Abstract
Signal transducer and activator of transcription 3 (STAT3) is an important oncogenic transcription factor residing in the cytoplasm in the resting cells. Upon stimulation, STAT3 is activated and translocated to the nucleus to regulate target genes. Although the canonical transcriptional function of STAT3 has been intensively studied, less is known about its cytoplasmic localization. In this study, by immunoprecipitation, microtubule cosedimentation, and immunofluorescence assays, we present the first evidence that cytoplasmic STAT3 interacts with both tubulin and microtubules. By using small-molecule inhibitor approaches, we further demonstrate that the localization of STAT3 on microtubules and its activation are independent of histone deacetylase 6 (HDAC6) activity. In addition, disruption of microtubule dynamics does not alter the activation and nuclear translocation of STAT3 in response to interleukin-6 treatment. These findings reveal that cytoplasmic STAT3 is physically associated with microtubules, whereas its activation and nuclear translocation are independent of microtubule dynamics, implicating that the association of STAT3 with microtubules might be involved in the regulation of noncanonical functions of STAT3 in the cytoplasm.
Introduction
Microtubules are important for a variety of cellular activities, including the maintenance of cell shape, cell division, cell motility, intracellular transport, and signal transduction. In cells, a number of proteins known as microtubule-associated proteins (MAPs) are localized to microtubules and regulate microtubule dynamics to fulfill their distinct functions (Gao et al., 2008; Sun et al., 2012; Tala et al., 2014a). Aberrant expression and localization of these MAPs are frequently associated with pathological diseases such as cancer and neurodegenerative diseases (Tala et al., 2014b; Xie et al., 2014).
Signal transducer and activator of transcription 3 (STAT3), the most pleiotropic member of the STAT family, resides largely in the cytoplasm in the resting cells (Boulton et al., 1995). In response to cytokines and growth factors, STAT3 is activated by tyrosine phosphorylation, resulting in its dimerization and nuclear translocation to regulate the transcription of genes involved in cell proliferation, survival, and differentiation (Walker et al., 2010, 2011). Besides its transcriptional function, STAT3 interacts with stathmin, a microtubule depolymerizing protein that sequesters free tubulin (Ng et al., 2006; Verma et al., 2009), suggesting a noncanonical role for STAT3 in the regulation of microtubule dynamics. However, the physical association of STAT3 with microtubules has not been characterized. This study was designed to test this possibility directly.
Materials and Methods
Materials
Paclitaxel, nocodazole, tubacin, tubastatin A, anti-α-tubulin antibody, and anti-acetylated α-tubulin antibody were obtained from Sigma-Aldrich. Anti-STAT3 antibody, anti-GFP antibody, and horseradish peroxidase-conjugated secondary antibody were purchased from Santa Cruz. Anti-phosphorylated STAT3 (Y705) antibody was purchased from Cell Signaling Technology. Fluorescein- and rhodamine-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories.
Cell culture and transfection
Jurkat cells were cultured in an RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and incubated at 37°C in a humidified atmosphere with 5% CO2. HeLa, Cos7, and 293T cells were cultured in a DMEM containing 10% FBS. Cells were transfected with GFP or GFP-tubulin by using polyethylenimine.
Microtubule cosedimentation assay
The microtubule cosedimentation assay was performed as described previously with minor changes (Sun et al., 2011). 5×106 HeLa cells were collected and homogenized, and the cell lysates were centrifuged at 4°C for 10 min at 12,000 rpm. The supernatants were incubated at 37°C for 30 min in the presence or absence of 20 μM paclitaxel and 1 mM GTP. Subsequently, the MME buffer (100 mM MES, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT, and 1% Triton) containing 20% glycerol and 20 μM paclitaxel was added and then centrifuged at 37°C for 1 h at 100,000 g. The supernatant and pellet fractions were collected for the detection of tubulin and STAT3.
Immunoprecipitation
293T cells transfected with GFP or GFP-tubulin were lyzed, and the cell lysates were incubated with anti-GFP agarose beads at 4°C for 4 h. The beads were centrifuged at 4°C for 30 s and then washed with a cell lysis buffer thrice. The beads were resuspended with a 50 μL cell lysis buffer and used for STAT3 detection.
Immunoblotting
Protein samples were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore), followed by blocking with Tris-buffered saline containing 0.2% Tween 20 for 1 h. The membranes were incubated sequentially with primary antibodies and horseradish peroxidase-conjugated secondary antibodies. Specific proteins were detected with an enhanced chemiluminescence detection reagent (Millipore) according to the manufacturer's protocol.
Immunofluorescence microscopy
HeLa and Cos7 cells were cultured on glass coverslips, and Jurkat cells were settled down on poly-lysine coated coverslips. Cells were fixed with 4% paraformaldehyde for 15 min at room temperature, perforated with 0.1% Triton X-100 for 20 min, and blocked with 2% bovine serum albumin in PBS for 40 min. Cells were then incubated with primary antibodies and fluorescein- or rhodamine-conjugated secondary antibodies. Nuclei were stained with DAPI. Images were captured using a Zeiss fluorescence microscope or a Leica TCS SP5 confocal microscope.
Statistical analysis
All data were derived from three independent experiments and presented as mean±SD. Student's t-test was performed for statistical analysis.
Results
Interaction of STAT3 with both tubulin and microtubules
To investigate the interaction of STAT3 with tubulin, we transfected GFP or GFP-tubulin to 293T cells to analyze whether endogenous STAT3 could be immunoprecipitated by GFP-tubulin. As shown in Figure 1A, STAT3 was detected in the GFP-tubulin immunoprecipitate, suggesting an association between STAT3 and tubulin. We then performed the microtubule cosedimentation assay to examine the interaction of STAT3 with microtubules. HeLa cells were homogenized, and the cell lysates were incubated at 37°C in the presence of paclitaxel and GTP to allow microtubules to polymerize. Microtubules and MAPs were pelleted by centrifugation. The majority of STAT3 was present in the pellet fraction when the experiment was performed in the presence of paclitaxel and GTP (Fig. 1B), suggesting a robust interaction of STAT3 with microtubules. By contrast, STAT3 was entirely present in the supernatant fraction and hard to detect in the pellet fraction when the experiment was performed in the absence of paclitaxel and GTP (Fig. 1B). Taken together, these data demonstrate that STAT3 can interact with both tubulin and microtubules in cells.
FIG. 1.
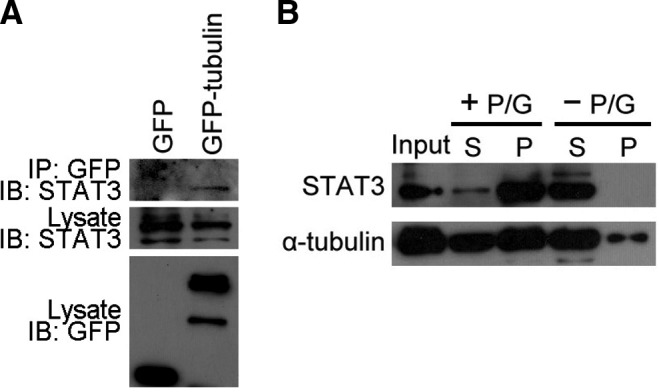
Immunoprecipitation and microtubule cosedimentation assays reveal that signal transducer and activator of transcription 3 (STAT3) interacts with both tubulin and microtubules. (A) 293T cells transfected with GFP or GFP-tubulin were lyzed and the cell lysates were incubated with anti-GFP agarose beads. The immunoprecipitate was analyzed by immunoblotting with the anti-STAT3 antibody, and cell lysates were analyzed with anti-STAT3 and anti-GFP antibodies. (B) HeLa cells were lyzed, and the lysates were incubated at 37°C for 30 min in the presence or absence of paclitaxel and GTP (P/G indicates paclitaxel and GTP). The microtubules and microtubule-associated proteins were pelleted by ultracentrifugation, and proteins in the pellet fraction (P) and the supernatant fraction (S) were examined by immunoblotting.
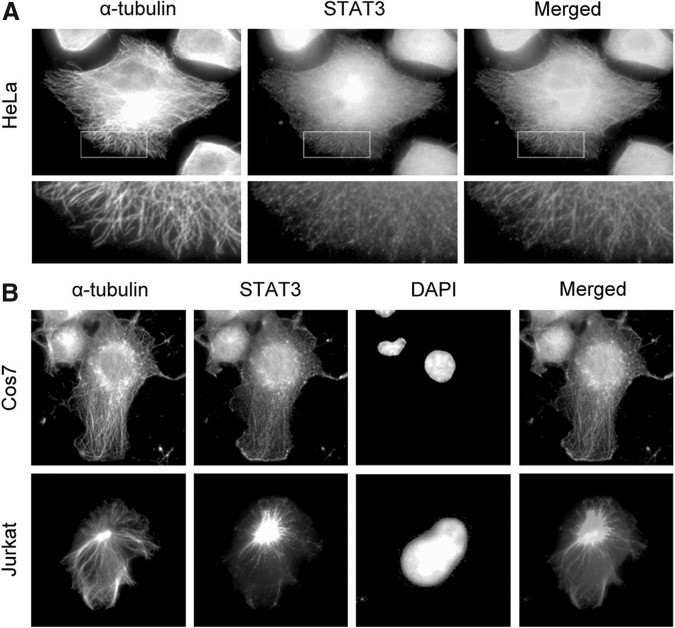
STAT3 partially colocalizes with microtubules in cells
We next investigated the localization of STAT3 in cells. By immunostaining α-tubulin and STAT3 in HeLa cells, we found that STAT3 was mainly localized in the cytoplasm in the resting cells (Fig. 2A). Interestingly, we observed a microtubule-like distribution pattern of STAT3, indicating a partial colocalization of STAT3 with microtubules (Fig. 2A). To investigate whether the microtubule colocalization pattern of STAT3 is cell-type dependent, we examined the localization of STAT3 in Cos7 and Jurkat cells. As shown in Figure 2B, STAT3 also colocalized with microtubules in these cells. In addition, STAT3 localized at the centrosome, the microtubule organizing center, in Jurkat cells (Fig. 2B).
FIG. 2.
Immunofluorescence microscopy shows that STAT3 partially colocalizes with microtubules in cells. (A) HeLa cells were stained with anti-α-tubulin and anti-STAT3 antibodies, and the boxed areas were magnified and shown in the lower panel. (B) Immunofluorescence microscopy of Cos7 and Jurkat cells stained with anti-α-tubulin and anti-STAT3 antibodies and DAPI.
Inhibition of HDAC6 activity increases tubulin acetylation without affecting STAT3 localization
Microtubules close to the centrosome are known to be relatively stable with hyperacetylation. Regarding the observation that endogenous STAT3 was concentrated at the centrosome in Jurkat cells, we examined whether the centrosomal localization of STAT3 is associated with the level of microtubule acetylation. Histone deacetylase 6 (HDAC6) is known to regulate microtubule dynamics through its action on microtubule acetylation (Li et al., 2011). By inhibition of HDAC6 activity with its selective inhibitor tubacin, we found that microtubule acetylation was remarkably increased (Fig. 3). Interestingly, the subcellular localization of STAT3 was not affected (Fig. 3). These results indicate that STAT3 localization is independent of HDAC6-mediated microtubule deacetylation.
FIG. 3.
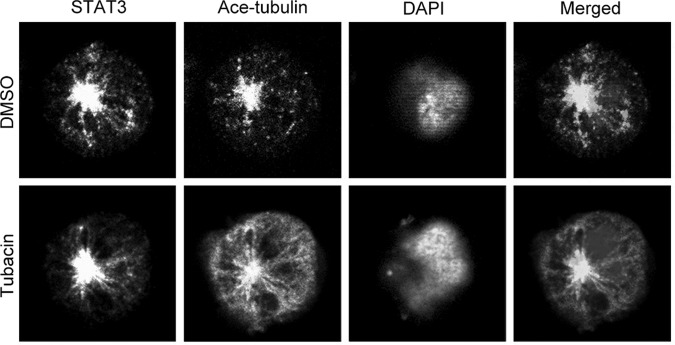
Inhibition of histone deacetylase 6 (HDAC6) activity increases tubulin acetylation without affecting STAT3 localization. Jurkat cells were treated with DMSO or HDAC6-selective inhibitor tubacin for 4 h, and then cells were stained with anti-STAT3 and anti-acetylated tubulin antibodies and DAPI.
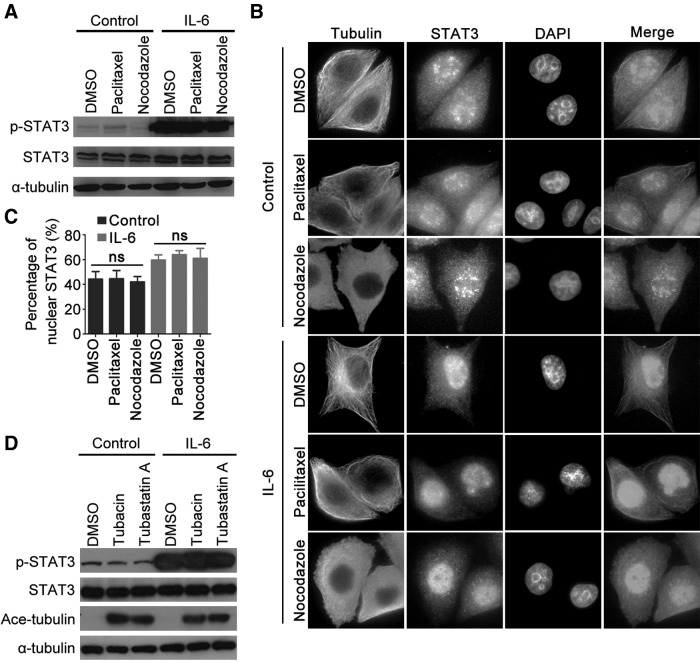
Microtubule dynamics or HDAC6 activity does not affect IL-6-induced STAT3 activation
Upon the stimulation of cytokines such as interleukin-6 (IL-6), STAT3 is activated and translocated to the nucleus to regulate the targeted genes (Zhong et al., 1994). Microtubule dynamics are thought to be involved in the activation of a number of transcription factors. To investigate whether STAT3 activation is modulated by microtubule dynamics, we treated cells with the microtubule-stabilizing agent paclitaxel and the microtubule-destabilizing agent nocodazole. The response of STAT3 signaling to IL-6 was examined by detecting the level of STAT3 phosphorylation. As shown in Figure 4A, IL-6 stimulation resulted in a significant increase of STAT3 phosphorylation, while paclitaxel and nocodazole had little effect on STAT3 activation. Consistently, alteration of the microtubule dynamics by either the microtubule-stabilizing agent paclitaxel or microtubule-destabilizing agent nocodazole had little effects on the IL-6-induced STAT3 nuclear translocation (Fig. 4B, C). We further examined whether HDAC6 is involved in IL-6-induced STAT3 activation. By using HDAC6-selective inhibitors, tubacin and tubastatin A, we found that microtubule acetylation was markedly elevated; however, tubacin and tubastatin A did not affect STAT3 phosphorylation in response to IL-6 (Fig. 4D). Taken together, these results suggest that IL-6-induced STAT3 activation is independent of microtubule dynamics and HDAC6 activity.
FIG. 4.
Inhibition of microtubule dynamics or HDAC6 activity does not alter STAT3 activation in response to interleukin-6 (IL-6) treatment. (A–C) HeLa cells were treated with DMSO, paclitaxel, or nocodazole for 1 h, followed by IL-6 treatment for 30 min. (A) Phosphorylated STAT3, total STAT3, and α-tubulin were examined by immunoblotting. (B) Cells were stained with anti-STAT3 and anti-α-tubulin antibodies and DAPI. (C) The percentage of nuclear STAT3 was measured by dividing the fluorescence intensity of STAT3 in the nucleus with that in the whole cell. (D) HeLa cells treated with DMSO, tubacin, or tubastatin A for 4 h were stimulated with IL-6 for 30 min. Phosphorylated STAT3, total STAT3, acetylated tubulin, and α-tubulin were examined by immunoblotting. ns, not significant (p≥0.05).
Discussion
The oncogenic transcription factor STAT3 has been considered as an important target for cancer therapy (Lavecchia et al., 2011). STAT3 is constitutively activated in several types of cancers, such as breast cancer, T-cell lymphomas, and liver cancer (Garcia et al., 1997; Mitchell and John, 2005; Wang et al., 2011). Besides its role in activating cancer-related genes, emerging evidence reveals that STAT3 interacts with stathmin to regulate microtubule dynamics and metastasis of cancer cells (Ng et al., 2006; Walker et al., 2011), implicating a potential association STAT3 with microtubules. However, whether cytoplasmic STAT3 colocalizes with microtubules remains undefined. In this study, we reveal that STAT3 interacts with both microtubules and tubulin, and immunostaining exhibits a partial localization of STAT3 with microtubules. Like many other transcription factors, STAT3 possesses a coiled-coil domain (Zhang et al., 2000), which is found in a variety of microtubule-binding proteins as well; for example, the coiled-coil motif in MAP7 domain-containing protein 3 (Mdp3) is essential for its interaction with tubulin and microtubules (Sun et al., 2011; Tala et al., 2014a). It is warranted to investigate in the future whether this motif is required for the interaction of STAT3 with microtubules.
Factors that regulate microtubule dynamics are thought to have potential effects on the localization of MAPs (Cassimeris and Spittle, 2001). In Jurkat cells, STAT3 is mainly concentrated in the centrosome region where microtubules are more stable and hyperacetylated. We found that the localization of STAT3 was not affected by HDAC6-mediated microtubule dynamics, and IL-6-induced STAT3 activation was independent of microtubule dynamics and HDAC6 activity as well, suggesting that STAT3 localization and transcriptional function were modulated by other mechanisms. Regarding the importance of microtubules in STAT3 transport, we speculated that STAT3 was recruited to the microtubules where it was activated and translocated to the nucleus to regulate biological behaviors. However, our further data revealed that disrupting the microtubule dynamics with paclitaxel or nocodazole did not affect the IL-6-induced STAT3 translocation to the nucleus. These evidences thus demonstrate that the activation and nuclear translocation of STAT3 are independent of microtubule dynamics, suggesting that the physical association of STAT3 with microtubules is not relevant to the canonical transcriptional functions of STAT3.
STAT3 is known to regulate microtubule dynamics through sequestration of the microtubule depolymerizing protein stathmin (Ng et al., 2006). Recent study reveals that the regulatory role of STAT3 in microtubule dynamics is critical for the lifecycle of hepatitis C virus (McCartney et al., 2013). In addition, mounting evidence demonstrates that microtubules are required and served as a track for the transport of various viruses to the nucleus (Martin and Helenius, 1991; Su et al., 2010; Rode et al., 2011). The transactivator of the transcription (Tat) protein of human immunodeficiency virus and Jembrana disease virus interacts robustly with microtubules and regulates microtubule dynamics (Xuan et al., 2007; Huo et al., 2011; Liu et al., 2014). Similarly, a robust interaction of STAT3 with microtubules was observed in this study. It would be interesting to investigate whether disrupting the interaction of STAT3 with microtubules would be a potential approach for the modulation of STAT3-associated diseases.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31171334 and 31170820) and the Tianjin Natural Science Foundation (13JCZDJC30300).
Disclosure Statement
No competing financial interests exist.
References
- Boulton T.G., Zhong Z., Wen Z., Darnell J.E., Jr., Stahl N., and Yancopoulos G.D. (1995). STAT3 activation by cytokines utilizing gp130 and related transducers involves a secondary modification requiring an H7-sensitive kinase. Proc Natl Acad Sci U S A 92,6915–6919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L., and Spittle C. (2001). Regulation of microtubule-associated proteins. Int Rev Cytol 210,163–226 [DOI] [PubMed] [Google Scholar]
- Gao J., Huo L., Sun X., Liu M., Li D., Dong J.T., et al. (2008). The tumor suppressor CYLD regulates microtubule dynamics and plays a role in cell migration. J Biol Chem 283,8802–8809 [DOI] [PubMed] [Google Scholar]
- Garcia R., Yu C.L., Hudnall A., Catlett R., Nelson K.L., Smithgall T., et al. (1997). Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ 8,1267–1276 [PubMed] [Google Scholar]
- Huo L., Li D., Sun L., Liu M., Shi X., Sun X., et al. (2011). Tat acetylation regulates its actions on microtubule dynamics and apoptosis in T lymphocytes. J Pathol 223,28–36 [DOI] [PubMed] [Google Scholar]
- Lavecchia A., Di Giovanni C., and Novellino E. (2011). STAT-3 inhibitors: state of the art and new horizons for cancer treatment. Curr Med Chem 18,2359–2375 [DOI] [PubMed] [Google Scholar]
- Li D., Xie S., Ren Y., Huo L., Gao J., Cui D., et al. (2011). Microtubule-associated deacetylase HDAC6 promotes angiogenesis by regulating cell migration in an EB1-dependent manner. Protein Cell 2,150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Li D., Sun L., Chen J., Sun X., Zhang L., et al. (2014). Modulation of Eg5 activity contributes to mitotic spindle checkpoint activation and Tat-mediated apoptosis in CD4-positive T-lymphocytes. J Pathol 233,138–147 [DOI] [PubMed] [Google Scholar]
- Martin K., and Helenius A. (1991). Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol 65,232–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney E.M., Helbig K.J., Narayana S.K., Eyre N.S., Aloia A.L., and Beard M.R. (2013). Signal transducer and activator of transcription 3 is a proviral host factor for hepatitis C virus. Hepatol 58,1558–1568 [DOI] [PubMed] [Google Scholar]
- Mitchell T.J., and John S. (2005). Signal transducer and activator of transcription (STAT) signalling and T-cell lymphomas. Immunology 114,301–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D.C., Lin B.H., Lim C.P., Huang G., Zhang T., Poli V., et al. (2006). Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J Cell Biol 172,245–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode K., Dohner K., Binz A., Glass M., Strive T., Bauerfeind R., et al. (2011). Uncoupling uncoating of herpes simplex virus genomes from their nuclear import and gene expression. J Virol 85,4271–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Qiao W., Guo T., Tan J., Li Z., Chen Y., et al. (2010). Microtubule-dependent retrograde transport of bovine immunodeficiency virus. Cell Microbiol 12,1098–1107 [DOI] [PubMed] [Google Scholar]
- Sun X., Li D., Yang Y., Ren Y., Li J., Wang Z., et al. (2012). Microtubule-binding protein CLIP-170 is a mediator of paclitaxel sensitivity. J Pathol 226,666–673 [DOI] [PubMed] [Google Scholar]
- Sun X., Shi X., Liu M., Li D., Zhang L., Liu X., et al. (2011). Mdp3 is a novel microtubule-binding protein that regulates microtubule assembly and stability. Cell Cycle 10,3929–3937 [DOI] [PubMed] [Google Scholar]
- Tala , Sun X., Chen J., Zhang L., Liu N., Zhou J., et al. (2014a). Microtubule stabilization by Mdp3 is partially attributed to its modulation of HDAC6 in addition to its association with tubulin and microtubules. PLoS One 9,e90932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tala , Xie S., Sun X., Sun X., Ran J., Zhang L., et al. (2014b). Microtubule-associated protein mdp3 promotes breast cancer growth and metastasis. Theranostics 4,1052–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma N.K., Dourlat J., Davies A.M., Long A., Liu W.Q., Garbay C., et al. (2009). STAT3-stathmin interactions control microtubule dynamics in migrating T-cells. J Biol Chem 284,12349–12362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S.R., Chaudhury M., and Frank D.A. (2011). STAT3 inhibition by microtubule-targeted drugs: dual molecular effects of chemotherapeutic agents. Mol Cell Pharmacol 3,13–19 [PMC free article] [PubMed] [Google Scholar]
- Walker S.R., Chaudhury M., Nelson E.A., and Frank D.A. (2010). Microtubule-targeted chemotherapeutic agents inhibit signal transducer and activator of transcription 3 (STAT3) signaling. Mol Pharmacol 78,903–908 [DOI] [PubMed] [Google Scholar]
- Wang H., Lafdil F., Wang L., Park O., Yin S., Niu J., et al. (2011). Hepatoprotective versus oncogenic functions of STAT3 in liver tumorigenesis. Am J Pathol 179,714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Dong B., Sun X., Tala , He X., Zhou J., et al. (2014). Identification of a cytoplasmic linker protein as a potential target for neovascularization. Atherosclerosis 233,403–409 [DOI] [PubMed] [Google Scholar]
- Xuan C., Qiao W., Gao J., Liu M., Zhang X., Cao Y., et al. (2007). Regulation of microtubule assembly and stability by the transactivator of transcription protein of Jembrana disease virus. J Biol Chem 282,28800–28806 [DOI] [PubMed] [Google Scholar]
- Zhang T., Kee W.H., Seow K.T., Fung W., and Cao X. (2000). The coiled-coil domain of Stat3 is essential for its SH2 domain-mediated receptor binding and subsequent activation induced by epidermal growth factor and interleukin-6. Mol Cell Biol 20,7132–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Wen Z., and Darnell J.E., Jr. (1994). Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264,95–98 [DOI] [PubMed] [Google Scholar]