Abstract
Ca2+ signals are thought to play important roles in plant growth and development, including key aspects of pollen tube growth and fertilization. The dynamics of a Ca2+ signal are largely controlled by influx (through channels) and efflux (through pumps and antiporters). The Arabidopsis genome encodes 14 Ca2+ pumps, 10 of which belong to a family of autoinhibited Ca2+ ATPases (ACA) that are predicted to be activated by Ca2+/calmodulin. Here, we show that isoform ACA9 is expressed primarily in pollen and localized to the plasma membrane. Three independent T-DNA [portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] gene disruptions of ACA9 were found to result in partial male sterility. Complementation was observed by using a ACA9-yellow fluorescence protein (YFP) fusion that displayed plasma membrane localization. Mutant aca9 pollen displayed a reduced growth potential and a high frequency of aborted fertilization, resulting in a >80% reduction in seed set. These findings identify a plasma membrane Ca2+ transporter as a key regulator of pollen development and fertilization in flowering plants.
Ca2+ is essential for eukaryotic cells, both as an important second messenger (signaling) as well as a structural component of enzymes and macromolecular complexes (“cellular nutrition”) (1). Although Ca2+ is essential, high concentrations in the cytoplasm can also be toxic. As a result, many different systems have evolved to sense, respond to, and regulate cytosolic Ca2+. Both plants and animals use a combination of ion pumps, antiporters, and uniporters to control cytoplasmic Ca2+ dynamics (2, 3), including a family of calmodulin-activated Ca2+-ATPase ion pumps: plasma membrane Ca2+ ATPases (PMCAs) in animals and autoinhibited Ca2+ ATPases (ACAs) in plants (4, 5). In mice, phenotypes resulting from knock-out mutants of different PMCAs include hearing and balance defects (for PMCA2), male sterility (for PMCA4), and lethality (for PMCA1) (6, 7). A multiplicity of gene-specific phenotypes support the expectation that Ca2+ ATPases have evolved to fulfill multiple cellular functions, some of which are essential for development of multicellular organisms.
In plants, Ca2+ has been implicated in many physiological processes, including tip growth by pollen tubes (8–17) and fertilization (18). Sexual reproduction in plants requires that pollen (the male gametophyte) undergoes directional growth to reach the embryo sac (the female gametophyte) and discharge two sperm cells into a receptive synergid cell (19). Whereas invasive experiments that block or alter Ca2+ signals have been shown to modulate pollen development and fertilization, genetic evidence supporting the role of Ca2+ transport has so far been lacking.
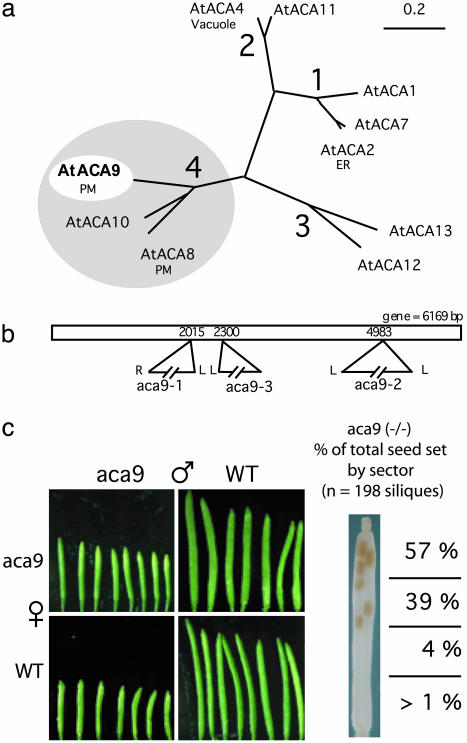
In Arabidopsis, there are 14 Ca2+ pumps from two distinct families (20) [10 ACAs and 4 endoplasmic reticulum-type Ca2+ ATPases (ECAs)]. Although the ACAs are most closely related to the animal plasma membrane Ca2+ ATPases, they are distinct in (i) having isoforms located in the endoplasmic reticulum and tonoplast, in addition to the plasma membrane and (ii) having their calmodulin-regulated autoinhibitor located at the N-terminal instead of C-terminal end (21–23). Based on amino acid similarity and intron positions, ACAs can be divided into 4 subfamilies (20), with ACA8, ACA9, and ACA10 constituting subfamily 4 (Fig. 1a). One member of this subfamily, ACA8, has previously been localized to the plasma membrane (23).
Fig. 1.
Diagrams showing relationship of ACA9 to other Ca2+ pumps in the P2B family and positions of T-DNA gene disruptions. (a) A sequence distance tree for the P2B Ca2+-ATPases in A. thaliana, based on ref. 20. PM, plasma membrane; ER, endoplasmic reticulum. (b) Map of T-DNA insertions in ACA9. T-DNA borders for left (L) and right (R) are shown. Numbers indicate the T-DNA insertion point, as reported as nucleotide position downstream from the start codon at position 1. The T-DNA insertions in aca9-2 and aca9-3 are inserted in exons whereas the T-DNA insertion in aca9-1 is inserted in an exon-intron boundary. (c) Siliques from selfed and reciprocal crosses between aca9 and WT plants. An enlarged view of a silique cleared with ethanol is shown to the right. The percentage of total seed set by sector is indicated (n = 198 siliques).
Here, we provide evidence that ACA9 functions in the pollen tube plasma membrane and is a key regulator of pollen tube growth and fertilization. Three independent gene disruptions of ACA9 were shown to result in a semisterile phenotype. This phenotype originates from two defects in the male gametophyte. First, mutant pollen tubes display a reduced growth rate and growth potential (i.e., length). Second, mutant pollen tubes that reach the ovule display a high frequency of aborted fertilization. Our results provide genetic evidence for the role of a Ca2+ transporter in pollen growth and fertilization.
Materials and Methods
Isolation of T-DNA Insertion Mutants. Mutants were identified by a PCR based screen of Arabidopsis thaliana (Wassilewskija-2) T-DNA [portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] insertion mutants generated at the University of Wisconsin Arabidopsis knockout facility (24). Three different aca9 mutants [aca9-1 (ssJH no. 108), aca9-2 (ssJH no. 109), and aca9-3 (ssMP no. 22)] were identified, all harboring a kanamycin-resistance marker within the T-DNA insertion. Plants homozygous for a T-DNA insertion were identified by the inability to amplify a WT band by using the ACA9-specific primers flanking the insert. T-DNA border fragments were amplified and sequenced for each insertion to verify the site of insertion.
Plasmid Constructs and Plant Transformation. Plants were transformed by using Agrobacterium (GV3101) and a floral dip method (25). For all plasmids derived using PCR, coding sequences were sequenced to ensure the absence of PCR mistakes.
ACA9-promoter::GUS construct (ps no. 533). A 1,171-bp sequence upstream of the ATG start codon of the ACA9 ORF was PCR amplified from Arabidopsis WT Ws-2 DNA and inserted into a pGreen vector (26) harboring a hygromycin resistance marker. Representative transgenic plant lines expressing this construct are ssJH nos. 468–470.
ACA9-promoter::ACA9-YFP construct (ps no. 580). An ACA9 genomic/cDNA hybrid sequence (At3g21180) was constructed from PCR-amplified fragments of cDNA and genomic DNA (ecotype Col) and inserted along with the ACA9 promoter sequence into a pGreen vector harboring a hygromycin resistance marker. T2 plants from six independent lines were imaged with similar results (plant lines ss nos. 471–473 and ss nos. 474–476 in aca9-1 and aca9-2 backgrounds, respectively).
ACA9-promoter::GFP construct (ps no. 536). The ACA9 promoter sequence was inserted into a pBIN vector (harboring a basta-resistance marker (27) and modified to express GFP fusions downstream of a 35s promoter (28). Representative transgenic plant lines expressing this construct are ss nos. 456, 460, and 464, in parental backgrounds corresponding to aca9-1, aca9-2, and Ws, respectively.
The ACA9-promoter::ACA8-GFP-TAP2 construct (ps no. 534) was based on the pGreen vector harboring a hygromycin resistance marker. Representative transgenic plant lines expressing this construct are ssJH nos. 480 and 483 in aca9-1 and aca9-2 backgrounds, respectively.
Pollen Germination. Pollen were germinated on an agar medium with 1% agar, 80 ppm boric acid, 5 mM CaCl2, 20% sucrose, and 10 ppm myo-inositol (pH 5.8), and incubated overnight at room temperature at 100% humidity.
Confocal Microscopy. Images of green and yellow fluorescent proteins (GFP and YFP) were collected on a Bio-Rad MRC1024 confocal system attached to a Zeiss Axiovert 100TV microscope using the ×63 objective (NA1.4), a 522-DF35 emission filter set, and a krypton-argon mixed gas laser for generating a 488-nm excitation line. Differential interference contrast (DIC) images were collected on the same system by using a single transmitted light detector. Pseudocolored images were merged by using Improvision (Lexington, MA) openlab software.
Aniline Blue Staining. Pistils were fixed in 10% acetic acid, 30% chloroform, and 60% ethanol for 2 h, incubated in 4 M NaOH for 10 min, washed three times with 50 mM potassium phosphate (pH 7.5), and then stained for 12 h with 0.05% aniline blue in 50 mM potassium phosphate (pH 7.5) (29). Pistils were squashed lightly between a slide and a coverslip, and photographed with a Leica DC 300F camera mounted on a Leica (Deerfield, IL) MZ FL III microscope using a 425/40-nm excitation and 475-nm long pass filter.
Real-Time PCR. Approximately 100 ng of total RNA extracted from Arabidopsis tissue was reverse transcribed by using either the ACA9 specific primer 5′-AAT TGC TAG TGG CCA GCT GAC-3′ or the Triose Phosphate Isomerase (At3g55440) primer 5′-GTC GAT AAA CTC AGG CTT A-3′. Real-time PCR reactions were run on a Roche Lightcycler using LightCycler FastStart DNA Master SYBR Green I dye (Roche), and the primer pairs 5′-TAG CCA AGA AAA TAA CGG TGA T-3′, 5′-TTG TTA TAA GAG GTT CCC TTC T-3′ for ACA9 and 5′-TTT CAC TGG TGA AGT GAG TG-3′,5′-GTC GAT AAA CTC AGG CTT TA-3′ for triose phosphate isomerase (normalization control). The software application q-gene (30) was used for the data analysis.
Complementation of Yeast Mutants. Constructs encoding a full-length ACA9 and truncated ACA9ΔN (lacking the first 93 aa) were engineered into a yeast expression vector (pYES2, Invitrogen) under the control of a galactose inducible promoter. Saccharomyces cerevisiae strain K616 (MATa, pmr1::HIS3, pmc1::TRP1, cnb1::LEU2, ura3) (31) was transformed with the plasmids pYES2-ACA9, pYES2-ACA9ΔN, and empty pYES2 vector by using the LiOAc/polyethylene glycol (PEG) methods (32). Likewise, the strain K601 (MATa, leu2, his3, ade2, trp1, ura3) was transformed with the empty pYES2 vector. The transformants were selected for uracil prototrophy by plating on yeast medium containing 2% glucose, 2% bacto-agar, 50 mM succinic acid (pH 5.5), 0.7% (wt/vol) yeast nitrogen base, 30 μg/ml adenine, 30 μg/ml each of amino acids his, leu, and trp, and 10 mM CaCl2 and incubating at 30°C for 3 days. Ura+ colonies were grown in a similar medium without agar at 30°C until the OD600 reached 0.5, and 7 μl of yeast culture was placed as drops on four different media: 2% glucose or galactose, 2% Bacto-agar, 50 mM succinic acid (pH 5.5), 0.7% (wt/vol) yeast nitrogen base, 30 μg/ml adenine, 30 μg/ml each of amino acids his, leu, and trp, and either 10 mM CaCl2 or 10 mM EDTA.
Calmodulin Overlay Assays. A 247-bp fragment of ACA9 encoding Met-40 to Ala-95 was amplified by PCR and cloned into plasmid pGEX-4T (Amersham Pharmacia Bioscience), leading to an N-terminal fusion with glutathione-S-transferase. The fusion protein was expressed in E. coli strain BL21(DE3)pLysS (Novagen) by standard procedures. Whole-cell lysate (50 μg of protein) was separated by SDS/PAGE and electroblotted onto an Immobilon-P membrane (Millipore). After renaturation of membrane-bound polypeptides by using a gradient of guanidine-HCl (4 M, 3 M, 1 M, and 0.1 M) in a Tris-saline buffer (25 mM NaCl/25 mM Tris·HCl, pH 7.5/1 mM DTT/5 mM MgCl2), the membrane was coated with 1% BSA in the same buffer with 0.04% Tween 20 but without guanidine-HCl, and subsequently probed with biotinylated bovine brain calmodulin (Calbiochem) according to the manufacturer's protocol in the presence of 1 mM CaCl2 or 5 mM EGTA.
Results
Gene Disruptions of ACA9 Result in Reduced Seed Set. To investigate the biological functions of Ca2+ transporters in plants, we identified plants harboring T-DNA disruptions in all 14 Arabidopsis Ca2+ pumps. In contrast to animals (7), no single pump was found to be essential under laboratory conditions, as shown by the ability to recover viable homozygous plants for each of the 14 gene disruptions. Nevertheless, a partial sterility phenotype was observed for disruptions of ACA9, as first noticed by a non-Mendelian segregation of homozygotes (4.8% -/- vs. expected 25%, n = 1899, P < 0.0001).
Three independent ACA9 mutants (aca9-1, aca9-2, and aca9-3, Fig. 1b) were identified by PCR screening of a collection of T-DNA-transformed plants (24). All homozygous aca9 plants produced shorter siliques, with 80% fewer seeds per silique (i.e., 5–8 seeds) (Fig. 1c). Seed counts varied under different culture conditions. In Copenhagen, typical yields were aca9 = 4.9 ± 0.5 and WT WS-2 = 56.2 ± 1.4; in San Diego, typical yields were aca9 = 7.8 ± 2.4 and WT WS-2 = 47.2 ± 2.7. A second feature of this phenotype was an unequal distribution of seeds within the silique, with 96% of the seeds located in the upper half (stigma end) (Fig. 1c Inset).
Evidence that the reduced seed set phenotype was the result of a gene disruption in ACA9 was confirmed by complementation. Mutant aca9-1 plants were transformed with ACA9 fused to YFP and expressed under the control of an ACA9 promoter. In second generation transgenic plant lines homozygous for the ACA9-YFP construct, seed set was restored to near WT levels (average seed set per silique = 40.6 ± 2.2, n = 3 transgenic lines). Thus, three independent gene disruptions and complementation all indicate that poor seed set results from a loss of ACA9 gene function (i.e., a “knockout”).
aca9 Pollen Tubes Show Defects in Growth and Fertilization. To determine the biological basis of the reduced seed set phenotype, we initiated reciprocal crosses between mutants and WT. When mutant pollen was used to fertilize a WT flower, we always obtained very poor seed set, comparable to a self-fertilized aca9 (-/-) plant. In contrast, when WT pollen was used to fertilize a mutant flower, full seed set was restored in some siliques (Fig. 1c Upper Right). To further test for any phenotypic contribution from the female gametophyte, we used WT pollen to fertilize a heterozygous aca9 mutant (i.e., 50% of the maternal parent ovules = aca9). By genotyping the progeny of this cross, we observed the expected 1:1 ratio of wt and aca9 alleles (F1 = 48% aca9 (-/+), n = 350, P < 0.0001). Together, these results indicate that the reduced seed phenotype results from a pollen defect alone, with no significant influence from the female gametophyte.
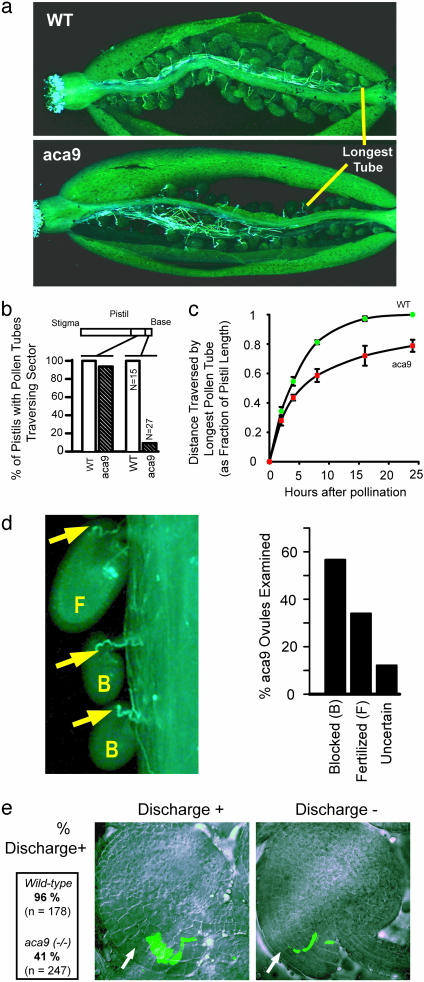
Mutant aca9 pollen were found to have two defects. First, they showed reduced growth potentials in vivo and in vitro. Aniline blue staining of pollen growing through pistils indicated that aca9 pollen tubes had a 90% reduced frequency of reaching the basal end of the pistil (Fig. 2 a and b). In a time course of pollen growth, a decreased growth rate became noticeable by around 5 h (Fig. 2c). This reduced growth potential is consistent with in vitro assays that show a roughly 75% length reduction in mutant pollen tubes compared with WT (after 24 h, 99.1 ± 20.1 μm vs. 378.1 ± 40.8 μm). This reduced growth potential partially explains the observed bias of successful fertilization at the stigma-end of the pistil (Fig. 1c Inset).
Fig. 2.
aca9 pollen tubes show defects in growth potential and interactions with ovules. (a) Examples of aca9 and WT pistils stained with aniline blue (29) to show positions of longest pollen tubes. (b) Fraction of pistils from WT and aca9 plants having pollen tubes that reach the basal third of the pistil vs. those that reach the bottom (i.e., last four seed positions), as revealed by aniline blue staining. (c) In vivo growth rate of WT and aca9 pollen tubes, as revealed by aniline blue staining. (d) Examples of aca9 pollen tubes where fertilization was successful (F, embryo has grown) or blocked (B, embryo unchanged). Arrows point at the pollen tube tip inside the ovule. Events were counted only in the upper half of the pistil where we previously determined that mutant pollen was capable of near normal growth potential. (Right) The percentage of blocked fertilization events was >50% in this region. (e) Examples of aborted pollen tube discharge (Discharge -) in comparison with successful fertilization reactions (Discharge +). Differential interference contrast (DIC) images are shown of ovules overlaid with fluorescent images of pollen tubes from aca9 (-/-) plants transformed with an ACA9-promoter::GFP construct. The fraction of pollen tubes showing successful discharge into a synergid cell (arrow) was scored by the release of cytosolic GFP into the large area occupied by the synergid, as previously established by Faure et al. (47). (Left) The fraction of pollen tubes showing successful discharge into a synergid cell in WT and aca9 (-/-) plants is summarized.
Second, mutant pollen also displayed a high frequency of aborted fertilization. This finding was first observed by aniline blue staining of maturing siliques (Fig. 2d). In cases where staining showed a pollen tube had reached an ovule, more than 50% of the ovules failed to develop an embryo. Because the aca9 defect seems to be restricted to the male gametophyte, we interpreted this aborted development as a failure to complete fertilization.
A defect in fertilization was corroborated by a microscopic examination of pollen-tube embryo-sac interactions, as monitored by the delivery of GFP from pollen tubes to synergids (Fig. 2e). A synergid cell represents the point of initial cell-cell contact between the male and female gametophyte. Upon contact, pollen tubes discharge their sperm cells into the synergid (in less than a minute) (33, 34). In contrast to WT fertilization, >50% of mutant pollen failed to discharge sperm, despite having reached the embryo-sac. This >50% abortion frequency correlated with that observed using aniline blue (Fig. 2d).
A prediction that arises from a high frequency of aborted fertilization is that a heterozygote aca9 mutant undergoing self-fertilization should show reduced seed set as a result of mutant pollen blocking the potential of WT pollen to fertilize available ovules (35). Consistent with this expectation, heterozygous plants exhibit a semidominant phenotype with a 25% reduction in seed set (in San Diego, 35.7 ± 2.9 seeds per silique, n = 290 siliques). Thus, despite having a reduced growth potential, aca9 pollen still effectively compete with WT pollen in growing and finding ovules, at least in the stigma-end half of the pistil.
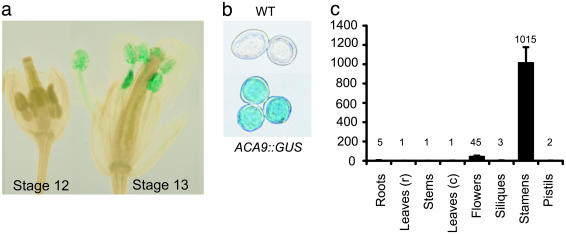
ACA9 Is Expressed Specifically in Pollen. To determine the expression pattern expected for ACA9 protein, a DNA fragment corresponding to 1,171 bp upstream of the ACA9 start codon was fused to the β-glucuronidase (GUS) reporter gene and transformed into Arabidopsis plants. GUS expression was observed primarily in pollen (Fig. 3 a and b). Expression was initiated between flower stages 12a to 14 [stages defined by Smyth et al. (36)]. There was no evidence for GUS expression in the pistil (including ovules and transmitting tract) before pollination.
Fig. 3.
ACA9 is preferentially expressed in pollen. Shown is X-Gluc staining showing expression of an ACA9-promoter::GUS fusion in flower anthers (a) and isolated pollen (b). Histochemical analysis of GUS activity was performed as described by Jefferson et al. (48). The same staining pattern was observed in the T2 generation of six independent transgenic lines. (c) Real-time PCR analysis of the expression of ACA9 in WT Arabidopsis tissues. The values are the normalized mean of three experiments ± SEM.
To confirm the GUS-reporter results, the expression pattern of ACA9 mRNA was evaluated by using real-time RT-PCR, using ACA9-specific primers. ACA9 mRNA was found primarily in stamens (Fig. 3c), at a level >500 times higher than in pistils. The low level expression detected in the pistils is likely due to contaminating pollen. This RT-PCR result confirms the GUS reporter analysis indicating that ACA9 is expressed in the male gametophyte, but not the female gametophyte. Finally, a publicly available oligo-based (Affy GeneChip) expression profiling analysis of pollen (http://ssbdjc2.nottingham.ac.uk/narrays/experimentbrowse.pl) revealed that ACA9 was the most highly expressed of all ACAs in mature pollen, including genes for known and expected plasma membrane Ca2+ pumps, such as ACA8 and ACA10 (e.g., ACA9 expression was 20-fold more than ACA8) (34). Significant ACA9 expression was observed starting at the tricellular pollen stage.
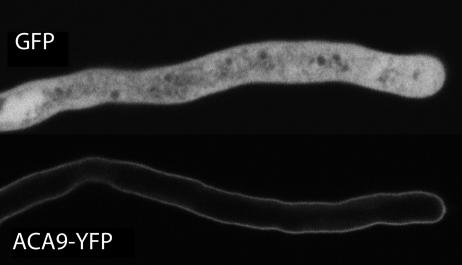
Subcellular Localization of ACA9. To determine the subcellular targeting potential of ACA9, an ACA9-YFP was expressed in stable transgenic Arabidopsis plants under the control of the ACA9 promoter. As a control, a GFP reporter was expressed in parallel under the same promoter. Pollen from transgenic plants was germinated on an agar-solidified medium and examined by confocal microscopy (Fig. 4). Consistent with a plasma membrane location, the ACA9-YFP fusion protein was always observed around the pollen tube perimeter. The same pattern of localization was observed in multiple transgenic lines, including those showing bright or barely detectable levels of expression. By contrast, GFP controls always showed strong fluorescence dispersed throughout the body of the cell. Because the ACA9-YFP fusion was able to complement an aca9 mutant, we conclude that at least a subset of the ACA9-YFP protein was localized to a functionally appropriate location, presumably the plasma membrane.
Fig. 4.
Confocal fluorescence microscopy showing plasma membrane localization of a YFP-tagged ACA9 expressed in pollen tubes. Plants were transformed with constructs harboring an ACA9-promoter::GFP construct (GFP) or an ACA9-promoter::ACA9::YFP (ACA9-YFP).
An independent line of evidence to support a plasma membrane function for ACA9 was obtained by showing that isoform ACA8 can also complement the aca9 mutant. ACA8 represents a well characterized plasma membrane Ca2+-ATPase (23) in the same ACA-subfamily as ACA9. ACA8 was fused to GFP and expressed under the ACA9 promoter. In second-generation transgenic plants, complementation was observed as a 2-fold (or more) increase in seed set in all 10 independent transgenic lines (average seed set per silique = 25.8).
ACA9 Functions as a Ca2+ Pump in Yeast Strain K616. Although ACA9 is closely related to well characterized Ca2+ pumps (21, 22), we confirmed a Ca2+ pump function for ACA9 by complementation of a yeast strain K616 that harbors a deletion of its own Ca2+ pumps, PMR1 and PMC1 (Fig. 5b). Expression of ACA9ΔN (ACA9 engineered with N-terminal truncation) allowed the K616 strain to grow on Ca2+-depleted medium. K616 cells normally fail to grow on Ca2+-depleted medium. For complementation, a functional Ca2+ pump activity is required for the secretory pathway to scavenge enough Ca2+ from the cytosol for proper function. However, because ACA9 is a plasma membrane pump, its ability to complement K616 presumably resulted either from (i) a transient activity as it moved through the yeast secretory pathway or (ii) a misaccumulation of the plant pump in the yeast endoplasmic reticulum, similar to that observed for the closely related plasma membrane calcium pump ACA8 (37). In either case, complementation provides genetic evidence that ACA9 can function as a high-affinity Ca2+ pump.
Fig. 5.
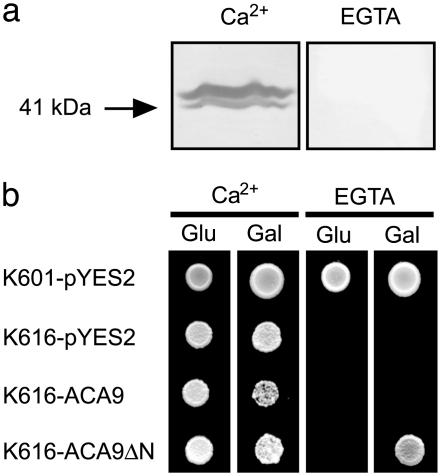
The N terminus of At-ACA9 binds calmodulin, and an N-terminally truncated version of ACA9 complements a yeast mutant devoid of Ca2+ pumps. (a) Overlay assay showing calcium-dependent binding of calmodulin to an N-terminal fragment of At-ACA9. CaM binding was observed with 1 mM Ca2+ whereas binding was abolished in the presence of 5 mM EGTA. GST alone did not bind CaM, either in the presence or in the absence of Ca2+ (results not shown). (b) Yeast strain K616 devoid of endogenous Ca2+ pumps transformed with empty vector (pYES2), full-length ACA9, or an N-terminal truncated version of ACA9 (ACA9ΔN) was grown on medium with 10 mM CaCl2 or 10 mM EGTA, and with either glucose (glu) or galactose (gal). Expression of the ACA9 genes was regulated by the galactose-inducible promoter of GAL1. K616 transformed with empty vector (pYES2) was used as a control.
Importantly, ACA9 complementation of yeast K616 was observed only when using a deregulated pump created by deleting 93 residues of the N-terminal domain that harbors a predicted calmodulin-regulated autoinhibitor. This result agrees with findings on other P2B ATPases that likewise are activated only when the autoinhibitor is removed (21, 22). To verify that the N-terminal regulatory domain of ACA9 harbors an expected calmodulin-binding site, the region Met-40 to Ala-95 of ACA9 was expressed as a GST fusion protein. In a calmodulin overlay assay (Fig. 5a), the fusion protein bound calmodulin in a Ca2+-dependent fashion. Thus, yeast complementation and calmodulin-binding studies both support the expectation that ACA9 is a calmodulin-activated calcium pump, similar to other P2B ATPases characterized from plants.
Discussion
Evidence presented here indicates that the poor seed set in an aca9 knockout mutant results from a partial male sterility (Fig. 1). This partial male sterility seems to have at least two underlying causes, including a reduced growth potential of mutant pollen and a high frequency of aborted fertilization at the point where pollen tubes reach the embryo sac (Fig. 2). Because ACA9 functions as a plasma membrane Ca2+ pump (Figs. 4 and 5), the cause of these pollen defects is expected to involve changes in Ca2+ signaling or nutrition. The identification of aca9 mutants with a disruption of a pollen-specific Ca2+ pump should provide a useful genetic tool to elucidate the role of Ca2+ dynamics in pollen tube growth and development.
The potential roles of Ca2+ in pollen have received considerable attention. For example, an oscillating tip-focused Ca2+ gradient in growing pollen tubes has been reported using fluorescent and luminescent Ca2+ indicators (9–11, 13, 38, 39). The Ca2+ gradient is usually seen only in growing pollen tubes, and manipulation of extracellular and intracellular Ca2+ concentrations using chelators and pharmacological agents correlates with an inhibition of growth (40–42). However, despite the vast amount of evidence indicating a connection between Ca2+ dynamics and pollen tube growth, our results on ACA9 provide genetic evidence to support a model in which Ca2+ signals are natural regulators of pollen tube growth and fertilization.
Whereas current models for pollen tip growth and fertilization already include Ca2+ influx channels at the plasma membrane, they have not included a specific function for a plasma membrane Ca2+ efflux pump. Evidence presented here indicates that a more complete model must include a Ca2+ pump (e.g., ACA9) as a necessary efflux pathway at the plasma membrane. Thus, we offer two models for considering the function of ACA9 in the pollen tube plasma membrane. First, ACA9 may provide a mechanism for regulating cytosolic Ca2+ homeostasis, thereby preventing the buildup of cytotoxic levels of Ca2+. Although we cannot exclude this “homeostasis” model, we consider it unlikely because expression profiling studies have provided evidence that pollen may express other Ca2+ efflux transporters (43) that, in the absence of ACA9, should be able to regulate Ca2+ homeostasis. In addition, yeast seem to regulate cytosolic Ca2+ levels without any plasma membrane localized Ca2+ pump (30). Thus, there is no precedent for expecting a plasma membrane Ca2+ pump to function as a master controller of Ca2+ homeostasis.
We therefore favor a second model in which ACA9 is part of a Ca2+ oscillator that functions in Ca2+ signaling at the plasma membrane (44). In this model, ACA9 provides a regulated efflux pathway that recycles the Ca2+ that enters through a plasma membrane channel back to its point of origin, thereby priming the system for a new Ca2+ spike. A defect in a plasma membrane oscillator may disrupt multiple signaling pathways that originate at the plasma membrane, including signals that feed-back regulate the oscillatory behavior of the tip-localized Ca2+ gradient during pollen tip growth. Thus, the disruption of an efflux component of a plasma membrane Ca2+ oscillator could easily result in both slower tip growth and impaired signaling interactions with the embryo sac, as documented here for aca9 mutants.
A feature of the aca9 phenotype is the apparent block in fertilization at the point of physical interaction between the pollen tube and embryo-sac/synergid. Currently, the best characterized mutations that disrupt this critical aspect of fertilization all correspond to gene functions expressed in the female gametophyte [e.g., sirene/feronia (34, 45)] rather than the male gametophyte (e.g., like aca9). The mutants sirene and feronia affect female gametophytic functions that somehow prevent the rupture of pollen tubes upon penetration of the synergid (i.e., no sperm cell discharge). This result may be due to the loss of a signal produced by the female gametophyte, most likely from the synergid cell itself (33). In contrast, the aca9 mutation results in a deficiency within the male gametophyte and potentially represents a defect in decoding or acting upon a signal originating from the female gametophyte. Although sirene/feronia and aca9 all show a defect in discharging sperm, the pollen tubes in the sirene/feronia mutant continue to grow into the synergid whereas the aca9 pollen tubes respond appropriately to a signal that stops further growth. Thus, aca9 and sirene/feronia show distinctly different phenotypes at the cellular level.
In conclusion, genetic evidence identifies ACA9 as a Ca2+ transporter that functions in the male gametophyte for normal pollen tube growth and the interaction between the pollen tube and embryo sac. This finding provides an example of a loss of function phenotype for an ACA-type Ca2+ pump in plants. As such, aca9/ACA9 provides a paradigm for exploring the structure and function of all 10 members of this gene family in Arabidopsis, as well as calmodulin-regulated Ca2+ pumps in non-plant systems. For example, a plasma membrane Ca2+ pump has been implicated in the viability of mammalian sperm (7, 46), the animal counterpart to the male gametophyte in plants. Thus, our results provide a genetic tool to begin dissecting the role of calmodulin-regulated Ca2+ pumps in plants and animals.
Acknowledgments
Assistance with confocal microscopy was provided by Malcolm Wood. Special thanks to Kathy Truong, Citlali Villalobos, and Greta Granstedt for technical assistance. This work was supported by grants from the Human Frontier Science Program Organization (to M.G.P. and J.F.H.), the European Union Framework Five Program (to M.G.P.), the Danish Agricultural and Veterinary Research Council (to M.G.P.), and the Department of Energy (DE-FG03-94ER20152) and the National Institutes of Health (to J.F.H.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PMCA, plasma membrane Ca2+ ATPase; ACA, autoinhibited Ca2+ ATPase; T-DNA, portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells; YFP, yellow fluorescence protein; GUS, β-glucuronidase.
References
- 1.White, P. J. & Broadley, M. R. (2003) Ann. Bot. 92, 487-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berridge, M. J., Lipp, P. & Bootman, M. D. (2000) Nat. Rev. Mol. Cell. Biol. 1, 11-21. [DOI] [PubMed] [Google Scholar]
- 3.Sanders, D., Pelloux, J., Brownlee, C. & Harper, J. F. (2002) Plant Cell 14, S401-S417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geisler, M., Axelsen, K. B., Harper, J. F. & Palmgren, M. G. (2000) Biochim. Biophys. Acta 1465, 52-78. [DOI] [PubMed] [Google Scholar]
- 5.Sze, H., Liang, F., Hwang, I., Curran, A. C. & Harper, J. F. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 433-462. [DOI] [PubMed] [Google Scholar]
- 6.Kozel, P. J., Friedman, R. A., Erway, L. C., Yamoah, E. N., Liu, L. H., Riddle, T., Duffy, J. J., Doetschman, T., Miller, M. L., Cardell, E. L., et al. (1998) J. Biol. Chem. 273, 18693-18696. [DOI] [PubMed] [Google Scholar]
- 7.Shull, G. E., Okunade, G., Liu, L. H., Kozel, P., Periasamy, M., Lorenz, J. N. & Prasada, V. (2003) Ann. N.Y. Acad. Sci. 986, 453-460. [DOI] [PubMed] [Google Scholar]
- 8.Holdaway-Clarke, T. L. & Hepler, P. K. (2003) New Phytol. 159, 539-563. [DOI] [PubMed] [Google Scholar]
- 9.Reiss, H. D. & Herth, W. (1978) Protoplasma 97, 373-377. [Google Scholar]
- 10.Rathore, K. S., Cork, R. J. & Robinson, K. R. (1991) Dev. Biol. 148, 612-619. [DOI] [PubMed] [Google Scholar]
- 11.Miller, D. D., Callaham, D. A., Gross, D. J. & Hepler, P. K. (1992) J. Cell Sci. 101, 7-12. [Google Scholar]
- 12.Malho, R., Read, N. D., Pais, M. S. & Trewavas, A. J. (1994) Plant J. 5, 331-341. [Google Scholar]
- 13.Pierson, E. S., Miller, D. D., Callaham, D. A., Shipley, A. M., Rivers, B. A., Cresti, M. & Hepler, P. K. (1994) Plant Cell 6, 1815-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malho, R. & Trewavas, A. J. (1996) Plant Cell 8, 1935-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor, L. P. & Hepler, P. K. (1997) Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 461-491. [DOI] [PubMed] [Google Scholar]
- 16.Franklin-Tong, V. E. (1999) Plant Cell 11, 727-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hepler, P. K., Vidali, L. & Cheung, A. Y. (2001) Annu. Rev. Cell Dev. Biol. 17, 159-187. [DOI] [PubMed] [Google Scholar]
- 18.Digonnet, C., Aldon, D., Leduc, N., Dumas, C. & Rougier, M. (1997) Development (Cambridge, U.K.) 124, 2867-2874. [DOI] [PubMed] [Google Scholar]
- 19.Wilhelmi, L. K. & Preuss, D. (1997) Plant Physiol. 113, 307-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baxter, I., Tchieu, J., Sussman, M. R., Boutry, M., Palmgren, M. G., Gribskov, M., Harper, J. F. & Axelsen, K. B. (2003) Plant Physiol. 132, 618-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisler, M., Frangne, N., Gomes, E., Martinoia, E. & Palmgren, M. G. (2000) Plant Physiol. 124, 1814-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harper, J. F., Hong, B., Hwang, I., Guo, H. Q., Stoddard, R., Huang, J. F., Palmgren, M. G. & Sze, H. (1998) J. Biol. Chem. 273, 1099-1106. [DOI] [PubMed] [Google Scholar]
- 23.Bonza, M. C., Morandini, P., Luoni, L., Geisler, M., Palmgren, M. G. & De Michelis, M. I. (2000) Plant Physiol. 123, 1495-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krysan, P. J., Young, J. C., Tax, F. & Sussman, M. R. (1996) Proc. Natl. Acad. Sci. USA 93, 8145-8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 26.Hellens, R. P., Edwards, E. A., Leyland, N. R., Bean, S. & Mullineaux, P. M. (2000) Plant Mol. Biol. 42, 819-832. [DOI] [PubMed] [Google Scholar]
- 27.Becker, D., Kemper, E., Schell, J. & Masterson, R. (1992) Plant Mol. Biol. 20, 1195-1197. [DOI] [PubMed] [Google Scholar]
- 28.Hong, B., Ichida, A., Wang, Y., Gens, J. S., Pickard, B. G. & Harper, J. F. (1999) Plant Physiol. 119, 1165-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, F. W. (1959) Stain Technol. 34, 125-128. [DOI] [PubMed] [Google Scholar]
- 30.Muller, P. Y., Janovjak, H., Miserez, A. R. & Dobbie, Z. (2002) Biotechniques 32, 1372-1379. [PubMed] [Google Scholar]
- 31.Cunningham, K. W. & Fink, G. R. (1994) J. Cell Biol. 124, 351-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker, D. M. & Guarente, L. (1991) Methods Enzymol. 194, 182-187. [DOI] [PubMed] [Google Scholar]
- 33.Higashiyama, T., Yabe, S., N., Nishimura, Y., Miyagishima, S., Kuroiwa, H. & Kuroiwa, T. (2001) Science 293, 1480-1483. [DOI] [PubMed] [Google Scholar]
- 34.Rotman, N., Rozier, F., Boavida, L., Dumas, C., Berger, F. & Faure, J. E. (2003) Curr. Biol. 13, 432-436. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu, K. K. & Okada, K. (2000) Development (Cambridge, U.K.) 127, 4511-4518. [DOI] [PubMed] [Google Scholar]
- 36.Smyth, D. R., Bowman, J. L. & Meyerowitz, E. M. (1990) Plant Cell 2, 755-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonza, M. C., Luoni, L. & De Michelis, M. I. (2004) Planta 218, 814-823. [DOI] [PubMed] [Google Scholar]
- 38.Holdaway-Clarke, T. L., Feijo, J. A., Hackett, G. R., Kunkel, J. G. & Hepler, P. K. (1997) Plant Cell 9, 1999-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Messerli, M. & Robinson, K. R. (1997) J. Cell Sci. 110, 1269-1278. [DOI] [PubMed] [Google Scholar]
- 40.Picton, J. M. & Steer, M. W. (1983) J. Cell Sci. 63, 303-310. [DOI] [PubMed] [Google Scholar]
- 41.Reiss, H. D. & Herth, W. (1979) Planta 145, 225-232. [DOI] [PubMed] [Google Scholar]
- 42.Reiss, H. D. & Herth, W. (1982) Planta 156, 218-225. [DOI] [PubMed] [Google Scholar]
- 43.Honys, D. & Twell, D. (2003) Plant Physiol. 132, 640-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harper, J. F. (2001) Trends Plant Sci. 6, 395-397. [DOI] [PubMed] [Google Scholar]
- 45.Huck, N., Moore, J. M., Federer, M. & Grossniklaus, U. (2003) Development (Cambridge, U.K.) 130, 2149-2159. [DOI] [PubMed] [Google Scholar]
- 46.Wennemuth, G., Babcock, D. F. & Hille, B. (2003) J. Gen. Physiol. 122, 115-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faure, J. E., Rotman, N., Fortune, P. & Dumas, C. (2002) Plant J. 30, 481-488. [DOI] [PubMed] [Google Scholar]
- 48.Jefferson, R. A. (1987) Plant Mol. Biol. Rep. 5, 387-405. [Google Scholar]