Abstract
Purpose
To determine the expression patterns of imprinted genes and their methylation status in aborted cloned porcine fetuses and placentas.
Methods
RNA and DNA were prepared from fetuses and placentas that were produced by SCNT and controls from artificial insemination. The expression of 18 imprinted genes was determined by quantitative real-time PCR (q-PCR). Bisulfite sequencing PCR (BSP) was conducted to determine the methylation status of PRE-1 short interspersed repetitive element (SINE), satellite DNA and H19 differentially methylated region 3 (DMR3).
Results
The weight, imprinted gene expression and genome-wide DNA methylation patterns were compared between the mid-gestation aborted and normal control samples. The results showed hypermethylation of PRE-1 and satellite sequences, the aberrant expression of imprinted genes, and the hypomethylation of H19 DMR3 occurred in mid-gestation aborted fetuses and placentas.
Conclusions
Cloned pigs generated by somatic cell nuclear transfer (SCNT) showed a greater ratio of early abortion during mid-gestation than did normal controls because of the incomplete epigenetic reprogramming of the donor cells. Altered expression of imprinted genes and the hypermethylation profile of the repetitive regions (PRE-1 and satellite DNA) may be associated with defective development and early abortion of cloned pigs, emphasizing the importance of epigenetics during pregnancy and implications thereof for patient-specific embryonic stem cells for human therapeutic cloning and improvement of human assisted reproduction.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-014-0320-2) contains supplementary material, which is available to authorized users.
Keywords: Imprinted gene, SCNT, Cloned pig, DNA methylation, PRE-1, Satellite DNA
Introduction
Therapeutic cloning, or somatic-cell nuclear transfer (SCNT), is used in stem cell research and has been used to create perfectly matched tissues. In this process, the nucleus is amenable to adequate remodeling and subsequent genomic reprogramming, although SCNT cloning remains inefficient. In humans, assisted reproductive technology (ART) embryos suffer a high ratio of early spontaneous abortions for a number of reasons. Swine are closely related to humans in terms of genome structure, and they also avoid some ethical problems. Moreover, pigs have been used as a good model for human assisted reproduction.
SCNT has been used to a clone wide variety of animals since Dolly was first cloned via a differentiated somatic donor cell; however, SCNT remains low efficiency and causes various developmental problems, such as abnormal fetal growth, enlarged placenta, perinatal death and an increased ratio of mid-gestation abortion [1]. The majority of SCNT embryos are lost during the different stages of pregnancy, including the death of embryos prior to implantation or early abortion during mid-gestation.
Recent studies demonstrated that abnormal gene expression and epigenetic modification are related to early abortion during mid-gestation and a low birth ratio of cloned animals [2–4]. Imprinted genes are specifically expressed by one of the two homologous chromosomes and contain parent-origin genetic information. Previous studies demonstrated that the majority of imprinted genes play important roles in regulating the normal development of the fetus and placenta, and the expression of these genes is regulated by the methylation status of the promoters [5, 6]. Numerous studies reported that aberrant DNA methylation occurs in cloned mammalian embryos [7, 8]. PRE-1, a short interspersed nucleotide element (SINEs), has 105 copies in porcine genome, whereas centromeric satellite DNA is a highly repetitive sequence in porcine heterochromatic region. The methylation levels of these two sequences could represent the genome-wide methylation status in swine [9, 10]. Insulin-like growth factor two (IGF2) is paternally expressed, whereas H19 is transcribed from the maternal allele. The expression of these two genes is regulated through the DMR locus of H19, which contains transcription factor 11-zinc finger protein (CTCF) binding sites within an imprinting control region (ICR) [11].
A growing body of evidence suggests that the epigenetic status may play a role in early abortion and other problems of human assisted reproduction [12]. In our previous study, porcine SCNT embryos underwent a greater ratio of early abortion during mid-gestation days 27 to 34 [13]. In this study, we attempt to determine the expression patterns of imprinted genes and the methylation status in aborted cloned porcine fetuses and placentas to achieve a better understanding of porcine SCNT and identify implications to improve human assisted reproduction.
Materials and methods
Sample collection
The porcine breed used in the experiment was Yorkshire. Fetal fibroblasts were used as the donor cells. The acquisition of donor cells and recipient oocytes, in vitro maturation (IVM), SCNT, artificial activation of reconstructed oocytes and embryo transfer were performed as previously described [14]. Unless indicated otherwise, the reagents were obtained from Sigma-Aldrich Co. (St. Louis, MO). Briefly, prepubertal porcine ovaries were collected from an abattoir and transported to the laboratory in a thermos filled with saline maintained at 30–35 °C. The oocyte–cumulus complexes (COCs) were aspirated from the ovarian follicles using a 10 ml disposable syringe with an 18-gauge needle and matured in TCM-199 (Gibco, BRL, Grand Island, NY, USA) supplemented with 0.1 % PVA, 3.05 mM D-glucose, 0.91 mM sodium pyruvate, 10 ng/ml epidermal growth factor (EGF), 0.57 mM cysteine, 0.5 μg/ml follicle-stimulating hormone (FSH), 0.5 μg/ml luteinizing hormone (LH), 75 μg/ml penicillin and 50 μg/ml streptomycin for 44 h 38.5 °C under 5 % (v/v) CO2. Following maturation, the oocytes were isolated from the COCs by pipetting up and down several times in a PVA-TL HEPES stock solution supplemented with 0.1 % hyaluronidase. Rounded oocytes with an extruded first polar body were selected for SCNT. The first polar body and the metaphase II plate in a small amount of surrounding cytoplasm were removed from the oocytes using a beveled glass pipette with a diameter of 25–30 μm in PBS micro drops supplemented with 7.5 μg/mL cytochalasin B (CB) and 10 % FBS. Confirmation of successful enucleation was observed by visualizing the karyoplast, while inside the pipette, under violet light. The donor nucleus was directly injected into the cytoplasm of the enucleated oocytes. Then, the reconstructed oocytes were pooled in a chamber filled with 0.3 M mannitol, 1.0 mM CaCl2 · 2H2O, 1.0 mM MgCl2 · 6H2O and 0.5 mM Hepes. 2 DC pulses of 1.2 kV/cm for 30 μs were subsequently administered using a BTX Electro Cell Manipulator 2001 (BTX, San Diego, CA, USA). After activation, the oocytes were cultured in PZM-3 medium supplemented with 7.5 μg/ml CB at 38.5 °C in a humidified atmosphere containing 5 % (v/v) CO2 for 4 h. After 1 day, the embryos were then transferred to the fallopian tubes of surrogate Yorkshire sows. Ultrasound examination was performed to verify the pregnancy of the sows after embryo transfer. Aborted cloned fetuses (N = 11) and placentas (N = 11) were collected on day 28 of pregnancy. Artificially inseminated sows were then anesthetized after 28 days of pregnancy, and the fetuses (N = 13) and placentas (N = 13) were collected from the uterine horn. All samples were weighed and stored in liquid nitrogen until further use.
The pig experiments were performed in accordance with the guidelines on the animal care and use of animals in research, which were approved by the Animal Care and Use Committee of Jilin University, Changchun, China.
Quantitative real-time PCR
Total RNA was isolated using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. DnaseI-treated RNA was reverse transcribed using the cDNA First Strand synthesis kit (Bioer Technology, Hangzhou, Zhejiang, China). Primers used for q-PCR are listed in Table S1. Quantitative gene expression analysis was performed according to the manufacturer’s instructions using a BIO-RAD iQ5 Multicolor Real-Time PCR Detection System with the BioEasy SYBR Green I Real Time PCR Kit (Bioer Technology, Hangzhou, Zhejiang, China). The reaction conditions were as follows: initial denaturation at 95 °C for 10 min, 40 cycles of denaturation at 95 °C for 10s, annealing at 58 °C for 15 s and extension at 72 °C for 30s. The melting protocol was from 55 to 95 °C in increments of 0.5 °C/5 s. All gene expression experiments were performed three times for the harvested samples. The gene expression is presented as the mean ± S.E.M. GAPDH was selected as the reference gene. The PCR products were identified by agarose gel (1 %) electrophoresis and DNA sequencing.
Bisulfite sequencing PCR
Genomic DNA was extracted with TIANamp Genomic DNA Kit (Tiangen, Beijing, China) and was treated with the CpGenome Turbo Bisulfite Modification Kit (Millipore, USA). The primers and PCR condition for the treated sequences of PRE-1, satellite region and H19 DMR3 were used as previously described [11, 15]. The PCR products were purified using a TIANgel Midi Purification Kit (Tiangen, Beijing, China) and cloned into the PGM-T vector (Tiangen, Beijing, China). Ten positive plasmid clones were sequenced at Tiangen Corporation.
Statistical analysis
The gene expression and methylation patterns were analyzed using the SPSS 16.0 software (SPSS Inc., Chicago, IL, USA). A probability of p < 0.05 was considered statistically significant. The methylation status was analyzed with the online programs methprimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) and BiQ Analyzer (http://biq-analyzer.bioinf.mpi-inf.mpg.de/tools/MethylationDiagrams/index.php).
Results
Weight comparison and imprinted gene expression analysis
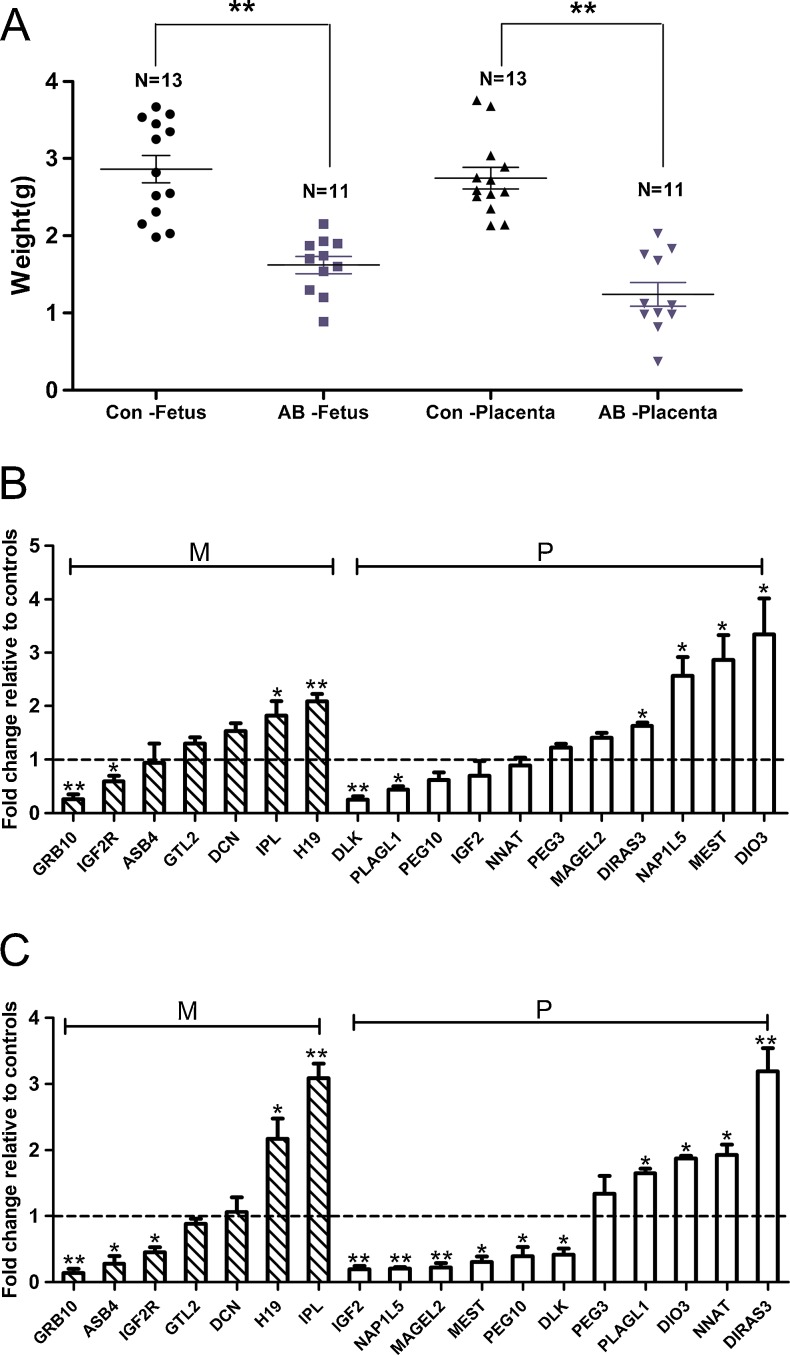
The weight of the aborted (n = 13) and control (n = 11) samples was recorded in this study. The results show significantly reduced weight for the aborted fetuses and placentas (Fig. 1A), indicating unsynchronized and retarded development in mid-gestation aborted cloned porcine fetuses and placentas, which suggests that developmental defects may be related to the mid-gestation abortion of porcine SCNTs.
Fig. 1.
Weight comparison and relative expression levels of 18 imprinted genes. A: Weight comparison between the aborted cloned pigs and controls. Con, control, AB, aborted. Different shapes represent different samples. N represents the number of collected fetuses or placentas. B: The relative mRNA levels of 18 imprinted genes in aborted cloned porcine fetuses analyzed by quantitative real-time PCR. C: The relative mRNA levels of 18 imprinted genes in aborted porcine placentas analyzed by quantitative real-time PCR. M, maternal, P, paternal. The hyphenated lines (set as 1) represent the average expression level of each gene compared to the controls. Three repeats were performed for each sample. The values are presented as the mean ± S.E.M. *p < 0.05, **p < 0.01
Although numerous studies reported that imprinted genes are important for early embryonic development, there is little knowledge concerning mid-gestation abortion. In this study, we examined the expression of 18 imprinted genes, which play important roles in embryonic development using q-PCR. Figure 1B and C demonstrate that H19, IPL, DIO3, MEST, NAP1L5 and DIRAS3 are significantly over-expressed (p < 0.05), whereas the expression of GRB10, IGF2R, DLK and PLAGL1 are significantly reduced in the aborted fetuses (p < 0.05). In the aborted placentas, the expression of IPL, H19, DIRAS3, NNAT, DIO3 and PLAGL1 are significantly elevated (p < 0.05), whereas the expression of GRB10, ASB4, IGF2R, IGF2, NAP1L5, MAGEL2, MEST, PEG10 and DLK are significantly down-regulated (p < 0.05). These data indicate that the abnormal expression of these 18 genes may play a critical role in the retarded development or early abortion of porcine SCNT fetuses and placentas.
DNA methylation status of PRE-1 and satellite DNA
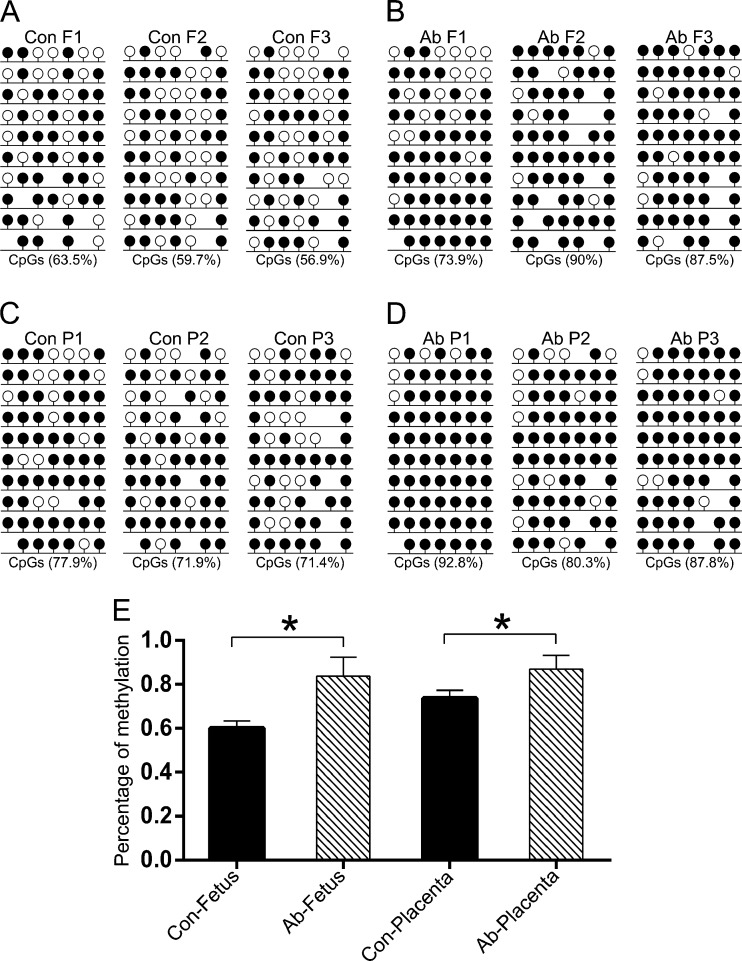
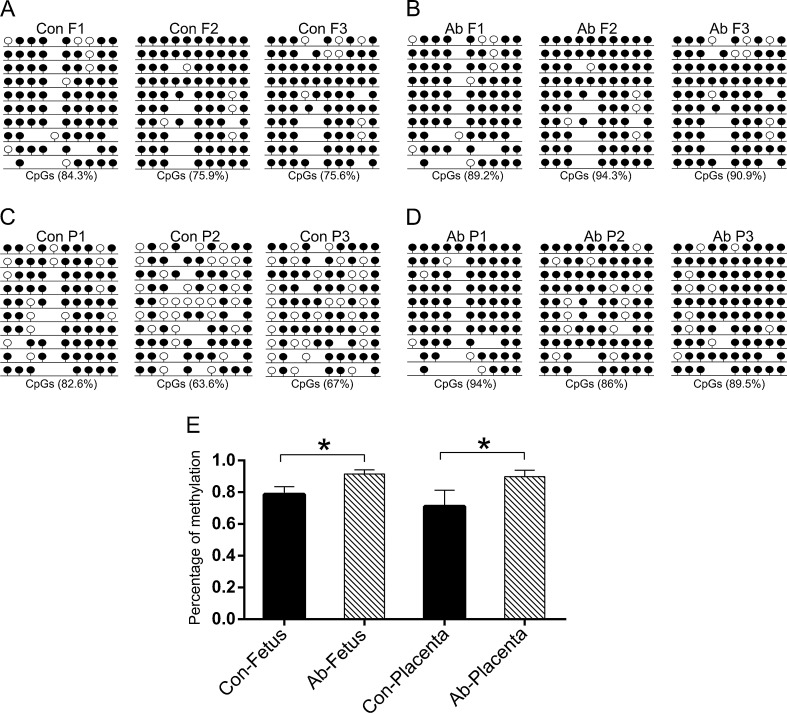
DNA methylation is a common epigenetic modification that plays an important role during gene transcription. To better understand the mechanism of DNA methylation in mid-gestation abortion SCNT embryos, we analyzed the methylation patterns of PRE-1 and satellite sequences using BSP. The results demonstrated that both PRE-1 (Fig. 2A-D) and satellite sequence (Fig. 3A-D) were hyper methylated in the aborted cloned fetuses and placentas compared to the controls (p < 0.05). Statistical analysis confirmed this result for the three aborted and control samples (Figs. 2 and 3E).
Fig. 2.
DNA methylation status of the PRE-1 element. The methylation profiles of PRE-1 in control fetuses (a), aborted fetuses (b), control placentas (c) and aborted placentas (d) analyzed by BSP. Con, control, Ab, aborted, F, fetus, P, placenta. Numbers 1, 2 and 3 represent each fetus or placenta. Unfilled (white) and filled (black) circles represent unmethylated and methylated CpGs, respectively. Horizontal lines with circles represent one separate sequenced clone. Lollipop diagrams were generated with the BIQ Analyzer software. The percentage under each figure represents the methylation proportion of the corresponding sample. E shows the statistical DNA methylation level of PRE-1 according to a, b, c and d. The values are presented as the mean ± SEM. *p < 0.05
Fig. 3.
DNA methylation status of satellite region. The methylation profiles of satellite region in control fetuses (a), aborted fetuses (b), control placentas (c) and aborted placentas (d) analyzed by BSP. E shows the statistical DNA methylation level of the satellite region according to a, b, c and d. The details are described in the legend for Fig. 2
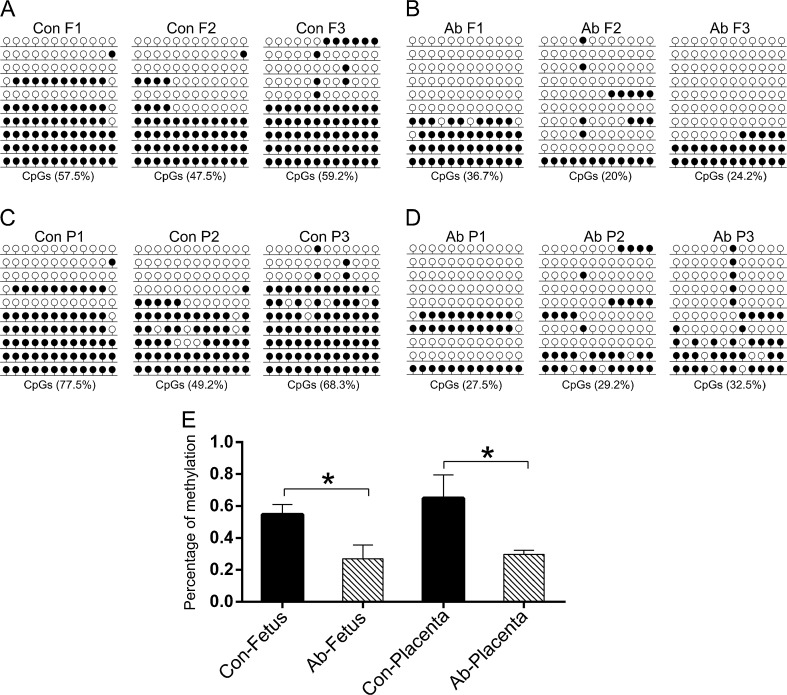
DNA methylation status of H19 DMR3
In this study, H19 was abnormally and significantly over-expressed, whereas IGF2 was down-regulated in aborted porcine fetuses and placentas. To determine the aberrant expression of H19 and Igf2 in the aborted samples, we analyzed the methylation patterns of H19 DMR3 using BSP. The sequencing data showed that the methylation patterns of H19 DMR3 were significantly demethylated (p < 0.05) in aborted samples compared to the controls. Statistical analysis confirmed this result for the three aborted and control samples (Fig. 4E). These results indicated the hypo methylated status of H19 DMR3 is related to the up-regulated expression of H19 and mid-abortion of SCNT fetuses.
Fig. 4.
DNA methylation status of H19 DMR3. The methylation profiles of H19 DMR3 in control fetuses (a), aborted fetuses (b), control placentas (c) and aborted placentas (d) analyzed by BSP. E shows the statistical DNA methylation level of H19 DMR3 according to a, b, c and d. The details are described in the legend for Fig. 2
Discussion
In this study, we found that the aborted cloned porcine fetuses and placentas had lower weights than the normal controls, suggesting the defective development of early SCNT cloned embryos, which has frequently been reported. A previous study found that placental overgrowth occurs in mice lacking the imprinted gene IPL [16]. In our study, the expression of IPL was 3 times higher in aborted cloned placentas than in controls; therefore, we hypothesized that the smaller size of the early aborted cloned porcine placentas may be partially due to the high expression of IPL. Furthermore, the smaller size of the fetus and the lack of relevant products may affect the recognition between mother and fetus, which may result in abortion.
It is generally accepted that incorrect epigenetic modifications during SCNT and altered imprinted gene expression are responsible for the low efficiency of SCNT cloned pigs. The majority of imprinted genes play roles in fetal growth and development, and both the maternal and paternal genomes are required for normal development. H19, IGF2, PEG3, GRB10 and IGF2R were previously found significantly differentially expressed in dead cloned piglets [17, 18]. These findings were confirmed by our present results, which revealed that the altered expression of imprinted genes was related to defective development and early abortion of SCNT cloned pigs. Recent reports also showed that aberrant imprinted gene expression is related to intrauterine growth restriction (IUGR) of human fetus [19]. This result is consistent with our finding in swine, demonstrating the similarity of developmental regulation of imprinted gene between pig and human.
Additionally, aberrantly expressed genes varied between the fetuses and placentas and showed more significant abnormalities in placentas. Studies demonstrated that imprinted genes control the development of the fetus by regulating the development of the placenta and the exchange of substances between the mother and fetus. Our experiment demonstrated that serious defects of the placenta might cause abnormal imprinted gene expression in the fetus. Recently, a study showed that DNA methylation of imprinted genes was higher in children conceived by intra-cytoplasmic sperm injection (ICSI) [12], which was not significantly relevant with the method of conception. Here, we postulate that the aberrant methylation levels of imprinted genes may result from oocyte reactivation, which leads to the early abortion of SCNT or ICSI embryos.
PRE-1 SINE and satellite sequences were aberrantly hyper methylated in early aborted porcine fetuses and placentas, suggesting that the expression of several important growth-related genes may be repressed. A previous study found typical demethylation processes in these repetitive sequences in pig preimplantation embryos [15]. Similar results were also found in mouse [20]. In cloned bovine embryos, these repeated regions of the donor genome exhibited aberrant methylation patterns [8]. In our study, the aborted cloned pigs showed aberrant high levels of methylation. Previous studies suggested that the in vitro culture medium affects imprinting methylation [21]. Further investigations are necessary to identify the molecular mechanisms of epigenetic reprogramming and methods to reduce abortion.
H19/Igf2 is a pair of growth-regulating imprinted genes, which are particularly important in early embryonic development. In this study, H19 was abnormally over-expressed, whereas IGF2 was down-regulated in the aborted fetuses and placentas. Furthermore, our results showed that H19 DMR3 was hypo methylated. Because H19 and IGF2 share common enhancers located downstream of H19, demethylation of DMR inhibits the expression of IGF2. A previous study showed that H19 and IGF2 are negatively correlated in naturally produced and somatically cloned mice [22], which displayed low expression of H19 and high expression of IGF2. Our findings confirmed the regulation of these two genes. In human IUGR fetuses, H19 is highly expressed, whereas IGF2 expression is reduced [19], which is identical to our findings in aborted cloned pigs.
SCNT may also be used to generate patient-specific embryonic stem cells for human therapeutic cloning. Our study revealed that genomic imprinting of SCNT embryos is incomplete, which may occur during nuclear transfer and be maintained through embryonic development. Additionally, many of the observed defects involve the placenta. Figure 1B and C show that paternally expressed genes have more significantly aberrant expression in placentas than in fetuses. This suggests that epigenetic reprogramming of paternal alleles is more susceptible to the in vitro culture medium or maturation than maternal alleles. For ART, we may need to pay greater attention to the sperm source. Further studies are necessary to identify the molecular mechanisms of epigenetic reprogramming of paternal alleles.
In conclusion, the altered expression of 18 imprinted genes, the hyper methylated patterns of repetitive regions and the demethylation of H19 DMR3 were observed in early aborted cloned porcine fetuses and placentas. Therefore, we hypothesize that the altered expression of imprinted genes is associated with abnormal DNA methylation levels, which is due to aberrant epigenetic reprogramming during SCNT, resulting in defective development and, ultimately, natural fetal abortion. Our study demonstrates the importance of epigenetic reprogramming during normal pig development and the reproductive similarities between swine and human. We also determined the imprinted gene expression profile of aborted SCNT cloned pigs, which has implications for therapeutic cloning and improving human assisted reproduction.
Electronic supplementary material
(DOC 50 kb)
Acknowledgments
We thank Xue Chen, Peiyan Hu for technical support at the Embryo Engineering Center. The National Natural Science Foundation of China (Grant No. 31201080 and 31272394) and a grant from the Changjiang scholars and innovative research team of universities (PCSIRT, No. IRT1248) supported this study.
Author contributions
Z.L. conceived of the study. Z.L. and D.W. designed the experiments. X.Z., Y.H., F.D. and Q.L. performed the experiments. X.Z. and Z.L. analyzed the data and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Capsule Our study demonstrated altered expression of imprinted genes and hypermethylation profile of the repetitive regions (PRE-1and satellite DNA) maybe associated with defective development and early abortion of cloned pigs, which emphases the importance of epigenetics during pregnancy and human assisted reproduction.
Xiaoyang Zhang and Dongxu Wang are authors contributed equally to this work.
Contributor Information
Xiaoyang Zhang, Email: recsy1126@163.com.
Dongxu Wang, Email: wang_dong_xu@163.com.
Yang Han, Email: hanyang_0008@163.com.
Feifei Duan, Email: 519482906@qq.com.
Qinyan Lv, Email: 1107334601@qq.com.
Zhanjun Li, Phone: +86-431-87836176, Email: lizj_1998@jlu.edu.cn.
References
- 1.Wilmut I, Beaujean N, de Sousa PA, et al. Somatic cell nuclear transfer. Nature. 2002;419(6907):583–6. doi: 10.1038/nature01079. [DOI] [PubMed] [Google Scholar]
- 2.Yang X, Smith SL, Tian XC, et al. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet. 2007;39(3):295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- 3.Chae JI, Lee KS, Kim DJ, et al. Abnormal gene expression in extraembryonic tissue from cloned porcine embryos. Theriogenology. 2009;71(2):323–33. doi: 10.1016/j.theriogenology.2008.07.025. [DOI] [PubMed] [Google Scholar]
- 4.Chae JI, Yu K, Cho SK, et al. Aberrant expression of developmentally important signaling molecules in cloned porcine extraembryonic tissues. Proteomics. 2008;8(13):2724–34. doi: 10.1002/pmic.200701134. [DOI] [PubMed] [Google Scholar]
- 5.Young LE, Fairburn HR. Improving the safety of embryo technologies: possible role of genomic imprinting. Theriogenology. 2000;53(2):627–48. doi: 10.1016/S0093-691X(99)00263-0. [DOI] [PubMed] [Google Scholar]
- 6.Jiang L, Jobst P, Lai L, et al. Expression levels of growth-regulating imprinted genes in cloned piglets. Cloning Stem Cells. 2007;9(1):97–106. doi: 10.1089/clo.2006.0041. [DOI] [PubMed] [Google Scholar]
- 7.Shi W, Dirim F, Wolf E, et al. Methylation reprogramming and chromosomal aneuploidy in in vivo fertilized and cloned rabbit preimplantation embryos. Biol Reprod. 2004;71(1):340–7. doi: 10.1095/biolreprod.103.024554. [DOI] [PubMed] [Google Scholar]
- 8.Kang YK, Koo DB, Park JS, et al. Aberrant methylation of donor genome in cloned bovine embryos. Nat Genet. 2001;28(2):173–7. doi: 10.1038/88903. [DOI] [PubMed] [Google Scholar]
- 9.Tajima K, Enishi O, Amari M, et al. PCR detection of DNAs of animal origin in feed by primers based on sequences of short and long interspersed repetitive elements. Biosci Biotechnol Biochem. 2002;66(10):2247–50. doi: 10.1271/bbb.66.2247. [DOI] [PubMed] [Google Scholar]
- 10.Harumi T, Kimura M, Yasue H. Survey on swine SINEs (PRE‐1) as candidates for SSCP markers in genetic linkage analysis. Anim Genet. 1995;26(6):403–6. doi: 10.1111/j.1365-2052.1995.tb02691.x. [DOI] [PubMed] [Google Scholar]
- 11.Park C-H, Kim H-S, Lee S-G, et al. Methylation status of differentially methylated regions at <i> Igf2/H19</i> locus in porcine gametes and preimplantation embryos. Genomics. 2009;93(2):179–86. doi: 10.1016/j.ygeno.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Whitelaw N, Bhattacharya S, Hoad G, Horgan GW, Hamilton M, Haggarty P. Epigenetic status in the offspring of spontaneous and assisted conception. Hum Reprod. 2014. doi:10.1093/humrep/deu094. [DOI] [PubMed]
- 13.Huang Y, Ouyang H, Yu H, et al. Efficiency of porcine somatic cell nuclear transfer - a retrospective study of factors related to embryo recipient and embryos transferred. Biol Open. 2013;2(11):1223–8. doi: 10.1242/bio.20135983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai L, Prather RS. Production of cloned pigs by using somatic cells as donors. Cloning & Stem Cells. 2003;5(4):233–41. doi: 10.1089/153623003772032754. [DOI] [PubMed] [Google Scholar]
- 15.Kang YK, Koo DB, Park JS, et al. Typical demethylation events in cloned pig embryos. Clues on species-specific differences in epigenetic reprogramming of a cloned donor genome. J Biol Chem. 2001;276(43):39980–4. doi: 10.1074/jbc.M106516200. [DOI] [PubMed] [Google Scholar]
- 16.Frank D, Fortino W, Clark L, et al. Placental overgrowth in mice lacking the imprinted gene Ipl. Proc Natl Acad Sci U S A. 2002;99(11):7490–5. doi: 10.1073/pnas.122039999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei Y, Zhu J, Huan Y, et al. Aberrant expression and methylation status of putatively imprinted genes in placenta of cloned piglets. Cell Reprogram. 2010;12(2):213–22. doi: 10.1089/cell.2009.0090. [DOI] [PubMed] [Google Scholar]
- 18.Su JM, Yang B, Wang YS, et al. Expression and methylation status of imprinted genes in placentas of deceased and live cloned transgenic calves. Theriogenology. 2011;75(7):1346–59. doi: 10.1016/j.theriogenology.2010.11.045. [DOI] [PubMed] [Google Scholar]
- 19.Amilcar Cordeiro APN, Filipa Carvalho, Carla Ramalho & Sofia Dória. Relevance of genomic imprinting in intrauterine human growth expression of CDKN1C, H19, IGF2, KCNQ1 and PHLDA2 imprinted genes. J Assist Reprod Genet. 2014. doi:10.1007/s10815-014-0278-0. [DOI] [PMC free article] [PubMed]
- 20.Mayer W, Niveleau A, Walter J, et al. Demethylation of the zygotic paternal genome. Nature. 2000;403(6769):501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 21.Market-Velker BA, Fernandes AD, Mann MR. Side-by-side comparison of five commercial media systems in a mouse model: suboptimal in vitro culture interferes with imprint maintenance. Biol Reprod. 2010;83(6):938–50. doi: 10.1095/biolreprod.110.085480. [DOI] [PubMed] [Google Scholar]
- 22.Hall JG. Genomic imprinting: review and relevance to human diseases. Am J Hum Genet. 1990;46(5):857–73. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 50 kb)