCNT buffalo (Bubalus bubalis) embryos in different culture medium and analysis of mRNA expression of insulin-like growth factors during embryogenesis. Reprod Domest Anim. 2010;45:786–95. doi: 10.1111/j.1439-0531.2009.01353.x. [DOI] [PubMed] [Google Scholar]
- 48.Chauhan MS, Singla SK, Palta P, Manik RS, Tomer OS. Development of in vitro produced buffalo (Bubalus bubalis) embryos in relation to time. Asian Aust J Anim Sci. 1998;11:398–403. [Google Scholar]
- 49.Bracket BG, Oliphant G. Capaciation of rabbit spermatozoa in vitro. Biol Reprod. 1975;12:260–74. doi: 10.1095/biolreprod12.2.260. [DOI] [PubMed] [Google Scholar]
- 50.Shuang T, Yongsheng W, Dong Z, Yajun G, Yefei M, Baoying Y, et al. Reprogramming donor cells with oocyte extracts improves in vitro development of nuclear transfer embryos. Anim Reprod Sci. 2009;115:1–9. doi: 10.1016/j.anireprosci.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 51.Shah RA, George A, Singh MK, Kumar D, Chauhan MS, Manik R, et al. Hand-made cloned buffalo (Bubalus bubalis) embryos: comparison of different media and culture systems. Cloning Stem Cells. 2008;10:435–42. doi: 10.1089/clo.2008.0033. [DOI] [PubMed] [Google Scholar]
- 52.Leese HJ. Quiet please, do not disturb hypothesis of embryo metabolism and viability. Bioessays. 2002;24:845–9. doi: 10.1002/bies.10137. [DOI] [PubMed] [Google Scholar]
- 53.Misica-Turner PM, Oback FC, Eichenlaub M, Well D, Oback B. Aggregating embryonic but not somatic nuclear transfer embryos increases cattle cloning efficiency. Biol Reprod. 2006;76:268–78. doi: 10.1095/biolreprod.106.050922. [DOI] [PubMed] [Google Scholar]
- 54.Beyhan Z, Forsberg EJ, Eilertsen KJ, Kent-First M, First NL. Gene expression in bovine nuclear transfer embryos in relation to donor cell efficiency in producing live offspring. Mol Reprod Dev. 2007;74:18–27. doi: 10.1002/mrd.20618. [DOI] [PubMed] [Google Scholar]
- 55.Kremenskoy M, Kremenska Y, Suzuki M, Imai K, Takahashi S, Hashizume K, et al. Epigenetic characterization of the CpG islands of bovine leptin and POU5F1 genes in cloned bovine fetuses. J Reprod Dev. 2006;52:277–85. doi: 10.1262/jrd.17100. [DOI] [PubMed] [Google Scholar]
- 56.Kang YK, Koo DB, Park JS, Choi YH, Chung AS, Lee KK. Aberrant methylation of donor genome in cloned bovine embryos. Nat Genet. 2001;28:173–7. doi: 10.1038/88903. [DOI] [PubMed] [Google Scholar]
- 57.Shiqiang J, Rong R, Qing L, Pengfei L, Huili G. Analysis of apoptosis and methyltransferase mRNA expression in porcine cloned embryos cultured in vitro. J Assist Reprod Genet. 2010;27:49–59. doi: 10.1007/s10815-009-9378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brison DR, Schultz RM. Apoptosis during mouse blastocyst formation: evidence for a role for survival factors including transforming growth factor alpha. Biol Reprod. 1997;56:1088–96. doi: 10.1095/biolreprod56.5.1088. [DOI] [PubMed] [Google Scholar]
- 59.Hardy K, Spanos S. Growth factor expression and function in the human and mouse pre-implantation embryo. J Endocrinol. 2002;172:221–36. doi: 10.1677/joe.0.1720221. [DOI] [PubMed] [Google Scholar]
- 60.Fahrudin M, Otoi T, Karja NW, Mori M, Murakami M, Suzuki T. Analysis of DNA fragmentation in bovine somatic nuclear transfer embryos using TUNEL. Reproduction. 2002;124:813. doi: 10.1530/rep.0.1240813. [DOI] [PubMed] [Google Scholar]
- 61.Kepp O, Rajalingam K, Kimmig S, Rudel T. BAK and BAX are non-redundant during infection- and DNA damage-induced apoptosis. Eur Mol Biol Organ J. 2007;26:825–34. doi: 10.1038/sj.emboj.7601533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zinkel S, Gross A, Yang E. BCL2 family in DNA damage and cell cycle control. Cell Death Differ. 2006;13:1351–9. doi: 10.1038/sj.cdd.4401987. [DOI] [PubMed] [Google Scholar]
- 63.Mundim TC, Ramos AF, Sartori R, Dode MA, Melo EO, Gomes LF, et al. Changes in gene expression profiles of bovine embryos produced in vitro, by natural ovulation, or hormonal superstimulation. Genet Mol Res. 2009;8:1398–407. doi: 10.4238/vol8-4gmr646. [DOI] [PubMed] [Google Scholar]
- 64.Miyamoto K, Nagai K, Kitamura N, Nishikawa T, Ikegami H, Binh NT. Identification and characterization of an oocyte factor required for development of porcine nuclear transfer embryos. Proc Nat Acad Sci. 2011;108:7040–5. doi: 10.1073/pnas.1013634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morita Y, Tsutsumi O, Hosoya I, Taketani Y, Oka Y, Kato T. Expression and possible function of glucose transporter protein GLUT1 during pre-implantation mouse development from oocytes to blastocysts. Biochem Biophys Res Commun. 1992;188:8–15. doi: 10.1016/0006-291X(92)92342-U. [DOI] [PubMed] [Google Scholar]
- 66.Chi MM, Pingsterhaus J, Carayannopoulos M, Moley KH. Decreased glucose transporter expression triggers BAX-dependent apoptosis in the murine blastocyst. J Biol Chem. 2000;275:40252–7. doi: 10.1074/jbc.M005508200. [DOI] [PubMed] [Google Scholar]
- 67.Kurosaka S, Eckardt S, McLaughlin KJ. Pluripotent lineage definition in bovine embryos by OCT4 transcript localization. Biol Reprod. 2004;71:1578–82. doi: 10.1095/biolreprod.104.029322. [DOI] [PubMed] [Google Scholar]
- 68.Donnison M, Pfeffer PL. Isolation of genes associated with developmentally competent bovine oocytes and quantitation of their levels during development. Biol Reprod. 2004;71:1813–21. doi: 10.1095/biolreprod.104.032367. [DOI] [PubMed] [Google Scholar]
- 69.Harvey AJ, Kind KL, Pantaleon M, Armstrong DT, Thompson JG. Oxygen–regulated gene expression in bovine blastocysts. Biol Reprod. 2004;71:1108–19. doi: 10.1095/biolreprod.104.028639. [DOI] [PubMed] [Google Scholar]
- 70.Rizos D, Ward F, Duffy P, Boland MP, Lonergan P. Consequences of bovine oocyte maturation, fertilization or early embryo development in vitro versus in vivo: implications for blastocyst yield and blastocyst quality. Mole Reprod Dev. 2002;61:234–48. doi: 10.1002/mrd.1153. [DOI] [PubMed] [Google Scholar]
- 71.Knijn HM, Wrenzycki C, Hendriksen PJ, Vos PL, Herrmann D, Van der Weijden GC, et al. Effects of oocyte maturation regimen on the relative abundance of gene transcripts in bovine blastocysts derived in vitro or in vivo. Reproduction. 2002;124:365–75. doi: 10.1530/rep.0.1240365. [DOI] [PubMed] [Google Scholar]
- 72.Park SH, Park SB, Kim NH. Expression of early developmental-related genes in bovine nuclear transfer and fertilized embryos. Zygote. 2003;11:355–60. doi: 10.1017/S0967199403002454. [DOI] [PubMed] [Google Scholar]
- 73.Han YM, Kang YK, Kon DB, Lee KK. Nuclear reprogramming of cloned embryos produced in vitro. Theriogenology. 2003;59:33–44. doi: 10.1016/S0093-691X(02)01271-2. [DOI] [PubMed] [Google Scholar]
- 74.Shi D, Lu F, Wei Y. Buffalos (Bubalus bubalis) cloned by nuclear transfer of somatic cells. Biol Reprod. 2007;77:285–91. doi: 10.1095/biolreprod.107.060210. [DOI] [PubMed] [Google Scholar]
- 75.Rodríguez-Alvarez L, Cox J, Tovar H, Einspanier R, Castro FO. Changes in the expression of pluripotency-associated genes during pre-implantation and peri-implantation stages in bovine cloned and in vitro produced embryos. Zygote. 2010;18:269–79. doi: 10.1017/S0967199409990323. [DOI] [PubMed] [Google Scholar]