Abstract
A myriad of structurally and functionally diverse non-coding RNAs (ncRNAs) have recently been implicated in numerous human diseases including cancer. Small nucleolar RNAs (snoRNAs), the most abundant group of intron-encoded ncRNAs, are classified into two families (box C/D snoRNAs and box H/ACA snoRNAs) and are required for post-transcriptional modifications of ribosomal RNA (rRNA). There is now a growing appreciation that nucleotide modifications on rRNA may impart regulatory potential to the ribosome, however the functional consequence of site-specific snoRNA-guided modifications remains poorly defined. Discovered almost 20 years ago, H/ACA snoRNAs are required for the conversion of specific uridine residues to pseudouridine on rRNA. Interestingly, recent reports indicate that the levels of subsets of H/ACA snoRNAs required for pseudouridine modifications at specific sites on rRNA are altered in several diseases, particularly cancer. In this review, we describe recent advances in understanding the downstream consequences of H/ACA snoRNA-guided modifications on ribosome function, discuss the possible mechanism by which H/ACA snoRNAs may be regulated, and explore prospective expanding functions of H/ACA snoRNAs. Furthermore, we will discuss the potential biological implication of alterations in H/ACA snoRNA expression in several human diseases.
Keywords: H/ACA snoRNA, pseudouridine, ribosomal RNA, RNA modifications, translational control, ribosomopathy, ribosome, dyskerin, post-transcriptional gene regulation, non-coding RNA, hematological abnormalities, cancer, dyskeratosis congenita, leukemia, lymphoma, multiple myeloma
The RNA component of the ribosome, ribosomal RNA (rRNA), undergoes numerous site-specific post-transcriptional nucleotide modifications, several of which are located within functionally important regions of the ribosome (1, 2). Two predominant types of rRNA modifications involve the addition of a methyl group to the 2′-hydroxyl group of a ribose residue (2′-O-methylation) and the isomerization of uridine to pseudouridine (Ψ), a process known as pseudouridylation. In eukaryotes, both types of modifications occur in the nucleolus and require hundreds of small nucleolar RNAs (snoRNAs) (3), ranging in length from 60–300 nucleotides, as well as multicomponent protein complexes, collectively referred to as small nucleolar ribonucleoprotein (snoRNP) complexes (4, 5). Importantly, snoRNP-guided nucleotide modifications are extremely conserved and present in two domains of life, archaea and eukaryotes (6). The highly conserved box C/D snoRNAs, first described in the late 1980’s (7), guide 2′-O-methylation at specific sites on rRNA (8) together with the methyltransferase fibrillarin (9–11). A different highly conserved snoRNP complex consisting of the pseudouridine synthase dyskerin, additional core proteins including NOP10, NHP2 and GAR1, and small ncRNAs known as box H/ACA snoRNAs, guide pseudouridine modifications at specific sites on rRNA (12–15). Although the precise function of distinct types of rRNA modifications is not fully understood, there is now a growing realization that the machinery required for site-specific rRNA modifications is necessary for normal development and is altered in numerous human diseases, particularly cancer (16–22). Together, these findings suggest an important yet perhaps unappreciated functional role of snoRNAs in cellular physiology that, when deregulated, may directly contribute to disease. For the purpose of this review, we will focus exclusively on H/ACA snoRNAs that guide pseudouridine modifications on rRNA. Although rRNA pseudouridine modifications were identified in the mid 1960’s, it was not until studies by the Kiss and Fournier laboratories that H/ACA snoRNAs were implicated in guiding pseudouridine modifications on rRNA (14, 15). These important findings have revolutionized our current understanding of the mechanism by which site-specific rRNA pseudouridylation occurs and have raised several questions as to why such a sophisticated mechanism underlying these modifications exists to modify distinct regions of rRNA. In the following sections, we will provide new insights into H/ACA snoRNA biology and their emerging role in human disease.
Architecture of H/ACA snoRNAs and their function in guiding rRNA pseudouridine modifications
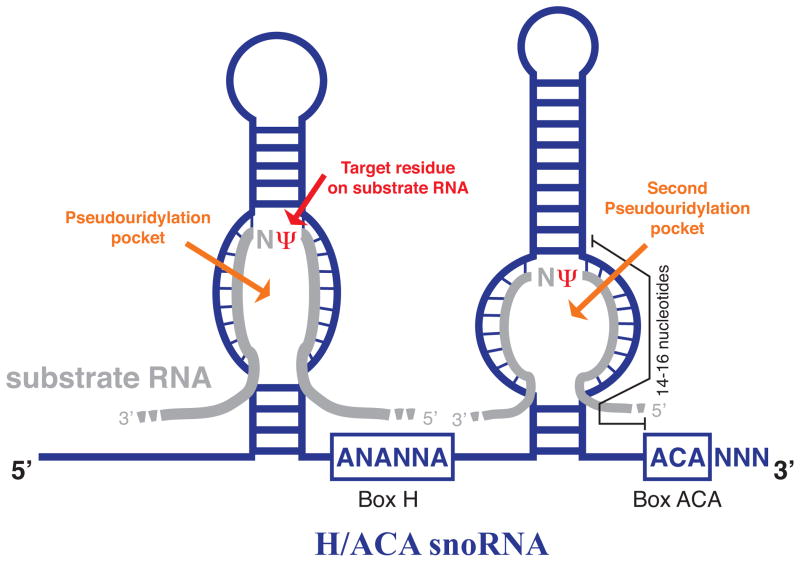
H/ACA snoRNAs are distinctly classified from the other major class of snoRNAs, C/D snoRNAs, according to both sequence and structural features (4). These evolutionarily conserved sequences include box H and box ACA motifs and structural elements such as hairpin structures and pseudouridylation pockets (4, 14, 23) (Figure 1). The secondary structure of H/ACA snoRNAs typically consists of 60 to 75 nucleotide-long hairpins that contain a region referred to as a pseudouridylation pocket, where isomerization of the target uridine residue on the substrate RNA occurs (24). As implied by the name, H/ACA snoRNAs are comprised of two conserved box motifs, the box H motif and the ACA triplet (Figure 1). Recently, it has been demonstrated in human cells that the pseudouridine synthase dyskerin, the enzyme responsible for converting uridine to pseudouridine within the H/ACA snoRNP, has a strong preferential association to the H box sequence of H/ACA snoRNAs (25). Indeed, these findings support the notion that the H box motif represents a protein recognition signal for H/ACA snoRNP-associated proteins and is required for H/ACA snoRNA stability as well as biogenesis of a functionally active H/ACA snoRNP complex (23). In addition, the ACA triplet, located approximately three nucleotides from the 3′ end of the RNA, also appears to be necessary for H/ACA snoRNA stability (4, 23). The selection of a uridine residue for modification on substrate RNA is achieved by base pairing of the H/ACA snoRNA with 3–10 nucleotides on either side of the target uridine, with the exception of one nucleotide adjacent to the target uridine (Figure 1). Several questions arise as to why such distinct secondary structures are required for the function of H/ACA snoRNAs in RNA-guided modifications. One possibility is that the unique spatial and structural arrangement between the guide H/ACA snoRNA and the substrate RNA is required for isomerization of target uridines.
Figure 1. Schematic secondary structure of a box H/ACA snoRNA.
Schematic representation of a box H/ACA snoRNA (blue) containing several evolutionarily conserved structural elements, including a box H (ANANNA) and box ACA motif and two pseudouridylation pockets. Pseudouridylation pockets are shown base pairing to the complementary sequence on substrate RNA (grey). The position of the target uridine modified to pseudouridine (Ψ) on substrate RNA is highlighted in red.
The H/ACA snoRNP protein components, together with H/ACA snoRNAs, are responsible for guiding up to 100 pseudouridine modifications on mammalian rRNAs; rRNA is thus often referred to as the canonical substrate of H/ACA snoRNAs. In eukaryotes, the base-pairing interaction between H/ACA snoRNAs and rRNAs allows the H/ACA snoRNP complex to isomerize distinct uridine residues to pseudouridine (15, 26). Each H/ACA snoRNP consists of a single guide H/ACA snoRNA, a protein complex comprised of the pseudouridine synthase dyskerin and other core snoRNP components NOP10, NHP2, and GAR1. The association of dyskerin, NOP10, and NHP2 with H/ACA snoRNAs appears to be essential for the biogenesis and formation of a catalytically active H/ACA snoRNP complex. For a comprehensive account of the structural and functional organization of H/ACA snoRNPs, please refer to (27–32). Interestingly, the same snoRNP protein complex, along with another class of H/ACA small RNAs, termed H/ACA small cajal body RNAs (scaRNAs), modify uridine residues on small nuclear RNAs (snRNA) that are required for RNA splicing (33, 34). In addition to a role in RNA-guided modifications, it has also been demonstrated that one H/ACA snoRNA, U17/E1, is required for the cleavage and processing of pre-rRNA with no detectable role in guiding rRNA pseudouridylation (35, 36). Although it has not been formally proven, it is also possible that additional H/ACA snoRNAs may be involved in nucleolytic processing of rRNA, or alternatively, that H/ACA snoRNA U17 may function in nucleolytic processing of additional classes of RNAs. Therefore, it is evident that while one specific H/ACA snoRNA is directly involved in the nucleolytic processing of rRNA, the most well characterized role for the overwhelming majority of H/ACA snoRNAs is in guiding pseudouridine modifications within the ribosome.
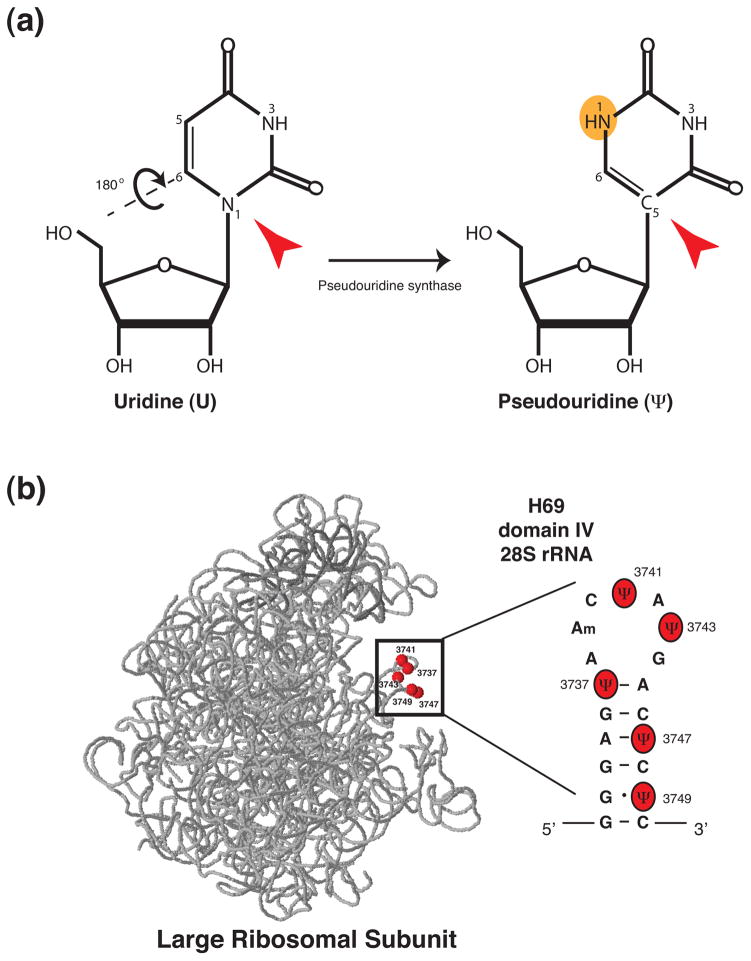
Does pseudouridine confer unique properties to rRNA that may modulate its structure and function? Interestingly, it has been proposed that the presence of an additional hydrogen bond donor site on pseudouridine (Figure 2a) confers unique properties to the RNA backbone, such as enhanced rigidity (37–39). Consistent with the hypothesis that pseudouridine residues on tRNA restrict the mobility of the RNA backbone, thus stabilizing the RNA (40), recent findings in human cells suggest that pseudouridine residues within 28S rRNA may play a conserved role in stabilizing rRNA (41). For example, pseudouridine residues within helix 69 (H69) of human 28S rRNA (Figure 2b) appear to affect RNA stability and structure within this functionally important region of the ribosome (41). Overall, it seems likely that specific conformational changes in the tertiary structure of rRNA, imposed by pseudouridine modifications, may stabilize rRNA, thereby potentially impacting the structure, protein composition, and function of the ribosome. Broadening our knowledge of the mechanisms governing H/ACA snoRNA expression and H/ACA snoRNP biogenesis will undoubtedly prove invaluable in understanding the role of pseudouridine modifications within the ribosome. Furthermore, understanding whether H/ACA snoRNA expression and function may be modulated in a cell and tissue specific manner will provide significant insights into the role of this abundant rRNA modification in maintaining normal cellular physiology—an area of research that, to date, remains very poorly explored.
Figure 2. Pseudouridine, an isomer of uridine, is implicated in stabilizing rRNA.
Pseudouridine (Ψ) is an isomer of uridine (U) and is the only nucleotide to possess a carbon-carbon (C-C) glycosidic bond (C5, highlighted with a red arrowhead). Isomerization of uridine to pseudouridine involves the detachment of the uracil base at position N1 (red arrowhead) and rotation (180°) through the N3-C6 axis. The newly synthesized pseudouridine possesses an additional hydrogen bond donor site, highlighted in orange. (b) 3D model of human 28S (light grey) and 5S rRNA (dark grey) (57) with the position of pseudouridine residues (red) within H69 of 28S rRNA highlighted (in box). Pseudouridine residues within H69 appear to play a conserved role in stabilizing rRNA (41).
Biogenesis and regulation of H/ACA snoRNPs involved in rRNA pseudouridine modifications
The biogenesis of catalytically active H/ACA snoRNPs is a complex process that requires the orchestrated expression, assembly, and nuclear transport of H/ACA snoRNP components, in addition to a number of chaperone proteins (32, 42). One of the first steps required for H/ACA snoRNP biogenesis is the coordinated expression and association of H/ACA snoRNAs with H/ACA snoRNP proteins, namely dyskerin, NHP2, and NOP10. Intriguingly, the majority of H/ACA snoRNA genes appears to lack detectable transcriptional regulatory elements and are located within intronic regions of protein coding genes that are often referred to as H/ACA snoRNA ‘host’ genes (43, 44). The apparent lack of a detectable, independent promoter element in the majority of H/ACA snoRNAs implies that their expression may inevitably be regulated by the transcription of their host gene. Indeed, human H/ACA snoRNAs are processed from the excised and debranched host gene intron by exonucleases (45), and the association of core H/ACA snoRNP proteins such as dyskerin with H/ACA snoRNA motifs define the termini of mature H/ACA snoRNAs (23). The binding of H/ACA snoRNP proteins to H/ACA snoRNAs also appears to be critical for their processing, stability, and nucleolar localization. One striking observation is that in contrast to C/D snoRNAs, processing of human H/ACA snoRNAs does not appear to be coupled with host gene splicing (46). Instead, the recognition of intronic H/ACA snoRNAs and assembly of pre-snoRNPs appears to occur during transcription elongation and, in the case of one H/ACA snoRNA (U64), correct processing and nuclear localization are dependent on RNA polymerase II transcription of the snoRNA precursor (46). Thus, it seems likely that the processing and assembly of H/ACA snoRNPs may be regulated by some components of the RNA polymerase II machinery. In addition to coupling the expression of H/ACA snoRNAs with their binding to protein components of H/ACA snoRNPs, several chaperone proteins (for example, NAF1 and SHQ1) are also implicated in modulating the stepwise assembly and nuclear targeting of mature H/ACA snoRNPs (47–49). Indeed, it appears that a hierarchy of component assembly must occur in order to achieve a functionally active H/ACA snoRNP complex in the nucleolus. A detailed account of H/ACA snoRNA processing and the stepwise assembly and biogenesis of H/ACA snoRNPs can be found in (32, 42).
An important and poorly understood question is whether the formation of a catalytically active H/ACA snoRNP is regulated in response to cellular cues, for instance upon increased demands for cell growth. Evidence suggesting that some steps in the biogenesis of H/ACA snoRNPs may indeed be regulated is supported by the observation that, in most cases, H/ACA snoRNA host genes encode proteins implicated in ribosome biogenesis and function (50). These findings suggest that H/ACA snoRNA expression and H/ACA snoRNP biogenesis may be innately regulated in response to increased demands for protein synthesis. In support of this hypothesis, it is interesting to note that several H/ACA snoRNA host genes including DKC1, encoding the pseudouridine synthase dyskerin, are direct targets of the well-characterized oncogene and transcription factor Myc (51, 52). As Myc plays an essential role in controlling cell growth and protein synthesis (52), it is reasonable to speculate that modulation of Myc transcriptional activity may also regulate H/ACA snoRNP biogenesis in order to accommodate increased demands for protein synthesis. Additionally, it remains poorly understood whether the biogenesis of catalytically active H/ACA snoRNPs, comprised of H/ACA snoRNAs that guide modifications at specific sites on rRNA, are differentially regulated for example, during development or upon oncogenic activation. Some evidence suggesting that H/ACA snoRNPs, comprised of distinct H/ACA snoRNAs, may be differentially modulated is supported by findings that H/ACA snoRNAs display a tissue-specific expression pattern and/or are variably expressed amongst different human tissues (53, 54). Additional findings that the expression of subsets of H/ACA snoRNAs may be modulated by components of the RNA polymerase II machinery also suggest that the biogenesis of distinct H/ACA snoRNPs targeting different sites on rRNA may be tightly regulated. For instance, a post-translational modification in the carboxy-terminal domain (CTD) of RNA polymerase II (RNAPII) in mammal cells facilitates the expression of distinct RNAs, including subsets of H/ACA snoRNAs known to guide pseudouridine modifications on 18S rRNA (e.g., H/ACA SNORA55 and SNORA74) (55). This fascinating observation opens up the exciting and intriguing possibility that H/ACA snoRNPs comprised of distinct H/ACA snoRNAs may differentially guide pseudouridine modifications on rRNA in mammalian cells. In light of recent findings that altered expression of specific subsets of H/ACA snoRNAs manifest in several hematological diseases and malignancies, it is of particular interest to understand whether the corresponding pseudouridine sites on rRNA, guided by these subsets of H/ACA snoRNAs, are also deregulated in human disease. In the next section, we will discuss recent advances in understanding the downstream functional outcomes of H/ACA snoRNA-guided pseudouridine modifications within the ribosome.
Role for H/ACA snoRNA-guided rRNA pseudouridine modifications in translational control
Ribosomes from organisms in all domains of life contain pseudouridine modifications, and in most cases, pseudouridine residues are located within conserved and functionally important regions of rRNA (56). Notably, the number of uridine residues converted to pseudouridine has increased throughout evolution (57), as has the complexity of the machinery necessary for performing pseudouridine modifications. These findings indicate that perhaps in higher organisms an increase in pseudouridine modifications provides an additional regulatory layer in modulating post-transcriptional gene expression mediated by the ribosome. Support for this hypothesis is evident from findings that an apparent lack of pseudouridine modifications within bacterial ribosomes does not adversely affect growth or the overall rate of protein synthesis (58), while in eukaryotes lack of rRNA pseudouridine modifications is not compatible with life (17, 59). Remarkably, approximately 8–10% of total uridine residues in human 28S and 18S rRNA are converted to pseudouridine (50), indicating that this process is highly selective for specific uridine residues. An outstanding question that arises is why are specific uridine residues selectively modified within rRNA? One clue as to the functional importance of pseudouridine modifications on rRNA is that pseudouridine residues tend to cluster within regions of rRNA critical for modulating ribosome activity, namely the decoding center and peptidyl transferase center (PTC). In fact, a role for rRNA pseudouridine modifications in affecting specific aspects of ribosome function is evident from findings in yeast, mouse, and human cells demonstrating that rRNA pseudouridine modifications influence translational fidelity, stop codon recognition, and ribosome-ligand interactions (60–63). Therefore, alterations in rRNA pseudouridylation levels may have profound effects on the ribosome’s ability to accurately and efficiently translate mRNA. These observations add to and support a growing body of evidence that structural components of the ribosome modulate specific aspects of protein synthesis.
In line with the observations that rRNA pseudouridylation plays a conserved role in regulating specific aspects of ribosome function, it is not surprising that a global decrease in rRNA pseudouridine modifications has no apparent overall effect on ribosome biogenesis or the global rate of protein synthesis (64). Indeed, deregulation of the pseudouridine synthase dyskerin, for instance, leads to defects in the translation of specific mRNAs (64–66). Importantly, mRNAs found to be sensitive to changes in rRNA pseudouridylation harbor cis-regulatory elements in their 5′ untranslated regions, such as internal ribosome entry site (IRES) elements. IRES elements are structured RNAs of variable length, originally identified in picornavirus RNAs, that directly engage with the 40S ribosomal subunit during translation initiation (67–69). Intriguingly, rRNA pseudouridine modifications play an evolutionarily conserved role in modulating the ability of ribosomes to bind to RNAs harboring IRES elements (60). IRES-dependent translation is an important RNA-based mode of translation initiation that effectively modulates gene expression during specific cellular events, for instance, mitosis, quiescence, hypoxia, nutrient deprivation, and apoptosis (69–71). Therefore, it is tempting to speculate that rRNA pseudouridine modifications may be particularly important for the temporal translation of distinct subsets of mRNAs. In line with this hypothesis, impairments in the translation of distinct mRNAs known to harbor IRES elements, including the tumor suppressor p53, have been identified upon dyskerin deregulation during oncogenic activation (66). Thus, pseudouridine modifications may impart a dynamic regulatory role to the ribosome, critical for control of gene expression at the translation level.
An interesting question arises as to whether distinct pseudouridine residues located at specific sites on rRNA guided by H/ACA snoRNAs may also modulate the translation of selective mRNAs in mammalian cells. For example, it is possible that modulating the pattern of rRNA pseudouridylation may provide an adaptive mechanism to control cell fate by regulating translation of specific mRNAs. This is a particularly important question to address in light of recent observations demonstrating that subsets of H/ACA snoRNAs are deregulated in several human diseases and is an avenue of H/ACA snoRNA biology that requires further investigation. Overall, although historically thought to exert a housekeeping function in the cytoplasm, emerging evidence suggests that ribosomes exhibit tremendous regulatory potential in modulating gene expression post-transcriptionally (72), and rRNA pseudouridylation appears to play an important role in this process.
Expanding cellular roles of H/ACA snoRNAs
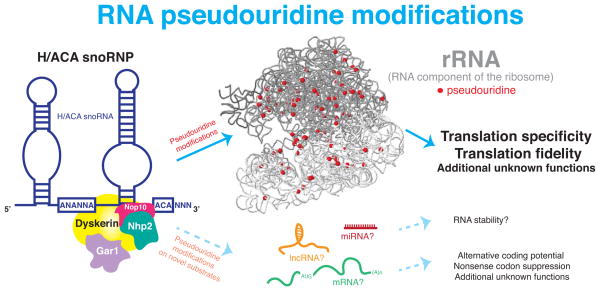
Pseudouridine modifications on rRNA mediated by H/ACA snoRNAs are emerging as evolutionarily conserved and important regulators of ribosome function. Intriguingly, the continuous identification of mammalian H/ACA snoRNA genes from both computational and experimental approaches (25, 73, 74) raise the exciting possibility that the number of novel rRNA target sites are rapidly expanding, and that non-canonical RNA substrates may also likely exist. Furthermore, tremendous advancements in small RNA deep sequencing have aided the identification of seemingly abundant and conserved small ncRNAs derived from H/ACA snoRNAs in eukaryotes (75, 76). Is it possible that H/ACA snoRNAs may have evolved additional regulatory roles in eukaryotes that may provide a new layer of complexity in gene expression control? Exploring the functional diversity of H/ACA snoRNAs in eukaryotes will undoubtedly ignite several new avenues of research and identify perhaps previously uncharacterized functions of H/ACA snoRNAs. In this section, we will discuss the likelihood that novel RNA substrates of H/ACA snoRNAs exist, and explore the potential significance of H/ACA snoRNA derivatives and ncRNAs with H/ACA snoRNA-like features in predominantly RNA-based cellular processes at the nexus of gene regulation (Figure 3 and 4).
Figure 3. Evolutionarily conserved and emerging novel substrates of H/ACA snoRNAs.
Schematic representation of the evolutionarily conserved and well-characterized role of H/ACA snoRNAs in rRNA modifications and potential novel substrates. rRNA pseudouridine modifications (red) guided by evolutionarily conserved H/ACA snoRNPs on human 18S and 28S rRNA (grey) (57) are shown. rRNA pseudouridine modifications play an important role in modulating specific aspects of ribosome function, particularly translation fidelity and specificity. Emerging evidence also suggests that additional RNA substrates (mRNAs, lncRNAs, and miRNAs) may be targeted for pseudouridine modifications by H/ACA snoRNPs.
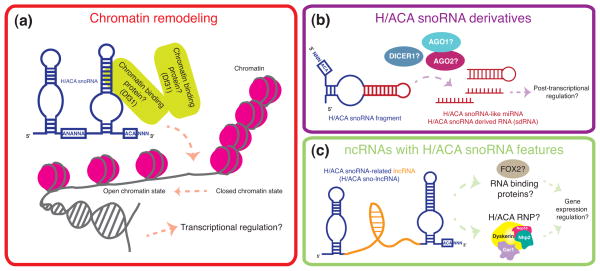
Figure 4. Potential novel and emerging roles of H/ACA snoRNAs.
Schematic representation of the expanding functions of (a) H/ACA snoRNAs, and emerging roles of (b) H/ACA snoRNA derivatives, and (c) ncRNAs with H/ACA snoRNA features. (a) Evidence that subsets of H/ACA snoRNAs are associated with chromatin, independent of H/ACA snoRNP proteins, indicates that H/ACA snoRNAs may play a role in chromatin remodeling. (b) The identification of H/ACA snoRNA derivatives, namely H/ACA snoRNA-like miRNA and H/ACA snoRNA-derived RNAs (sdRNA) suggests a potential role for H/ACA snoRNA derivatives in post-transcriptional gene regulation. (c) H/ACA snoRNA-related lncRNAs may play a role in transcriptional and post-transcriptional gene expression regulation.
New RNA substrates for pseudouridine modifications
Since its discovery in 1957 (77), pseudouridine has, to date, only been identified on non-coding regulatory RNAs, namely tRNA, rRNA, and snRNA. However, increasing evidence indicates that uridine residues on non-canonical RNA substrates, which may include mRNAs and lncRNAs, may also be subject to isomerization. For example, emerging evidence is revealing that human H/ACA snoRNAs, known to guide pseudouridine modifications on rRNA, are predicted to guide modifications at specific uridine residues on distinct mRNAs. These predictions raise several interesting questions regarding the putative role of pseudouridine modifications on mRNA. Importantly, a study by Karijolich and colleagues has uncovered that targeted pseudouridylation of UAA, UAG, or UGA stop codons by a synthetic H/ACA snoRNA leads to highly specific recognition of pseudouridylated stop codons by aminoacyl-tRNAs in yeast (78). This unexpected role for pseudouridine in nonsense codon suppression imparts an enormous capacity of pseudouridine to alter the coding potential of mRNAs (78, 79). Although not yet identified on cellular mRNAs, it is clear that mRNA pseudouridylation may have the ability to post-transcriptionally alter the genetic code and greatly diversify the human proteome (Figure 3). In addition to hypotheses that H/ACA snoRNAs with defined rRNA targets may guide modifications on novel RNA substrates, it is also possible that orphan H/ACA snoRNAs, so-called because their putative RNA targets are not known (80), may also guide isomerization of select uridine residues on rRNA and non-canonical RNA substrates, some of which may include mRNAs. Likewise, it is also conceivable that a recently identified class of H/ACA small RNAs termed AluACA RNAs (81) may participate in pseudouridylation of novel RNA substrates.
Potential role for H/ACA snoRNAs in chromatin remodeling
In addition to the well-characterized role of H/ACA snoRNAs in rRNA pseudouridine modifications from archaea to eukaryotes (82), recent evidence opens up the possibility that H/ACA snoRNAs may also play a role in chromatin biology in eukaryotes. Findings that C/D and H/ACA snoRNAs are enriched in chromatin-associated RNAs (caRNAs) from human and drosophila cells (83), independent of snoRNP proteins suggest a conserved and previously uncharacterized role for H/ACA snoRNAs in chromatin remodeling (Figure 4a). For example, in human fibroblasts several H/ACA snoRNAs (U64, U23, and ACA44) that guide modifications on rRNA, were found to be associated with chromatin. Although the precise function of H/ACA snoRNAs on chromatin was not assessed, the association of C/D snoRNAs with a drosophila chromatin binding protein (Df31) was shown to play a role in maintaining open chromatin structure (83). Whether this unanticipated function occurs in human cells and whether H/ACA snoRNAs can directly modulate chromatin structure remains to be investigated.
H/ACA snoRNA-derived RNAs (sdRNAs) and snoRNA-like miRNAs
Recently, a number of small RNAs containing snoRNA features have been identified in eukaryotes. One class of small RNAs ranging in length from 20–24 nucleotides, supposedly derived from H/ACA snoRNAs, has been designated H/ACA snoRNA-derived RNAs (sdRNAs) and snoRNA-like miRNAs (75, 76, 84). Interestingly, computational analyses indicate that these RNAs are located within and may be processed from H/ACA snoRNA and orphan H/ACA snoRNA genomic regions (84). These findings suggest that H/ACA snoRNAs, involved in rRNA modifications, may serve as precursors of novel small RNAs. Surprisingly, H/ACA sdRNAs and snoRNA-like miRNAs appear to be regulated by or associated with components of the RNAi pathway, such as DICER1 (75), AGO1 and AGO2 (76) (Figure 4b). Although the function of H/ACA snoRNA-derived small RNAs has not been assessed, one snoRNA-like miRNA derived from an H/ACA scaRNA (and designated ACA45 sRNA), was found to play a role in post-transcriptional gene silencing in a similar manner to miRNAs (76). Together these findings provide a previously uncharacterized connection between H/ACA snoRNAs, components of the RNA silencing machinery, and miRNAs. It remains to be addressed whether H/ACA sdRNAs and/or snoRNA-like miRNAs merely represent non-functional degradation products of H/ACA snoRNAs or whether they hold novel regulatory potential in vivo.
snoRNA-related lncRNAs (sno-lncRNAs)
In addition to the H/ACA snoRNA-derived small RNAs described above, human lncRNAs whose ends correspond to sequences of C/D snoRNAs or H/ACA snoRNAs have been identified and termed snoRNA-related lncRNAs (sno-lncRNAs) (85). Interestingly, it appears that sno-lncRNAs are synthesized from introns imbedded between two snoRNAs (for example, H/ACA snoRNAs ACA5 and ACA5C) and are comprised of 5′ and 3′ sequences that correspond to the flanking snoRNAs. Although the potential function of H/ACA sno-lncRNA remains unknown, analysis of C/D sno-lncRNAs indicated that they bind to fox family splicing regulators, thereby possibly modulating some aspects of RNA splicing (85). Whether H/ACA sno-lncRNAs function similarly to lncRNAs (86), associate with H/ACA snoRNP proteins to modify RNA, or possess novel functions remains to be addressed (Figure 4c).
In summary, although the function of H/ACA snoRNA derivatives and ncRNAs with H/ACA snoRNA features remains completely unknown, based on the recent discoveries described above, it is reasonable to envision that these ncRNAs may hold novel regulatory potential in modulating chromatin state or controlling gene expression in a manner similar to that described for miRNAs and lncRNAs (Figure 4). Is it possible that H/ACA snoRNA derivatives and ncRNAs with H/ACA snoRNA features may have evolved to provide an additional regulatory step in decoding the genetic template in higher eukaryotes? As these hypothetical regulatory ncRNAs have not yet been identified in eubacteria or archaea, it is possible that they may provide an additional step in controlling gene expression in multicellular organisms such as mammals, where temporal and spatial gene regulation is critical during development and, when deregulated, may lead to human disease.
Deregulation of H/ACA snoRNAs in human disease
ncRNAs control critical cellular processes and are emerging as key players in human disease (87, 88). Increasing studies suggest that H/ACA snoRNAs that guide pseudouridine modifications at distinct sites on rRNA are frequently altered in hematological disorders and solid tumors (Table 1). Although their contribution to disease remains largely unexplored, one of the most intriguing and puzzling aspects of H/ACA snoRNA deregulation in disease is that only specific subsets of H/ACA snoRNAs appear to be altered. Moreover, the observation that deregulation of H/ACA snoRNA expression in several malignancies is often independent of host gene transcription (89–91) reinforces the notion that this in not simply a global phenomenon and, rather, indicates that H/ACA snoRNA deregulation may be directly implicated in disease. Furthermore, findings that mutations in several genes encoding protein components of H/ACA snoRNPs have been identified in cancer and congenital bone marrow failure syndromes (19, 92–100) suggest that functional perturbations of H/ACA snoRNAs may, in fact, contribute to disease. In this section, we will discuss and provide our perspective on the potential functional consequences of alterations in H/ACA snoRNA expression in human disease.
Table 1.
Illustrative list of H/ACA snoRNAs commonly altered in human disease
| Name | Locus | Substrate | Expression Levels | Disease | Genetic or epigenetic alteration Alteration | Disease | Reference |
|---|---|---|---|---|---|---|---|
| SNORA15 | 7p11 | 18S rRNA | Decrease | Acute Myeloblastic Leukemia | 20 | ||
| Acute Lymphoblastic Leukemia | 20 | ||||||
| Peripheral T-Cell Lymphoma | 21 | ||||||
| X-linked dyskeratosis congenita | 102 | ||||||
| SNORA24 | 4q26 | 18S rRNA | Decrease | Acute Myeloblastic Leukemia | 20 | ||
| Acute Lymphoblastic Leukemia | 20 | ||||||
| Chronic Lymphocytic Leukemia | 91 | ||||||
| Peripheral T-Cell Lymphoma | 21 | ||||||
| X-linked dyskeratosis congenita | 102 | ||||||
| SNORA41 | 2q33 | 18S rRNA | Decrease | Acute Myeloblastic Leukemia | Nucleotide substitution and deletion | Endometrial Cancer | 20, 103 |
| Acute Lymphoblastic Leukemia | 20 | ||||||
| SNORA48 | 17p13 | 28S rRNA | Decrease | Acute Myeloblastic Leukemia | Nucleotide substitution | Breast Cancer | 20, 103 |
| Acute Lymphoblastic Leukemia | 20 | ||||||
| Multiple Myeloma | 89 | ||||||
| X-linked dyskeratosis congenita | 102 | ||||||
| SNORA70C | 9q33 | 18S rRNA | Decrease | Chronic Lymphocytic Leukemia | CpG island hypermethylation | Leukemia cell line | 91, 105 |
| Colorectal cancer cell line | Colorectal cancer cell line | 105 | |||||
| SNORA71C | 20q11 | 18S rRNA | Decrease | Chronic Lymphocytic Leukemia | Gene deletion | Myelofibrosis | 91, 104 |
| Nucleotide substitution | Head and Neck Cancer | 103 | |||||
| SNORA71D | 20q11 | 18S rRNA | Decrease | Acute Myeloblastic Leukemia | Gene deletion | Myelofibrosis | 20, 104 |
| Peripheral T-Cell Lymphoma | Nucleotide substitution | Colorectal Cancer | 21, 103 | ||||
| SNORA74A | 5q31 | 28S rRNA | Increase | Chronic Lymphocytic Leukemia | 91 | ||
| U3 snRNA | Multiple Myeloma | 89 | |||||
| Prostate Cancer | 109 | ||||||
| SNORA42 | 1q22 | 18S rRNA | Increase | Non-Small Cell Lung Cancer | Gene amplification | Non-Small Cell Lung Cancer | 110, 90 |
| Prostate Cancer | 109 | ||||||
| X-linked dyskeratosis congenita | 102 | ||||||
| SNORA64 | 16p13 | 28S rRNA | Increase | Multiple Myeloma | 89 | ||
| Prostate Cancer | 109 | ||||||
| X-linked dyskeratosis congenita | 102 |
Hematological disorders
Alterations in specific subsets of H/ACA snoRNAs have been reported predominantly in hematological disorders, in particular acute myeloblastic and lymphoblastic leukemia, chronic lymphocytic leukemia, T-cell lymphoma, multiple myeloma and the cancer susceptibility and bone marrow failure syndrome X-linked dyskeratosis congenita (X-DC) (20, 21, 89, 91, 101, 102). Intriguingly, as opposed to a global perturbation in H/ACA snoRNA expression in these diseases, only specific subsets of H/ACA snoRNAs appear to be altered. Importantly, the levels of most H/ACA snoRNAs found altered in hematological disorders, as well as many other diseases, appear to be decreased compared to control cells. For instance H/ACA SNORA15 and SNORA24 are two examples of H/ACA snoRNAs that are commonly downregulated in disease (Table 1). In some cases, changes in H/ACA snoRNA expression do not correlate with host gene levels, suggesting that host gene transcriptional changes (89, 91) are uncoupled from and cannot account for alterations in H/ACA snoRNA expression. Therefore, it seems likely that alterations in specific subsets of H/ACA snoRNAs may occur post-transcriptionally and may directly affect H/ACA snoRNP biogenesis and function. Intriguingly, nucleotide deletions or substitutions reported in H/ACA snoRNAs (103, 104) are located within structurally and functionally important regions of H/ACA snoRNAs that may in fact significantly alter the predicted structure of H/ACA snoRNAs (Figure 5) and may hinder their association with H/ACA snoRNP proteins as well as their ability to guide modifications on substrate RNAs. It is also interesting to note that epigenetic alterations involving deletion and hypermethylation of distinct H/ACA snoRNA loci have also been reported in cancer cells, including those of hematological origin (105). Collectively, these findings strongly indicate that deregulated expression of distinct subsets of H/ACA snoRNAs is emerging as a common feature of disease, particularly in those originating from hematopoietic tissue.
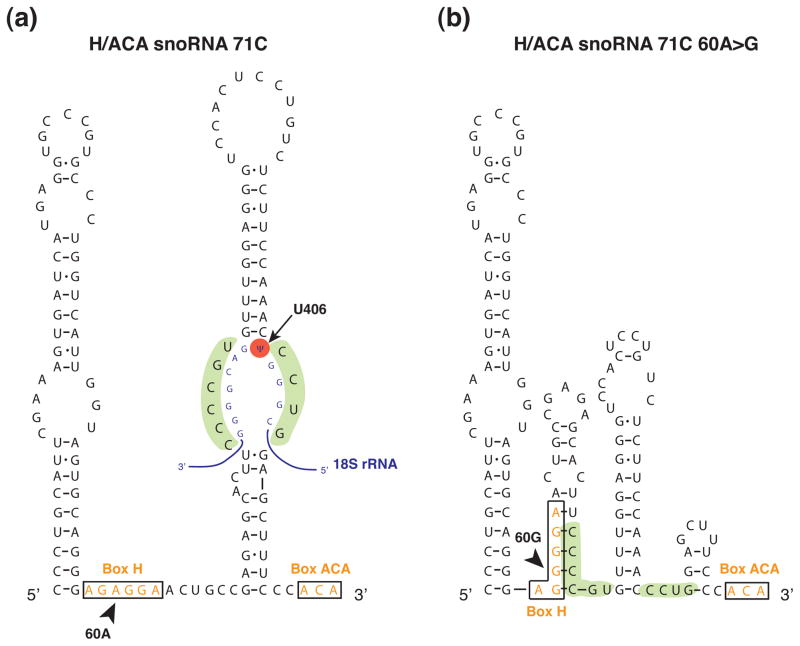
Figure 5. Predicted effect of a single nucleotide substitution on H/ACA snoRNA secondary structure.
Sequence and predicted secondary structure of SNORA71C (a) and a SNORA71C variant found in head and neck cancer, SNORA71 60A>G (103) (b). The position of the substituted nucleotide is indicated with an arrowhead. The nucleotide substitution appears to alter the predicted structure of the pseudouridylation pocket within SNORA71C (highlighted in green) and may likely inhibit base pairing to human 18S rRNA (blue, with position of pseudouridine highlighted in red). The box H and box ACA elements are boxed and shown in orange. Secondary structure predictions were obtained using RNAfold (113) and visual inspection.
One evolutionarily conserved functional outcome of deregulated H/ACA snoRNA expression and biogenesis is impairment in rRNA pseudouridine modifications. Consistent with these findings, alterations in H/ACA snoRNA expression also lead to a functional perturbation in H/ACA snoRNA-guided pseudouridylation at specific sites on rRNA in hematological diseases (102). For example, decreased expression of H/ACA SNORA15 results in a corresponding reduction in pseudouridine modifications at nucleotide U1367 on 18S rRNA in X-DC patient cells. X-DC, a congenital disorder characterized by a wide range of defects, some of which include bone marrow failure, skin abnormalities, hematopoietic malignancies, and pulmonary fibrosis (106) is caused by mutations in DKC1, the gene encoding the evolutionarily conserved pseudouridine synthase dyskerin. DKC1 mutations appear to affect the expression and function of unique subsets of H/ACA snoRNAs in modifying rRNA in X-DC (102) and are consistent with an evolutionarily conserved role of dyskerin and additional H/ACA snoRNP components in modulating H/ACA snoRNA expression and function (12, 107, 108). Although the downstream functional contribution of perturbations in H/ACA snoRNAs and site-specific rRNA pseudouridylation in disease remains relatively unexplored, it is likely that defects in the expression of specific subsets of H/ACA snoRNAs may lead to heterogeneous pools of ribosomes harboring unique differences in rRNA pseudouridine modifications. As mentioned previously, such heterogeneity in ribosome nucleotide modifications may have profound functional implications for translational regulation. Although not formally proven, it is tempting to speculate that a decrease in pseudouridine modifications at distinct sites on rRNA in cells of hematopoietic origin may impinge on the translation of specific mRNAs, such as those encoding important mediators of cell fate, including tumor suppressors and regulators of stem cell differentiation. Importantly, the enzymatic activity of dyskerin in catalyzing pseudouridine modifications on RNA was shown to rescue, to a large extent, hematopoietic stem cell differentiation defects in primary CD34+ hematopoietic progenitor cells from a patient harboring a DKC1 promoter mutation (102). These findings illuminate an important requirement for H/ACA snoRNA-guided RNA pseudouridine modifications in stem cell differentiation and may provide one explanation for why H/ACA snoRNA deregulation is frequently reported in hematological disorders. Given that alterations in H/ACA scaRNAs involved in spliceosomal snRNA modifications are also perturbed in some hematological disorders, it cannot be excluded that defective pseudouridine modifications on additional RNA substrates may also contribute to disease. Similarly, it is also possible that, although functionally uncharacterized, emerging and potential non-canonical functions of H/ACA snoRNAs, H/ACA snoRNA derivatives, and ncRNAs with H/ACA snoRNA features (Figure 4) may also be deregulated in diseases where alterations in H/ACA snoRNAs have been reported.
Solid tumors
It is also emerging that specific subsets of H/ACA snoRNAs are altered in solid tumors of distinct histological origin including prostate (109) and lung (110) (Table 1). Several mechanisms have been identified that may lead to dysfunction of H/ACA snoRNAs in solid tumors, including H/ACA snoRNA nucleotide deletions and substitutions (103). Importantly, the position of these nucleotide deletions and substitutions may have a dramatic impact on the structure, stability, and function of H/ACA snoRNAs as predicted for a nucleotide substitution of one H/ACA snoRNA found in head and neck cancer (Figure 5). Furthermore, CpG island hypermethylation associated with transcriptional silencing of H/ACA snoRNAs in cancer cell lines from colon and renal tumors (105) have also been reported. Intriguingly, although the overwhelming majority of H/ACA snoRNAs altered in disease appear to be decreased, a small subset of H/ACA snoRNAs appear to be increased, most noticeably H/ACA snoRNA42. H/ACA snoRNA42 guides a pseudouridine modification on 18S rRNA and is commonly increased in a number of solid tumors as well as in X-DC patient cells. Interestingly, X-DC patients have a high incidence of solid tumors, particularly head and neck cancer (111). Therefore, it is possible that, although uncommon, increased expression of unique H/ACA snoRNAs may also contribute to disease. For instance, H/ACA snoRNA42 is found significantly upregulated in Non-Small Cell Lung Cancer (NSCLC), amongst other cancers (Table 1). SNORA42 is encoded within a chromosomal region commonly amplified in NSCLC, and high SNORA42 expression in NSCLC patients correlates with poor survival (90). Importantly, gain and loss of function studies suggest that increased H/ACA snoRNA42 expression may be pro-tumorigenic in the lung (90). However, whether the function of H/ACA snoRNA42 in modifying rRNA is increased and may directly contribute to NSCLC in vivo remains to be determined. Likewise, elucidating whether the observed deregulation of additional H/ACA snoRNAs in solid tumors can directly promote tumorigenesis or is merely a secondary effect due to changes in proliferation or host gene transcription requires further investigation. As mentioned earlier, it seems likely that direct perturbations of distinct subsets of H/ACA snoRNAs in solid tumors may lead to the production of ribosomes harboring unique patterns of rRNA pseudouridine modifications. Is it possible that alterations in the pattern of rRNA pseudouridine modifications may contribute to tumorigenesis by modulating specific aspects of ribosome functions? An emerging role for rRNA post-transcriptional modifications in modulating translational specificity and accuracy in cancer is supported by findings that rRNA ribose methylation plays a key role in regulating ribosome activity in breast cancer cells (18, 112). Collectively, these studies illuminate a functionally important role of H/ACA snoRNA-guided rRNA modifications in cellular physiology that, when deregulated, may directly contribute to human disease.
Conclusion
The contribution of the non-coding genome to human disease is a rapidly expanding and important avenue of research, particularly in light of findings that the expression and function of several classes of ncRNAs are frequently altered in diseases such as cancer and developmental and neurological disorders (87). There is now an emerging realization that snoRNAs, whose origins lie in archaea (82), do not simply exhibit housekeeping roles in the production of ribosomes, but instead have evolved to regulate accurate and efficient translation by modulating post-transcriptional rRNA modifications (63). Thus, the function of H/ACA snoRNAs may be similar to primordial RNA-based mechanisms of gene expression regulation present in an ‘RNA world’. Furthermore, it seems likely that H/ACA snoRNA-mediated translational regulation may play a particularly important role in cellular physiology and cell fate decisions. For example, in multicellular organisms it is possible that increased numbers of rRNA pseudouridine modifications guided by H/ACA snoRNAs may constitute an additional layer of complexity in post-transcriptional gene regulation that may occur in a cell and/or tissue specific manner. Broadening our understanding of the mechanisms by which site-specific H/ACA snoRNA-guided rRNA modifications modulate ribosome activity in the context of normal cellular physiology will undoubtedly advance our knowledge of the contribution of deregulated H/ACA snoRNA expression towards hematological disorders and solid tumors. Such advances may allow for the development of novel therapeutic interventions for diseases associated with H/ACA snoRNA dysfunction. Furthermore, recent findings that H/ACA snoRNA expression signatures distinguish specific subtypes of T-cell lymphoma (21) raises the exciting prospects that H/ACA snoRNAs may represent important diagnostic and/or prognostic markers for human disease. Clearly, this is just the tip of the iceberg in understanding how alterations of specific H/ACA snoRNAs may contribute to human disease. An exciting avenue for future studies lies in dissecting the precise downstream functional consequences of deregulated H/ACA snoRNA expression towards human disease and developing therapeutic strategies to counteract perturbations of H/ACA snoRNAs.
Acknowledgments
We are grateful to Craig Stumpf for helpful discussion and to Christine Milentis for editing this manuscript. We apologize to those whose work we were unable to cite. This work was supported by the NIH (R01DK098057 and R01DK098057-07S1 to D.R.) and the Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation (M.M.). D.R. is a Leukemia and Lymphoma Society Research Scholar.
References
- 1.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends in biochemical sciences. 2002;27(7):344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 2.Maden BE. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annual review of biochemistry. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 4.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86(5):823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 5.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. The EMBO journal. 2001;20(14):3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3(3):397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 7.Tyc K, Steitz JA. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. The EMBO journal. 1989;8(10):3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85(7):1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 9.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt EC. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. The EMBO journal. 1991;10(3):573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, et al. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103(3):457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 11.Reichow SL, Hamma T, Ferre-D’Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic acids research. 2007;35(5):1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watkins NJ, Gottschalk A, Neubauer G, Kastner B, Fabrizio P, et al. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4(12):1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Gene Dev. 1998;12(4):527–537. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89(5):799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 15.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89(4):565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 16.Newton K, Petfalski E, Tollervey D, Caceres JF. Fibrillarin is essential for early development and required for accumulation of an intron-encoded small nucleolar RNA in the mouse. Molecular and cellular biology. 2003;23(23):8519–8527. doi: 10.1128/MCB.23.23.8519-8527.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J, Navarrete S, Jasinski M, Vulliamy T, Dokal I, et al. Targeted disruption of Dkc1, the gene mutated in X-linked dyskeratosis congenita, causes embryonic lethality in mice. Oncogene. 2002;21(50):7740–7744. doi: 10.1038/sj.onc.1205969. [DOI] [PubMed] [Google Scholar]
- 18.Marcel V, Ghayad SE, Belin S, Therizols G, Morel AP, et al. p53 Acts as a Safeguard of Translational Control by Regulating Fibrillarin and rRNA Methylation in Cancer. Cancer Cell. 2013;24(3):318–330. doi: 10.1016/j.ccr.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, et al. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19(1):32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 20.Valleron W, Laprevotte E, Gautier EF, Quelen C, Demur C, et al. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2012;26(9):2052–2060. doi: 10.1038/leu.2012.111. [DOI] [PubMed] [Google Scholar]
- 21.Valleron W, Ysebaert L, Berquet L, Fataccioli V, Quelen C, et al. Small nucleolar RNA expression profiling identifies potential prognostic markers in peripheral T-cell lymphoma. Blood. 2012;120(19):3997–4005. doi: 10.1182/blood-2012-06-438135. [DOI] [PubMed] [Google Scholar]
- 22.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nature reviews Cancer. 2012;12(2):84–88. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- 23.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Gene Dev. 1997;11(7):941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 24.Bortolin ML, Ganot P, Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. The EMBO journal. 1999;18(2):457–469. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kishore S, Gruber AR, Jedlinski DJ, Syed AP, Jorjani H, Zavolan M. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biology. 2013;14(5) doi: 10.1186/gb-2013-14-5-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiss AM, Jady BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol. 2004;24(13):5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rashid R, Liang B, Baker DL, Youssef OA, He Y, et al. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Molecular cell. 2006;21(2):249–260. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443(7109):302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 29.Duan J, Li L, Lu J, Wang W, Ye K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Molecular cell. 2009;34(4):427–439. doi: 10.1016/j.molcel.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Hamma T, Reichow SL, Varani G, Ferre-D’Amare AR. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat Struct Mol Biol. 2005;12(12):1101–1107. doi: 10.1038/nsmb1036. [DOI] [PubMed] [Google Scholar]
- 31.Hamma T, Ferre-D’Amare AR. The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. The Journal of biological chemistry. 2010;285(2):805–809. doi: 10.1074/jbc.R109.076893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harbor symposia on quantitative biology. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 33.Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2’-O-methylation and pseudouridylation guide RNAs. The EMBO journal. 2002;21(11):2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terns M, Terns R. Noncoding RNAs of the H/ACA family. Cold Spring Harbor symposia on quantitative biology. 2006;71:395–405. doi: 10.1101/sqb.2006.71.034. [DOI] [PubMed] [Google Scholar]
- 35.Morrissey JP, Tollervey D. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Molecular and cellular biology. 1993;13(4):2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atzorn V, Fragapane P, Kiss T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Molecular and cellular biology. 2004;24(4):1769–1778. doi: 10.1128/MCB.24.4.1769-1778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Newby MI, Greenbaum NL. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. P Natl Acad Sci USA. 2002;99(20):12697–12702. doi: 10.1073/pnas.202477199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z, Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic acids research. 2014;42(5):3492–3501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49(5):341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 40.Yarian CS, Basti MM, Cain RJ, Ansari G, Guenther RH, et al. Structural and functional roles of the N1- and N3-protons of psi at tRNA’s position 39. Nucleic acids research. 1999;27(17):3543–3549. doi: 10.1093/nar/27.17.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sumita M, Desaulniers JP, Chang YC, Chui HM, Clos L, 2nd, Chow CS. Effects of nucleotide substitution and modification on the stability and structure of helix 69 from 28S rRNA. RNA. 2005;11(9):1420–1429. doi: 10.1261/rna.2320605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol. 2002;14(3):319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 43.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109(2):145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 44.Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94(2):83–88. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Gene Dev. 1995;9(11):1411–1424. doi: 10.1101/gad.9.11.1411. [DOI] [PubMed] [Google Scholar]
- 46.Richard P, Kiss AM, Darzacq X, Kiss T. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Molecular and cellular biology. 2006;26(7):2540–2549. doi: 10.1128/MCB.26.7.2540-2549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. Journal of Cell Biology. 2006;173(2):207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grozdanov PN, Roy S, Kittur N, Meier UT. SHQ1 is required prior to NAF1 for assembly of H/ACA small nucleolar and telomerase RNPs. Rna-a Publication of the Rna Society. 2009;15(6):1188–1197. doi: 10.1261/rna.1532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walbott H, Machado-Pinilla R, Liger D, Blaud M, Rety S, et al. The H/ACA RNP assembly factor SHQ1 functions as an RNA mimic. Gene Dev. 2011;25(22):2398–2408. doi: 10.1101/gad.176834.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic acids research. 2006;34(Database issue):D158–162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alawi F, Lee M. DKC1 is an evolutionarily conserved c-Myc target. Faseb Journal. 2007;21(6):A1155–A1155. [Google Scholar]
- 52.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nature Reviews Cancer. 2010;10(4):301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 53.Cavaille J, Buiting K, Kiefmann M, Lalande M, Brannan CI, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. P Natl Acad Sci USA. 2000;97(26):14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castle JC, Armour CD, Lower M, Haynor D, Biery M, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One. 2010;5(7):e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sims RJ, Rojas LA, Beck D, Bonasio R, Schuller R, et al. The C-Terminal Domain of RNA Polymerase II Is Modified by Site-Specific Methylation. Science. 2011;332(6025):99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lane BG, Ofengand J, Gray MW. Pseudouridine and O2’-methylated nucleosides. Significance of their selective occurrence in rRNA domains that function in ribosome-catalyzed synthesis of the peptide bonds in proteins. Biochimie. 1995;77(1–2):7–15. doi: 10.1016/0300-9084(96)88098-9. [DOI] [PubMed] [Google Scholar]
- 57.Piekna-Przybylska D, Decatur WA, Fournier MJ. The 3D rRNA modification maps database: with interactive tools for ribosome analysis. Nucleic acids research. 2008;36(Database issue):D178–183. doi: 10.1093/nar/gkm855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ofengand J. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS letters. 2002;514(1):17–25. doi: 10.1016/s0014-5793(02)02305-0. [DOI] [PubMed] [Google Scholar]
- 59.Jiang W, Middleton K, Yoon HJ, Fouquet C, Carbon J. An essential yeast protein, CBF5p, binds in vitro to centromeres and microtubules. Molecular and cellular biology. 1993;13(8):4884–4893. doi: 10.1128/mcb.13.8.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, et al. rRNA Pseudouridylation Defects Affect Ribosomal Ligand Binding and Translational Fidelity from Yeast to Human Cells. Molecular cell. 2011;44(4):660–666. doi: 10.1016/j.molcel.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Molecular cell. 2003;11(2):425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 62.Baudin-Baillieu A, Fabret C, Liang XH, Piekna-Przybylska D, Fournier MJ, Rousset JP. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic acids research. 2009;37(22):7665–7677. doi: 10.1093/nar/gkp816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McMahon M, Bellodi C, Ruggero D. The ‚ÄúFifth‚Äù RNA Nucleotide: A Role for Ribosomal RNA Pseudouridylation in Control of Gene Expression at the Translational Level. In: Dinman JD, editor. Biophysical approaches to translational control of gene expression. Springer; New York: 2013. pp. 253–288. [Google Scholar]
- 64.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, et al. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312(5775):902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 65.Montanaro L, Calienni M, Bertoni S, Rocchi L, Sansone P, et al. Novel dyskerin-mediated mechanism of p53 inactivation through defective mRNA translation. Cancer Res. 2010;70(11):4767–4777. doi: 10.1158/0008-5472.CAN-09-4024. [DOI] [PubMed] [Google Scholar]
- 66.Bellodi C, Kopmar N, Ruggero D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010;29(11):1865–1876. doi: 10.1038/emboj.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5’ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. Journal of virology. 1988;62(8):2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334(6180):320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 69.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15(13):1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 70.Lewis SM, Holcik M. IRES in distress: translational regulation of the inhibitor of apoptosis proteins XIAP and HIAP2 during cell stress. Cell Death Differ. 2005;12(6):547–553. doi: 10.1038/sj.cdd.4401602. [DOI] [PubMed] [Google Scholar]
- 71.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6(4):318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 72.Xue SF, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Bio. 2012;13(6):355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gu AD, Zhou H, Yu CH, Qu LH. A novel experimental approach for systematic identification of box H/ACA snoRNAs from eukaryotes. Nucleic acids research. 2005;33(22):e194. doi: 10.1093/nar/gni185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schattner P, Barberan-Soler S, Lowe TM. A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA. 2006;12(1):15–25. doi: 10.1261/rna.2210406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15(7):1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, et al. A human snoRNA with microRNA-like functions. Molecular cell. 2008;32(4):519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 77.Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. The Journal of biological chemistry. 1957;227(2):907–915. [PubMed] [Google Scholar]
- 78.Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474(7351):395–398. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fernandez IS, Ng CL, Kelley AC, Wu G, Yu YT, Ramakrishnan V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500(7460):107–110. doi: 10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vitali P, Royo H, Seitz H, Bachellerie JP, Huttenhofer A, Cavaille J. Identification of 13 novel human modification guide RNAs. Nucleic acids research. 2003;31(22):6543–6551. doi: 10.1093/nar/gkg849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jady BE, Ketele A, Kiss T. Human intron-encoded Alu RNAs are processed and packaged into Wdr79-associated nucleoplasmic box H/ACA RNPs. Gene Dev. 2012;26(17):1897–1910. doi: 10.1101/gad.197467.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Terns MP, Terns RM. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 2002;10(1–2):17–39. [PMC free article] [PubMed] [Google Scholar]
- 83.Schubert T, Pusch MC, Diermeier S, Benes V, Kremmer E, et al. Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin. Molecular cell. 2012;48(3):434–444. doi: 10.1016/j.molcel.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 84.Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol. 2009;5(9):e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, et al. Long noncoding RNAs with snoRNA ends. Molecular cell. 2012;48(2):219–230. doi: 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 86.Baker M. Long noncoding RNAs: the search for function. Nature Methods. 2011;8(5):379–383. [Google Scholar]
- 87.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 88.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. The Journal of pathology. 2010;220(2):126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 89.Ronchetti D, Todoerti K, Tuana G, Agnelli L, Mosca L, et al. The expression pattern of small nucleolar and small Cajal body-specific RNAs characterizes distinct molecular subtypes of multiple myeloma. Blood Cancer J. 2012;2:e96. doi: 10.1038/bcj.2012.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mei YP, Liao JP, Shen J, Yu L, Liu BL, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31(22):2794–2804. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ronchetti D, Mosca L, Cutrona G, Tuana G, Gentile M, et al. Small nucleolar RNAs as new biomarkers in chronic lymphocytic leukemia. Bmc Med Genomics. 2013;6 doi: 10.1186/1755-8794-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rahman N. Realizing the promise of cancer predisposition genes. Nature. 2014;505(7483):302–308. doi: 10.1038/nature12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, et al. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Gene Dev. 2011;25(1):11–16. doi: 10.1101/gad.2006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Walne AJ, Vulliamy T, Marrone A, Beswick R, Kirwan M, et al. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Human Molecular Genetics. 2007;16(13):1619–1629. doi: 10.1093/hmg/ddm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vulliamy T, Beswick R, Kirwan M, Marrone A, Digweed M, et al. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc Natl Acad Sci U S A. 2008;105(23):8073–8078. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Knight SW, Heiss NS, Vulliamy TJ, Aalfs CM, McMahon C, et al. Unexplained aplastic anaemia, immunodeficiency, and cerebellar hypoplasia (Hoyeraal-Hreidarsson syndrome) due to mutations in the dyskeratosis congenita gene, DKC1. Br J Haematol. 1999;107(2):335–339. doi: 10.1046/j.1365-2141.1999.01690.x. [DOI] [PubMed] [Google Scholar]
- 97.Bellodi C, Krasnykh O, Haynes N, Theodoropoulou M, Peng G, et al. Loss of function of the tumor suppressor DKC1 perturbs p27 translation control and contributes to pituitary tumorigenesis. Cancer Res. 2010;70(14):6026–6035. doi: 10.1158/0008-5472.CAN-09-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2011;39(Database issue):D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Teittinen KJ, Laiho A, Uusimaki A, Pursiheimo JP, Gyenesei A, Lohi O. Expression of small nucleolar RNAs in leukemic cells. Cell Oncol (Dordr) 2012 doi: 10.1007/s13402-012-0113-5. [DOI] [PubMed] [Google Scholar]
- 102.Bellodi C, McMahon M, Contreras A, Juliano D, Kopmar N, et al. H/ACA Small RNA Dysfunctions in Disease Reveal Key Roles for Noncoding RNA Modifications in Hematopoietic Stem Cell Differentiation. Cell Rep. 2013;3(5):1493–1502. doi: 10.1016/j.celrep.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brecqueville M, Rey J, Devillier R, Guille A, Gillet R, et al. Array comparative genomic hybridization and sequencing of 23 genes in 80 patients with myelofibrosis at chronic or acute phase. Haematologica. 2014;99(1):37–45. doi: 10.3324/haematol.2013.091454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ferreira HJ, Heyn H, Moutinho C, Esteller M. CpG island hypermethylation-associated silencing of small nucleolar RNAs in human cancer. RNA biology. 2012;9(6):881–890. doi: 10.4161/rna.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dokal I. Dyskeratosis congenita in all its forms. Br J Haematol. 2000;110(4):768–779. doi: 10.1046/j.1365-2141.2000.02109.x. [DOI] [PubMed] [Google Scholar]
- 107.Gu BW, Bessler M, Mason PJ. A pathogenic dyskerin mutation impairs proliferation and activates a DNA damage response independent of telomere length in mice. Proc Natl Acad Sci U S A. 2008;105(29):10173–10178. doi: 10.1073/pnas.0803559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Alawi F, Lin P. Dyskerin is Required for Tumor Cell Growth Through Mechanisms that are Independent of its Role in Telomerase and only Partially Related to its Function in Precursor rRNA Processing. Mol Carcinogen. 2011;50(5):334–345. doi: 10.1002/mc.20715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, et al. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31(8):978–991. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 110.Liao JP, Yu L, Mei YP, Guarnera M, Shen J, et al. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer. 2010;9 doi: 10.1186/1476-4598-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenita. Blood. 2009;113(26):6549–6557. doi: 10.1182/blood-2008-12-192880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Basu A, Das P, Chaudhuri S, Bevilacqua E, Andrews J, et al. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular Internal Ribosome Entry Sites. Molecular and cellular biology. 2011 doi: 10.1128/MCB.05804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Denman RB. Using RNAFOLD to predict the activity of small catalytic RNAs. Biotechniques. 1993;15(6):1090–1095. [PubMed] [Google Scholar]