Abstract
Key signaling pathways (such as phosphoinositide 3-kinase, Myc, and RAS) act as sensors of energy, stress, and nutrient availability and integrate these inputs to directly control ribosomeproduction and gene expression at the translational level. This activity is normally directly coupled to cell growth, division, and survival. However, it remains poorly understood the extent to which changes in ribosome number and nucleolar integrity downstream of these key signaling pathways contribute to their oncogenic activity. Emerging studies provide interesting insight into how deregulations in RNA polymerase I activity may lead to tumorigenesis and suggest that new drugs targeting ribosomal DNA transcription may hold great promise for the treatment of cancer.
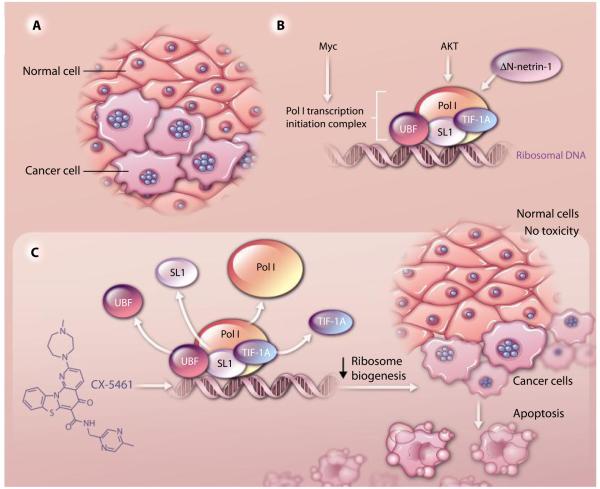
Imagine being a scientist in the 1800s. Although cancer was already recognized as a malignant disease, how would you distinguish a cancer cell from a normal one without modern-day molecular biology? Remarkably, the Italian pathologist Giuseppe Pianese described 200 years ago the first distinguishing features of cancer cells using only a simple light microscope (Fig. 1A). The earliest hall-marks of cancer cells that he observed were enlarged nucleoli, the cellular domain of ribosome production (1). Indeed, increases in the number and size of nucleoli have continued to be a useful prognostic marker for tumor development up to the present day.
Fig. 1.
The role of Pol I and the nucleolus in tumor formation and therapeutic intervention. (A) A reproduction of a drawing from (1). Pianese described prominent nuclear structures (blue circles) in a mammary gland carcinoma which he termed “Kernkörperchen” (literal translation: “nuclear corpuscles”) (1), which we now know are nucleoli. He raised several questions regarding the abundance of these corpuscles as well as their altered size in only specific cells within the tumor (presumably advanced cancer cells), concluding with “What is the fate of Kernkörperchen?” (B) Upstream oncogenic signaling pathways such as Myc and PI3K-AKT-mTOR—as well as ΔN-netrin-1, which is present only in cancer cells—regulate rDNA transcription by modulating the abundance or the activity of Pol I transcription machinery (or both). (C) The novel small-molecule CX-5461 targets the Pol I transcription complex and may represent a new treatment for cancer. CX-5461, which induces apoptosis in cancer cells with no cytotoxic effects in normal cells, has illuminated in part how tumors can be “addicted” to the increased numbers of ribosomes required to sustain cancer cell growth and division.
These early studies speak to an important relation between the cell cycle, ribosome production, and nucleoli morphology. Ribosome biogenesis and global protein synthesis are tightly and dynamically regulated to accommodate the demands triggered by growth (increases in cell size) of a cell, a prerequisite for accurate cell division (2). Indeed, increased ribosome production manifests in increased nucleolar number and size, such that highly proliferative cells have more and larger nucleoli compared with quiescent cells (3, 4). However, several key questions related to these observations remain to be addressed: Does an increase in ribosome biogenesis play a causal role in cellular transformation? If so, what are the underlying mechanisms? Is nucleolar integrity an important determinant of cellular transformation? Recent studies have shed light on some aspects of these unresolved questions and provided an unprecedented understanding of the balance between nucleolar activity and cancer development while unraveling a novel therapeutic window to target ribosomal RNA (rRNA) synthesis (5–7).
Ribosome biogenesis is a highly regulated process that requires the coordinated activity of all three RNA polymerases (Pol I, II, and III) along with a vast cohort of small nucleolar RNAs, transcription factors, and nonribosomal proteins that promote the transcription, processing, and modification of rRNA. Mature rRNA is assembled with 79 ribosomal proteins into the 40S and 60S ribosomal subunits that compose the functional ribosome (8). There has been a growing realization that the expression and activity of many of the components involved in ribosome production and translational control are directed by signal transduction pathways, which are often deregulated in cancer (9). Two of the best-studied examples, the proto-oncogene Myc and the PI3K (phosphoinositide 3-kinase)-AKT-mTOR (mammalian target of rapamycin) signaling pathway, are exquisite regulators of the translation machinery (10). Both Myc and PI3K-AKT-mTOR augment the protein synthetic capacity of cancer cells, which is a major determinant of their oncogenic activity (11, 12). Myc and PI3K-AKT-mTOR also directly regulate ribosome biogenesis (Fig. 1B). For instance, Myc promotes Pol II and Pol III–mediated transcription of ribosomal proteins and 5S rRNA in the nucleoplasm (13, 14). Myc also increases transcription of rDNA in the nucleolus by directing the assembly of the Pol I preinitiation complex or by enhancing the expression of rDNA transcription factors such as UBF (upstream binding factor), SL1 (selectivity factor 1), TIF-1A (transcription initiation factor 1A), and POLR1B (polymerase I polypeptide B) (15–17). The kinase mTOR modulates the activity of the transcription initiation factor TIF-1A, which associates with Pol I to mediate rRNA synthesis (18). Although Myc and PI3K-AKT-mTOR regulate many aspects of ribosome biogenesis, whether and how this directly translates to tumorigenesis is still poorly understood.
Employing a unique pharmacologic approach, Chan et al. uncovered that the protein kinase AKT, in addition to its well-documented role in controlling translation initiation, also modulates ribosome biogenesis at multiple steps, specifically by promoting Pol I loading during rDNA transcription initiation, rDNA transcription elongation, and rRNA processing (Fig. 1B) (6). In striking contrast to the majority of research centered on AKT’s ability to modulate translational control by activating its downstream target the mTOR kinase complex 1 (mTORC1), Chan et al. found that the PI3K-AKT-mTOR pathway controls ribosome biogenesis at least in part through an mTORC1-independent mechanism. They showed that acute and short-term inhibition of AKT in cell lines results in decreased rDNA transcription rates with minimal effect on mTORC1 activity at these early time points. One feature required for an oncogene to induce cellular transformation is its intrinsic ability to promote cell growth or division (or both) independent from mitogenic signals such as growth factors (19, 20). In this context, Chan et al. showed that over-expression of AKT results in increased ribosome biogenesis, ribosome number, and cell growth, even in growth factor–deprived cells. This suggests that AKT-mediated rRNA synthesis is a direct effect and active participant in the AKT-mediated oncogenic program.
New findings from Chan et al. now also show that PI3K-AKT-mTOR and Myc co-operate to promote ribosome biogenesis. Unraveling the mechanisms that underlie this important synergistic activity will have far-reaching implications. Indeed, concomitant hyperactivation of these two oncogenic pathways is associated with increased disease severity and resistance to therapy (21, 22). It remains unknown whether there may be a graded biological response to increases in rRNA production toward cell growth, proliferation, or translation. It would be of great importance to assess whether the synergistic effects between Myc and the PI3K-AKT-mTOR pathway on ribosome biogenesis might explain at least in part the observed severity and resistance to therapy in tumors with both oncogenic lesions.
Do increases in Pol I–mediated rDNA transcription underlie Myc oncogenic potential, and, if so, could Pol I itself be a druggable target? Oncogenic Myc has historically been considered undruggable, and therapeutically targeting Pol I activity to induce synthetic lethality in the context of increased activation of Myc might be a huge leap forward in the design of effective cancer treatments. Bywater et al. employ complementary genetic and pharmacologic approaches to address this outstanding question (5). Specifically, the authors utilize the Eμ-Myc mouse model in which Myc is overexpressed in the B cell compartment. Eμ-Myc mice develop lymphoma and recapitulate many aspects of human Burkitt’s lymphoma, a disease characterized by translocation of Myc in 100% of cases. In agreement with previous findings, Eμ-Myc B cells display increased expression of Pol I components, including the key activator of Pol I, UBF; the Pol I initiation factor RRN3 (also known as TIF-1A); and POLR1B, one of the largest subunits of Pol I. The increased abundance of these activators of Pol I is already evident in premalignant cells, which suggests that increased rates of rDNA transcription are not merely a consequence of cellular transformation, but rather are directly regulated by Myc and, therefore, may contribute to tumor initiation. Hannan’s group showed that restoring Pol I activity to that found in normal cells by an RNA interference technique triggers cell death of Eμ-Myc lymphoma cells. Importantly, they also demonstrated the therapeutic efficacy of CX-5461, the first selective small-molecule inhibitor of Pol I (created by Cylene Pharmaceuticals), in B cell lymphoma (Fig. 1C). In allograft experiments, the authors determined that, remarkably, CX-5461 selectively induces cell death in Eμ-Myc malignant cells while allowing normal B cells to grow and proliferate. CX-5461 also significantly prolonged the survival of tumor-bearing mice. Altogether, these findings strongly suggest that CX-5461, which is scheduled to enter phase I clinical trials soon, holds great promise as a small-molecule therapy.
Can CX-5461 be employed as a novel treatment for possibly many human cancers that display augmented ribosome numbers? This might reflect a means to target a universally important feature of cancer cells: a rewiring of the translational machinery to meet the increased translational demands necessary to promote and sustain their growth and division. In essence, compared with normal cells, cancer cells may be “addicted” to increases in ribosome biogenesis and number. Bywater et al. showed that a surprising prerequisite for therapeutic efficacy of CX-5461 is not only increased Pol I activity but also the presence of the tumor suppressor p53. Unexpectedly, CX-5461 treatment leads to a rapid increase in p53 abundance and activity, which presumably mitigates the cytotoxicity of this drug. The effect of CX-5461 on p53 abundance may be attributed to the activation of a putative nucleolar surveillance pathway in which perturbations of nucleolar integrity result in increased amounts of cellular p53. Several mechanisms by which p53 may be trigged by nucleolar “stress” have been proposed (23), one of which has been in part tested by Bywater et al. The authors showed that in Eμ-Myc lymphoma cells treated with CX-5461, ribosomal proteins L5 and L11 exert an “extra-ribosomal” function by binding to MDM2, the ubiquitin ligase for p53; this leads to p53 accumulation. These are interesting findings; however, much remains to be further understood, including the extent of CX-5461 therapeutic efficacy in human cancers, as well as whether additional p53-independent mechanisms may underlie the striking cytotoxic response in cancer cells.
Besides the “usual suspects” such as Myc and the PI3K-AKT-mTOR oncogenic path-way, regulators of ribosome biogenesis are also emerging as putative contributors to cellular transformation (7, 24, 25). For example, Delloye-Bourgeois et al. made the surprising discovery that a specific isoform of netrin-1 (ΔN-netrin-1), a protein essential for proper axon guidance during neuronal development, is localized in the nucleolus only in human tumor cells and not in normal tissues (Fig. 1B) (7). Netrin-1 has been previously implicated in tumorigenesis (26). Delloye-Bourgeois et al. showed that in cancer cells, ΔN-netrin-1 is produced from an alternative promoter and has a nucleolar localization signal at the C terminus responsible for its recruitment to the nucleolus. They also demonstrated that this isoform of netrin-1 is a member of the rRNA transcription complex in the nucleolus, which directly interacts with the rRNA promoter to increase 45S rRNA synthesis; this leads to the production of more ribosomes. Delloye-Bourgeois et al. showed that the nucleolar localization of ΔN-netrin-1 is required to promote cell proliferation in vitro, as well as oncogenic potential in vivo in xenograft experiments in chick embryos.
Together, these emerging studies not only shed new light on a critical role for Pol I, ribosome biogenesis, and cancer development (Fig. 1), but they also contribute to our understanding of how ribosome production is controlled by specific oncogenic signals. These studies also raise new questions regarding the mechanisms by which ribosome numbers impinge on cancer etiology. A critical unresolved question is whether translation of the cancer genome is directly influenced by increases in Pol I activity and ribosome numbers. For example, a qualitative change in ribosome production may alter the dynamics and specificity in translational control of classes of mRNAs with unique regulatory elements—for example, in their 5′ untranslated regions—which are sensitive to changes in ribosome numbers. Posttranscriptional control of the cancer genome may play an unexpectedly important role in the etiology of cancer and reflects a new frontier in cancer research. The surprising selectivity of agents that target ribosome production, which kill cancer cells but not normal cells, reveals new avenues for cancer therapeutics and further illuminates how ribosome synthesis might be controlled in the first place. Historically, the mechanisms controlling ribosome abundance in normal cells during development, differentiation, and cell cycle progression have remained poorly understood. However, extensive buffering of ribosome numbers may allow for greater tolerance of their reduction in normal cells. In contrast, a greater demand for ribosome production in cancer cells may herald a new therapeutic window. Ongoing research is coming full circle from identifying the nucleolus as the earliest noted marker of cancer cells hundreds of years ago to using this unique identifier as a meaningful distinguishing feature to selectively eradicate cancer cells. “Viva la differenza!”—as Giuseppe Pianese would say!
Acknowledgments
I would like to thank M. Barna and K. Tong for critical reading and editing. Funding: This work is supported by NIH R01 CA140456 and the Leukemia and Lymphoma Society Scholar Award.
References and Notes
- 1.Pianese G. Beitrag zur histologie und aetiologie der carcinoma. histologische und experimentelle untersuchungen. Beitr. Pathol. Anat. Allgem. Pathol. 1896;142:1–193. [Google Scholar]
- 2.Hall MN, Raff M, Thomas G. Cell Growth: Control of Cell Size. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; NY: 2004. [Google Scholar]
- 3.Dubben HH. Different nucleolar antigen expression in resting and proliferating human lymphocytes as studied by fluorescence microscopy and flow cytometry. Cell Tissue Kinet. 1990;23:89–97. doi: 10.1111/j.1365-2184.1990.tb01335.x. [DOI] [PubMed] [Google Scholar]
- 4.Derenzini M, Trerè D, Pession A, Govoni M, Sirri V, Chieco P. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J. Pathol. 2000;191:181–186. doi: 10.1002/(SICI)1096-9896(200006)191:2<181::AID-PATH607>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 5.Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, Huser N, Proffitt C, Bliesath J, Haddach M, Schwaebe MK, Ryckman DM, Rice WG, Schmitt C, Lowe SW, Johnstone RW, Pearson RB, McArthur GA, Hannan RD. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell. 2012;22:51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan JC, Hannan KM, Riddell K, Ng PY, Peck A, Lee RS, Hung S, Astle MV, Bywater M, Wall M, Poortinga G, Jastrzebski K, Sheppard KE, Hemmings BA, Hall MN, Johnstone RW, McArthur GA, Hannan RD, Pearson RB. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Sci. Signal. 2011;4:ra56. doi: 10.1126/scisignal.2001754. [DOI] [PubMed] [Google Scholar]
- 7.Delloye-Bourgeois C, Goldschneider D, Paradisi A, Therizols G, Belin S, Hacot S, Rosa-Calatrava M, Scoazec J-Y, Diaz J-J, Bernet A, Mehlen P. Nucleolar localization of a netrin-1 isoform enhances tumor cell proliferation. Sci. Signal. 2012;5:ra57. doi: 10.1126/scisignal.2002456. [DOI] [PubMed] [Google Scholar]
- 8.Xue S, Barna M. Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 10.Ruggero D. Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 2012 doi: 10.1101/cshperspect.a012336. 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- 14.Ruggero D. The role of Myc-induced protein synthesis in cancer. Cancer Res. 2009;69:8839–8843. doi: 10.1158/0008-5472.CAN-09-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arabi A, Wu S, Ridderstråle K, Bierhoff H, Shiue C, Fatyol K, Fahlén S, Hydbring P, Söderberg O, Grummt I, Larsson LG, Wright AP. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 16.Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 17.Poortinga G, Hannan KM, Snelling H, Walkley CR, Jenkins A, Sharkey K, Wall M, Brandenburger Y, Palatsides M, Pearson RB, McArthur GA, Hannan RD. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J. 2004;23:3325–3335. doi: 10.1038/sj.emboj.7600335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoakum GH, Lechner JF, Gabrielson EW, Korba BE, Malan-Shibley L, Willey JC, Valerio MG, Shamsuddin AM, Trump BF, Harris CC. Transformation of human bronchial epithelial cells transfected by Harvey ras oncogene. Science. 1985;227:1174–1179. doi: 10.1126/science.3975607. [DOI] [PubMed] [Google Scholar]
- 20.Wheeler EF, Rettenmier CW, Look AT, Sherr CJ. The v-fms oncogene induces factor independence and tumorigenicity in CSF-1 dependent macrophage cell line. Nature. 1986;324:377–380. doi: 10.1038/324377a0. [DOI] [PubMed] [Google Scholar]
- 21.Clegg NJ, Couto SS, Wongvipat J, Hieronymus H, Carver BS, Taylor BS, Ellwood-Yen K, Gerald WL, Sander C, Sawyers CL. MYC cooperates with AKT in prostate tumorigenesis and alters sensitivity to mTOR inhibitors. PLoS ONE. 2011;6:e17449. doi: 10.1371/journal.pone.0017449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zafarana G, Ishkanian AS, Malloff CA, Locke JA, Sykes J, Thoms J, Lam WL, Squire JA, Yoshimoto M, Ramnarine VR, Meng A, Ahmed O, Jurisca I, Milosevic M, Pintilie M, van der Kwast T, Bristow RG. Copy number alterations of c-MYC and PTEN are prognostic factors for relapse after prostate cancer radiotherapy. Cancer. 2012;118:4053–4062. doi: 10.1002/cncr.26729. [DOI] [PubMed] [Google Scholar]
- 23.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibaragi S, Yoshioka N, Kishikawa H, Hu JK, Sadow PM, Li M, Hu GF. Angiogenin-stimulated rRNA transcription is essential for initiation and survival of AKT-induced prostate intraepithelial neoplasia. Mol. Cancer Res. 2009;7:415–424. doi: 10.1158/1541-7786.MCR-08-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saporita AJ, Chang HC, Winkeler CL, Apicelli AJ, Kladney RD, Wang J, Townsend RR, S. Michel L, Weber JD. RNA helicase DDX5 is a p53-independent target of ARF that participates in ribosome biogenesis. Cancer Res. 2011;71:6708–6717. doi: 10.1158/0008-5472.CAN-11-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazelin L, Bernet A, Bonod-Bidaud C, Pays L, Arnaud S, Gespach C, Bredesen DE, Scoazec J-Y, Mehlen P. Netrin-1 controls colorectal tumorigenesis by regulating apoptosis. Nature. 2004;431:80–84. doi: 10.1038/nature02788. [DOI] [PubMed] [Google Scholar]