Abstract
New neurons are generated in adult mammalians and may contribute to repairing the brain after injury. Here, we show that the number of new neurons in the dentate gyrus of adult rats increased in cerebral ischemic stroke and correlated with activation of the cAMP-response-element-binding protein (CREB). Inhibition of endogenous CREB by expression of a dominant-negative mutant of CREB (CREB-S133A or CREB-R287L) blocked ischemia-induced neurogenesis in the dentate gyrus of adult rats, whereas expression of constitutively active CREB, VP16-CREB, increased the number of new neurons. Thus, our findings provide roles and regulatory mechanisms for CREB in adult neurogenesis and possibly suggest a practical strategy for replacing dead neurons in brain injury.
Progenitor cells in the adult dentate gyrus can proliferate and differentiate into mature neurons when maintained in culture medium containing growth factors in vitro (1) or when grafted into the adult brain in vivo (2), suggesting that residential neuronal progenitors are capable of responding to environmental factors in the adult host (2, 3). Consistent with this hypothesis, several recent studies show that neurogenesis in the dentate gyrus of adults is regulated by stress (4), exercise (5), and learning (6, 7).
There is also precedent for neuronal injury's modifying the fate of immature precursor cells. An earlier report showed that the number of BrdUrd-labeled cells in the dentate gyrus of the gerbil is increased on the day after transient global ischemia (8). Consistent with this finding, our studies, as well as others, show that focal cerebral ischemia increases the number of newly generated neurons that migrate from the subgranule zone into the granule cell layer of the dentate gyrus in adult rats (9, 10). A more recent study extended these findings to demonstrate that activation of endogenous progenitors after transient forebrain ischemia leads to massive regeneration of pyramidal neurons in the CA1 area of the hippocampus (11). These results have been interpreted as evidence for the direct migration of neuronal precursors toward injured areas, possibly to trigger brain repair (11).
Because neurons that die in adulthood can be replaced by neurons of the same class (12–14), it is crucial to determine what signaling molecules promote the production of replacement neurons. Several signals control the proliferation, differentiation, and survival of endogenous progenitors (15, 16). In this study, we examined cAMP-response-element-binding protein (CREB) in regulation of adult neurogenesis and found that CREB activation is responsible for recruiting new neurons into the dentate circuits of adult rats that have been subjected to cerebral ischemic stroke.
Materials and Methods
Focal Cerebral Ischemia and Stereotaxic Operation. In our preliminary studies, we found that adult male rats after focal cerebral ischemia revealed greater increase in the number of BrdUrd-labeled cells than did female rats (Fig. 5, which is published as supporting information on the PNAS web site). Therefore, adult male (300–350 g) Sprague–Dawley rats from the breeding colony at the University of Calgary were used in this study. Anesthesia was induced in animals with ketamine (100 mg/kg, i.p.) and xylazine (5 mg/kg, i.p.). Focal cerebral ischemia was induced by intraluminal middle cerebral artery (MCA) occlusion, as described previously (10). Briefly, a 4-0 surgical nylon monofilament with a rounded tip was introduced into the left internal carotid through the external carotid stump and advanced 20–21 mm past the carotid bifurcation. The filament was left in place for 90 min and then withdrawn. Sham-operated animals were treated identically, except that the MCAs were not occluded after the neck incision. Body temperature was maintained at 37 ± 1°C until the animal had recovered from surgery. The local cerebral blood flow was monitored on the front parietal cortex of the occluded side by using a Perimed PF5050 (Järfälla, Sweden) multichannel laser Doppler flowmetry (LDF). Activated virus particles were infused (2 μl at 0.2 μl/min) into each side of the dentate gyrus, as described previously (10, 17, 18). The injection site was 2.2 mm posterior to the bregma, 1.9 mm lateral to the midline, and 2.9 mm below the dura.
Mutagenesis and Viral Gene Expression Vectors. Construction of the shutoff-deficient Semliki Forest virus (pSFV) gene expression vectors and packaging of recombinant virions were described previously (10, 17, 18). Dominant-negative mutants of CREB, CREB-S133A and CREB-R287L, were generated from wild-type CREB by using the QuikChange site-directed mutagenesis kit (Stratagene). Mutagenesis was verified by sequencing. The construct containing VP16-CREB was a gift from E. Kandel (Columbia University, New York). A cDNA encoding CREB-S133A, CREB-R287L, or VP16-CREB was then inserted directly into the SFV vector pSFV-GFP to produce pSFV-CREB-S133A-GFP, pSFV-CREB-R287L-GFP, or pSFV-VP16-CREB-GFP constructs, respectively.
cAMP-Response Element (CRE)-Binding Assay and Western Blotting. Hippocampal neurons infected with virus were homogenized by using 100 μl of buffer A per sample. Buffer A contained 10 mM Hepes, pH 7.9/1.5 mM MgCl2/10 mM KCl/1 mM DTT/1 mM PMSF/0.1% (wt/vol) Nonidet P-40. The samples were incubated on ice for 15 min and then were centrifuged at 1,000 × g at 4°C for 10 min; the supernatant was discarded. The pellet was resuspended in 500 μl of buffer A without Nonidet P-40 and was centrifuged at 1,000 × g for 10 min; the supernatant was discarded. The pellet was resuspended in 100 μl of TransAm lysis buffer (Active Motif, Carlsbad, CA) containing DTT and a protease inhibitor mixture. The samples were rocked at 4°C for 30 min and then were microcentrifuged for 10 min at 14,000 × g at 4°C; the supernatant (nuclear extract) was collected. Protein concentrations were determined by using the BCA Protein Assay kit (Pierce). CRE binding was examined by using the NE-PER kit (Pierce). Dentate nuclear extract (10 μg) was incubated at 25°C for 30 min in the presence of the 32P-labeled CRE double-stranded DNA (dsDNA) oligonucleotide (5′-AGAGATTGCCTGGACGTCAGAGCTAG-3′). The mixtures were then immunoprecipitated with anti-VP16 (2 μg, Santa Cruz Biotechnology). Radioactivity of the immunoprecipitates was determined by scintillation counting.
To examine pCREB-S133 levels, the extracts (500 mg of protein) were incubated with nonspecific IgG (2 μg, control for nonspecific binding) or polyclonal mouse anti-CREB (2 μg, Santa Cruz Biotechnology) overnight at 4°C, followed by the addition of 40 μl of Protein G-Sepharose (Sigma) for 3 h at 4°C. Immunoprecipitates were washed four times with lysis buffer, denatured with SDS sample buffer, and separated by 12% SDS/PAGE. Proteins were transferred onto nitrocellulose membranes by using a Bio-Rad Miniprotein III System wet transfer unit overnight at 4°C. Transfer membranes were then incubated with blocking solution [5% nonfat dried milk dissolved in TBST buffer, pH 7.5 (10 mM Tris·HCl/150 mM NaCl/0.1% Tween 20)] for 1 h at room temperature, washed three times, and incubated with rabbit anti-pCREB-S133 (1:1,000; Chemicon) for 1 h at room temperature. Membranes were washed three times with TBST buffer and incubated with secondary antibodies for 1 h, followed by washing four times. Signal detection was performed with an enhanced chemiluminescence kit (Amersham Biosciences) and quantitated by using a GS-710 Calibrated Image Densitometer (Bio-Rad).
BrdUrd Labeling and Immunocytochemistry. Animals were treated i.p. with 50 mg/kg BrdUrd (Sigma) two times with an 8-h interval between injections. This dose was chosen because it was used in our recent studies (10). In our preliminary experiments, we tested the relationship between BrdUrd dose and cell labeling in the dentate gyrus and treated rats with four different doses of BrdUrd (two injections at a dose of 50 mg/kg or a single injection at a dose of 75, 150, or 300 mg/kg). Animals were then killed 1 d after the injection of BrdUrd to estimate the number of BrdUrd-labeled cells. Our data showed that ischemia-induced increase in the number of BrdUrd-labeled cells is independent of BrdUrd dose (Fig. 6, which is published as supporting information on the PNAS web site). BrdUrd injection took place either 1 d after or 1 week before infection with virus vectors, as indicated in the figures. Animals were then killed 1 d or 4 weeks after the injection of BrdUrd to estimate the number of BrdUrd-labeled cells. BrdUrd staining was described previously (10, 17). The sections were heated (85°C for 5 min) in antigen-unmasking solution (Vector Laboratories), incubated in 2 M HCl (30°C for 30 min), rinsed in 0.1 M boric acid (pH 8.5) for 10 min, incubated in 1% H2O2 in PBS for 30 min, and blocked in 3% normal goat serum/0.3% (wt/vol) Triton X-100/0.1% BSA in PBS (room temperature for 1 h), followed by incubation with rat monoclonal anti-BrdUrd (1:200; Accurate Chemicals) at 4°C overnight. Subsequently, the sections were developed with the ABC kit (Vector Laboratories). For fluorescence imaging, the sections were incubated with the affinity-purified goat anti-rat Cy3 secondary antibody (1:200; Chemicon). These sections were then incubated with rabbit anti-glial fibrillary acidic protein (GFAP, 1:1,000; Sigma) or rabbit anti-TUC-4 (1:5,000; Chemicon). The sections were then incubated with goat anti-rabbit Cy5 (1:100; Chemicon). Sections were rinsed, dried, and coverslipped by using DAKO fluorescence mounting medium. Control sections were processed with omission of the primary antisera. Labeling was imaged with a confocal laser-scanning microscope (LSM-GB200; Olympus, Melville, NY) and analyzed with image-pro plus software (Media Cybernetics, Silver Spring, MD). Images were taken at 0.5-μm intervals through the region of interest, and optical stacks of 6–10 images were produced for the figures.
Cell Counting. An experimenter (D.Y.Z.) coded all slides from the experiments before quantitative analysis. Stereological analysis of BrdUrd-positive cells in every sixth section in a series of 30-μm coronal sections (beginning at a random starting position) was performed by another experimenter (J.S.W) who was unaware of the experimental conditions of each sample. The analysis was conducted by using a modified version of the optical dissector method on peroxidase-stained sections on coded slides (19). Labeled cells were counted, excluding those in the outermost focal plane to avoid counting cell caps. The total volume of the granule cell layer was determined with stereo investiga-tor software (MicroBrightfield, Williston, VT) by summing the traced granule cell areas for each section multiplied by the distance between sampled sections. For purposes of comparison, we also examined the number of BrdUrd-labeled cells in the subventricular zone (SVZ), a region lining the wall of the labeled ventricles and known to produce cells in adulthood (20). For this analysis, we counted the number of BrdUrd-labeled cells in the SVZ present on every 10th coronal section. Fluorescence labeling was analyzed by sampling every section from the experimental animals with a fluorescence microscope as described previously (10, 17, 18).
Results
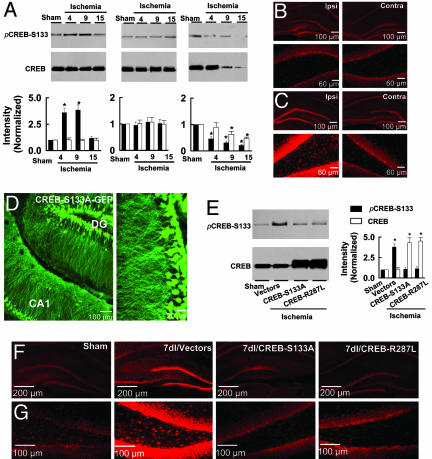
Focal Cerebral Ischemia Activates CREB in the Dentate Gyrus. To determine CREB activity, we examined phosphorylated CREB at Ser-133 (pCREB-S133), an activated form of CREB (21). In this study, the nuclear extracts were prepared from microdissected dentate gyrus of rats that had been subjected to focal cerebral ischemia or a sham operation. Western blots of the extracts showed a substantial increase of pCREB-S133 in the ipsilateral dentate gyrus after ischemic insult; the peak level of pCREB-S133 occurred in rats 4 d after ischemia (4dI; ≈3.9-fold of sham controls) (Fig. 1A). The elevated pCREB-S133 remained in 9dI rats and then declined to control level in 15dI rats (Fig. 1 A). In contrast to the dentate gyrus, in the CA1 area of the hippocampus pCREB-S133 levels were reduced after ischemic insult (Fig. 1 A). Consistent with the Western blots, staining of the hippocampal sections with anti-pCREB-S133 antibody demonstrated that the number of pCREB-S133-labeled nuclei was markedly increased in the ipsilateral dentate gyrus of 4dI rats, compared with sham controls (Fig. 1 B and C). However, in the contralateral dentate gyrus, CREB activity was unchanged at any time point after focal cerebral ischemia, showing that ischemic insult activates CREB only in the ipsilateral dentate gyrus.
Fig. 1.
Expression of dominant-negative mutants of CREB blocks ischemia-induced activation of CREB in the dentate gyrus. (A) Focal cerebral ischemia activates CREB in the dentate gyrus. (Upper) Nuclear extracts (500 mg of proteins) from microdissected ipsilateral dentate gyrus (Left), contralateral dentate gyrus (Center), and ipsilateral area CA1 (Right) of sham-operated, 4dI, 9dI, and 15dI rats were immunoprecipitated by anti-CREB and then probed with anti-pCREB-S133 or anti-CREB antibodies. (Lower) Anti-pCREB-S133 (filled bars) and anti-CREB (open bars) were normalized to respective sham-operated (defined as 1.0). Data are shown as mean ± SEM (n = 5, P < 0.01, Student's t test). (B and C) Ipsilateral (Ipsi) and contralateral (Contra) hippocampus (Upper) and dentate gyrus (Lower) from sham-operated (B) or 4dI (C) rats were stained with anti-pCREB-S133 antibody. Similar results were observed in each of four experiments. (D) A representative fluorescence image of a hippocampal section from a rat 1 d after infection with pSFV-CREB-S133A-GFP vectors (Left) with a selected area shown at increased magnification (Right). DG, dentate gyrus. (E Left) Nuclear extracts (500 mg) from ipsilateral dentate gyrus of sham-operated or 7dI rats infected with pSFV-GFP (control) vectors or pSFV-CREB-S133A-GFP or pSFV-CREB-R287L-GFP vectors were immunoprecipitated with anti-CREB and blotted with anti-pCREB-S133 or anti-CREB, as indicated. (Right) Anti-pCREB-S133 (filled bars) and anti-CREB (open bars) were normalized to respective sham-operated (defined as 1.0). Data are shown as mean ± SEM (n = 5, P < 0.01, Student's t test). (F and G) Hippocampus (F) and dentate gyrus (G) from sham-operated or 7dI rats infected with pSFV-GFP (control) vectors or pSFV-CREB-S133A-GFP or pSFV-CREB-R287L-GFP vectors were stained with anti-pCREB-S133 antibody. Similar results were observed in each of four experiments.
Inhibition of Endogenous CREB by the Expression of Dominant-Negative Mutants of CREB in the Dentate Gyrus. To block ischemia-induced activation of CREB, we constructed a dominant-negative CREB, CREB-S133A (22), which was inserted to pSFV vectors tagged with enhanced GFP (pSFV-CREB-S133A-GFP). The pSFV-CREB-S133A-GFP infectious particles (2 μl of >1010 infectious units per ml) were injected into each side of the rat hippocampus. CREB-S133A-GFP fusion proteins were observed in most neurons 24 h after the infection (Fig. 1D). Subsequently, we examined pCREB-S133 levels and observed no increase of pCREB-S133 intensities in the dentate gyrus expressing CREB-S133A at any time after ischemic injury (Fig. 1 E–G). As controls, pSFV vectors lacking CREB-S133A or expressing wild-type CREB were infused into the hippocampus. Neither control altered CREB activity (Fig. 1 E–G). A second CREB mutant, CREB-R287L, was also constructed by changing Arg-287 to Leu to disrupt the binding of endogenous CREB to targeted genes (23). Consistent with the results for CREB-S133A, CREB-R287L expression suppressed CREB activation in the dentate gyrus of rats after cerebral ischemic stroke (Fig. 1 E–G).
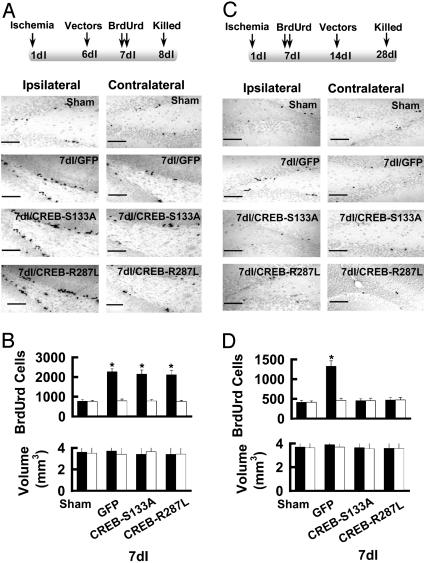
Inhibition of CREB Suppresses Newborn Cell Survival in the Dentate Gyrus. Our previous data show that the number of new cells in the dentate gyrus begins to increase 3 d after focal cerebral ischemia and reaches a peak in 7dI rats (10). This enhanced neurogenesis could result from increased precursor cell survival/proliferation and/or differentiation (8–10). To examine which of these events is affected by CREB dominant-negative mutants, we first assessed cell proliferation by treating 7dI animals twice with BrdUrd with an 8-h interval between injections; on the following day, the animals were killed to analyze BrdUrd labeling of dividing cells (Fig. 2A). Specific comparisons showed that 7dI rats infected with pSFV-CREB-S133A or pSFV-CREB-R287L vectors had similar proliferation to that of 7dI rats infected with pSFV-CREB-GFP or pSFV-GFP (control vectors, n = 6; Fig. 2B). No significant difference in the granule cell layer volume was observed between groups. These data indicate that inhibition of CREB did not alter progenitor cell proliferation in adult rats.
Fig. 2.
Inhibition of CREB blocks ischemia-induced production of new neurons in the dentate gyrus. (A Upper) Schematic representation of the experimental arrangement. (Lower) Photomicrographs of BrdUrd-labeled nuclei in the dentate gyrus of sham-operated and 7dI rats 1 d after the injection of BrdUrd. (Scale bar = 200 μm.) (B) Total number of BrdUrd-labeled cell per dentate gyrus (Upper) and granule cell layer volume (Lower) from ipsilateral (filed bars) and contralateral (open bars) dentate gyrus are summarized (mean ± SEM, n = 6; *, P < 0.01, compared with sham-operated). (C Upper) Schematic representation of the experimental arrangement. (Lower) Photomicrographs of BrdUrd-labeled nuclei in the dentate gyrus of sham-operated and 7dI rats 4 weeks after the injection of BrdUrd. (Scale bar = 200 μm.) (D) Total number of BrdUrd-labeled cells per dentate gyrus (Upper) and granule cell layer volume (Lower) from ipsilateral (filed bars) and contralateral (open bars) dentate gyrus are summarized (mean ± SEM, n = 6; *, P < 0.01, compared with sham-operated).
Next, we determined whether activation of CREB after ischemia increased the survival of neural precursor cells. We infected 7dI animals with pSFV-CREB-S133A-GFP, pSFV-CREB-R287L, or control (pSFV-CREB-GFP or pSFV-GFP) vectors 1 week after the BrdUrd injections. BrdUrd-labeled neurons in the dentate gyrus were then examined 3 weeks later (4 weeks after the injection of BrdUrd; Fig. 2C). When CREB was inhibited by the expression of either CREB-S133A or CREB-R287L, the number of BrdUrd-labeled nuclei in the ipsilateral dentate gyrus of 7dI rats was reduced (463 ± 37 cells for CREB-S133A, 446 ± 52 cells for CREB-R287L; Fig. 2D) compared with that in 7dI rats infected with control vectors (1,318 ± 137 cells for GFP; Fig. 2D).
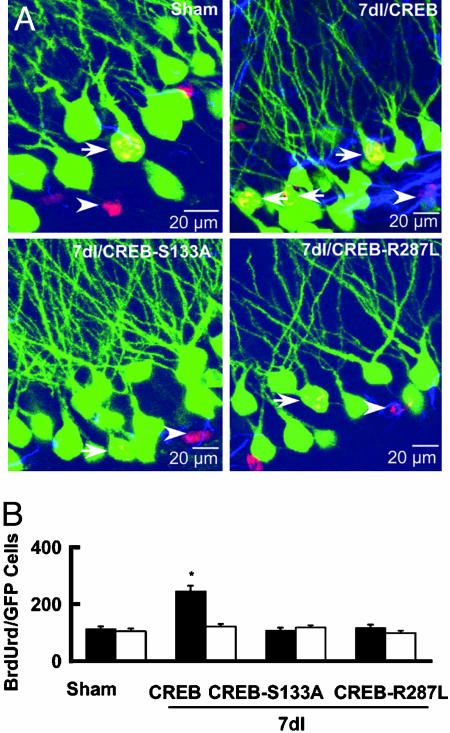
To determine neuronal identity of BrdUrd-labeled cells, we examined colocalization of BrdUrd and GFP and observed a number of BrdUrd-labeled neurons expressing GFP (BrdUrd/GFP cells, n = 7; Fig. 3) in the granule cell layer that had the morphological characteristics of a granule cell (a round or oval cell body ≈20 μm in diameter with long dendrites extending toward the molecular layer) in 7dI rats infected with control (pSFV-CREB-GFP) vectors (249 ± 38 BrdUrd/GFP cells per hippocampus, n = 7; Fig. 3A). In contrast, few BrdUrd/GFP neurons were detected in the dentate gyrus of 7dI rats infected with CREB-S133A-GFP (116 ± 16 BrdUrd/GFP cells per hippocampus, n = 7) or CREB-R287L-GFP vectors (108 ± 19 BrdUrd/GFP cells per hippocampus, n = 7, P < 0.01; Fig. 3B). Furthermore, the number of BrdUrd/GFP cells did not differ between sham-operated rats (109 ± 22 BrdUrd/GFP cells per hippocampus, n = 6) and 7dI rats infected with either CREB-S133A or CREB-R287L vectors (P > 0.1), and the percentage of new cells that differentiated into glia was similar between groups (Fig. 3B), suggesting that CREB activation was specific to the survival of precursor cells that differentiate into new neurons in adult rats.
Fig. 3.
Focal cerebral ischemia increases the number of granule neurons, but not glia, in the dentate gyrus. (A) Confocal laser-scanning fluorescence images of BrdUrd-positive (red) cells expressing GFP (green) or glial fibrillary acidic protein (GFAP; blue) in the dentate gyrus of sham-operated and 7dI rats infected with pSFV-CREB-GFP or pSFV-CREB-S133A-GFP or pSFV-CREB-R287L-GFP vectors 4 weeks after the injection of BrdUrd. Arrows and arrowheads indicate BrdUrd/GFP and BrdUrd/GFAP cells, respectively. (B) The numbers of BrdUrd/GFP granule cells (filled bars) and BrdUrd/GFAP colabeled glia (open bars) per dentate gyrus are summarized (mean ± SEM, n = 7; *, P < 0.01).
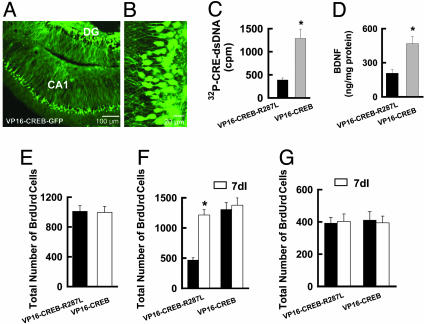
Expression of Constitutively Active CREB, VP16-CREB, in the Dentate Gyrus. To determine whether CREB activation is sufficient to induce neurogenesis in the dentate gyrus of adult rats, we expressed the constitutively active fusion protein VP16-CREB (24, 25), using pSFV-VP16-CREB-GFP vectors (Fig. 4 A and B). For this study, pSFV-VP16-CREB-R287L-GFP was used as a control. To examine whether VP16-CREB exhibited activity, we used an antibody to VP16 to precipitate VP16-CREB-GFP or VP16-CREB-R287L-GFP from the nuclear extracts of neurons infected with pSFV-VP16-CREB-GFP or pSFV-VP16-CREB-R287L-GFP, respectively (Fig. 4C). A CRE-binding assay showed that VP16-CREB-GFP, but not VP16-CREB-R287L-GFP, bound to a CRE oligonucleotide probe (n = 4; Fig. 4C). Subsequently, we analyzed brain-derived neurotrophic factor (BDNF) expression and observed that BDNF levels in the dentate gyrus doubled after expression of VP16-CREB (n = 4, P < 0.01, compared with control; Fig. 4D), indicating that VP16-CREB was functional for inducing CRE-dependent gene expression in vivo.
Fig. 4.
Expression of VP16-CREB increases the number of new neurons in the dentate gyrus of adult rats. (A) A representative fluorescence image of a hippocampal section from a rat 1 d after infection with pSFV-VP16-CREB-GFP vectors. DG, dentate gyrus. (B) High magnification of the image in A shows dentate granule neurons. (C) Nuclear extracts from neurons infected with VP16-CREB, but not VP16-CREB-R287L, vectors bound to 32P-labeled CRE oligonucleotides (mean ± SEM, n = 4; *, P < 0.01). dsDNA, double-stranded DNA. (D) Expression of VP16-CREB, but not VP16-CREB-R287L, induced expression of BDNF (mean ± SEM, n = 4; *, P < 0.01). (E) Neither VP16-CREB nor VP16-CREB-R287L altered proliferating activity. Proliferating cells in the dentate gyrus were labeled 1 d after the injection of BrdUrd. (F) Expression of VP16-CREB, but not VP16-CREB-R287L, increased the number of new neurons in sham-operated rats. Data are shown as mean ± SEM (n = 6; *, P < 0.01, ANOVA followed by Neuman–Keuls post hoc test). New neurons in the dentate gyrus were labeled 4 weeks after the injection of BrdUrd. Note that expression of VP16-CREB produced no further increase of new neurons in 7dI rats. (G) Neither VP16-CREB nor VP16-CREB-R287L altered survival of BrdUrd-labeled cells in the subventricular zone (SVZ). New cells in the SVZ were labeled 4 weeks after the injection of BrdUrd.
CREB Activation Increases New Neurons in the Dentate Gyrus. To address the consequences of expressing VP16-CREB on progenitor cell proliferation, we investigated BrdUrd labeling and observed that proliferating cells in the dentate gyrus of rats infected with VP16-CREB-GFP vectors did not differ from those infected with VP16-CREB-R287L-GFP vectors (Fig. 4E). However, the total number of new neurons (BrdUrd-positive cells 4 weeks after labeling) was significantly greater in animals infected with VP16-CREB (1,262 ± 166 cells, n = 6) than control (493 ± 78 cells, n = 6, P < 0.01; Fig. 4F). Expression of VP16-CREB in the dentate gyrus of 7dI rats produced no further increase of BrdUrd-labeling (1,379 ± 158 cells for VP16-CREB and 1,264 ± 141 cells for VP16-CREB-R287L, respectively, n = 6, P > 0.1; Fig. 4F). Thus, these results provide further evidence that CREB activation is responsible for ischemia-induced neurogenesis. CREB regulation of neurogenesis seems to be specific to the dentate gyrus because neither focal cerebral ischemic insult nor expression of VP16-CREB altered the survival of BrdUrd-labeled cells in the subventricular zone (SVZ; Fig. 4G).
Discussion
In this article, we expressed two dominant-negative mutants of CREB, CREB-S133A and CREB-R287L, in the hippocampus of adult rats using pSFV-based gene expression vectors. Our data show that inhibition of endogenous CREB did not alter progenitor cell proliferation but blocked ischemia-induced survival of neuronal precursors in the dentate gyrus. Further, we constructed a constitutively active CREB, VP16-CREB, and observed that its expression in the dentate gyrus of adult rats increased the number of newly generated granule neurons. Specific comparison showed that the increased number of new neurons induced by VP16-CREB was similar to that induced by ischemic injury. Thus, our results show that CREB activation fulfills a necessary and sufficient condition for ischemia-induced neurogenesis in the dentate gyrus of adult rats.
Our data demonstrate that the number of newly generated granule neurons doubles in the ipsilateral dentate gyrus of rats in response to focal cerebral ischemia. Previous studies indicate that the number of new cells in the dentate gyrus of normal adult rats increases between 2 h and 1 week after BrdUrd labeling and then declines dramatically by 2 weeks after DNA synthesis, because of dividing cell death (6, 26). Thus, enhanced neurogenesis in cerebral ischemic stroke could result from increased proliferation and/or survival of neural precursor cells (8–10). In the present study, our data demonstrate that inhibition of CREB did not change proliferating activity but reduced the survival rate of precursor cells, thereby suggesting that CRE transcription is one of the critical requisites for neuronal precursor cell survival but not their proliferation. This notion was further supported by our observations that the number of new granule neurons increased upon expression of VP16-CREB, during the time when many newborn cells typically die, whereas its expression did not alter progenitor cell proliferation in the dentate gyrus of adult rats. Consistent with our results, chronic treatment of adult rats with fluoxetine, which increases the level of pCREB-S133 in the dentate gyrus, enhances the survival of immature neurons in vivo (27). Another study on this issue reported that pCREB-S133 is present in immature neurons and suggests that pCREB-S133 may be involved in their survival/maturation (28). Together, our data indicate that CREB activation is critical for ischemia-induced neurogenesis in adult rats because it facilitates the survival of neuronal precursor cells.
Our previous studies demonstrate that the biochemical events underlying ischemia-induced neurogenesis in the dentate gyrus are thought to involve activation of N-methyl-d-aspartate (NMDA) receptors (10). There are also extensive data that CREB in neuronal cultures is activated by stimulation of NMDA receptors (29). Thus, the most parsimonious explanation of our findings is that, after focal cerebral ischemia, CREB is activated through NMDA receptor-dependent events. CREB regulation of neural precursor survival may involve expression of BDNF that provides a growth environment for the residential precursor cells in the dentate gyrus. This hypothesis is supported by earlier reports that dietary restriction stimulates the expression of BDNF that correlated with increased neurogenesis in the dentate gyrus of adult rats (30, 31). CREB activation and its ability to facilitate the survival of neuronal precursor cells may also be responsible for experience-dependent changes of dentate neurogenesis (32). For example, hippocampus-associative learning tasks that require activation of CREB enhance the survival, but not the proliferation, of neuronal precursor cells in the dentate gyrus (6). CREB-dependent expression of BDNF is elevated by exercise as well as by spatial learning (33). Thus, CREB activation induces BDNF expression, thereby enhancing the survival of new neurons that facilitate the encoding of new memories.
Supplementary Material
Acknowledgments
This work was supported by Canadian Institute of Health Research Grant MOP-67164, Heart and Stroke Foundation of Canada Grant M2C47765, the Alberta Heritage Foundation for Medical Research Grant 20010519, the Canada Foundation for Innovation Grant JV0730206 (to Y.M.L.), and by the Alberta Foundation of Innovation and Science.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CRE, cAMP-response element; CREB, CRE-binding protein; SFV, Semliki Forest virus; ndI, n days after ischemia; BDNF, brain-derived neurotrophic factor.
References
- 1.Reynolds, B. A. & Weiss, S. (1992) Science 255, 1707-1710. [DOI] [PubMed] [Google Scholar]
- 2.Suhonen, J. O., Peterson, D. A., Ray, J. & Gage, F. H. (1996) Nature 383, 624-627. [DOI] [PubMed] [Google Scholar]
- 3.Kempermann, G., Kuhn, H. G. & Gage, F. H. (1997) Nature 386, 493-495. [DOI] [PubMed] [Google Scholar]
- 4.Gould, E., Tanapat, P., McEwen, B. S., Flugge, G. & Fuchs, E. (1998) Proc. Natl. Acad. Sci. USA 95, 3168-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Praag, H., Kempermann, G. & Gage, F. H. (1999) Nat. Neurosci. 2, 266-270. [DOI] [PubMed] [Google Scholar]
- 6.Gould, E., Beylin, A., Tanapat, P., Reeves, A. & Shors, T. J. (1999) Nat. Neurosci. 2, 260-265. [DOI] [PubMed] [Google Scholar]
- 7.Shors, T. J., Miesegaes, G., Beylin, A., Zhao, M., Rydel, T. & Gould, E. (2001) Nature 410, 372-376. [DOI] [PubMed] [Google Scholar]
- 8.Liu, J., Solway, K., Messing, R. O. & Sharp, F. R. (1998) J. Neurosci. 18, 7768-7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin, K., Minami, M., Lan, J. Q., Mao, X. O., Batteur, S., Simon, R. P. & Greenberg, D. A. (2001) Proc. Natl. Acad. Sci. USA 98, 4710-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu, D. Y., Liu, S. H., Sun, S. H. & Lu, Y. M. (2003) J. Neurosci. 23, 223-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakatomi, H., Kuriu, T., Okabe, S., Yamamoto, S., Hatano, O., Kawahara, N., Tamura, A., Kirino, T. & Nakafuku, M. (2002) Cell 110, 429-441. [DOI] [PubMed] [Google Scholar]
- 12.Kirn, J. R. & Nottebohm, F. (1993) J. Neurosci. 13, 1654-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snyder, E. Y., Yoon, C., Flax, J. D. & Macklis, J. D. (1997) Proc. Natl. Acad. Sci. USA 94, 11663-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magavi, S. S., Leavitt, B. R. & Macklis, J. D. (2000) Nature 405, 951-955. [DOI] [PubMed] [Google Scholar]
- 15.Menard, C., Hein, P., Paquin, A., Savelson, A., Yang, X. M., Lederfein, D., Barnabe-Heider, F., Mir, A. A., Sterneck, E., Peterson, A. C., et al. (2002) Neuron 36, 597-610. [DOI] [PubMed] [Google Scholar]
- 16.Barnabe-Heider, F. & Miller, F. D. (2003) J. Neurosci. 23, 5149-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, S., Wang, J., Zhu, D., Fu, Y., Lukowiak, K. & Lu, Y. M. (2003) J. Neurosci. 23, 732-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, J., Liu, S., Fu, Y., Wang, J. H. & Lu, Y. M. (2003) Nat. Neurosci. 6, 1039-1047. [DOI] [PubMed] [Google Scholar]
- 19.Gundersen, H. J., Bagger, P., Bendtsen, T. F., Evans, S. M., Korbo, L., Marcussen, N., Moller, A., Nielsen, K., Nyengaard, J. R., Pakkenberg, B., et al. (1988) Acta Pathol. Microbiol. Immunol. Scand. 96, 857-881. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn, H. G., Dickinson-Anson, H. & Gage, F. H. (1996) J. Neurosci. 16, 2027-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez, G. A. & Montminy, M. R. (1989) Cell 59, 675-680. [DOI] [PubMed] [Google Scholar]
- 22.Riccio, A., Ahn, S., Davenport, C. M., Blendy, J. A. & Ginty, D. D. (1999) Science 286, 2358-2361. [DOI] [PubMed] [Google Scholar]
- 23.Reusch, J. E., Colton, L. A. & Klemm, D. J. (2000) Cell Biol. 20, 1008-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barco, A., Alarcon, J. M. & Kandel, E. R. (2002) Cell 108, 689-703. [DOI] [PubMed] [Google Scholar]
- 25.Pittenger, C., Huang, Y. Y., Paletzki, R. F., Bourtchouladze, R., Scanlin, H., Vronskaya, S. & Kandel, E. R. (2002) Neuron 34, 447-462. [DOI] [PubMed] [Google Scholar]
- 26.Cameron, H. A., Woolley, C. S., McEwen, B. S. & Gould, E. (1993) Nat. Neurosci. 56, 337-344. [DOI] [PubMed] [Google Scholar]
- 27.Malberg, J. E., Eisch, A. J., Nestler, E. J. & Duman, R. S. (2000) J. Neurosci. 20, 9104-9110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa, S., Kim, J. E., Lee, R., Chen, J., Fujioka, T., Malberg, J., Tsuji, S. & Duman, R. S. (2002) J. Neurosci. 22, 9868-9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bading, H., Ginty, D. D. & Greenberg, M. E. (1993) Science 260, 181-186. [DOI] [PubMed] [Google Scholar]
- 30.Lee, J., Duan, W., Long, J. M., Ingram, D. K. & Mattson, M. P. (2000) J. Mol. Neurosci. 15, 99-108. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J., Seroogy, K. B. & Mattson, M. P. (2002) J. Neurochem. 80, 539-547. [DOI] [PubMed] [Google Scholar]
- 32.Tully, T., Bourtchouladze, R., Scott, R. & Tallman, J. (2003) Nat. Rev. Drug Discov. 2, 267-277. [DOI] [PubMed] [Google Scholar]
- 33.Tokuyama, W., Okuno, H., Hashimoto, T., Xin, L. Y. & Miyashita, Y. (2000) Nat. Neurosci. 3, 1134-1142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.