Abstract
Purpose
To characterize each chromosome’s risk for being involved in embryonic aneuploidy.
Methods
This is a retrospective cohort study conducted at a single, academic center. The cohort consisted of 15,169 consecutive trophectoderm biopsies which then underwent comprehensive chromosome screening utilizing validated real-time polymerase chain reaction (RT-PCR) or single nucleotide polymosphism (SNP) array platforms. Analysis was done to determine probability of aneuploidy by chromosome, changes in that risk with increasing maternal age, and in relationship of aneuploidy to chromosomal structure as classified by prior cytogenetic literature.
Results
The highest prevalence of imbalances leading to aneuploidy was seen for chromosomes 13, 15, 16, 18, 19, 21, and 22. While elevated in all age groups, there was a disproportionate rise in aneuploidy rates for these chromosomes with increasing maternal age. When classic cytogenetic karyotype groups were compared, the overall smaller groups D, E, and G were associated with the highest rates. Similarly, when grouped based upon structure, acrocentric chromosomes exhibited the highest rates of aneuploidy, followed by the metacentric chromosomes, with the lowest prevalence of error in those with submetacentric structures.
Conclusions
The highest rates of chromosomal aneuploidy were found in chromosomes known to be involved in clinically detectable, abnormal pregnancies, not just simply implantation failure. The rate of aneuploidy in these chromosomes rises disproportionately with age when compared to the other chromosomes which may provide information about chromosomal susceptibility to aging. The biological structure groupings did show varied aneuploidy rates which may provide insight into the biology of aneuploidy.
Keywords: Aneuploidy, CCS, Chromosomal abnormalities, Genetic diagnosis
Introduction
The contribution of embryonic aneuploidy to the inefficiency of human reproduction is now well established [1, 2]. While significant at all ages, the prevalence of aneuploidy rises dramatically with increasing maternal age [3]. We have shown previously that characterization of rates of aneuploidy at the embryonic level which results in a diagnosis of either euploid or aneuploid is important for many reasons including patient counseling, active management of ART cycles including decisions regarding transfer order or the need for aneuploidy screening, and to create a reference standard against which future aneuploidy assays can be measured [3]. While these characterizations at the embryonic level are important, further characterization of the nature of aneuploidy at the individual chromosomal level at the embryonic stage of development may be helpful when exploring the biological mechanisms underlying this pathological process.
The emphasis on specific chromosomes can be seen in the early forms of preimplantation genetic screening (PGS) which screened for chromosomes 13, 18, 21, X and Y, the chromosome abnormalities compatible with viable pregnancies [4]. Several others, such as 14, 15, 16, and 22, were added given these were seen most frequently when spontaneously aborted fetuses were examined [5–8]. In fact, data compiled from chromosomal evaluation of spontaneous abortions confirm the importance of evaluating the full complement of chromosomes.
In several large series which reported on karyotype analysis of spontaneous abortions, trisomies were identified for each of the autosomes, although 1 and 19 were rare. These sentinel studies could only characterize those errors which would allow development of a clinically recognized pregnancy capable of producing sufficient viable cellular material to allow cytogenetic analysis following clinical pregnancy loss [9, 10]. Thus while the ability to assess all 24 chromosomes provides a broader assessment of the nature of human chromosomal errors, significant limits prevent extrapolation back to the point of embryonic development and as such provide only a “filtered” view of the nature of those errors.
To date there has not been a comprehensive look at individual chromosomes in the general infertility population at the blastocyst level. The largest sample to date evaluated 1,046 trophectoderm biopsies for aneuploidy using array comparative genomic hybridization (aCGH) [11]. However, this sample set came from only 396 patients, 56 % of whom were referred for genetic testing due to advanced maternal age with the average age being 39.6 years. Of the remaining, 27 % had recurrent implantation failure, and 17 % had repeated miscarriages. Other studies have also been limited to patients with advanced maternal age [12] or recurrent implantation failure [13].
Several other studies, while providing some insight into the aneuploidy at the blastocyst level, suffer from small numbers [14] or combine known aneuploid embryos and multiple platforms, including FISH and aCGH, for chromosomal analysis [15]. Some have relied upon methods which are combined with fluorescent in situ hybridization (FISH) [12, 16] which has since been shown less accurate than other platforms when evaluating blastocysts [17, 18] and thus can be difficult to rely upon when attempting to completely describe aneuploidy in the infertile population at the blastocyst stage.
Comprehensive characterization of the rates of individual chromosomes at different stages of development, such as embryonic versus miscarriage specimen, is important when interpreting the body of literature on aneuploidy. When attempting to understand more about the biological mechanisms responsible for aneuploidy, it would be informative to know about the rates of abnormalities across the entire genetic complement and how these rates change with age in the general infertility population (i.e. not just in those with recurrent pregnancy loss or single gene cases). Do all chromosomes errors increase over time proportionally or are certain chromosomes disproportionately represented? Do error rates differ when classified according to classic cytogenetic groups based upon chromosomal size (A-G) and structure (acrocentric, metacentric, and submetacentric)? Patterns which emerge from the answers to these questions may improve contemporary understanding of the nature of embryonic aneuploidy and contribute to subsequent investigations regarding the specific mechanisms that lead to these abnormalities or strategies to prevent them.
This is a large-scale analysis of comprehensive chromosome screening (CCS) results utilizing validated real-time polymerase chain reaction (RT-PCR) or single nucleotide polymosphism (SNP) array platforms from blastocysts looking specifically at the chromosomal level in a general IVF population. Additionally, it includes a comparison of data obtained via these platforms utilizing classic cytogentic classifications in order to help put into context a large body of literature detailing aneuploidy in the miscarriage specimen, much of which was done utilizing karyotype analysis. The study seeks to characterize the nature of aneuploidy at the specific chromosomal level, as opposed the whole embryonic level, by describing the probability of aneuploidy by chromosome, each chromosome’s specific susceptibility to aging, as well as an analysis of physical structure as it relates to chromosomal errors.
Materials and methods
The nature of each chromosome’s contribution to embryonic aneuploidy was characterized by determination of the following: 1. The overall probability of aneuploidy by chromosome for the population as a whole and within various age groups 2. The relative impact of maternal age on the prevalence of specific chromosome abnormalities; and 3. The difference in aneuploidy rates when chromosomes were grouped by established cytogenetic structural classification schemes. The retrospective review was conducted under IRB approval, protocol # 20021333.
Population
The population studied has been previously characterized when detailing aneuploidy at the embryonic, rather than the individual chromosomal, level [3]. In an effort to achieve a representative sample of patients seeking infertility treatment, all blastocysts undergoing CCS of trophectoderm biopsies that were submitted to the RMA Genetics laboratory for analysis from 2010 to 2013 were included. These samples included both clinical cases in which patients elected to undergo CCS as well as patients enrolled in clinical studies who may not have had a traditional indication for aneuploidy screening. The population includes the full breadth of clinical patients.
Standard regimens for controlled ovarian hyperstimulation were employed using purified urinary FSH or recombinant FSH and LH activity in the form of low-dose hCG or human menopausal gonadotropins (hMG) along with GnRH agonist (long down-regulation or microdose flare) or GnRH antagonist to prevent a premature LH surge. Monitoring of IVF cycles were per practice routine. Oocyte maturation was induced with recombinant hCG (typically 500 mcg) or purified urinary hCG (typically 10,000 IU) or with GnRH agonist (leuprolide acetate 2 mg in 2 doses 12 h apart) ±1,500 IU of hCG when 2 or 3 follicles reached or exceeded 17–18 mm or when the follicular cohort was deemed to be mature by the patient’s primary physician.
Transvaginal oocyte aspiration was performed approximately 36 h later. Cumulus stripping occurred after retrieval. Insemination of mature oocytes was performed by intracytoplasmic sperm injection (ICSI) or by conventional insemination. Embryos were then cultured with sequential media with Quinns Advantage™ (CooperSurgical Inc., Trumbull, CT) followed by BlastAssist™ (Origio Inc., Malov, Denmark).
Expanded blastocysts, equivalent to Gardner blastocele expansion score of 3 to 6, were biopsied for CCS. Trophectoderm biopsies were obtained using laser dissection. Five to six trophectoderm cells were obtained from the portion of the embryo opposite the inner cell mass.
All biopsies were reviewed and the following information was collected: 1. The ploidy result by chromosome of the genetic analysis; 2. The age of the female producing the oocyte resulting in the embryo that was biopsied. There were no inclusion or exclusion criteria beyond having those pieces of information available.
Assays
The trophectoderm biopsies were placed into lysis buffer using a previously established protocol and then submitted for evaluation. The samples were analyzed either via real-time quantitative polymerase chain reaction (RT-PCR) or single nucleotide polymosphism (SNP) array platforms using an established 24 chromosome assay which has been specifically validated for trophectoderm biopsies [19–21]. During the period of time trophectoderm biopsies were obtained, the genetics laboratory began utilizing qPCR rather than SNP array due to the shorter time in which results were available. Prior to use, cross-validation of qPCR with the extensively validated SNP array platform was performed ensuring equivalent results between platforms [17, 20]. All assay were run and diagnoses made as published previously.
Study design and data analysis
The results of each biopsy individually were categorized as being euploid or aneuploid. Amongst the embryos that were aneuploid, some had a single chromosome involved while others had multiple chromosomes involved. Thus, any difference in the number of aneuploid chromosomes and the number of aneuploid biopsy samples reflects the fact that some embryos were aneuploid for more than one chromosome. All analyses of aneuploidy took place on the individual chromosome level.
First, the full genetic complement across all 22 autosomes and both sex chromosomes was characterized. This was done by dividing the total number of errors for each chromosome by the number of embryos evaluated. The same method was used to examine the probability of monosomy and trisomy by chromosome. The diagnosis of monsomy and trisomy from trophectoderm biopsies were made utilizing previously published methods [17, 20]. Chromosome X and Y were evaluated together as sex chromosome abnormalities since the one inappropriately present or absent cannot be determined by simply characterizing the ploidy status.
Then, in order to provide more information on the biological effect aging has on aneuploidy, the individual chromosomes based up on the maternal age at the time of trophectoderm biopsy were grouped into the age groups for reporting by the Society for Assisted Reproductive Technology (<35, 35–37, 38–40, 41–42, 43+). The lowest age group was further divided into <26, 26–30, and 31–34 based upon prior characterization of aneuploidy at the embryonic level [3]. The prevalence was calculated by looking at the total number of errors per chromosome and dividing the number of embryos that were evaluated, irrespective of embryo transfer order.
Next, evaluation of relative changes over time was undertaken to determine if there is a disproportionate change in prevalence of certain chromosomes over others with aging. Given that the lowest aneuploidy prevalence in this dataset was ages 26–30 this was assigned a relative prevalence of 1. Using this as an index group we looked at the relative change in other age groups for each chromosome. Thus, if the increase in aneuploidy was simply age related increase one would expect to see a flat line for each age group across all chromosomes.
Finally, chromosomal structure was evaluated by placing results into previously established cytogenetic structural classification. This was done because much of the prior literature detailing aneuploidy at the miscarriage specimen level was done utilizing karyotypes. Additionally, chromosome size and configuration may impact the mechanisms involved with synapsis, recombination, and longitudinal separation in the diplotene phase, the error rate for chromosomes with different structures. Since the centromere impacts the interactions with the spindle fibers and the mechanisms involved in chromosomal separation, the chromosomes were compared after being grouped according to their structure relative to their centromere location. They were categorized as acrocentric (chromosomes 13, 14, 15, 21 and 22), metacentric [1, 3, 16, 19, 20], and submetacentric (2, 4–12, 17, 18, and X).
Additionally, results were classified utilizing the classic karyotype structural groups. These morphologic classifications (Groups A-G) generally correlate with size and configuration in most classes. Although they are not a true structural designation, they facilitate comparisons with prior literature and classification schema. Group A included chromosomes 1–3, B chromosomes 4–5, C chromosomes 6–12, D chromosomes 13–15, E chromosomes 16–18, F chromosomes 19–20, and G chromosomes 21–22.
Given that the groups for both the karyotype criteria and structural types were not equally sized, which would give some groups more chances of having errors than others, the error rate per chromosome was calculated within each group. The group with the lowest error rate was assigned a relative rate of 1 and the other groups were then compared using a one-way ANOVA test. These two groupings were subsequently stratified by age group for analysis.
In all cases, an alpha error less than 0.05 was considered significant.
Results
There were 15,169 biopsy results from 2,701 patients in 3,292 cycles which constituted 362,056 chromosomes evaluated. There were 9,001 euploid biopsy (59.3 %) results and 6,168 aneuploid biopsy (40.7 %) results from these samples. The aneuploid biopsies resulted in 9,166 distinct chromosomal abnormalities (either monsomy or trisomy). Ages of the female patients ranged from 22 to 49 years.
The indications for CCS have been described [3] and included routine infertility care, recurrent pregnancy loss, single gene cases, and family balancing. Routine fertility care represents patients who were offered and underwent CCS aneuploidy screening as a means to increase pregnancy rates, decrease loss rates [22–24], and decrease transfer order and did not carry the other diagnoses listed. In patients under age 26, the most common indication was routine infertility care (61.4 %) followed by single-gene preimplantation genetic diagnosis (PGD) cases (36.4 %) and recurrent pregnancy loss (2.3 %). For the remainder of the age groups, routine infertility, defined as those seeking aneuploidy screening as a way of increasing pregnancy rates and decreasing miscarriages, represented nearly 80 % or more of the cases. Recurrent loss as an indication was highest in patients 43 years and older and represented 16.4 % of cases. Family balancing represented a minority of cases for all age groups as did single-gene PGD cases with the exception of the patients <26 years old, as indicated above. Overall, routine infertility care represented 85.0 % of cases followed by recurrent pregnancy loss at 11.0 %, single gene cases at 3.1 %, and family balancing at 0.9 %.
Aneuploidy by chromosome
The proportions of biopsies demonstrating aneuploidy for each individual chromosome are shown in Fig. 1a. The probability of a particular chromosome exhibiting monosomy and trisomy are shown in Fig. 1b. The ratio of monosomies and trisomies approaches unity for most chromosomes. The most frequently observed aneuploidy was seen for chromosomes 13, 15, 16, 18, 19, 21, and 22 (P < 1*10−6). This group of 7 chromosomes was more frequently observed to be aneuploidy when compared to the other 16 chromosomes combined as a group (RR = 1.8). This is of importance since these most commonly abnormal chromosomes are often involved in clinically detectable, abnormal pregnancies, not just simply failed implantation.
Fig. 1.
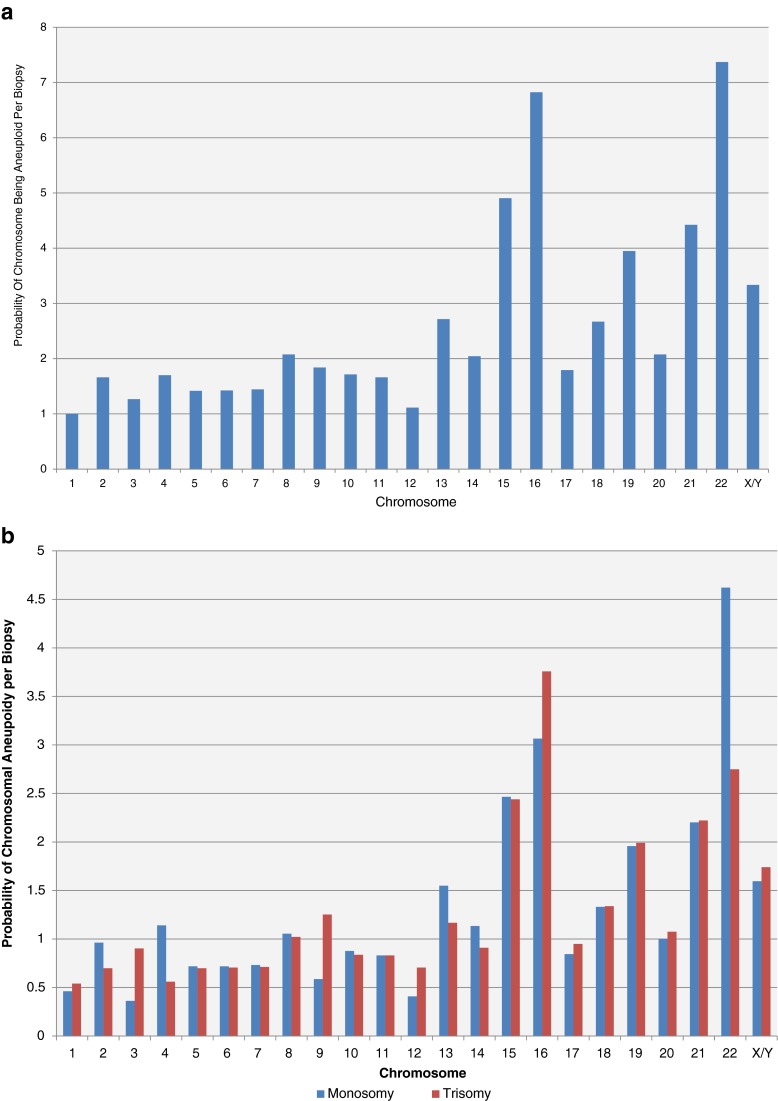
The probability (%) of a chromosome being aneuploid per biopsy is depicted (a). The most common errors are seen in chromosomes 13, 15, 16, 18, 19, 21, and 22. The probability (%) of a chromosome being monosomic or trisomic per biopsy is also shown (b)
Aneuploidy by chromosome and age
After examining monosomy and trisomy separately, the combined errors by chromosome were analyzed by age group. The lowest risk group was ages 26–30. This age group was thus designated to provide an estimate of the basal risk for aneuploidy for each chromosome and assigned a relative risk of 1. The relative risk of aneuploidy by chromosome was evaluated by age group in order to take into account the difference of prevalence of errors seen at baseline and highlight disproportional relative increases in error rates (Fig. 2).
Fig. 2.
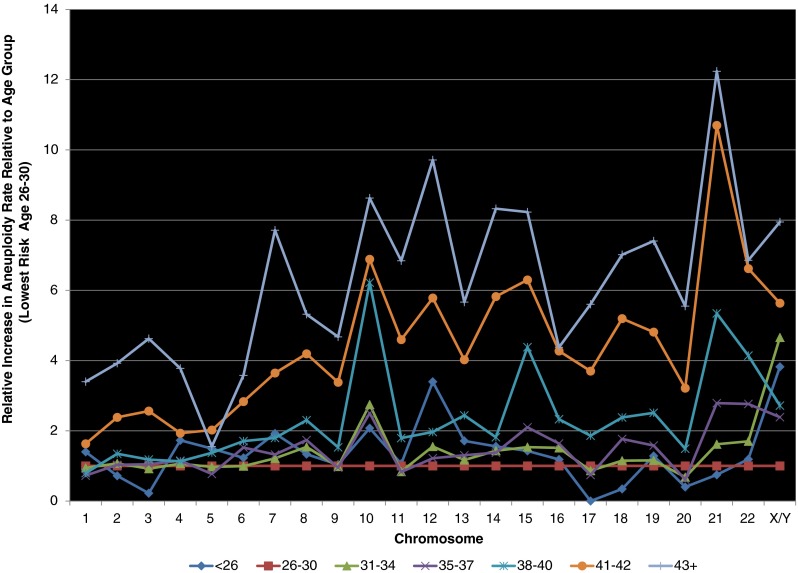
The combined errors by chromosome, including both monosomy and trisomy, was analyzed by SART age groups with the lowest age group divided into less than 26, 26–30, and 31–34. These shows mean centered changes in chromosome error rates by age group. There is a disproportionate relative increase in chromosomes 13, 15, 16, 19, 21 and 22
Not surprisingly, each age group for women age 35 and greater shows an increase in aneuploidy across all chromosomes. There was a disproportionate rise in Chromosomes 13, 15, 16, 18, 19, 21, and 22 seen in the older age groups (P < 1*10−6) and this group comprised 35.3 % of all aneuploidy. These chromosomes are more commonly associated with successful implantation and clinical abnormalities beyond simply failed implantation. While uniparental disomy must also be considered, particularly in chromosome 15, the phenomenon has been shown to be rare utilizing validated platforms and is unlikely to have altered these results [25]. Given the fact these chromosomes are commonly involved in clinically-recognized, abnormal pregnancies, it suggests the clinical risk rises disproportionately to the risk of aneuploidy with age.
Aneuploidy by chromosome classification
In order to better characterize the biology involved in aneuploidy, the chromosomes were categorized based upon classic karyotyping criteria (Groups A-G) and by structural characteristics (metacentric, submetacentric, and acrocentric). Table 1 summarized the number of errors seen across these groups.
Table 1.
Number of errors by chromosome and designation by classic karyotyping group and structure
| Number of errors (total and %) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | Structure | Group | <35 | 35-37 | 38-40 | 41-42 | 43+ | |||||
| 1 | metacentric | A | 70 | 2.6 % | 27 | 1.5 % | 25 | 1.1 % | 16 | 1.0 % | 14 | 1.8 % |
| 2 | submetacentric | A | 92 | 3.4 % | 50 | 2.8 % | 55 | 2.3 % | 34 | 2.2 % | 21 | 2.7 % |
| 3 | metacentric | A | 65 | 2.4 % | 41 | 2.3 % | 39 | 1.6 % | 27 | 1.8 % | 20 | 2.5 % |
| 4 | submetacentric | B | 103 | 3.8 % | 58 | 3.2 % | 48 | 2.0 % | 28 | 1.8 % | 21 | 2.7 % |
| 5 | submetacentric | B | 87 | 3.2 % | 36 | 2.0 % | 54 | 2.3 % | 30 | 2.0 % | 8 | 1.0 % |
| 6 | submetacentric | C | 66 | 2.5 % | 54 | 3.0 % | 51 | 2.2 % | 31 | 2.0 % | 14 | 1.8 % |
| 7 | submetacentric | C | 70 | 2.6 % | 42 | 2.3 % | 48 | 2.0 % | 32 | 2.1 % | 27 | 3.4 % |
| 8 | submetacentric | C | 100 | 3.7 % | 68 | 3.8 % | 76 | 3.2 % | 48 | 3.1 % | 23 | 2.9 % |
| 9 | submetacentric | C | 92 | 3.4 % | 48 | 2.7 % | 65 | 2.7 % | 48 | 3.1 % | 26 | 3.3 % |
| 10 | submetacentric | C | 68 | 2.5 % | 42 | 2.3 % | 88 | 3.7 % | 31 | 2.0 % | 31 | 3.9 % |
| 11 | submetacentric | C | 68 | 2.5 % | 34 | 1.9 % | 62 | 2.6 % | 57 | 3.7 % | 31 | 3.9 % |
| 12 | submetacentric | C | 57 | 2.1 % | 25 | 1.4 % | 34 | 1.4 % | 31 | 2.0 % | 22 | 2.8 % |
| 13 | acrocentric | D | 120 | 4.5 % | 73 | 4.1 % | 115 | 4.9 % | 69 | 4.5 % | 35 | 4.4 % |
| 14 | acrocentric | D | 95 | 3.5 % | 54 | 3.0 % | 60 | 2.5 % | 65 | 4.3 % | 36 | 4.6 % |
| 15 | acrocentric | D | 170 | 6.3 % | 141 | 7.9 % | 248 | 10.5 % | 124 | 8.1 % | 61 | 7.8 % |
| 16 | metacentric | E | 329 | 12.2 % | 217 | 12.1 % | 260 | 11.0 % | 165 | 10.8 % | 64 | 8.1 % |
| 17 | submetacentric | E | 77 | 2.9 % | 36 | 2.0 % | 76 | 3.2 % | 53 | 3.5 % | 30 | 3.8 % |
| 18 | submetacentric | E | 99 | 3.7 % | 89 | 5.0 % | 101 | 4.3 % | 77 | 5.0 % | 39 | 5.0 % |
| 19 | metacentric | F | 156 | 5.8 % | 118 | 6.6 % | 158 | 6.7 % | 106 | 6.9 % | 61 | 7.8 % |
| 20 | metacentric | F | 91 | 3.4 % | 40 | 2.2 % | 82 | 3.5 % | 62 | 4.1 % | 40 | 5.1 % |
| 21 | acrocentric | G | 120 | 4.5 % | 130 | 7.2 % | 210 | 8.9 % | 148 | 9.7 % | 63 | 8.0 % |
| 22 | acrocentric | G | 258 | 9.6 % | 263 | 14.7 % | 332 | 14.0 % | 193 | 12.6 % | 72 | 9.1 % |
| X/Y | submetacentric | C | 235 | 8.7 % | 108 | 6.0 % | 83 | 3.5 % | 52 | 3.4 % | 28 | 3.6 % |
| Total errors | 2,688 | 1,794 | 2,370 | 1,527 | 787 | |||||||
| Number of biopsies | 6,819 | 3,659 | 3,084 | 1,203 | 404 | |||||||
Karyotyping criteria
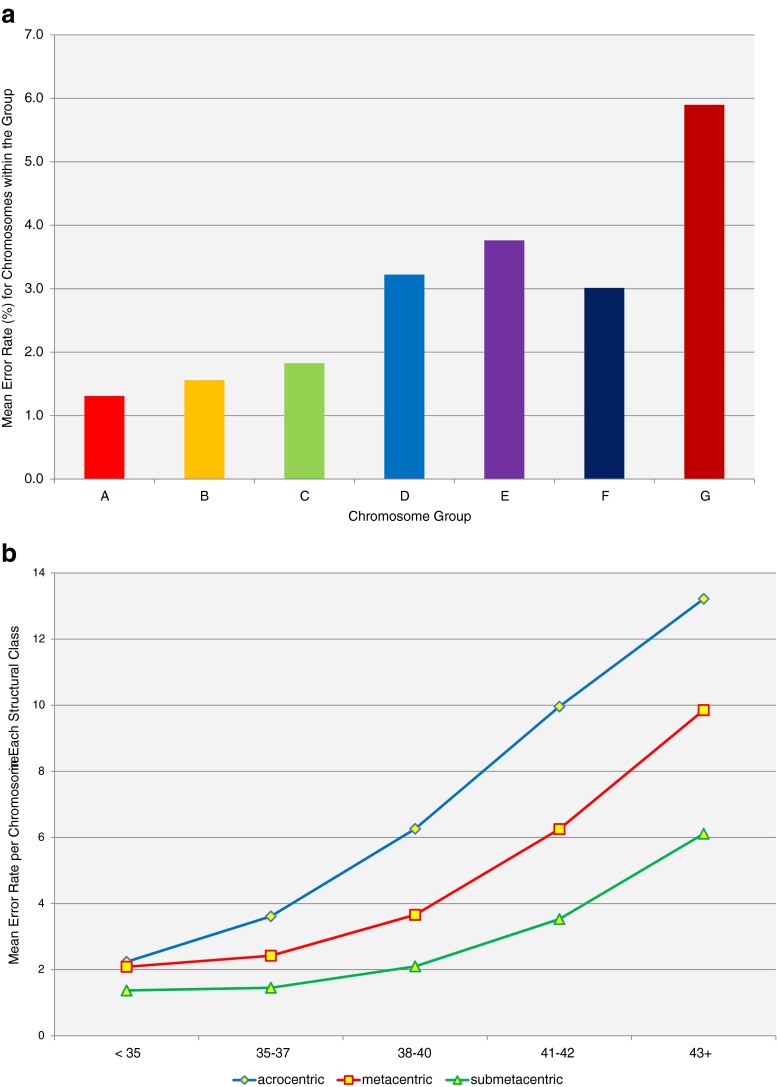
The chromosomal mean error rates by karyotype classification are shown in Fig. 3. There was a clear difference (p = 0.02) in error rates across different groups. Groups D, E, and G were associated with the highest error rate. The evaluation of karyotype groups stratified by age group also revealed clear differences in errors by chromosome group with increasing age.
Fig. 3.
Error rates per chromosome karyotype group (a), calculated by taking the number of total errors within a group and dividing by the number of chromosomes represented within the group. Groups D, E, and G were associated with the highest error rate. When error rates were looked at by chromosome type (b), acrocentric followed by metacentric were most likely to exhibit chromosomal errors
Chromosome structure
Chromosomal error rates by structure type are shown in Fig. 3b. Submetacentric chromosomes had the lowest rate of error and were given the relative value of 1 for comparison of error rates. Metacentric were 1.6 times and acrocentric 2.3 times more likely to have errors (p = 0.02). There was also an age related increase in the proportion of errors that occur with acrocentric chromosomes relative to other types of chromosomes.
Discussion
This data represents the largest systematic report of individual chromosome aneuploidy at the blastocyst stage in the general IVF population to date. It has implications in patient counseling, gaining a better understanding of chromosomal susceptibility to aging, and the biology of aneuploidy at the chromosome level. Additionally, it places chromosome aneuploidy rate in the context of historical classification schemes—karyotype classifications and chromosome structure—in order to make comparisons with prior literature.
In terms of patient counseling, it is important to recognize that the most common aneuploidies are seen in chromosomes 22, 16, 15, 21 and 19, in that order. This is consistent with prior literature in a study where patients were referred for aneuploidy screening specifically for advanced age [11]. This data adds to the literature in that it represents a larger sample of patients with a broad distribution of ages that were undergoing aneuploidy screening as part of their routine infertility care, not because of advanced age.
When analyzed by age group it was discovered that chromosomes 13, 15, 16, 18, 19, 21, and 22 aneuploidy rates rise disproportionately with age at the blastocyst stage. Thus, not only are patients more likely to have genetically abnormal embryos with increasing age, they are also more likely to have abnormal embryos that are more likely to be involved with clinically detectable, abnormal pregnancies. Thus, it will not just simply manifest as increased failed implantation; they will be more likely to have pregnancies which require prolonged management and thus would possibly increase time to live birth.
Of interest is the comparisons made between rates of aneuploidy seen at the embryonic level and those seen in products of conception. When comparing the relative contributions to trisomy of products of conception, we see relatively low levels of the large autosomes as compared to small, primarily acrocentric chromosomes (chromosomes 13, 14, 15, 21 and 22) as well as chromosome 16 [9, 10]. At the embryonic level we see these large autosomes represented with more frequency, although, the relative contribution of the most common chromosomes, namely 13, 15, 16, 18, 19, 21, and 22, remain high in both embryos and products of conception.
In addition to illuminating the distribution of aneuploidy across chromosomes, the data highlight differences in rates of aneuploidy when chromosomes are grouped by structural cytogenetic classification systems. This was seen with both the classic karyotype groupings as well as the metacentric, submetacentric, and acrocentric designations. While these groupings are morphologic in nature and do not necessarily indicate specific function, it is of great biological interest that differences exist between these groups.
The observation that the most common chromosomes which exhibited aneuploidy comprised both the short and medium acrocentric chromosomes is of note. Additionally, these particular chromosomes exhibited disproportionately higher rates of aneuploidy with advancing age. This suggests that perhaps the process of crossing over during pachytene and separation during diplotene puts these particular chromosomes at risk [26]. Moreover, the biologic machinery governing this process appears to be susceptible to the aging process, associated with less fidelity over time. Further studies of chromosomal susceptibility to aneuploidy may better elucidate the mechanism behind the differences seen.
These findings not only allow for better patient counseling and understanding of the biology, but also are a valuable tool as a reference standard as new assays are tested for CCS. To date there is not a large report of aneuploidy which specifically analyzes aneuploidy rates per chromosome across a large cohort of embryos using validated modern day platforms. These data accomplish this and allow for comparisons going forward as new technology is evaluated.
There are several limitations to this study. First it is a retrospective review of results from infertile couples who elected to use clinical CCS screening. While the previously reported demographics of the population closely match those of the overall population of patients undergoing care in this center, it is not a prospective cross sectional assessment and may contain some selection biases. Additionally, the number of embryos which each patient contributed was not uniform. Thus, individual higher responders with a large number of blastocysts are over represented compared to low responders who may have only a single embryo to contribute to the study population. Additionally, embryos are tested at the blastocyst stage and thus those that arrested in culture were not assessed. Embryonic mosaicism must always be considered and it is important to note that mosaicism would only be detected when approximately 50 % of the cells biopsied for analysis possessed aneuploidy [17, 18]. Thus, mosaicism will remain a limit with any trophectoderm biopsy study. Finally, factors such as paternal age, socioeconomic status, drug exposure, smoking, alcohol use, and familial factors may have contributed to age-related aneuploidy.
While the data presented here provide important information in terms of patient counseling and describes the biology of aneuploidy, they do not provide direct answers to the pathophysiologic questions raised. However, this data is an important first step. To date, much of our understanding of early aneuploidy is based upon cytogenetic analysis of products of conception. Furthermore, what data does exist on embryonic aneuploidy rates of individual chromosomes have been produced by platforms which have yet to undergo rigorous validation. This data serves to further our understanding of the rates of specific chromosome aneuploidy at the embryonic level.
While the findings are of great interest, understanding why we see disproportionate rises in aneuploidy rates in specific chromosomes with age or why certain structural groupings have higher rates of aneuploidy remains a question to be answered in future investigations. As investigators explore differences in chromosomal centric indices, percentage of haploid content, and further compare cytogenetic properties such as qh regions and banding polymorphisms, frequency and distribution of chiasmata and DNA base size, among many other things, this data serves as the most complete and reliable demographic overview of individual chromosome aneuploidy at the blastocyst level published to date.
Footnotes
Capsule Chromosomes 13, 15, 16, 18, 19, 21, and 22 had the highest aneuploidy rates, and that risk rose disproportionately with age. Structurally, acrocentric chromosomes were most likely to be aneuploid.
References
- 1.Hassold T, Hunt P. To ERR (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 2.Fragouli E, Wells D. Aneuploidy in the human blastocyst. Cytogenet Genome Res. 2011;133:149–159. doi: 10.1159/000323500. [DOI] [PubMed] [Google Scholar]
- 3.Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, et al. The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril. 2014;101(3):656–663.e1. doi: 10.1016/j.fertnstert.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Ambartsumyan G, Clark AT. Aneuploidy and early human embryo development. Hum Mol Genet. 2008;17:R10–R15. doi: 10.1093/hmg/ddn170. [DOI] [PubMed] [Google Scholar]
- 5.Staessen C, Platteau P, Van Assche E, Michiels A, Tournaye H, Camus M, et al. Comparison of blastocyst transfer with or without preimplantation genetic diagnosis for aneuploidy screening in couples with advanced maternal age: a prospective randomised controlled trial. Hum Reprod. 2004;19:2849–2858. doi: 10.1093/humrep/deh536. [DOI] [PubMed] [Google Scholar]
- 6.Fritz MA. Perspectives on the efficacy and indications for preimplantation genetic screening: where are we now? Hum Reprod. 2008;23:2617–2621. doi: 10.1093/humrep/den400. [DOI] [PubMed] [Google Scholar]
- 7.Mastenbroek S, Twisk M, van Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, et al. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:9–17. doi: 10.1056/NEJMoa067744. [DOI] [PubMed] [Google Scholar]
- 8.Hardarson T, Hanson C, Lundin K, Hillensjo T, Nilsson L, Stevic J, et al. Preimplantation genetic screening in women of advanced maternal age caused a decrease in clinical pregnancy rate: a randomized controlled trial. Hum Reprod. 2008;23:2806–2812. doi: 10.1093/humrep/den217. [DOI] [PubMed] [Google Scholar]
- 9.Menasha J, Levy B, Hirschhorn K, Kardon NB. Incidence and spectrum of chromosome abnormalities in spontaneous abortions: new insights from a 12-year study. Genet Med. 2005;7:251–263. doi: 10.1097/01.GIM.0000160075.96707.04. [DOI] [PubMed] [Google Scholar]
- 10.Hassold TJ, Jacobs PA. Trisomy in man. Annu Rev Genet. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- 11.Fragouli E, Alfarawati S, Spath K, Jaroudi S, Sarasa J, Enciso M, et al. The origin and impact of embryonic aneuploidy. Hum Genet. 2013;132(9):1001–1013. doi: 10.1007/s00439-013-1309-0. [DOI] [PubMed] [Google Scholar]
- 12.Capalbo A, Bono S, Spizzichino L, Biricik A, Baldi M, Colamaria S, et al. Sequential comprehensive chromosome analysis on polar bodies, blastomeres and trophoblast: insights into female meiotic errors and chromosomal segregation in the preimplantation window of embryo development. Hum Reprod Oxf Engl. 2013;28(2):509–518. doi: 10.1093/humrep/des394. [DOI] [PubMed] [Google Scholar]
- 13.Fragouli E, Katz-Jaffe M, Alfarawati S, Stevens J, Colls P, Goodall N, et al. Comprehensive chromosome screening of polar bodies and blastocysts from couples experiencing repeated implantation failure. Fertil Steril. 2010;94(3):875–887. doi: 10.1016/j.fertnstert.2009.04.053. [DOI] [PubMed] [Google Scholar]
- 14.Fragouli E, Alfarawati S, Daphnis DD, Goodall NN, Mania A, Griffiths T, et al. Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: scientific data and technical evaluation. Hum Reprod. 2011;26:480–490. doi: 10.1093/humrep/deq344. [DOI] [PubMed] [Google Scholar]
- 15.Fragouli E, Lensi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod. 2008;23:2596–2608. doi: 10.1093/humrep/den287. [DOI] [PubMed] [Google Scholar]
- 16.Daphnis DD, Fragouli E, Economou K, Jerkovic S, Craft IL, Delhanty JD, et al. Analysis of the evolution of chromosome abnormalities in human embryos from day 3 to 5 using CGH and FISH. Mol Hum Reprod. 2008;14:117–125. doi: 10.1093/molehr/gam087. [DOI] [PubMed] [Google Scholar]
- 17.Treff NR, Levy B, Su J, Northrop LE, Tao X, Scott R. SNP microarray-based 24 chromosome aneuploidy screening is significantly more consistent than FISH. Mol Hum Reprod. 2010;16:583–589. doi: 10.1093/molehr/gaq039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Northrop LE, Treff NR, Levy B, Scott RT. SNP microarray-based 24 chromosome aneuploidy screening demonstrates that cleavage-stage FISH poorly predicts aneuploidy in embryos that develop to morphologically normal blastocysts. Mol Hum Reprod. 2010;16:590–600. doi: 10.1093/molehr/gaq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott RT, Upham KM, Forman EJ, Zhao T, Treff NR. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: a randomized and paired clinical trial. Fertil Steril. 2013;100(3):624–630. doi: 10.1016/j.fertnstert.2013.04.039. [DOI] [PubMed] [Google Scholar]
- 20.Treff NR, Su J, Tao X, Levy B, Scott RT. Accurate single cell 24 chromosome aneuploidy screening using whole genome amplification and single nucleotide polymorphism microarrays. Fertil Steril. 2010;94:2017–2021. doi: 10.1016/j.fertnstert.2010.01.052. [DOI] [PubMed] [Google Scholar]
- 21.Treff NR, Tao X, Ferry KM, Su J, Taylor D, Scott RT. Development and validation of an accurate quantitative real-time polymerase chain reaction-based assay for human blastocyst comprehensive chromosomal aneuploidy screening. Fertil Steril. 2012;97:819–824. doi: 10.1016/j.fertnstert.2012.01.115. [DOI] [PubMed] [Google Scholar]
- 22.Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, et al. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fertil Steril. 2013;100(1):100–107. doi: 10.1016/j.fertnstert.2013.02.056. [DOI] [PubMed] [Google Scholar]
- 23.Forman EJ, Hong KH, Franasiak JM, Scott RT., Jr Obstetrical and neonatal outcomes from the BEST Trial: single embryo transfer with aneuploidy screening improves outcomes after in vitro fertilization without compromising delivery rates. Am J Obstet Gynecol. 2014;210(2):157.e1–6. doi: 10.1016/j.ajog.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 24.Forman EJ, Hong KH, Treff NR, Scott RT. Comprehensive chromosome screening and embryo selection: moving toward single euploid blastocyst transfer. Semin Reprod Med. 2012;30:236–242. doi: 10.1055/s-0032-1311526. [DOI] [PubMed] [Google Scholar]
- 25.Gueye N-A, Devkota B, Taylor D, Pfundt R, Scott RT, Jr, Treff NR. Uniparental disomy in the human blastocyst is exceedingly rare. Fertil Steril. 2014;101(1):232–236. doi: 10.1016/j.fertnstert.2013.08.051. [DOI] [PubMed] [Google Scholar]
- 26.Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet. 2007;16(Spec No. 2):R203–R208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]