Abstract
Over the past decade, our understanding of T cell activation, differentiation and function has markedly expanded, providing a greater appreciation of the signals and pathways that regulate these processes. It has become clear that evolutionarily conserved pathways that regulate stress responses, metabolism, autophagy and survival have crucial and specific roles in regulating T cell responses. Recent studies suggest that the metabolic pathways involving MYC, hypoxia-inducible factor 1α (HIF1α), AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) are activated upon antigen recognition and that they are required for directing the consequences of T cell receptor engagement. The purpose of this Review is to provide an integrated view of the role of these metabolic pathways and of canonical T cell signalling pathways in regulating the outcome of T cell responses.
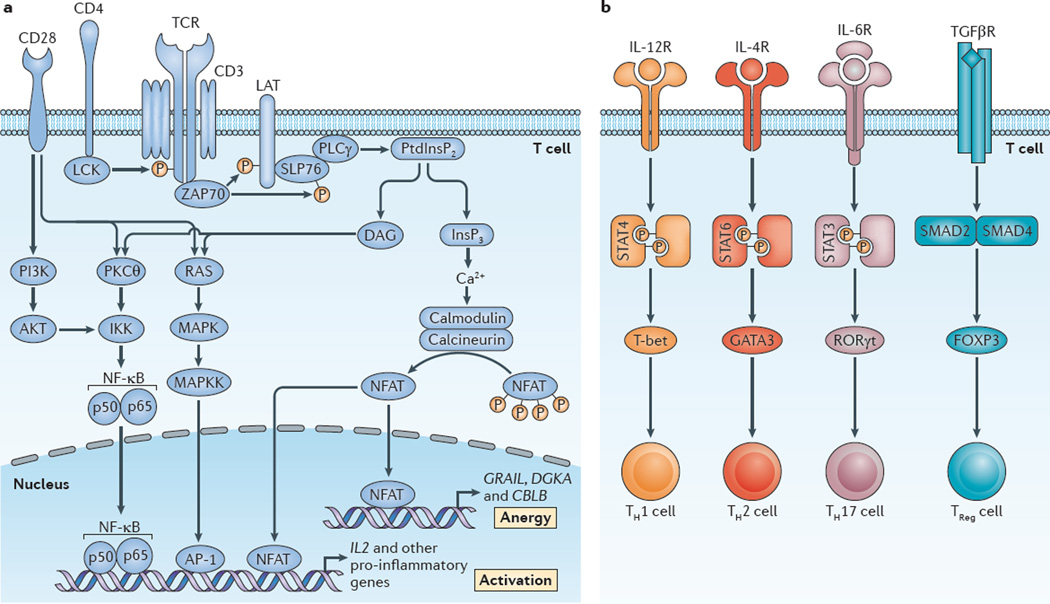
T cell receptor (TCR) engagement by peptide–MHC complexes initiates a multitude of signalling programmes that prepare the cell for differentiation, proliferation and effector function. The canonical signalling pathways that lead to activation-induced transcription are mediated by nuclear factor-κB (NF-κB), activator protein 1 (AP-1) and nuclear factor of activated T cells (NFAT). These three pathways collaborate to promote the expression of effector molecules that are crucial for T cell function1–7 (FIG. 1a). It is generally thought that TCR-induced signalling only leads to T cell activation when it occurs in the context of a second co-stimulatory signal, such as the ligation of CD28 (REF. 8). The precise pathways that mediate CD28-induced co-stimulation have not been completely elucidated. However, one such model posits that TCR-induced NFAT activation leads to T cell anergy, whereas in the context of co-stimulation, NFAT and AP-1 collaborate to promote full T cell activation3. Likewise, CD28 signalling leads to the activation of phosphoinositide 3-kinase (PI3K) and the subsequent activation of mammalian target of rapa-mycin (mTOR)9. In addition to co-stimulation, further signals from the microenvironment influence the outcome of TCR ligation. For example, specific cytokines are required to promote the differentiation of naive CD4+ T cells into various T helper (TH) cell subsets (FIG. 1b). Thus, immuno-logical inputs in the form of antigen recognition, co-stimulatory ligand engagement and cytokine stimulation guide the outcome of T cell activation and differentiation.
Figure 1. Canonical T cell signalling pathways: signal 1 and signal 2.
a | Signal 1 (T cell receptor (TCR) engagement) in the setting of signal 2 (co-stimulation; depicted as CD28) leads to full T cell activation122. This is facilitated by the activation of three canonical transcription factors — nuclear factor-κB (NF-κB), activator protein 1 (AP-1) and nuclear factor of activated T cells (NFAT)6,7,123,124. This, in turn, leads to the expression of multiple cytokines, chemokines and cell surface receptors, all of which promote T cell activation and proliferation3. Alternatively, TCR recognition alone (in the absence of co-stimulation) leads to an ‘off’ signal in the form of T cell anergy5,125. Under these conditions, NFAT is activated in the absence of full AP-1 activation, which leads to the expression of genes such as diacylglycerol kinase-α (DGKA) and the E3 ubiquitin-protein ligases CBLB and GRAIL (which encodes gene related to anergy in lymphocytes; also known as RNF128), which inhibit full T cell activation3. b | Upon T cell activation, cytokines in the T cell microenvironment determine the outcome of antigen recognition with regard to effector T cell differentiation51. As shown for CD4+ T cells, interleukin-12 (IL-12), IL-4 and IL-6 activate signal transducer and activator of transcription 4 (STAT4), STAT6 and STAT3, respectively. This leads to the expression of T-bet, GATA-binding protein 3 (GATA3) and retinoic acid receptor-related orphan receptor-γt (RORγt), which facilitates the generation of T helper 1 (TH1), TH2 and TH17 cells. Alternatively, transforming growth factor-β (TGFβ) signalling through SMAD2-SMAD4 promotes the expression of forkhead box P3 (FOXP3) and the generation of regulatory T (TReg) cells. DAG, diacylglycerol; IKK, inhibitor of NF-κB kinase; InsP3, inositol-1,4,5-trisphosphate; LAT, linker for activation of T cells; MAPK, mitogen-activated protein kinase; MAPKK, MAPK kinase; PI3K, phosphoinositide 3-kinase; PKCθ, protein kinase Cθ; PLCγ, phospholipase Cγ; PtdInsP2, phosphatidylinositol-4,5-bisphosphate; SLP76, SH2 domain-containing leukocyte protein of 76 kDa (also known as LCP2); ZAP70, ζ-chain-associated protein kinase of 70 kDa.
Recently, the signalling pathways that control cellular metabolism have been shown to have a crucial role in dictating the outcome of T cell activation. Overall, this requirement for the coordination of T cell metabolism and T cell function reflects two important features of the T cell response: the ability of low frequency, antigen-specific naive T cells to rapidly increase in number in response to a pathogen, and their ability to generate long-lived memory T cells or regulatory T (TReg) cells that can modulate immune responses. In this Review, we aim to integrate the metabolic pathways with the canonical T cell signalling pathways to provide a comprehensive view of the pathways that regulate T cell immunity. This reveals potential new pharmacological targets for enhancing or inhibiting specific T cell responses.
Regulation of cellular metabolism
Cellular metabolism provides the means by which cells store and use macromolecules that are necessary for growth and for the generation of energy. Depending on nutrient availability and external or intracellular cues, cells can use different substrates and distinct pathways to produce energy. Likewise, cellular metabolism is dictated by the specific function of a cell. Glycolysis is a metabolic pathway by which the catabolism of six-carbon sugars (glucose) produces a net sum of two molecules of ATP and two of pyruvate from each molecule of glucose10. In the presence of oxygen, pyruvate derivatives enter the tricarboxylic acid cycle (TCA cycle) and promote the oxidative phosphorylation of energy inter mediates in the mitochondrial matrix to generate a total of ~30 ATP molecules (TABLE 1). If oxygen is unavailable, the two molecules of pyruvate that are generated from glyco lysis can be converted to lactate, which dramatically reduces the ATP yield but still provides an energy source for the cell10. In response to environmental cues, there are specific drivers of cellular metabolism that regulate the expression of enzymes that are crucial for various metabolic processes.
Table 1.
A summary of metabolic pathways and molecules
| Metabolic process | Description | Substrate(s) | Crucial components |
|---|---|---|---|
| Glycolysis |
|
Glucose |
|
| TCA cycle | A series of enzyme-catalysed chemical reactions that result in the reduction of NAD+ molecules, which can be substrates of the electron transport chain during oxidative phosphorylation | Acetyl-CoA (synthesized from sugars, lipids or amino acids) |
|
| Oxidative phosphorylation | Oxidation of energy intermediates during the electron transport chain establishes a proton gradient across the mitochondrial inner membrane, which drives ATP synthesis |
|
|
| Fatty acid oxidation | Catabolism of fatty acids into acetyl-CoA, which can be further broken down in the TCA cycle for ATP synthesis in the electron transport chain | Fatty acids |
|
| Glutaminolysis |
|
Glutamine |
|
| Fatty acid synthesis | Anabolic process leading to the generation of fatty acids from acetyl-CoA and malonyl-CoA precursors |
|
|
ACC1, acetyl-CoA carboxylase 1; CD98, a heterodimeric glutamine exchanger comprising SLC3A2 and SLC7A5; CoA, coenzyme A; CPT1A, carnitine palmitoyl-transferase 1A; GLUT, glucose transporter; PDK1, pyruvate dehydrogenase kinase 1; SREBPs, sterol regulatory element-binding proteins; TCA cycle, tricarboxylic acid cycle.
Glycolysis is promoted by the upregulation of MYC, which is a basic helix–loop–helix leucine zipper transcription factor (TABLE 2). MYC promotes the expression of glucose transporter type 1 (GLUT1; also known as SLC2A1), pyruvate kinase, lactate dehydrogenase A (LDHA) and hexokinase 2, which are required for glucose uptake and for the rate-limiting steps of glycolysis11,12. In addition, MYC promotes the expression of both glutaminase and glutamine transporters13, and further promotes glutaminolysis by transcriptionally repressing the microRNAs miR-23a and miR-23b, which allows for the increased expression of glutaminase14. Furthermore, MYC has also been found to have a role in promoting mitochondrial biogenesis15.
Table 2.
Regulators of metabolism are also regulators of T cell differentiation and function
| Regulator | Description | Metabolic function | T cell function |
|---|---|---|---|
| MYC | Transcription factor |
|
|
| HIF1α | Transcription factor | Under hypoxic conditions, regulates gene expression necessary for survival in low oxygen primarily by promoting metabolic switch to glycolysis | |
| AMPK | Serine/threonine kinase |
|
|
| mTOR | Serine/threonine kinase | Regulates cell growth, proliferation, survival and metabolic gene expression, resulting in enhanced glycolysis and lipid biosynthesis |
AMPK, AMP-activated protein kinase; FOXP3, forkhead box P3; HIF1α, hypoxia-inducible factor 1α; mTOR, mammalian target of rapamycin; RORγt, retinoic acid receptor-related orphan receptor-γt; TCR, T cell receptor; TH cell, T helper cell; TReg cell, regulatory T cell.
Glycolysis is also regulated by hypoxia-inducible factor 1α (HIF1α), which is a heterodimeric basic helix–loop–helix and Per–Arnt–Sim (PAS) domain-containing transcription factor that, during hypoxia, binds to cis-acting hypoxia-response elements and leads to the transcription of numerous genes that are important for cell survival in low oxygen conditions16 (TABLE 2). Not surprisingly, these genes include those encoding enzymes that are required for the glycolytic pathway17. In addition, HIF1α promotes the expression of GLUT1 (REF. 18) and enforces ATP synthesis by glycolysis, rather than oxidative phosphorylation, by upregulating pyruvate dehydrogenase kinase 1 (PDK1), which is an enzyme that inhibits the entry of pyruvate into the TCA cycle19,20.
HIF1α expression is not only regulated by oxygen levels but also depends on external cues that are integrated by mTOR activity21. mTOR is an evolutionarily conserved serine/threonine kinase that integrates a diverse array of environmental cues to regulate growth, survival and proliferation22 (TABLE 2). mT O R is present in two distinct protein complexes — mTOR complex 1 (mTORC1) and mTORC2 — that each have unique downstream targets and functions. Activation of mTORC1 occurs by growth factor stimulation of PI3K, which initiates a signalling cascade that results in the inhibitory phosphorylation of the mTORC1 repressor tuberous sclerosis 2 (TSC2; also known as tuberin) by the kinase AKT23. In addition to growth factors, amino acids also activate mTORC1 and this leads to recruitment of mTOR to the lysosomal surface where it can interact with, and become activated by, its activator RAS homologue enriched in brain (RHEB)24–26. The mechanisms that regulate mTORC2 activation are less clear than those for mTORC1. However, it is known that growth factor stimulation enhances mTORC2 activity and recent studies have implicated a role for the association of the mTORC2 complex with ribosomes in promoting its activation27.
The activity of mTORC1 enhances HIF1α expression at both the transcriptional and translational level, and thereby stimulates glycolysis and glucose transport28. The importance of HIF1α in mediating mTORC1-enhanced glycolysis is illustrated by the observation that small interfering RNA (siRNA)-mediated inhibition of Hif1a expression in cells that express constitutively active mTORC1 (Tsc2−/− cells) abrogates the expression of the glycolytic factors GLUT1, phosphofructokinase 1 and PDK1 (REF. 28). Interestingly, a recent report suggests that MYC activity is, in part, regulated by mTORC2 (REF. 29). It was observed that mTORC2 activity leads to the acetylation of forkhead box protein O1 (FOXO1), which initiates the release of MYC from a suppressive miR-34c–dependent network29.
Although MYC, HIF1α and mTOR signalling promote an increased metabolic output by cells, other regulators promote energy conservation during times of limited resources. One such regulator is AMP-activated protein kinase (AMPK), which is a heterotrimeric serine/threonine kinase complex that monitors cellular energy levels (TABLE 2). The binding of AMP or ADP to AMPK induces its phosphorylation and activation by upstream kinases30,31. AMPK activation enhances glucose uptake and, at the same time, inhibits glucose, glycogen and fatty acid synthesis31. This occurs through the phosphoryl-ation and inhibition of acetyl-CoA carboxylase 1 (ACC1) and the inhibition of the lipogenic transcription factor sterol regulatory element-binding protein 1 (SREBP1; also known as SREBF1)32. In addition, AMPK promotes fatty acid oxidation through the phosphorylation and inhibition of ACC2. This results in the enhanced expression of carnitine palmitoyltransferase 1A (CPT1A), which is the rate-limiting factor in mitochondrial lipid uptake10. AMPK also enhances mitochondrial bio genesis and oxidative metabolism by promoting the transcriptional activity of peroxisome proliferator-activated receptor-γ co-activator 1α (PGC1α; also known as PPARGC1A)31. Thus, AMPK regulates cell metabolism to limit energy expenditure and replenish ATP production. AMPK activity can also diminish mTORC1 signalling through the phosphorylation of TSC2 and regulatory-associated protein of mTOR (RAPTOR; also known as RPTOR), which is a crucial component of mTORC1 (REFS 33,34). Under conditions of prolonged energy deprivation (starvation), AMPK promotes autophagy) by phos-phorylating and activating the serine/threonine protein kinase Unc-51-like kinase 1 (ULK1)35. Thus, AMPK shuts down energy-demanding synthetic pathways but promotes mechanisms that generate energy — such as glycolysis, oxidative phosphorylation and autophagy — as a means of deriving substrates from within the cell. By contrast, deficiency of the AMPK activator liver kinase 1 (LKB1; also known as STK11), and therefore loss of AMPK activation, promotes enhanced glucose and glutamine metabolism through the mTORC1-dependent upregulation of HIF1α during normoxic conditions36.
Interestingly, although mTORC1 activity has been shown to increase glycolysis through regulation of HIF1α, mTORC1 activity can also promote oxidative phosphorylation. This occurs through the increased interaction of the transcriptional repres-sor yin and yang 1 (YY1) with PGC1α, which induces the expression of mitochondrial genes37. In addition, mTORC1 promotes lipid biosynthesis by enhancing the transcription and translation of SREBP1 (REF. 28) but limits fatty acid oxidation through the inhibition of CPT1A38. The activity of mTORC1 has also been shown to promote nucleotide synthesis. This process occurs through the activation of ribosomal protein S6 kinase β1, which post-translationally regulates de novo pyrimidine synthesis39,40.
T cells have a specialized metabolism
Most cells use oxidative phosphorylation to maximize ATP production but activated T cells (and cancer cells) mainly generate ATP through glycolysis41,42. This use of glycolysis in the presence of oxygen was first described by Otto Warburg for cancer cells and it is therefore referred to as the Warburg effect43. Although glycolysis provides less ATP than oxidative phosphorylation, it has been proposed that avoiding oxidative phosphorylation allows for the generation of substrates that are required for the synthesis of amino acids, nucleic acids and lipids, all of which are vital for proliferation44. Of note, a recent report has challenged the necessity of Warburg physiology in T cell proliferation, instead suggesting that glycolysis is required to release translational inhibition of the mRNA that encodes the effector cytokine interferon-γ (IFNγ)45. Nonetheless, glucose uptake is essential for glyco lysis and enhanced cell surface expression of GLUT1 is a crucial aspect of TCR-induced T cell activation46. In this regard, it has been shown that CD28 signalling upregulates the expression of glucose transporters47. Similarly, the uptake and metabolism of the amino acid glutamine is essential for T cell activation, as glutamine deprivation blocks T cell proliferation and cytokine production11,48. Glutamine oxidation can lead to the generation of α-ketoglutarate, which is a key intermediate of the TCA cycle and which, in turn, provides substrates for the generation of various macromolecules10. Furthermore, T cells require fatty acid metabolism for their proliferation and function. Cholesterol synthesis is essential for membrane biogenesis and T cells that are deficient in SREBPs (owing to a T cell-specific deletion of SREBP cleavage-activating protein (SCAP)) have diminished proliferative capacity and reduced antiviral responses49. Along these lines, the expression of SREBP1 is enhanced following TCR activation through the suppression of the liver X receptor signalling pathway50. Thus, T cell activation induces metabolic changes that allow for processes that are necessary to promote proliferation and cytokine secretion.
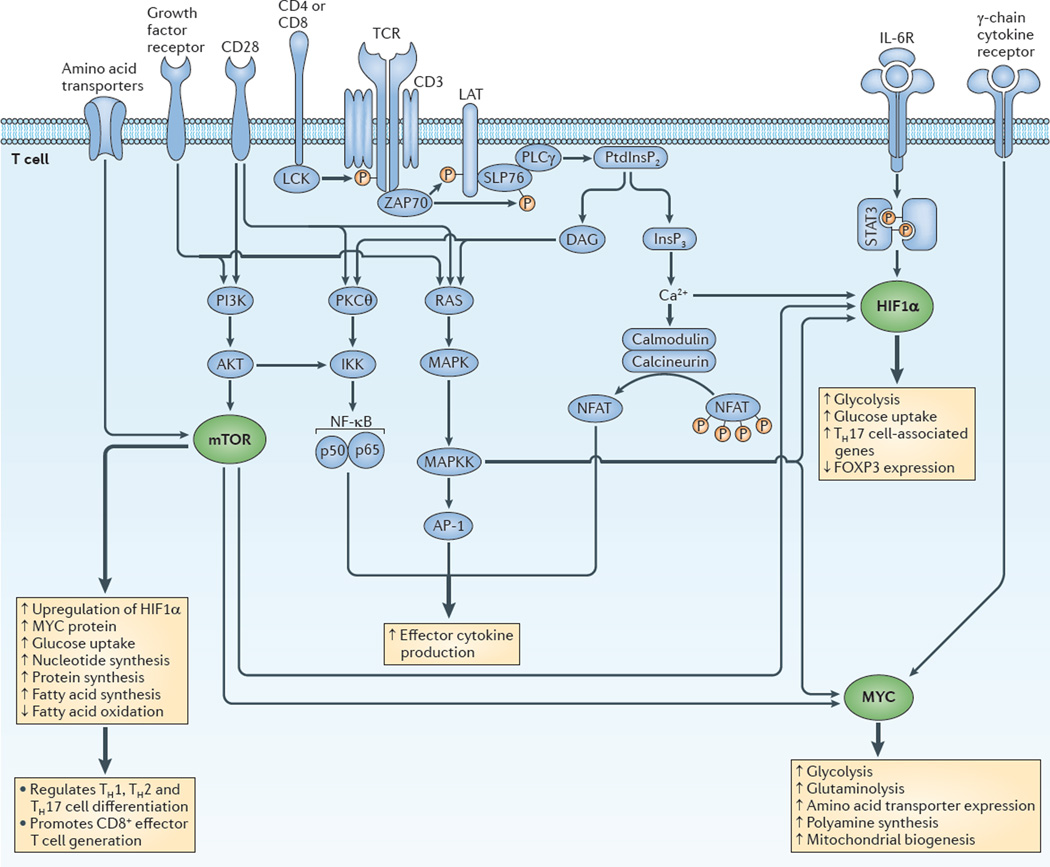
Although the considerations that are described above reflect the metabolic needs of T cells during activation and robust proliferation, it has become clear that different T cell subsets have unique metabolic needs. Thus, T cell activation and fate are linked to specific metabolic programmes that support selective T cell functions. In this regard, immunological signals such as co-stimulatory ligands, cytokines and antigen promote metabolism. Likewise, metabolic signals such as nutrient availability, hypoxia and growth factors regulate immune function. From a signalling perspective, central roles for the energy sensor AMPK and the evolutionarily conserved PI3K family member mTOR in integrating immuno-logical and metabolic pathways have emerged (TABLE 2). Similarly, the transcription factors MYC and HIF1α are central to promoting the expression of the genes that are required for metabolic programmes that support T cell responses (TABLE 2). Focusing on the metabolic and immunological roles of these molecules provides an integrative picture of the signalling pathways that regulate T cell activation, differentiation and function. In the following sections, we discuss the roles of these metabolic factors in influencing CD4+ and CD8+ effector T cell differentiation (FIG. 2), CD8+ memory T cell generation and CD4+ TReg cell function (FIG. 3).
Figure 2. Integrating immunological and metabolic signalling programmes to promote effector T cell generation and function.
The figure shows the coordinated integration of canonical T cell signalling (blue) and metabolic regulators (green) to promote the generation and function of effector T cells. In this perspective, hypoxia-inducible factor 1α (HIF1α) and MYC are just as integral to T cell effector generation as nuclear factor of activated T cells (NFAT), activator protein 1 (AP-1) and nuclear factor-κB (NF-κB). Similarly, mammalian target of rapamycin (mTOR) signalling is as crucial in effector T cell activation and differentiation as the activation of mitogen-activated protein kinase (MAPK), protein kinase Cθ (PKCθ) and calcineurin. Thicker arrows indicate the activation of metabolic programmes. Thinner arrows indicate signalling cascades. DAG, diacylglycerol; FOXP3, forkhead box P3; GLUT, glucose transporter; IKK, inhibitor of NF-κB kinase; IL-6R, interleukin-6 receptor; InsP3, inositol-1,4,5-trisphosphate; LAT, linker for activation of T cells; MAPKK, MAPK kinase; PI3K, phosphoinositide 3-kinase; PKCθ, protein kinase Cθ; PLCγ, phospholipase Cγ; PtdInsP2, phosphatidylinositol-4,5-bisphosphate; SLP76, SH2 domain-containing leukocyte protein of 76 kDa (also known as LCP2); STAT, signal transducer and activator of transcription; TCR, T cell receptor; TH cell, T helper cell; ZAP70, ζ-chain-associated protein kinase of 70 kDa.
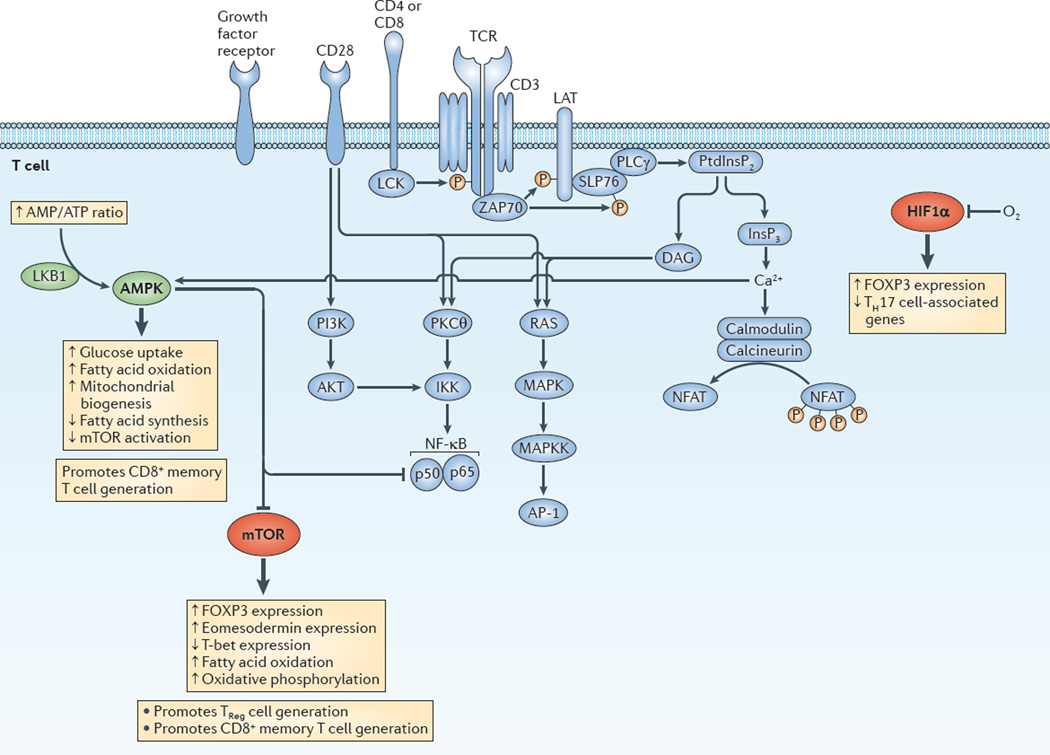
Figure 3. Integrating immunological and metabolic signalling programmes to promote CD8+ memory and CD4+ regulatory T cell generation.
This figure depicts the integration of the canonical T cell signalling pathways (blue) and metabolic regulators (green for activated and red for inhibited). AMP-activated protein kinase (AMPK) activation promotes metabolic programmes that enhance the generation of memory and regulatory T (TReg) cells. Alternatively, it is the inhibition of mammalian target of rapamycin (mTOR) and hypoxia-inducible factor 1α (HIF1α) activation that promotes the generation of CD8+ memory or CD4+ regulatory TReg cells. From this perspective, memory T cells and TReg cells share similar metabolic requirements. Thicker arrows indicate the downstream consequences of AMPK activation, and of the inhibition of mTOR and HIF1α. AP-1, activator protein 1; DAG, diacylglycerol; FOXP3, forkhead box P3; IKK, inhibitor of NF-κB kinase; InsP3, inositol-1,4,5-trisphosphate; LAT, linker for activation of T cells; LKB1, liver kinase B1; MAPK, mitogen-activated protein kinase; MAPKK, MAPK kinase; NFAT, nuclear factor of activated T cells; NF-κB, nuclear factor-κB; PI3K, phosphoinositide 3-kinase; PKCθ, protein kinase Cθ; PLCγ, phospholipase Cγ; PtdInsP2, phosphatidylinositol-4,5-bisphosphate; SLP76, SH2 domain-containing leukocyte protein of 76 kDa (also known as LCP2); TCR, T cell receptor; TH cell, T helper cell; ZAP70, ζ-chain-associated protein kinase of 70 kDa.
Metabolic regulation of CD4+ TH cell lineages
CD4+ T cells differentiate into distinct helper cell lineages following TCR engagement and cytokine stimulation51 (FIG. 1b). To determine whether TH cell subsets have distinct metabolic needs, the metabolic profiles of stimulated TH1, TH2 and TH17 cells have been assessed52. This study showed that all CD4+ TH cell subsets upregu-late GLUT1 expression upon TCR activation and have elevated glycolytic rates52. Thus, TH1, TH2 and TH17 cells all use glycolysis upon activation.
Recent studies have elucidated a role for MYC in establishing the metabolic profile that is required for effective T cell proliferation. T cell activation induces protein-level expression of both MYC and HIF1α within 2 hours of stimulation11, and MYC expression levels are highest in proliferating lymphocytes53. However, MYC — but not HIF1α — is required for upregulating the expression of the glycolytic machinery and the substrates that are essential for glutamine metabolism. Deletion of Myc abrogates the ability of activated T cells to undergo glycolysis and to initiate the catabolism of glutamine11. Furthermore, MYC deficiency diminishes the expression of the glu-tamine exchanger CD98 (a heterodimer of SLC3A2 and SLC7A5), which reduces mTORC1 activity. The absence of MYC in T cells markedly inhibits activation-induced glutaminolysis, and the subsequent generation of nucleotides and polyamines that is necessary for proliferation11.
Although HIF1α is not required for CD4+ T cell proliferation or interleukin-2 (IL-2) production11,54, several groups have shown that is has an important role in the generation and function of TH17 cells54,55. HIF1α expression is highly induced under TH17 cell-polarizing conditions during T cell activation. This upregulation of HIF1α expression is dependent on signal transducer and activator of transcription 3 (STAT3) and, importantly, occurs even under normoxic conditions55. Furthermore, HIF1α promotes TH17 cell differentiation by directly inducing the transcription of the gene that encodes retinoic acid receptor-related orphan receptor-γt (RORγt), and by cooperating with RORγt and the histone acetyltransferase p300 (also known as EP300) to drive the transcription of TH17 cell-associated genes55. In addition, it was found that polarizing T cells in vitro under conditions of 5% oxygen promotes TH17 cell differentiation in an mTORC1–HIF1α-dependent manner56. Under TH17 cell-polarizing conditions, HIF1α promotes the transcription of the genes encoding the rate-limiting enzymes of glycolysis, such as hexo kinase 2, glucose-6-phosphate isomerase, pyruvate kinase and LDHA, as well as GLUT1 (REF. 54).
HIF1α expression has also been linked to the maintenance of TH17 cells57. By studying T cells from patients with inflammation, it was observed that TH17 cells resemble long-lived effector memory cells57. Indeed, HIF1α was shown to have an important role in maintaining the expression of high levels of anti-apoptotic genes in TH17 cells57. In addition to HIF1α, the maintenance of TH17 cells has been linked to the upregulation of T cell factor 7 (TCF7; also known as TCF1) and lymphoid enhancer-binding factor 1 (LEF1), which are targets of the WNT–β-catenin pathway that are expressed at high levels in stem cells58. Interestingly, work that has been carried out in neural stem cells has shown that HIF1α positively regulates the expression of TCF7 and LEF1 (REF. 59). Although further work will be necessary to support a role for HIF1α in promoting the expression of TCF7 and LEF1 in lymphocytes, these data suggest that HIF1α expression may induce stem cell-like properties in TH17 cells. Thus, HIF1α coordinates immunological programmes — such as RORγt expression and forkhead box P3 (FOXP3) degradation (see below) — with metabolic programmes (for example, the upregulation of the glycolytic machinery and inhibitors of apoptosis) to promote the development of TH17 cells.
Of note, one group has shown increased T cell activation and IFNγ production in T cells that are deficient for the alternatively spliced isoform of HIF1α known as I.1 (REF. 60). As IL-17 has been shown to inhibit IFNγ production, it has been proposed that the increase in IFNγ in these mice is due to decreased IL-17 production55. Nonetheless, follow-up studies have revealed that deletion of the HIF1α isoform I.1 in T cells enhances immunity in a model of bacterial infection61. Therefore, the precise role of HIF1α in TH1 and TH2 cell differentiation and function remains to be determined.
Dissecting the mTOR pathway has revealed a crucial role for mTOR in the regulation of CD4+ T cell lineage differentiation. T cell-specific deletion of Mtor results in the abrogation of TH1, TH2 and TH17 cell differentiation62. Instead, stimulation of mTOR-deficient CD4+ T cells induces the accumulation of FOXP3+ TReg cells62. Furthermore, a specific deletion of Rheb (leading to the loss of mTORC1) in T cells results in the loss of TH1 and TH17 cell differentiation, although TH2 cell generation is unaffected63. By contrast, T cells that lack rapamycin-insensitive companion of mTOR (RICTOR), and thus lack mTORC2, are readily skewed towards TH1 or TH17 cell lineages (depending on which Cre recombinase is used) but they fail to differentiate into TH2 cells63,64. In addition, RICTOR-deficient mice are resistant to TH2 cell-mediated diseases63,65. Thus, mTORC1 is required for TH1 and TH17 cell differentiation, and mTORC2 is necessary for TH2 cell development. Although the metabolic profiling of these cells is currently an area of active investigation, we propose that mTOR regulates the metabolic potential of these cells to influence T cell differentiation. Of note, recent papers demonstrate that deletion of the mTORC1 component RAPTOR prevents the generation of TH1, TH2, TH17 and TReg cells66,67. This is different from Rheb−/− mice, in which there are only defects in TH1 and TH17 cell differentiation63. Thus, RHEB-dependent mTORC1 signalling seems to have more selective effects on immune cells. We speculate that the differences that have been observed between Raptor−/−T cells and Rheb−/− T cells are due to markedly enhanced or unopposed mTORC2 activity in Raptor−/− T cells, as many of the defects that are seen in these mice are not observed in mTOR-deficient T cells62. Nonetheless, the differences between the Raptor−/− and Rheb−/− T cells provide an opportunity to define mTORC1-dependent processes that selectively regulate T cell differentiation.
It should be pointed out that, in contrast to these two studies63,64 on the role of RHEB and RAPTOR in T cells, there is a report suggesting that RAPTOR (and therefore mTORC1) is not required for TH1 and TH2 cell differentiation and is only crucial for TH17 cell differentiation by enhancing the nuclear accumulation of RORγt68. An explanation for these discrepant findings remains to be elucidated. We speculate that the inconsistent results may be related to the enhanced expansion of a CD4−IFNγ+ cell population that can rapidly proliferate in cell cultures after magnetic isolation of RAPTOR-deficient CD4+ T cells (J.D.P., unpublished observations).
Other studies have used LKB1-deficient CD4+ T cells (which display a loss of AMPK activation) to investigate the roles of the AMPK and mTOR pathways in TH cell differentiation. LKB1-deficient CD4+ T cells show elevated production of IFNγ and IL-17, and have an enhanced propensity to differentiate into TH1 or TH17 cells69. LKB1 deficiency also results in enhanced glucose uptake with elevated protein-level expression of GLUT1 and hexokinase 2, which indicates that AMPK activation represses glycolysis69. In addition, LKB1-deficient T cells have increased expression of mTORC1 gene targets compared with wild-type cells, which further supports a crucial role for mTOR in TH1 and TH17 cell differentiation69.
The metabolic link to CD8+ effector T cell function
CD8+ effector T cells rely heavily on glycolysis to support their metabolic needs during their rapid proliferation in response to infection70. Inhibition of glycolysis during the activation of naive CD8+ T cells abrogates effector cell generation71. For example, the glucose analogue 2-deoxy-d-glucose inhibits glycolysis and downregulates the expression of mRNAs encoding the CD8+ T cell effector proteins IFNγ and perforin72,73. Interestingly, IL-2 production is unperturbed by 2-deoxy-d-glucose treatment.
MYC has been shown to be crucial for T cell proliferation and its requirement in T cell activation has been further highlighted by studies examining CD8+ T cell function in Myc+/− mice. CD8+ T cells that lack one copy of the Myc gene show impaired activation, as determined by the abrogated upregulation of CD44 (REF. 74). In addition, studies of HIF1β-deficient T cells have provided an insight into the role of HIF1β in CD8+ T cell effector differentiation and function75. Upon initial activation, HIF1β-deficient T cells readily take up glucose and initiate glycolysis. This is in contrast to MYC-deficient CD8+ T cells, which lack the ability to initiate activation and glycolysis11. However, in response to IL-2, HIF1β-deficient CD8+ T cells fail to sustain GLUT1 levels and they have reduced expression of the key rate-limiting glyco-lytic enzymes, such as hexokinase 2, pyruvate kinase, phospho fructokinase 1 and LDHA75. Concomitantly, the HIF1β-deficient CD8+ T cells have reduced expression of effector molecules (namely, perforin and granzymes), but proliferation, IFNγ production and T-bet expression remain intact. Of note, these data indicate that the induction of HIF1β depends on mTORC1 activity and thus, mTOR is a crucial regulator of the glycolytic machinery that is upregulated by HIF1β expression75. Interestingly, this study also showed that mTOR activation in cytotoxic T lymphocytes (CTLs) occurs independently of AKT activation. This suggests that CTLs may use an alternative signalling pathway for the activation of mTORC1 (REF. 75).
Consistent with this study is a recent report that examines the function of Von Hippel–Lindau disease tumour suppressor (VHL)-deficient CD8+ T cells, which have enhanced expression of HIF1α76. Compared with wild-type T cells, VHL-deficient T cells have enhanced effector activity and they more potently reject tumours. Interestingly, although such cells expressed increased levels of effector molecules — such as perforin and granzymes — the overexpression of HIF1α also resulted in the increased expression of inhibitory molecules, such as cytotoxic T lymphocyte antigen 4 (CTLA4) and lymphocyte activation gene 3 protein (LAG3)76.
In addition, branched chain amino acids activate the mTOR pathway, as well as providing the building blocks for protein synthesis. A recently defined feature of T cell activation is the increased cell surface expression of the neutral amino acids transporter solute carrier family 7 member 5 (SLC7A5; also known as LAT1). Deletion of SLC7A5 in T cells markedly inhibits clonal expansion and effector cell differentiation77. Similarly, T cell-specific deletion of Raptor abrogates CD8+ T cell effector function (including IFNγ production) and proliferation in response to infection66. Furthermore, RAPTOR deficiency results in the downregulation of glycolytic transcripts and MYC protein, and the generation of transcripts that are important in lipid synthesis and oxidative phosphorylation66. Thus, the RAPTOR–mTORC1 pathway coordinates metabolic programmes that are important for T cell activation and function.
AMPK is activated by an increase in the AMP/ATP ratio, as well as following TCR engagement. Interestingly, the activation of AMPK following antigen recognition requires the activation of calcium/calmodulin-dependent protein kinase kinases (CaMKKs) but this is not necessary for the activation of AMPK by an increase in the AMP/ATP ratio. These results suggest that in lymphocytes, AMPK activation in response to antigen anticipates ATP depletion even in the presence of adequate nutrients78. Nonetheless, CD8+ T cells that lack expression of the catalytic α1-subunit of AMPK (AMPKα1) are activated, proliferate and secrete cytokines to an extent that is similar to wild-type T cells79,80. Thus, AMPK activation is dispensable for T cell activation in the presence of adequate nutrients. However, metabolic stress due to glucose deprivation induces enhanced cell death in AMPKα1-deficient T cells80. Similarly, T cells that are deficient in tuberous sclerosis 1 protein homologue (TSC1; also known as hamartin) have increased mTOR activation and show increased apoptosis as a result of abnormal mito-chondrial potential and the increased production of reactive oxygen species81–83.
Memory and TReg cells are metabolically alike
It has been established that glucose uptake and a high glyco lytic rate are required for effective CD4+ and CD8+ T cell responses, but both peripherally derived TReg cells and CD8+ memory T cells do not primarily use glycolysis for energy generation and instead rely on fatty acid metabolism52,84. Compared with effector T cells, CD8+ memory T cells have enhanced mitochondrial spare respiratory capacity, which provides the extra energy storage that is necessary to promote survival85. Memory T cells must also respond rapidly following antigen re challenge. In this regard, it was found that memory T cells have a greater mitochondrial mass compared with naïve T cells86. Consequently, following antigen rechallenge, effector memory T cells more extensively use oxidative phosphorylation and glycolysis compared with activated naive T cells86,87. Furthermore, a recent study suggests that memory CD8+ T cells use an AKT-dependent, rapa mycin-insensitive metabolic programme that facilitates rapid activation-induced glycolysis87.
A role for mTOR in regulating the differentiation of CD8+ effector and memory T cells was revealed by treating mice with low doses of rapamycin during infection with lymphocytic choriomeningitis virus (LCMV)88. It was found that mTOR inhibition markedly enhanced the generation of memory T cells. Given that rapamycin is used as an immunosuppressive agent, these results seem counterintuitive. However, it was shown that rapamycin treatment mitigates the expression of T-bet and enhances the expression of eomeso-dermin, which is a transcription factor that is associated with memory T cell differentiation89. Furthermore, in a model of homeostatic proliferation-induced memory, rapamycin administration abrogates the requirement of IL-15 signalling for the upregulation of eomesodermin to promote a memory response90. Consistent with these studies is a recent report showing that treatment of mice with a 4-1BB aptamer–Raptor-specific siRNA — which targets Raptor to an aptamer that binds the CD8+ T cell co-stimulatory molecule 4-1BB (also known as CD137 and TNFRSF9) — led to diminished mTORC1 activity in CD8+ T cells and the generation of an enhanced memory response91.
In addition to coordinating transcription factors that are associated with effector and memory T cell generation, it is clear that mTOR regulates CD8+ T cell differentiation by guiding metabolic programmes. Pearce et al.84 defined the necessity of fatty acid metabolism in CD8+ memory T cell generation. They observed that CD8+ T cells that lack tumour necrosis factor (TNF) receptor-associated factor 6 (TRAF6) have impaired memory cell generation owing to defects in fatty acid metabolism. Treatment of Traf6−/− cells with metformin (which activates AMPK) or rapamycin restores fatty acid oxidation and consequently rescues memory T cell generation. In a similar system, culturing LCMV-specific T cells with rapamycin before adoptive transfer into infected mice leads to a marked increase in the frequency of long-lived memory cells92. However, the inhibition of oxidative phosphorylation using oligomycin reduced the survival advantage of the rapamycin-treated cells92. In addition, although AMPK deficiency does not affect CD8+ CTL activity, it has been found that AMPK is required for CD8+ memory T cell generation, as AMPKα1-deficient CD8+ T cells fail to mount an effective secondary response to an in vivo infection80.
As discussed, HIF1α has been shown to be crucial for promoting TH17 cell differentiation but some studies suggest that it has the opposite effect on TReg cell development. This inhibition of TReg cell development was shown to occur through HIF1α-mediated degradation of the FOXP3 protein during TH17 cell development via proline hydroxylation and subsequent ubiquitylation55. Such studies are consistent with other work showing that HIF1α expression is strongly induced in CD4+ T cells under TH17 cell-polarizing conditions, whereas its expression is mitigated under conditions that promote CD4+ TReg cell development54. Consistent with these findings, the deletion of Hif1a favours the generation of TReg cells54,55. Furthermore, blocking glycolysis inhibits TH17 cell development but promotes TReg cell differentiation54.
However, in contrast to these studies, other groups have suggested that HIF1α promotes TReg cell differen-tiation93,94. For example, it has been shown that hypoxia can enhance the expression of FOXP3 in Jurkat T cells in a HIF1α-dependent manner93. Similarly, another report showed a marked increase in FOXP3 expression under hypoxic conditions94. This upregulation was HIF1α dependent and was mediated by the direct binding of HIF1α to the FOXP3 promoter. HIF1α also promotes optimal TReg cell function, as HIF1α-deficient TReg cells fail to provide protection in an in vivo colitis model94. Although the precise mechanisms that account for the positive and negative roles of HIF1α in TReg cells are yet to be delineated, it has been suggested that HIF1α — similarly to IFN-regulatory factor 4 (IRF4), B lymphocyte-induced maturation protein 1 (BLIMP1; also known as PRDM1), GATA-binding protein 3 (GATA3) and B cell lymphoma 6 (BCL-6) — might have an intrinsic role in both effector and TReg cell differentiation and function94.
As indicated above, a crucial role for mTOR in CD4+ T cell differentiation was identified by studying mice in which Mtor expression was selectively deleted in T cells62. It was found that under TH1, TH2 and TH17 cell-polarizing conditions, the mTOR-deficient T cells failed to differentiate into the respective CD4+ effector T cell subsets and instead became FOXP3+ TReg cells62. These findings are consistent with previous studies showing that rapamycin promotes the generation of TReg cells both in vitro and in vivo95–97, as well as other studies suggesting that activation of the AKT–mTOR pathway through sphingosine-1-phosphate impedes the development of thymus-derived TReg cells during thymic generation and instead favours the generation of TH1 cells98. Furthermore, activation of the AKT–mTOR pathway by overexpression of a constitutively active form of AKT inhibits TReg cell genera-tion99. These findings are consistent with observations that TReg cells rely less on glycolysis and more on fatty acid metabolism52.
Interestingly, several reports cite the necessity of mTORC1 activity in promoting TReg cell function67,100. However, this seems to be at odds with the findings that Mtor deletion and rapamycin treatment promote TReg cell function. To reconcile these observations, we have proposed that decreased mTOR activity promotes the generation of ‘memory’ TReg cells and that high mTOR activity may be necessary for promoting the function of ‘effector’ TReg cells101. Such a model is consistent with the observation that strong TCR engagement in the presence of transforming growth factor-β (TGFβ) promotes TReg cell generation, even though it also promotes increased mTOR activity (J.D.P., unpublished observations).
Targeting metabolism for immunoregulation
It is well established that potent inhibition of T cells can be achieved by blocking the activation of NFAT, NF-κB and AP-1. Indeed, the robust ability of the cal-cineurin inhibitors cyclosporine A and FK506 to inhibit T cell responses has revolutionized transplantation102. However, blocking NFAT activation also inhibits tolerance and the activation of TReg cells103. Thus, more selective pharmacological agents are needed to specifically target effector T cell responses. As HIF1α, MYC, AMPK and mTOR have crucial and selective roles in defining T cell function and fate, they represent novel and specific targets for immune modulation.
As an example, a recent drug screen identified the cardiac glycoside digoxin as an inhibitor of HIF1α104. Interestingly, digoxin has been shown to be a potent inhibitor of TH17 cell differentiation105,106. Digoxin treatment selectively inhibits TH17 cell generation without affecting the differentiation of other effector T cell subsets. Furthermore, digoxin — as well as other RORγt-specific inhibitors — mitigates TH17 cell-mediated autoimmune disease in mice105,107. These studies showed that digoxin blocks RORγt activity. However, in light of the ability of digoxin to inhibit HIF1α, it is possible that the abrogation of HIF1α-induced gene expression might also be contributing to the reduced TH17 cell response. Similarly, direct targeting of MYC could be a potent immunosuppressive strategy. Indeed, it has been shown that inhibitors of bromo domain and extra-terminal domain (BET) protein and MYC protein can suppress CD4+ T cell-mediated cytokine production and autoimmunity108.
Targeting AMPK as a means of regulating T cell function has also been studied. The AMPK agonist 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR) has been used to decrease disease severity in mouse models of acute and chronic dextran sulphate sodium-induced colitis109. This was associated with decreased TH1 and TH17 cell responses109. In another study, AICAR treatment mitigated disease severity in experimental autoimmune encephalomyelitis (EAE)110. This effect was associated with a reduction in the levels of TH1 cell-associated cytokines (IFNγ and TNF) and an increase in the expression of IL-4 and IL-10 (REF. 110). Furthermore, compared with control-treated mice, treatment with another AMPK activator, metformin, reduced GLUT1 expression and increased the percentage of airway-infiltrating TReg cells in a mouse model of asthma52. Likewise, metformin has been shown to decrease the TH17 cell response and mitigate disease severity in EAE111. Overall, these data suggest that targeted activation of AMPK may prove to be a potent strategy for treating inflammatory diseases. Additionally, metformin treatment enhances memory T cell generation in mice84 and therefore AMPK activation might also have a role in potentiating vaccine efficacy.
Targeting mTOR has proven to be an effective means of suppressing immune responses. Indeed, the mTORC1 inhibitor rapamycin has been used to prevent transplant rejection102,112. Interestingly, the mechanism of action of rapamycin was initially thought to be related to its ability to inhibit T cell proliferation9. In fact, rapa mycin is a relatively poor inhibitor of proliferation and the effectiveness of this agent is most probably owing to its ability to inhibit effector T cell metabolism, inhibit effector T cell differentiation and promote TReg cell differ-entiation113. Along these lines, it should be noted that whereas rapamycin was initially thought to only inhibit mTORC1 activity, it is clear that — particularly under conditions of prolonged exposure — rapamycin can also inhibit mTORC2 activity114. This might explain the ability of rapamycin to promote the generation of TReg cells. Alternatively, given that rapamycin can promote the generation of memory T cells, investigators are exploring the use of rapamycin to enhance vaccine responses. Indeed, this strategy has proven effective in a non-human primate model of vaccinia virus vaccination115. Whereas rapamycin (and other rapalogues) inhibit mTOR activity by sterically blocking the formation of the mTOR complex, mTOR kinase inhibitors have also been developed more recently. These agents are designed to inhibit the activity of both mTORC1 and mTORC2 (REFS. 116, 117 ). These inhibitors are designed to become new cancer therapies but their use in regulating immune responses should also be explored.
Future perspectives
In this Review, we have demonstrated that metabolic signalling programmes are integral to T cell activation, differentiation and function. Thus, whereas classical immunotherapies target ubiquitous pathways of T cell activation, we propose a more selective means of regulating immune responses by targeting specific metabolic signalling programmes. As such, we believe that selective metabolic inhibitors might prove to be clinically useful immunomodulators. In the case of immunosuppression, such agents would have the advantage of inhibiting effector T cell function but also enhancing TReg cell function. For example, whereas calcineurin inhibitors (such as cyclosporine A and FK506) block the generation of TReg cells, mTOR inhibition does not118. To this end, treating liver transplant recipients with siro-limus (an mTOR inhibitor), rather than FK506, increases the number of liver FOXP3+ TReg cells119. Likewise, a calci neurin inhibitor-free regimen (using sirolimus) has been developed to promote stable mixed donor chimer-ism after non-myeloablative allogeneic haematopoietic stem cell transplantation for adult sickle cell disease120. Furthermore, there seems to be promise in the strategy of directly inhibiting glycolysis. In mouse models, 2-deoxy-d-glucose has been shown to prevent the development of EAE and to promote TReg cell generation52,54, as well as promoting CD8+ memory T cell generation71. Likewise, inhibitors of glutamine metabolism — which are being developed as anticancer agents — might turn out to be potent inhibitors of effector T cell responses121. Therefore, future work should not be focused on indiscriminate switching on or off of T cell responses but rather on modulating T cell responses depending on what immune mechanism is required. This therapeutic approach may harness the most potent response and minimize undesired effects.
Acknowledgements
The authors thank F. Pan for critical review of this manuscript. The authors’ work is supported by the US National Institutes of Health (grant R01AI077610-01A2). The authors apologize to those colleagues whose work has not been cited owing to space constraints.
Glossary
- Glucose transporter type 1 (GLUT1)
A unidirectional transporter that facilitates the transport of glucose across the plasma membrane
- Lactate dehydrogenase A (LDHA)
An enzyme that catalyses the conversion of pyruvate to lactate
- Hexokinase 2
An enzyme that initiates the first reaction of glycolysis by phosphorylating glucose to produce glucose-6-phosphate
- Pyruvate dehydrogenase kinase 1 (PDK1)
An enzyme that phosphorylates and inactivates pyruvate dehydrogenase, thereby inhibiting the catalysis of pyruvate to acetyl-CoA and preventing the initiation of the tricarboxylic acid cycle
- Phosphofructokinase 1
A rate-limiting enzyme of glycolysis that requires ATP to convert fructose-6-phosphate into fructose-1,6-bisphosphate
- Carnitine palmitoyltransferase 1A (CPT1A)
A rate-limiting mitochondrial enzyme that is necessary for fatty acid oxidation. CPT1A catalyses the transfer of the acyl group of long-chain fatty acids to acylcarnitine, which allows for its transport from the cytosol to the mitochondria
- Autophagy
An evolutionarily conserved process in which acidic double-membrane-bound vacuoles sequester intracellular contents (such as damaged organelles and macromolecules) and target them for degradation through fusion with secondary lysosomes
- α-ketoglutarate
A key intermediate of the tricarboxylic acid cycle that can be derived from glutaminolysis
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Gerondakis S, Fulford TS, Messina NL, Grumont RJ. NF-κB control of T cell development. Nature Immunol. 2014;15:15–25. doi: 10.1038/ni.2785. [DOI] [PubMed] [Google Scholar]
- 2.Srikanth S, Gwack Y. Orai1-NFAT signalling pathway triggered by T cell receptor stimulation. Mol. Cells. 2013;35:182–194. doi: 10.1007/s10059-013-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macian F, et al. Transcriptional mechanisms underlying lymphocyte tolerance. Cell. 2002;109:719–731. doi: 10.1016/s0092-8674(02)00767-5. [DOI] [PubMed] [Google Scholar]
- 4.Schwartz RH. Models of T cell anergy: is there a common molecular mechanism? J. Exp. Med. 1996;184:1–8. doi: 10.1084/jem.184.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fields PE, Gajewski TF, Fitch FW. Blocked Ras activation in anergic CD4+ T cells. Science. 1996;271:1276–1278. doi: 10.1126/science.271.5253.1276. [DOI] [PubMed] [Google Scholar]
- 6.Kane LP, Andres PG, Howland KC, Abbas AK, Weiss A. Akt provides the CD28 costimulatory signal for up-regulation of IL-2 and IFN-γbut not TH2 cytokines. Nature Immunol. 2001;2:37–44. doi: 10.1038/83144. [DOI] [PubMed] [Google Scholar]
- 7.Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nature Rev. Immunol. 2013;13:227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powell JD, Lerner CG, Schwartz RH. Inhibition of cell cycle progression by rapamycin induces T cell clonal anergy even in the presence of costimulation. J. Immunol. 1999;162:2775–2784. [PubMed] [Google Scholar]
- 10. MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. This is an outstanding and comprehensive review of metabolism in T cells.
- 11. Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. This is a seminal paper on the role of MYC in controlling T cell metabolism.
- 12.Osthus RC, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 13.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb. Perspect. Med. 2013;3:a014217. doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li F, et al. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol. Cell. Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl Acad. Sci. USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semenza GL. Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harb. Symp. Quant. Biol. 2011;76:347–353. doi: 10.1101/sqb.2011.76.010678. [DOI] [PubMed] [Google Scholar]
- 18.Iyer NV, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1 -mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell. Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell. Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 21.Sun Q, et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc. Natl Acad. Sci. USA. 2011;108:4129–4134. doi: 10.1073/pnas.1014769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J. Cell Sci. 2013;126:1713–1719. doi: 10.1242/jcs.125773. This is an outstanding and comprehensive review of mTOR signalling and function.
- 24.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nature Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 28. Duvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. This exhaustive work defines the regulation of metabolism by mTOR. Although it is a primary research article, it reads like a review.
- 29.Masui K, et al. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell. Metab. 2013;18:726–739. doi: 10.1016/j.cmet.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardie DG. AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 2011;25:1895–1908. doi: 10.1101/gad.17420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan HX, Xiong Y, Guan KL. Nutrient sensing, metabolism, and cell growth control. Mol. Cell. 2013;49:379–387. doi: 10.1016/j.molcel.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munday MR. Regulation of mammalian acetyl-CoA carboxylase. Biochem. Soc. Trans. 2002;30:1059–1064. doi: 10.1042/bst0301059. [DOI] [PubMed] [Google Scholar]
- 33.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 34.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faubert B, et al. Loss of the tumor suppressor LKB1 promotes metabolic reprogramming of cancer cells via HIF-1α. Proc. Natl Acad. Sci. USA. 2014;111:2554–2559. doi: 10.1073/pnas.1312570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 38.Deberardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J. Biol. Chem. 2006;281:37372–37380. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- 39.Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science. 2013;339:1323–1328. doi: 10.1126/science.1228792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robitaille AM, et al. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science. 2013;339:1320–1323. doi: 10.1126/science.1228771. [DOI] [PubMed] [Google Scholar]
- 41.Bental M, Deutsch C. Metabolic changes in activated T cells: an NMR study of human peripheral blood lymphocytes. Magn. Reson. Med. 1993;29:317–326. doi: 10.1002/mrm.1910290307. [DOI] [PubMed] [Google Scholar]
- 42. Jones RG, Thompson CB. Revving the engine: signal transduction fuels T cell activation. Immunity. 2007;27:173–178. doi: 10.1016/j.immuni.2007.07.008. Along with other articles by these authors, this review is paradigm shifting by proposing that metabolism is not dictated by activation but rather that metabolism guides the activation and function of T cells.
- 43.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 44.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. This paper demonstrates that aerobic glycolysis is required for T cell effector function but not for proliferation or survival.
- 46.Jacobs SR, et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J. Immunol. 2008;180:4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Frauwirth KA, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. This is one of the first papers demonstrating a crucial role for immunological signals in regulating cellular metabolism.
- 48.Carr EL, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kidani Y, et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nature Immunol. 2013;14:489–499. doi: 10.1038/ni.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bensinger SJ, et al. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Michalek RD, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. This is an early and important study in terms of defining the differences in metabolism between different T cell subsets.
- 53.Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur. J. Immunol. 2008;38:342–349. doi: 10.1002/eji.200737972. [DOI] [PubMed] [Google Scholar]
- 54.Shi LZ, et al. HIF1 α-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and TR cells. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dang EV, et al. Control of TH17/TReg balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. References 54 and 55 comprehensively examine the role of HIF1 a in regulating TH17 and TR cell differentiation.
- 56.Ikejiri A, et al. Dynamic regulation of TH17 differentiation by oxygen concentrations. Int. Immunol. 2012;24:137–146. doi: 10.1093/intimm/dxr111. [DOI] [PubMed] [Google Scholar]
- 57.Kryczek I, et al. Human TH17 cells are long-lived effector memory cells. Sci. Transl. Med. 2011;3:104ra100. doi: 10.1126/scitranslmed.3002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muranski P, et al. Th17 cells are long lived and retain a stem cell-like molecular signature. Immunity. 2011;35:972–985. doi: 10.1016/j.immuni.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mazumdar J, et al. O2 regulates stem cells through Wnt/β-catenin signalling. Nature Cell Biol. 2010;12:1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lukashev D, et al. Cutting edge: hypoxia-inducible factor 1 a and its activation-inducible short isoform I.1 negatively regulate functions of CD4+ and CD8+ T lymphocytes. J. Immunol. 2006;177:4962–4965. doi: 10.4049/jimmunol.177.8.4962. [DOI] [PubMed] [Google Scholar]
- 61.Georgiev P, et al. Genetic deletion of the HIF-1a isoform I.1 in T cells enhances antibacterial immunity and improves survival in a murine peritonitis model. Eur. J. Immunol. 2013;43:655–666. doi: 10.1002/eji.201242765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Delgoffe GM, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. This is the first paper to use genetic tools to demonstrate the crucial role of mTOR in regulating T cell differentiation.
- 63.Delgoffe GM, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nature Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee K, et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heikamp EB, et al. The AGC kinase SGK1 regulates TH1 and TH2 differentiation downstream of the mTORC2 complex. Nature Immunol. 2014;15:457–464. doi: 10.1038/ni.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang K, et al. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1 -mediated metabolic reprogramming. Immunity. 2013;39:1043–1056. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zeng H, et al. mTORC1 couples immune signals and metabolic programming to establish TR cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. References 63–67 demonstrate the ability of mTORC1 and mTORC2 signalling complexes to selectively regulate CD4+ T cell differentiation.
- 68.Kurebayashi Y, et al. PI3K–Akt-mTORC1-S6K1/2 axis controls Th17 differentiation by regulating Gfi1 expression and nuclear translocation of RORγ. Cell Rep. 2012;1:360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 69.MacIver NJ, et al. The liver kinase B1 is a central regulator of T cell development, activation, and metabolism. J. Immunol. 2011;187:4187–4198. doi: 10.4049/jimmunol.1100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frauwirth KA, Thompson CB. Regulation of T lymphocyte metabolism. J. Immunol. 2004;172:4661–4665. doi: 10.4049/jimmunol.172.8.4661. [DOI] [PubMed] [Google Scholar]
- 71.Sukumar M, et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 2013;123:4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cham CM, Gajewski TF. Glucose availability regulates IFN-γ production and p70S6 kinase activation in CD8+ effector T cells. J. Immunol. 2005;174:4670–4677. doi: 10.4049/jimmunol.174.8.4670. References 71 and 72 demonstrate the requirement of glycolysis for T cell effector function by using an inhibitor of glycolysis, 2-deoxy-d-glucose. Reference 71 indicates that inhibition of glycolysis in activated CD8+ T cells promotes the differentiation of long-lived memory cells.
- 73.Cham CM, Driessens G, O’Keefe JP, Gajewski TF. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. Eur. J. Immunol. 2008;38:2438–2450. doi: 10.1002/eji.200838289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bianchi T, Gasser S, Trumpp A, MacDonald HR. c-Myc acts downstream of IL-15 in the regulation of memory CD8 T-cell homeostasis. Blood. 2006;107:3992–3999. doi: 10.1182/blood-2005-09-3851. [DOI] [PubMed] [Google Scholar]
- 75. Finlay DK, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J. Exp. Med. 2012;209:2441–2453. doi: 10.1084/jem.20112607. This study demonstrates that mTORC1 activity controls glucose uptake and glycolysis by regulating the expression of HIF1α.
- 76.Doedens AL, et al. Hypoxia-inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nature Immunol. 2013;14:1173–1182. doi: 10.1038/ni.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sinclair LV, et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nature Immunol. 2013;14:500–508. doi: 10.1038/ni.2556. This is an elegant study demonstrating the upregulation of an amino acid transporter upon TCR engagement and reports the functional consequences of deleting this gene.
- 78.Tamas P, et al. Regulation of the energy sensor AMP-activated protein kinase by antigen receptor and Ca2+ in T lymphocytes. J. Exp. Med. 2006;203:1665–1670. doi: 10.1084/jem.20052469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mayer A, Denanglaire S, Viollet B, Leo O, Andris F. AMP-activated protein kinase regulates lymphocyte responses to metabolic stress but is largely dispensable for immune cell development and function. Eur. J. Immunol. 2008;38:948–956. doi: 10.1002/eji.200738045. [DOI] [PubMed] [Google Scholar]
- 80.Rolf J, et al. AMPKα1: A glucose sensor that controls CD8 T-cell memory. Eur. J. Immunol. 2013;43:889–896. doi: 10.1002/eji.201243008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang L, et al. TSC1/2 signaling complex is essential for peripheral naive CD8+ T cell survival and homeostasis in mice. PLoS ONE. 2012;7:e30592. doi: 10.1371/journal.pone.0030592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nature Immunol. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Brien TF, et al. Regulation of T-cell survival and mitochondrial homeostasis by TSC1. Eur. J. Immunol. 2011;41:3361–3370. doi: 10.1002/eji.201141411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pearce EL, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. van der Windt GJ, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. References 84 and 85 are seminal studies by the same group that define the role of metabolism in regulating CD8+ T cell memory.
- 86.van der Windt GJ, et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc. Natl Acad. Sci. USA. 2013;110:14336–14341. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gubser PM, et al. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nature Immunol. 2013;14:1064–1072. doi: 10.1038/ni.2687. [DOI] [PubMed] [Google Scholar]
- 88.Araki K, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. References 88 and 89 demonstrate the ability of rapamycin to enhance CD8+ T cell memory generation.
- 90.Li Q, et al. A central role for mTOR kinase in homeostatic proliferation induced CD8+ T cell memory and tumor immunity. Immunity. 2011;34:541–553. doi: 10.1016/j.immuni.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Berezhnoy A, Castro I, Levay A, Malek TR, Gilboa E. Aptamer-targeted inhibition of mTOR in T cells enhances antitumor immunity. J. Clin. Invest. 2014;124:188–197. doi: 10.1172/JCI69856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He S, et al. Characterization of the metabolic phenotype of rapamycin-treated CD8+ T cells with augmented ability to generate long-lasting memory cells. PLoS ONE. 2011;6:e20107. doi: 10.1371/journal.pone.0020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ben-Shoshan J, Maysel-Auslender S, Mor A, Keren G, George J. Hypoxia controls CD4+CD25+ regulatory T-cell homeostasis via hypoxia-inducible factor-1α. Eur. J. Immunol. 2008;38:2412–2418. doi: 10.1002/eji.200838318. [DOI] [PubMed] [Google Scholar]
- 94.Clambey ET, et al. Hypoxia-inducible factor-1α-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc. Natl Acad. Sci. USA. 2012;109:E2784–2793. doi: 10.1073/pnas.1202366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Battaglia M, Stabilini A, Roncarolo MG. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood. 2005;105:4743–4748. doi: 10.1182/blood-2004-10-3932. [DOI] [PubMed] [Google Scholar]
- 96.Kang J, Huddleston SJ, Fraser JM, Khoruts A. De novo induction of antigen-specific CD4 + CD25 + Foxp3 + regulatory T cells in vivo following systemic antigen administration accompanied by blockade of mTOR. J. Leukoc. Biol. 2008;83:1230–1239. doi: 10.1189/jlb.1207851. [DOI] [PubMed] [Google Scholar]
- 97.Kopf H, de la Rosa GM, Howard OM, Chen X. Rapamycin inhibits differentiation of Th17 cells and promotes generation of FoxP3+ T regulatory cells. Int. Immunopharmacol. 2007;7:1819–1824. doi: 10.1016/j.intimp.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu G, Yang K, Burns S, Shrestha S, Chi H. The S1P1-mTOR axis directs the reciprocal differentiation of TH1 and TReg cells. Nature Immunol. 2010;11:1047–1056. doi: 10.1038/ni.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J. Exp. Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Procaccini C, et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Powell JD, Heikamp EB, Pollizzi KN, Waickman AT. A modified model of T-cell differentiation based on mTOR activity and metabolism. Cold Spring Harb. Symp. Quant. Biol. 2013 doi: 10.1101/sqb.2013.78.020214. http://dx.doi.org/10.1101/sqb.2013.78.020214. [DOI] [PMC free article] [PubMed]
- 102.Powell JD, Zheng Y. Dissecting the mechanism of T-cell anergy with immunophilin ligands. Curr. Opin. Investig. Drugs. 2006;7:1002–1007. [PubMed] [Google Scholar]
- 103.Bopp T, et al. NFATc2 and NFATc3 transcription factors play a crucial role in suppression of CD4+ T lymphocytes by CD4+ CD25+ regulatory T cells. J. Exp. Med. 2005;201:181–187. doi: 10.1084/jem.20041538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang H, et al. Digoxin and other cardiac glycosides inhibit HIF-1α synthesis and block tumor growth. Proc. Natl Acad. Sci. USA. 2008;105:19579–19586. doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Huh JR, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORγt activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. This study uses digoxin, which is a small-molecule compound that specifically inhibits the transcription of the gene that encodes RORγt. Digoxin can inhibit both mouse and human TH17 cell differentiation, which suggests that it may be a novel therapeutic agent for autoimmune diseases.
- 106.Fujita-Sato S, et al. Structural basis of digoxin that antagonizes RORγt receptor activity and suppresses Th17 cell differentiation and interleukin (IL)-17 production. J. Biol. Chem. 2011;286:31409–31417. doi: 10.1074/jbc.M111.254003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xiao S, et al. Small-molecule RORγt antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 2014;40:477–489. doi: 10.1016/j.immuni.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bandukwala HS, et al. Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proc. Natl Acad. Sci. USA. 2012;109:14532–14537. doi: 10.1073/pnas.1212264109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bai A, et al. Novel anti-inflammatory action of 5-aminoimidazole-4-carboxamide ribonucleoside with protective effect in dextran sulfate sodium-induced acute and chronic colitis. J. Pharmacol. Exp. Ther. 2010;333:717–725. doi: 10.1124/jpet.109.164954. [DOI] [PubMed] [Google Scholar]
- 110.Nath N, et al. 5-aminoimidazole-4-carboxamide ribonucleoside: a novel immunomodulator with therapeutic efficacy in experimental autoimmune encephalomyelitis. J. Immunol. 2005;175:566–574. doi: 10.4049/jimmunol.175.1.566. [DOI] [PubMed] [Google Scholar]
- 111.Nath N, et al. Metformin attenuated the autoimmune disease of the central nervous system in animal models of multiple sclerosis. J. Immunol. 2009;182:8005–8014. doi: 10.4049/jimmunol.0803563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Touzot M, Soulillou JP, Dantal J. Mechanistic target of rapamycin inhibitors in solid organ transplantation: from benchside to clinical use. Curr. Opin. Organ. Transplant. 2012;17:626–633. doi: 10.1097/MOT.0b013e32835a4be2. [DOI] [PubMed] [Google Scholar]
- 113.Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol. Rev. 2012;249:43–58. doi: 10.1111/j.1600-065X.2012.01152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 115.Turner AP, et al. Sirolimus enhances the magnitude and quality of viral-specific CD8+ T-cell responses to vaccinia virus vaccination in rhesus macaques. Am. J. Transplant. 2011;11:613–618. doi: 10.1111/j.1600-6143.2010.03407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gao W, et al. Contrasting effects of cyclosporine and rapamycin in de novo generation of alloantigen-specific regulatory T cells. Am. J. Transplant. 2007;7:1722–1732. doi: 10.1111/j.1600-6143.2007.01842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Levitsky J, et al. Systemic immunoregulatory and proteogenomic effects of tacrolimus to sirolimus conversion in liver transplant recipients. Hepatology. 2013;57:239–248. doi: 10.1002/hep.25579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hsieh MM, et al. Allogeneic hematopoietic stem-cell transplantation for sickle cell disease. N. Engl. J. Med. 2009;361:2309–2317. doi: 10.1056/NEJMoa0904971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen JQ, Russo J. Dysregulation of glucose transport, glycolysis, TCA cycle and glutaminolysis by oncogenes and tumor suppressors in cancer cells. Biochim. Biophys. Acta. 2012;1826:370–384. doi: 10.1016/j.bbcan.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 122.Schwartz RH. Costimulation of T lymphocytes: the role of CD28, CTLA-4, and B7/BB1 in interleukin-2 production and immunotherapy. Cell. 1992;71:1065–1068. doi: 10.1016/s0092-8674(05)80055-8. [DOI] [PubMed] [Google Scholar]
- 123.Loh C, et al. Calcineurin binds the transcription factor NFAT1 and reversibly regulates its activity. J. Biol. Chem. 1996;271:10884–10891. doi: 10.1074/jbc.271.18.10884. [DOI] [PubMed] [Google Scholar]
- 124.Chen FE, Ghosh G. Regulation of DNA binding by Rel/NF-κB transcription factors: structural views. Oncogene. 1999;18:6845–6852. doi: 10.1038/sj.onc.1203224. [DOI] [PubMed] [Google Scholar]
- 125.Jenkins MK, Schwartz RH. Antigen presentation by chemically modified splenocytes induces antigen-specific T cell unresponsiveness in vitro and in vivo. J. Exp. Med. 1987;165:302–319. doi: 10.1084/jem.165.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]