Abstract
Birds are the most species-rich class of tetrapod vertebrates and have wide relevance across many research fields. We explored bird macroevolution using full genomes from 48 avian species representing all major extant clades. The avian genome is principally characterized by its constrained size, which predominantly arose because of lineage-specific erosion of repetitive elements, large segmental deletions, and gene loss. Avian genomes furthermore show a remarkably high degree of evolutionary stasis at the levels of nucleotide sequence, gene synteny, and chromosomal structure. Despite this pattern of conservation, we detected many non-neutral evolutionary changes in protein-coding genes and noncoding regions. These analyses reveal that pan-avian genomic diversity covaries with adaptations to different lifestyles and convergent evolution of traits.
With ~10,500 living species (1), birds are the most species-rich class of tetrapod vertebrates. Birds originated from a theropod lineage more than 150 million years ago during the Jurassic and are the only extant descendants of dinosaurs (2, 3). The earliest diversification of extant birds (Neornithes) occurred during the Cretaceous period. However, the Neoaves, the most diverse avian clade, later underwent a rapid global expansion and radiation after a mass extinction event ~66 million years ago near the Cretaceous-Paleogene (K-Pg) boundary (4, 5). As a result, the extant avian lineages exhibit extremely diverse morphologies and rates of diversification. Given the nearly complete global inventory of avian species, and the immense collected amount of distributional and biological data, birds are widely used as models for investigating evolutionary and ecological questions (6, 7). The chicken (Gallus gallus), zebra finch (Taeniopygia guttata), and pigeon (rock dove) (Columba livia) are also important model organisms in disciplines such as neuroscience and developmental biology (8). In addition, birds are widely used for global conservation priorities (9) and are culturally important to human societies. A number of avian species have been domesticated and are economically important. Farmed and wild water birds are key players in the global spread of pathogens, such as avian influenza virus (10).
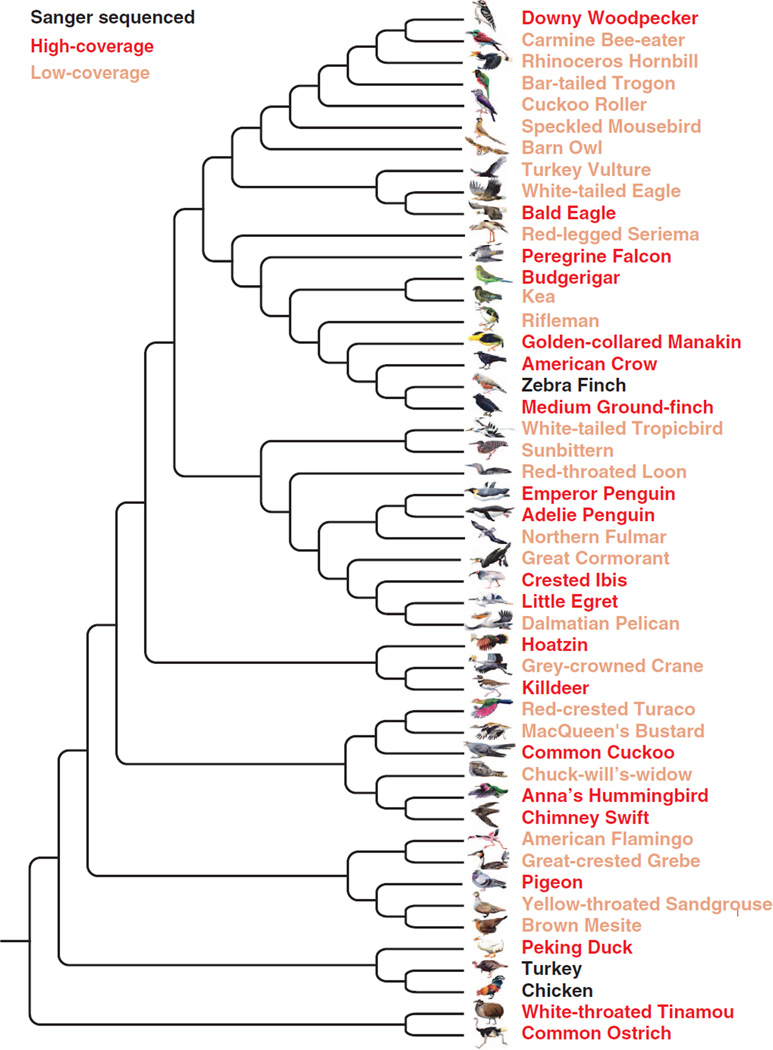
Despite the need to better understand avian genomics, annotated avian genomic data was previously available for only a few species: the domestic chicken, domestic turkey (Meleagris gallopavo) and zebra finch (11–13), together with a few others only published recently (14–16). To build an understanding of the genetic complexity of birds and to investigate links between their genomic variation and phenotypic diversity, we collected and compared genome sequences of these and other avian species (48 species total), representing all 32 neognath and two of the five palaeognath orders (Fig. 1) (17), thus representing nearly all of the major clades of living birds (5).
Fig. 1. Avian family tree and genomes sequenced.
The phylogenomic relationships of the 48 avian genomes from (5), with Sanger-sequenced (black), high-coverage (dark red), and low-coverage (light red) genomes denoted.
Results
Sequencing, assembly, and annotation
We used a whole-genome shotgun strategy to generate genome sequences of 45 new avian species (18), including two species representing two orders within the infraclass Paleognathae [common ostrich (Struthio camelus) and white-throated tinamou (Tinamus guttatus)], the other order within Galloanserae [Peking duck (Anas platyrhynchos)], and 41 species representing 30 neoavian orders (table S1) (19). In combination with the three previously published avian genomes (11–13), the genome assemblies cover 92% (34 of 37) of all avian orders (the three missing orders belong to the Paleognathae) (17). With the exception of the budgerigar (Melopsittacus undulatus), which was assembled through a multiplatform (Illumina/GS-FLX/PacBio) approach (20), all other new genomes were sequenced and assembled with Illumina (San Diego, CA) short reads (Fig. 1) (18). For 20 species, we produced high (>50×) coverage sequences from multiple libraries, with a gradient of insert sizes and built full-genome assemblies. For the remaining 25 species, we generated low (~30×) coverage data from two insert-size libraries and built less complete but still sufficient assemblies for comparative genome analyses. These de novo (18) genome assemblies ranged from 1.05 to 1.26 Gb, which is consistent with estimated cytology-based genome sizes (21), suggesting near complete genome coverage for all species. Scaffold N50 sizes for high-coverage genomes ranged from 1.2 to 6.9 Mb, whereas those for lower-coverage genomes were ~48 kb on average (table S2). The genomes of the ostrich and budgerigar were further assembled with optical maps, increasing their scaffold N50 sizes to 17.7 and 13.8 Mb, respectively (20, 22).
We annotated the protein-coding sequences using a homology-based method for all genomes, aided by transcriptome sequencing for some species (18). To avoid systematic biases related to the use of different methods in annotations of previously published avian genomes, we created a uniform reference gene set that included all genes from the chicken, zebra finch, and human (23). This database was used to predict protein gene models in all avian genomes and American alligator (Alligator mississippiensis) (24). All high-coverage genomes were predicted to contain ~15,000 to 16,000 transposable element-free protein-coding genes [table S3 and annotation files in (19)], similar to the chicken genome (~15,000). Despite the fragmented nature of the low-coverage genomes leading to ~3000 genes likely missing or partially annotated, it was still possible to predict 70 to 80% of the entire catalog of avian genes.
Broad patterns of avian genome evolution
Although many fishes and some amphibians have smaller genomes than birds, among amniotes, birds have the smallest (21). The genomes of mammals and nonavian reptiles typically range from 1.0 to 8.2 Gb, whereas avian genomes range from 0.91 in the black-chinned hummingbird (Archilochus alexanderi) to a little over 1.3 Gb in the common ostrich (21). A number of hypotheses have been proposed for the smaller avian genome size (25–28). Here, we document key events that have likely contributed to this smaller genome size.
The proliferation and loss of transposable elements (TEs) may drive vertebrate genome size evolution (29–31). Consistent with the zebra finch and galliformes genomes (11–13, 32), almost all avian genomes contained lower levels of repeat elements (~4 to 10% of each genome) (table S4) than in other tetrapod vertebrates (for example, 34 to 52% in mammals) (33).The sole outlier was the downy woodpecker (Picoides pubescens), with TEs representing ~22% of the genome, derived mainly from species-specific expansion of LINE (long interspersed elements) type CR1 (chicken repeat 1) transposons (fig. S1). In contrast, the average total length of SINEs (short interspersed elements) in birds has been reduced to ~1.3 Mb, which is ~10 to 27 times less than in other reptiles [12.6 Mb in alligator; 34.9 Mb in green sea turtle (Chelonia mydas)], suggesting that a deficiency of SINEs occurred in the common ancestor of birds.
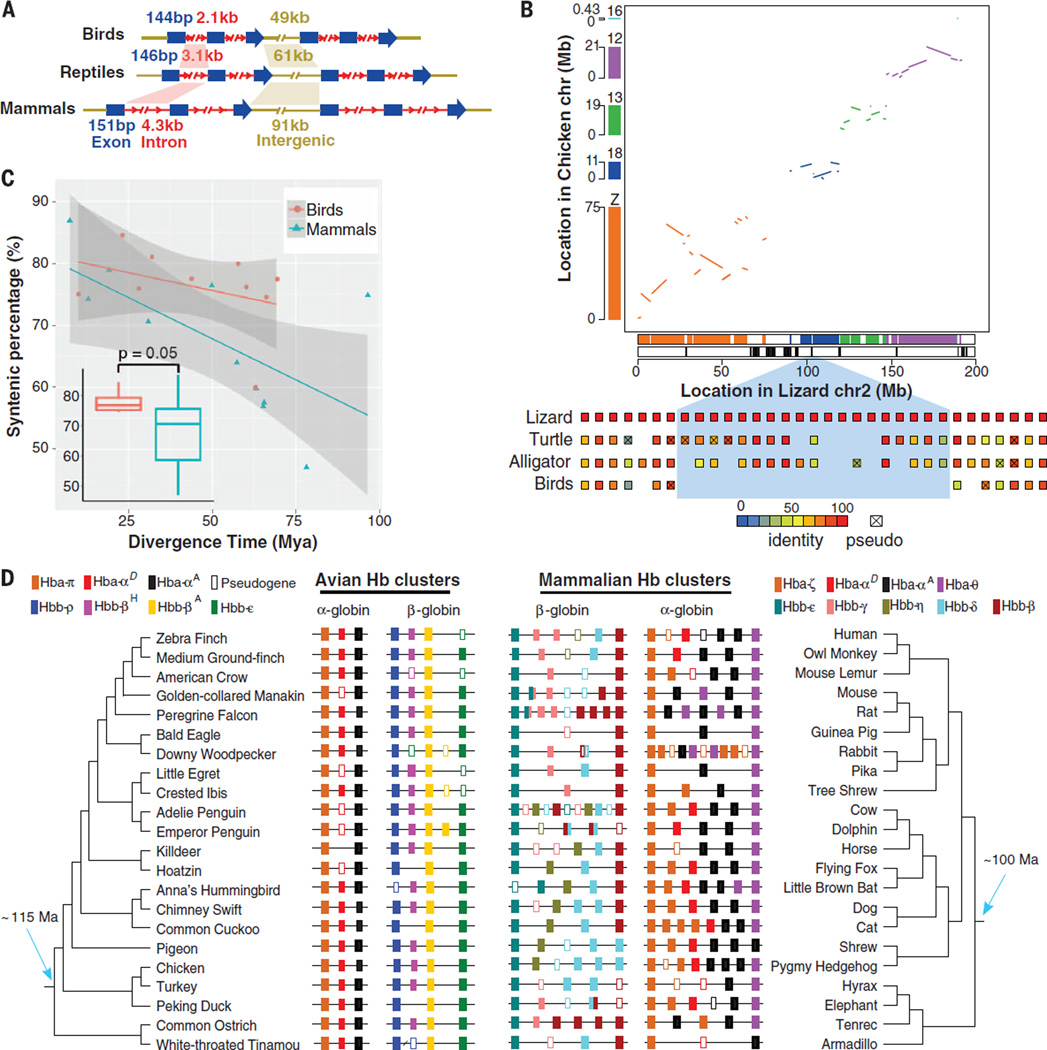
We compared the average size of genomic elements of birds with 24 mammalian and the three nonavian reptile genomes. Avian protein-coding genes were on average 50 and 27% shorter than the mammalian and reptilian genes, respectively (Fig. 2A). This reduction is largely due to the shortening of introns and reduced intergenic distances that resulted in an increased gene density (Fig. 2A). Such genomic contraction has also evolved convergently in bats (fig. S11), the only flying mammalian group. The condensed genomes may represent an adaptation tied to rapid gene regulation required during powered flight (34, 35).
Fig. 2. Genome reduction and conservation in birds.
(A) Comparison of average size of introns, exons, and intergenic regions within avian, reptilian, and mammalian genomes. (B) Synteny plot and large segmental deletions between green anole chromosome 2 and multiple chicken chromosomes. Colored bars and lines indicate homologous blocks between two species; black bars indicate location of large avian-specific segmental deletions, which are enriched at the breakpoints of interchromosome rearrangements. (Bottom) An example of a large segmental deletion in birds (represented by ostrich genes). Homologous genes annotated in each species are shown in small boxes. The color spectrum represents the percent identity of homologous genes with the green anole. (C) Distribution of gene synteny percentages identified for phylogenetically independent species pairs of various divergence ages. Dots indicate the percentage of genes remaining in a syntenic block in pairwise comparisons between two avian or mammalian species. Box plots indicate that the overall distributions of the synteny percentages in birds and mammals are different (P value was calculated by using Wilcoxon rank sum test with phylogenetically independent species pairs). (D) Chromosomal organization of the α- and β-globin gene clusters in representative avian and mammalian taxa. These genes encode the α- and β-type subunits of tetrameric (α2β) hemoglobin isoforms that are expressed at different ontogenetic stages. In; the case of the α-like globin genes, birds and mammals share orthologous copies of the αD- and αA-globin genes. Likewise, the avian π-globin and the mammalian ζ-globin genes are 1:1 orthologs. In contrast, the genes in the avian and mammalian β-globin gene clusters are derived from independent duplications of one or more β-like globin genes that were inherited from the common ancestor of tetrapod vertebrates (90, 91).
To further investigate whether avian genome size reduction is due to a lineage-specific reduction in the common avian ancestor of birds or expansion in other vertebrates (36), we performed ancestral state reconstructions of small [<100 base pairs (bp)] deletion events across an alignment of four representative well-assembled avian and three reptile genomes (18) and found that the avian ancestral lineage experienced the largest number of small deletion events—about twice the number in the common ancestor of birds and crocodiles (fig. S12). In contrast, many fewer small deletion events occurred in modern avian lineages (fig. S12).
We next created a gene synteny map between the highest-quality assembled avian genome (ostrich) and other reptile genomes to document lineage-specific events of large segmental deletions (18). We detected 118 syntenic blocks, spanning a total of 58 Mb, that are present in alligator and turtle genomes but lost in all birds (table S8). In contrast, ~8× and ~5× fewer syntentic blocks were missing in alligator (14 blocks, 9 Mb) and turtle (27 blocks, 8 Mb) relative to green anole, respectively, confirming the polarity of genome size reduction in birds (table S8).The large segmental losses in birds were skewed to losses from chr2 and chr6 of the green anole (fig. S13). Two of the green anole’s 12 pairs of microchromosomes, LGd and LGf, were completely missing in birds, with no homologous genes found within the avian genomes. Most of these lost segments were located at the ends of chromosomes or close to the centrosomes (fig. S13). Furthermore, lost segments were enriched at apparent breakpoints of the avian microchromosomes (Fig. 2B and fig. S13). These findings imply that the large segmental losses may be a consequence of chromosomal fragmentation events in the common ancestor of birds giving rise to additional microchromosomes in modern birds.
The large segmental deletions in birds contain at least 1241 functional protein-coding genes (table S9), with each lost segment containing at least five contiguous genes. The largest region lost in birds was a 2.1-Mb segment of the green anole chr2, which contains 28 protein-coding genes (Fig. 2B). Overall, at least 7% of the green anole macrochromosomal genes were lost through segmental deletions in birds. Although gene loss is a common evolutionary process, this massive level of segmental deletion has not been previously observed in vertebrates. Over 77% of the 1241 genes present in the large segmentally deleted regions have at least one additional paralog in the green anole genome, a level higher than the overall percentage of genes with paralogs in the green anole genome or avian genomes (both at ~70%). This suggests that birds may have undergone functional compensation in their paralogous gene copies, reducing selection against the loss of these segmental regions. We predict that the loss of functions associated with many genes in the avian ancestor may have had a profound influence on avian-specific traits (table S11).
Conservative mode of genome evolution
With ~2/3 of avian species possessing ~30 pairs of microchromosomes, the avian karyotype appears to be distinctly conserved because this phenotype is not a general feature of any other vertebrate group studied to date (37). We assessed the rates of avian chromosomal evolution among the 21 more fully assembled genomes (scaffold N50 > 1 Mb) (table S2) (18). From the alignment of chicken with the other 20 avian genomes, plus green anole and Boa constrictor (38), we identified homologous synteny blocks (HSBs) and 1746 evolutionary breakpoint regions (EBRs) in different avian lineages and then estimated the expected number of EBRs (18) and the rates of genomic rearrangements, using a phylogenetic total evidence nucleotide tree (TENT) as a guide (5). We excluded the turkey genome after detecting an unusually high fraction of small lineage-specific rearrangements, suggesting a high number of local misassemblies. Of the 18 remaining non–Sanger-sequenced genomes (table S2), the estimated rate of chimeric scaffolds that could lead to false EBRs was ~6% (39).
The average rate of rearrangements in birds is ~1.25 EBRs per million years; however, bursts of genomic reorganization occurred in several avian lineages (fig. S15). For example, the origin of Neognathae was accompanied by an elevated rate of chromosome rearrangements (~2.87 EBRs per million years). Intriguingly, all vocal learning species [zebra finch, medium-ground finch (Geospiza fortis), American crow (Corvus brachyrhynchos), budgerigar, and Anna’s hummingbird (Calypte anna)] had significantly higher rates of rearrangements than those of close vocal nonlearning relatives [golden-collared manakin (Manacus vitellinus), peregrine falcon (Falco peregrinus) and chimney swift (Chaetura pelagica)] [phylogenetic analysis of variance F statistic (F) = 5.78, P = 0.0499] and even higher relative to all vocal nonlearning species (F = 15.03, P = 0.004). This may be related to the larger radiations these clades experienced relative to most other bird groups. However, the golden-collared manakin, which belongs to suboscines (vocal non-learners) that have undergone a larger radiation than parrots and hummingbirds, has a low rearrangement rate.
We next compared microsynteny (local gene arrangements), which is more robust and accurate than macrosynteny analyses for draft assemblies (18). We compared with eutherian mammals, which are approximately the same evolutionary age as Neoaves and whose genome assemblies are of similar quality. We examined the fraction of orthologous genes identified from each pair of two-avian/mammalian genomes, on the basis of syntenic and best reciprocal blast matches (18). Birds have a significantly higher percentage of synteny-defined orthologous genes than that of mammals (Fig. 2C). The fraction of genes retained in syntenic blocks in any pairwise comparison was linearly related with evolutionary time, by which the overall level of genome shuffling in birds was lower than in mammals over the past ~100 million years (Fig. 2C). This suggests a higher level of constraint on maintaining gene synteny in birds relative to mammals.
The apparent stasis in avian chromosome evolution suggests that birds may have experienced relatively low rates of gene gain and loss in multigene families. We examined the intensively studied gene families that encode the various α- and β-type subunits of hemoglobin, the tetrameric protein responsible for blood oxygen transport in jawed vertebrates (40). In amniotes, the α- and β-globin gene families are located on different chromosomes (40) and experienced high rates of gene turnover because of lineage-specific duplication and deletion events (41). In birds, the size and membership composition of the globin gene families have remained remarkably constant during ~100 million years of evolution, with most examined species retaining an identical complement (Fig. 2D). Estimated gene turnover rates (λ) of α- and β-globin gene families were over twofold higher in mammals than birds (λ = 0.0023 versus 0.0011, respectively). Much of the variation in the avian α-globin gene family was attributable to multiple independent inactivations of the αD-globin gene (Fig. 2D), which encodes the α-chain subunit of a hemoglobin isoform (HbD) expressed in both embryonic and definitive erythrocytes (42). Because of uniform and consistent differences in oxygen-binding properties between HbD and the major adult-expressed hemoglobin isoform, HbA (which incorporates products of αA-globin) (42), the inactivations of αD-globin likely contribute to variation in blood-oxygen affinity, which has important consequences for circulatory oxygen transport and aerobic energy metabolism. Overall, the globin gene families illustrate a general pattern of evolutionary stasis in birds relative to mammals.
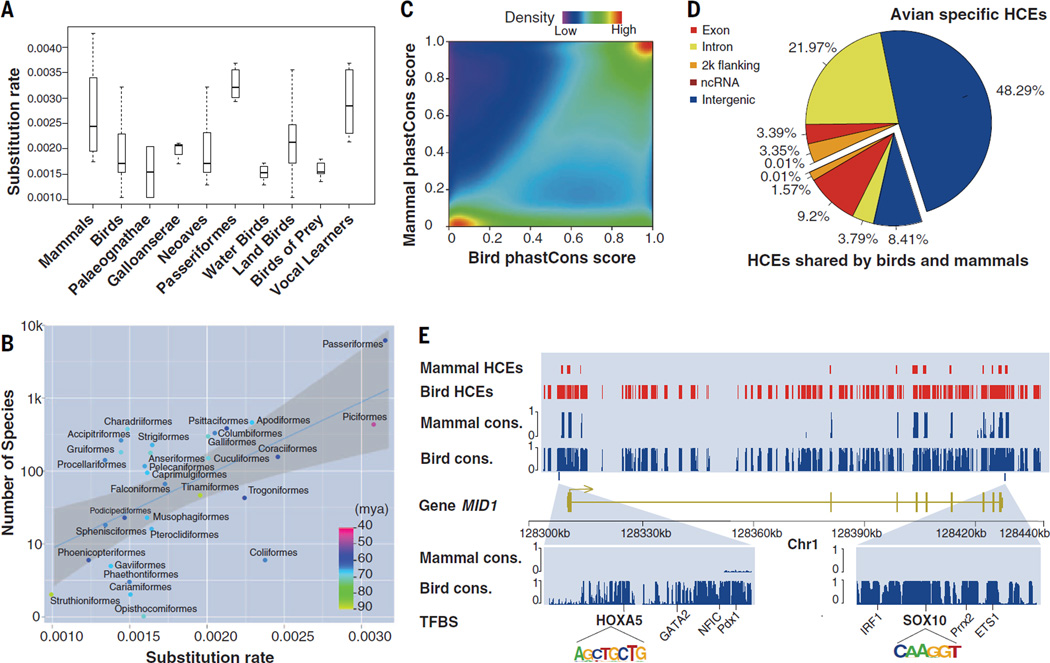
Genomic nucleotide substitution rates vary across species and are determined through both neutral and adaptive evolutionary processes (43).We found that the overall pan-genomic background substitution rate in birds (~1.9 × 10−3 substitutions per site permillion years) was lower than in mammals (~2.7 × 10−3 substitution per site per million years) (Fig. 3A). However, the substitution rate estimates also exhibited interordinal variation among birds (Fig. 3A). There was a positive correlation between the substitution rate and the number of species per order [coefficient of determination (R2) = 0.21, P = 0.01, Pearson’s test with phylogenetically independent contrasts] (Fig. 3B and fig. S19), evidencing an association with rates of macroevolution (44). For example, Passeriformes, the most diverse avian order, exhibited the highest evolutionary rate (~3.3 × 10−3 substitutions per site per million years), almost two times the average of Neoaves (~2 × 10−3 substitutions per site per million years, Fig. 3A). Landbirds exhibited an average higher substitution rate than that of waterbirds (landbirds, ~2.2 × 10−3 substitutions per site per million years; waterbirds, ~1.6 × 10−3 substitutions per site per million years), which is consistent with the observation that landbirds have greater net diversification rates than those of waterbirds (7). Among the landbirds, the predatory lineages exhibited slower rates of evolution (~ 1.6 × 10−3 substitutions per site permillion years), similar to that of waterbirds. Moreover, the three vocal learning landbird lineages (parrots, songbirds, and hummingbirds) are evolving faster than are nonvocal learners (Fig. 3A). Overall, our analyses indicate that genome-wide variation in rates of substitution is a consequence of the avian radiation into a wide range of niches and associated phenotypic changes.
Fig. 3. Evolutionary rate and selection constraints.
(A) Substitution rate in each lineage was estimated by the comparison of fourfold degenerate (4d) sites in coding regions, in units of substitutions per site per million years. Waterbirds and landbirds are defined in (5). (B) Correlation between average substitution rates and number of species within different avian orders. Divergence times were estimates from (5).The fit line was derived from least square regression analysis, and the confidence interval was estimated by “stat_smooth” in R. The units of the x axis are numbers of substitutions per site per million years. The correlation figure with phylogenetically independent contrasts is provided in the supplementary materials. (C) Density map for comparison of conservation levels between pan-avian and pan-mammalian genomes, on the basis of the homologous genomic regions between birds and mammals. Conservation levels were quantified by means of PhastCons basewise conservation scores. (D) HCEs found in both mammalian and avian genomes (smaller pie piece) and those that are avian-specific (larger pie piece). (E) MID1 contains abundant avian-specific HCEs in the upstream and downstream regulatory regions. Many regulatory motif elements are identified in these avian-specific HCEs. Cons., conservation level.
Selective constraints on functional elements
Conservation of DNA sequences across distantly related species reflects functional constraints (45). A direct comparison of 100-Mb orthologous genomic regions revealed more regions evolving slower than the neutral rate among birds (Fig. 3C) than mammals (46), which is consistent with the slower rate of avian mitochondrial sequence evolution (47). We predicted 3.2 million highly conserved elements (HCEs) at a resolution of 10 bp or greater spanning on average 7.5% of the avian genome, suggesting a strong functional constraint in avian genomes. Functional annotations revealed that ~12.6% of these HCEs were associated with protein-coding genes, whereas the majority of the remaining HCEs were located in intron and intergenic regions (Fig. 3, D and E). These HCEs enabled us to identify 717 new protein-coding exons and 137 new protein-coding genes, with 77% of the latter supported by the deep transcriptome data (table S17). Deep transcriptome sequencing also enabled us to annotate 5879 candidate long noncoding RNA (lncRNA) genes, of which 220 overlapped HCEs with a coverage ratio of >50% (table S18) (18).
Because HCEs may have different functions in different lineages, we separated the HCEs into two categories: bird-specific and amniote HCEs (shared by birds and mammals). Among the bird-specific HCEs, we identified 13 protein-coding genes that were highly conserved in birds but divergent in mammals (table S19).One of the most conserved was the sperm adhesion gene, SPAM1, which mediates sperm binding to the egg coat (48). This gene, however, was under positive selection driven by sperm competition in mammalian species (49). Noncoding HCEs play important roles in the regulation of gene expression (50); thus, we compared the transcription factor binding sites in the ENCODE project (51) with the HCEs and found that the avian-specific HCEs are significantly associated with transcription factors functioning in metabolism (table S20), whereas amniote core HCEs are enriched with transcription factors functioning in signal regulation, stimulus responses, and development (table S21).
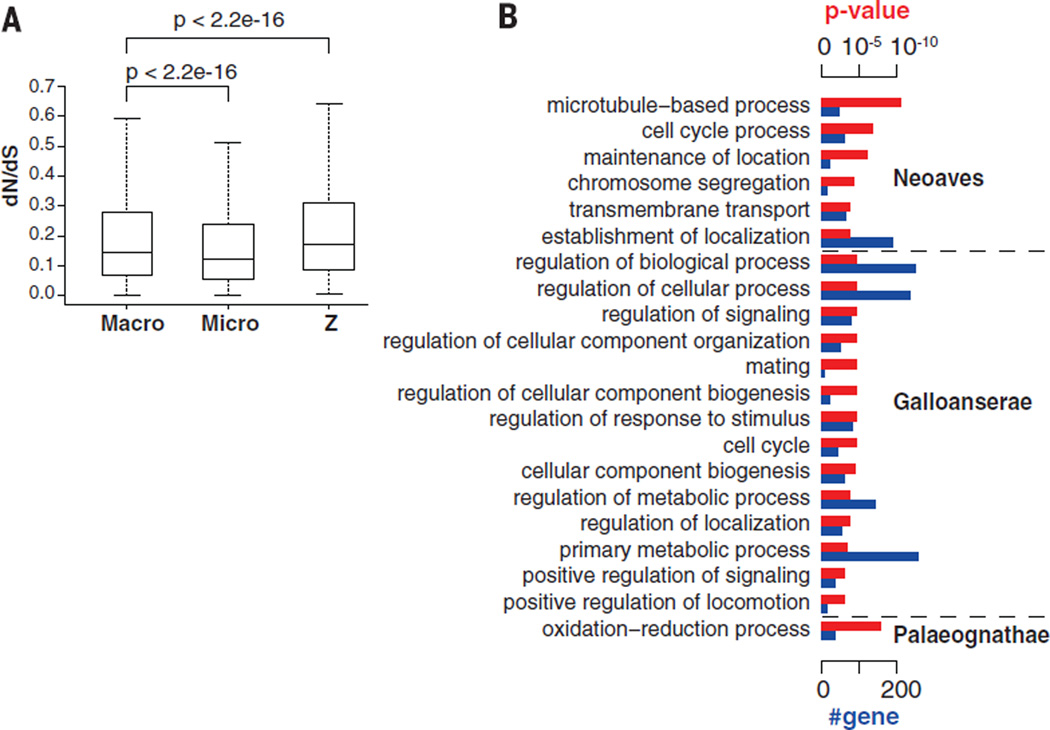
To investigate evolutionary constraints on gene regions, we calculated dN/dS [the ratio of the number of nonsynonymous substitutions per non-synonymous site (dN) to the number of synonymous substitutions per synonymous site (dS)] for 8295 high-quality orthologs. Consistent with the fast-Z sex chromosome hypothesis (52), the evolutionary rate of Z-linked genes was significantly higher than autosome genes (Fig. 4A). This is most likely driven by the reduction of effective population size (Ne) of Z-linked genes—because the Ne of Z chromosome is only 3/4 of that of autosomes—as well as by male sexual selection (52). Furthermore, consistent with the fast-macro hypothesis, the overall rate of macro-chromosomal genic evolution is higher than that of microchromosomes (Fig. 4A), which is probably due to differences in the recombination rates and genic densities between macro- and microchromosomes in birds (53).
Fig. 4. Selection constraints on genes.
(A) Box plot for the distribution of dN/dS values of genes on avian macrochromosomes, microchromosomes, and the Z chromosome. P values were calculated with Wilcoxon rank sum tests. (B) GO categories in Neoaves, Galloanserae, and Palaeognathae showing clade-specific rapid evolutionary rates. Red bars, P value of significance; blue bars, number of genes in each GO.
We also examined the dN/dS ratio of each avian Gene Ontology (GO) category for comparison with mammals and within birds. Those involved in development (such as spinal cord development and bone resorption) are evolving faster in birds, and those involved in the brain function (such as synapse assembly, synaptic vesicle transport, and neural crest cell migration) are evolving faster in mammals (tables S23 and S24). Genes involved in oxidoreductase activity were relatively rapidly evolving in the Palaeognathae clade that contains the flightless ratites (Fig. 4B and table S25). The fast evolving GOs in the Galloanserae participate in regulatory functions (Fig. 4B and table S26). In Neoaves, genes involved in microtubule-based processes were the fastest evolving (Fig. 4B and table S27). We speculate that these differences could be caused by relaxed selective constraints or positive selection in different lineages.
Genotype-phenotype convergent associations: Evolution of vocal learning
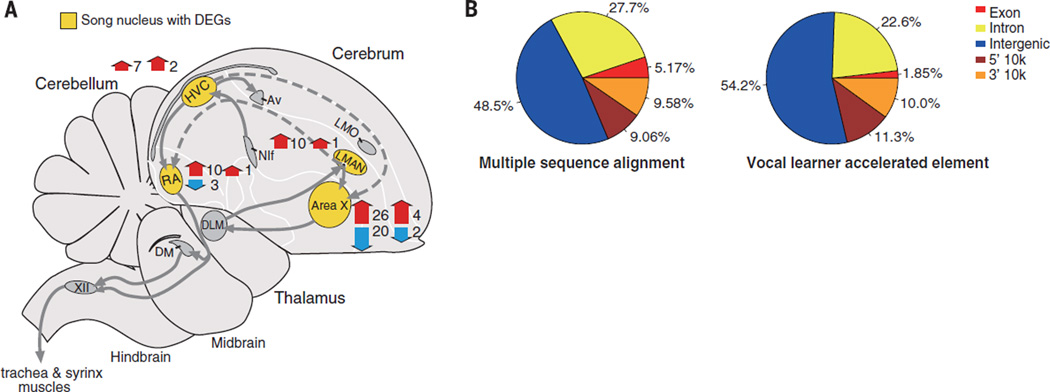
With the availability of genomes representing all major modern avian lineages and their revised phylogenetic relationships (5), it becomes possible to conduct genome-wide association studies across species with convergent traits. We focused on vocal learning, which given our phylogenetic analyses is inferred as having evolved independently, either twice, in hummingbirds and the common ancestor of songbirds and parrots, or three times (5, 54). All three groups have specialized song-learning forebrain circuits (song nuclei) not found in vocal nonlearners (Fig. 5A) (55).
Fig. 5. Convergent molecular changes among vocal learning birds.
(A) Songbird brain diagram showing the specialized forebrain song-learning nuclei (yellow) that controls the production (HVC and RA) and acquisition (LMAN and Area X) of learned song (55). Gray arrows indicate connections between brain regions; red and blue (thick) arrows indicate relative numbers of genes with increased or decreased specialized expression in zebra finch song nuclei and with convergent accelerated coding sequences (left numbers of 66 total) or convergent amino acid substitutions (right numbers of 6). Genes expressed in more than one song nucleus are counted multiple times. RA, robust nucleus of the arcopallium; LMAN, lateral magnocellular nucleus of the anterior nidopallium; Area X, Area X of the striatum; and HVC, a letter-based name. (B) Classification of vocal learner-specific accelerated elements, compared with the background alignment of 15 avian species.
Analyses of 7909 orthologous protein-coding genes with available amino acid sites in all three vocal-learning and control vocal nonlearning groups revealed convergent accelerated dN/dS for 227 genes in vocal learners (table S28). Of these, 73% (165) were expressed in the songbird brain (physically cloned mRNAs), and of these, 92% (151) were expressed in adult song-learning nuclei, which is much higher than the expected 60% of brain genes expressed in song nuclei (56). About 20% (33) were regulated by singing, which is twice the expected 10% (56). In addition, 41% of the song nuclei accelerated genes showed differential expression among song nuclei [expected 20% (56)], and 0.7 to 9% [0.7 to 4.3%expected (57)] showed specialized expression compared with the surrounding brain regions (table S28) (58). GO analyses of the accelerated differentially expressed song nuclei genes revealed 30 significant functionally enriched gene sets, which clustered into four major categories, including neural connectivity, brain development, and neural metabolism (fig. S25). For an independent measure of convergence, we developed an approach that scans for single amino acid substitutions common to species with a shared trait, controlling for phylogenetic relationships (18). Of the 7909 genes, 38 had one to two amino acid substitutions present only in vocal learners (table S31). At least 66% of these were expressed in the songbird brain, including in the song nuclei [58%; 20%expected (56)]. Two genes (GDPD4 and KIAA1919) showed convergent accelerated evolution on the amino acid sites specific to vocal learners (table S31).
To identify accelerated evolution in noncoding sequences in vocal learners, we scanned the genome alignment using phyloP (18, 59).We used a more limited sampling of vocal nonlearning species closely related to the vocal learners (table S32) because of the relatively faster evolutionary rate of noncoding regions. We scanned the entire genome alignment and found 822 accelerated genomic elements specifically shared by all three vocal learning groups (table S33). These convergent elements were skewed to intergenic regions in vocal learners relative to the background average accelerated elements across species (Fisher’s exact test, P < 2.2 × 10–16) (Fig. 5B). Of these elements, 332 were associated with 278 genes (within 10 kb 5′ or 3′ of the nearest gene), of which a high proportion (76%) was expressed in the brain; almost all of those (94%, 198 genes) expressed in one or more song nuclei, 20% were regulated by singing (10% expected), 51% (20% expected) showed differential expression among song nuclei, and 2 to 15% [0.7 to 4.3% expected, based on (56)] had specialized expression relative to the surrounding brain regions, including the FoxP1 gene involved in speech (table S34 and figs. S27 to S32). Overall, these analyses show a 2- to 3.5-fold enrichment of accelerated evolution in regulatory regions of genes differentially expressed in vocal learning brain regions. In contrast, there was very little overlap (2.5%) of genes with convergent accelerated noncoding changes and convergent accelerated amino acid changes, indicating two independent targets of selection for convergent evolution.
Evolution of ecologically relevant genes
We also investigated candidate genes that underlie traits relevant to avian ecological diversity. Although these analyses should be approached with caution given the phenotypic and ecological plasticity within major avian lineages, we examined genes putatively associated with major skeletal and tissue changes for the capacity for powered flight, feeding modification such as loss of teeth, the advanced visual system found in some lineages, and sexual and reproductive systems.
Evolution of the capacity for flight
Skeletal systems
The evolution of flight involved a series of adaptive changes at the morphological and molecular levels. One of the key requirements for flight is a skeleton that is both strong and lightweight. In both birds and nonavian theropods, this evolved through the fusion and elimination of some bones and the pneumatization of the remaining ones (60). Of 89 genes involved in ossification (table S36), 49 (~55%) showed evidence of positive selection in birds, which is almost twice as high as in mammals (31 genes, ~35%). For birds, most of these are involved in the regulation of remodeling and ossification-associated processes, or bone development in general, and those with the highest values for global dN/dS (>0.5) were obtained for AHSG (α-2-HS-glycoprotein), which is associated with bone mineral density, and P2RX7 (P2× purinoceptor 7), which is associated with bone homeostasis. The variation in the extension of pneumatization in avian post-cranial bones has been associated with the variation in body size and foraging strategies (61). Therefore, selection of these genes may explain variation in the levels of bone pneumatization in birds because the genes involved in the process of maintaining trabeculae within bones likely depends on the intrinsic network of genes participating in bone resorption and mineralization. These results suggest that most structural differences in bone between birds and mammals may be a result of bone remodeling and resorption (table S37).
Pulmonary structure and function
The increased metabolism associated with homeothermy and powered flight requires an efficient gas exchange process during pulmonary ventilation. Because of functional integration of ventilation and locomotion, birds evolved a volume-constant lung and a rigid trunk region, whereas mammals evolved a changing-volume lung, often coupled to locomotory flexion of the lumbar region (62). In contrast to the pulmonary alveola of the mammalian lung, the avian lung has a honeycomb-like structure incorporating a flow-through system with small air capillaries (63). We found five genes that function in mammalian lung development that were lost in the avian ancestor (table S11).
Feathers
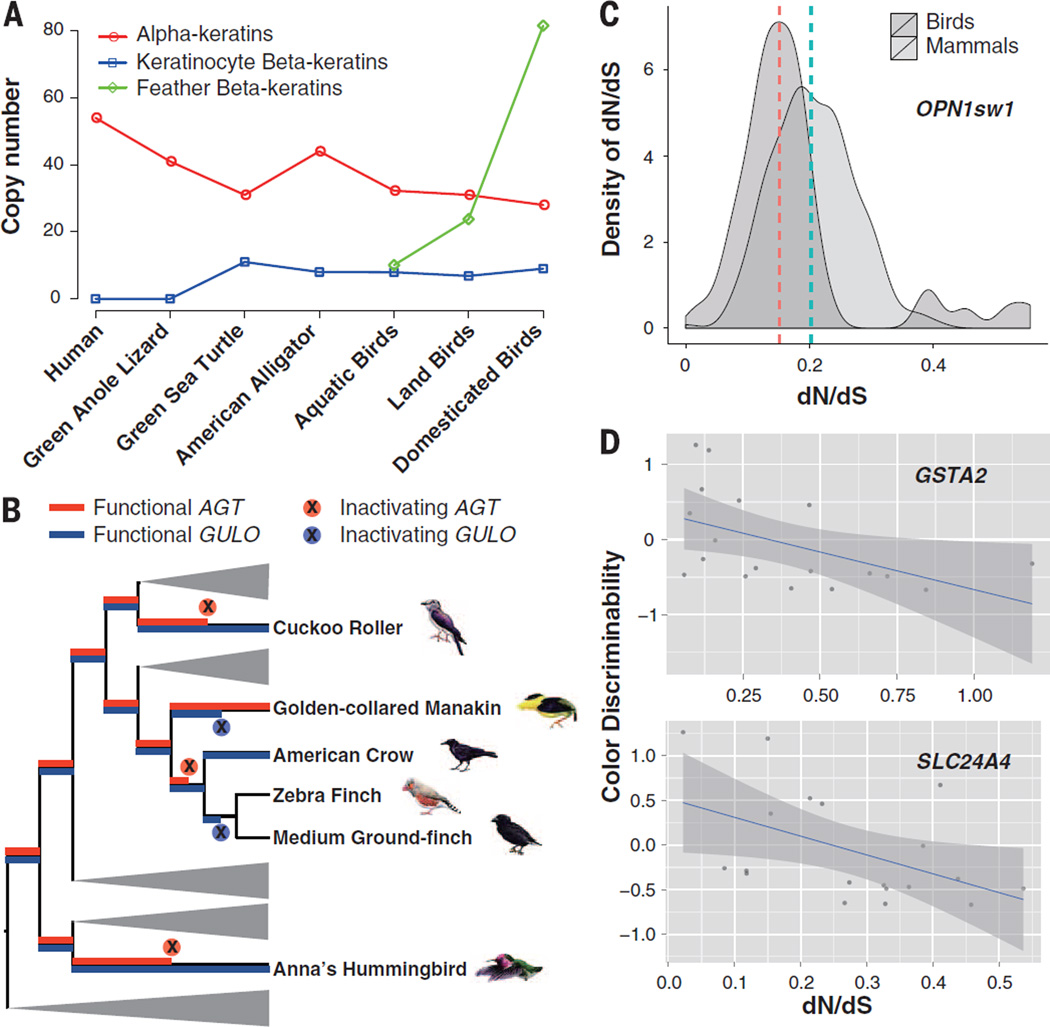
The evolution and subsequent morphological diversification of feathers have shaped avian physiology, locomotion, mate choice, and ecological niches (64). Feathers are composed of α- and β-keratins (65), the latter of which are structural proteins found only in the epidermal appendages of birds and other reptiles. The α-keratin gene family has contracted in birds relative to reptiles (except turtle) and mammals (0.7-fold change), whereas the β-keratin gene family has expanded (1.96-fold change) relative to reptiles (Fig. 6A and table S39). The avian β-keratins form six clusters, with all major avian lineages possessing members from each avian cluster (fig. S33), indicating that avian β-keratin diversity was present in the basal avian lineage. Of these, the feather β-keratin subfamily is avian-specific and comprises over 56% of the genes, whereas the remaining avian β-keratin subfamilies (claw, scale, and keratinocyte β-keratin subfamilies) are found in turtles and crocodiles (Fig. 6A and fig. S33). The mean number of keratinocyte β-keratins is similar across bird groups and their two closest living reptile relatives (turtle and alligator), suggesting copy number conservation since their common ancestor (Fig. 6A). In contrast, aquatic/semi-aquatic birds have a relatively low mean number of feather β-keratins compared with that of land birds, with land birds having more than double the number, and among them several domesticated land birds (zebra finch, chicken, pigeon, and budgerigar) having more than 8 times (Fig. 6A). Although the later observation is concordant with the hypothesis that domestication may increase the recombination rate at β-keratin loci (66, 67), domestic turkey and Peking duck did not exhibit this trend. Overall, these findings indicate that feather compositional adaptations are associated with different avian lifestyles.
Fig. 6. Genetic changes associated with ecological adaptations.
(A) Copy numbers of α- and β-keratins in humans, reptiles, and birds, including in aquatic birds, land birds, and domesticated birds. Definitions of aquatic and landbirds are provided in (5). (B) Pseudogenization events of the diet-related genes AGT and GULO along the avian phylogeny. (C) Density distribution of dN/dS values of the OPN1sw1 gene for mammals (median, 0.21) and birds (median, 0.16). (D) dN/dS values of two plumage color–related genes (GSTA2 and SLC24A4) show negative correlation with the color discriminability values (log transformation applied). The correlation figures with phylogenetically independent contrasts are provided in the supplementary materials.
Evolution of genes related to diet
Edentulism
The evolution of birds also had major consequences with regard to their feeding strategies and diets, with changes at the structural, biochemical, and sensory levels (among others). One of the most immediately obvious avian-specific traits is edentulism, the phenotype of being toothless. Edentulismis thought to have evolved independently in multiple theropod lineages (68). However, although most phylogenetic analyses suggest that teeth were lost in the common ancestor of modern birds (69), several studies have recovered dentate taxa (Hesperornis and Ichthyornis) from the Mesozoic inside of crown Neornithes, suggesting that tooth loss could have occurred independently (70). A scan of avian genomes for molecular fossils of tooth-specific genes recovered remnants of enamel and dentin formation genes in all species examined [table 1 in (71)]. Frameshift mutations and whole-exon deletions were widespread in all investigated tooth genes. The vast majority of debilitating mutations were not shared, but all species shared unambiguous deletions in protein-coding exons of enamel-specific genes (ENAM, AMEL, AMBN, MMP20, and AMTN) and one dentin-specific gene (DSPP). This shared pattern of pseudogenization across living birds supports the hypothesis that the common ancestor of modern birds lacked mineralized teeth (69).
Diet-related enzymes
Birds have evolved an extraordinary diversity of dietary specializations. The glyoxylate detoxifying enzyme alanine/glyoxylate aminotransferase (AGT) represents a candidate for study (72). We recovered complete AGT genes from 22 avian genomes (table S42), of which five exhibit pseudogenized forms in their MTS region (Fig. 6B and fig. S34). MTS function was lost in three unrelated avian orders, which is consistent with multiple independent dietary transitions during avian evolution. Detection of positively selected amino acids at 137 Q (dN/dS = 2.153) and 378 R (dN/dS = 2.153) in all birds provided additional support for diet-related adaptation in AGT (positions according to human AGT; posterior probability > 99%; P < 0.0001).
Vitamin C (Vc) is an important nutrient co-factor in a range of essential metabolic reactions. Loss of the ability to synthesize Vc has occurred in humans, Guinea pigs, and some bats. All species that do not synthesize Vc exhibit a pseudogenized gene for l-gulonolactone oxidase (GULO), an enzyme essential for catalyzing the last step of Vc synthesis (73). Genomic mining revealed GULO pseudogenization in two oscines (medium ground-finch and zebra finch) and the suboscine golden-collared manakin (Fig. 6B and fig. S35). In contrast, intact GULO was recovered from the third oscine species, American crow, and the basal passerine rifleman (Acanthisitta chloris) (table S43). Similar to mammals (74), this pseudogenization was caused by the loss of different exons and lethal mutations. We also found purifying selection has dominated GULO evolution, from the ancestral amniote node (dN/dS = 0.096) to ancestral birds (dN/dS = 0.133) and mammals (dN/dS = 0.355), suggesting conservation of the ability to synthesize Vc both before and after avian divergence. However, both the American crow and rifleman exhibited nonsynonymous changes in GULO at one order of magnitude higher than the average (fig. S36), a sign of potentially harmful mutations (75).
Rhodopsin/opsins and vision
Birds exhibit what is possibly the most advanced vertebrate visual system, with a highly developed ability to distinguish colors over a wide range of wavelengths. In contrast to mammals, which have relatively few photoreceptor classes, almost all birds studied to date have retained an ancestral tetrapod set of cones hypothesized to play a role in reproduction and feeding (76). Vertebrate visual opsins are classified into five genes in two families: rhodopsin (RH1) and conopsins (RH2, OPN1sw1, OPN1sw2, andOPN1lw). In most avian genomes, we detected higher numbers of opsin genes than in mammalian genomes, which lacked OPN4x (77), RH2, and either OPN1sw1 (Monotremata) or OPN1sw2 (Theria). All avian genomes contained RH1 and RH2, and most high-coverage genomes contained two to three of the remaining three conopsin genes (table S44), supporting that ancestral avian vision is tetrachromatic. Penguins were one of the exceptions, with both species exhibiting only three classes of functional opsins, and thus are trichromatic, which is in line with retinal examination (78). This is likely due to their aquatic lifestyle and is consistent with observations of marine mammals that also appear to have lost one, or even both, cone pigment (or pigments) (76).
Signs of strong positive selection were detected in the branch leading to the passerine group Passerida (represented by the medium ground-finch and zebra finch) (fig. S38), which corroborates that the shift from violet sensitive SWS1 cones in this clade was adaptive (79). Excluding these species, dN/dS values for OPN1sw1 were lower in birds than in mammals (Fig. 6C). Optimal color discrimination requires an even distribution of spectral sensitivities (80), which is more easily disturbed with an increasing number of cone classes. Hence, stabilizing selection on spectral sensitivity should be stronger in birds than in mammals, and the dN/dS values are consistent with this prediction. Besides two transmembrane regions (II and VII) encompassing previously identified spectral tuning amino acids in the SWS1 conopsin, we found markedly positive selection in region IV, strongly suggesting that there is one or more unknown amino acid sites important to spectral tuning of this ultraviolet-sensitive cone (fig. S40).
Sex-related and reproductive traits
Reproduction-related genes
Unlike other reptiles, almost all birds develop only a single functional ovary, on the left side (81), as a result of the evolutionary loss of the right ovary during the transition from nonavian theropods to birds (82). It has been hypothesized that this loss represents an adaptation to reduce weight during flight (82). We found that two genes related with ovary development (MMP19 and AKR1C3) have been lost in birds. MMP19, a matrix metalloprotease gene, functions during the follicular growth and the ovulation process (83), and the enzyme AKR1C3 catalyzes the conversion of androstenedione to testosterone and has been associated with polycystic ovary syndrome (84).
We analyzed a range of other genes related to reproduction, under the hypothesis that some of them may have been direct targets of the morphological and behavioral adaptations related to sexual selection in birds. Reproduction genes in Drosophila, humans, and marine invertebrates evolve faster than do nonreproduction genes (49). We chose 89 genes that may be involved in spermatogenesis (table S46) and six involved in oogenesis (table S47). We found that 19 out of 46 avian species show significantly accelerated evolution (lineage-specific dN/dS ratio) of spermatogenesis genes relative to the genomic background (table S48). In contrast, only the carmine bee-eater (Merops nubicoides) and Peking duck showed significantly accelerated evolution in oogenesis genes (table S49). These results suggest that male birds are the dominant targets of sexual selection, which drives rapid evolution of spermatogenesis genes via sperm competition (85).
Plumage color
We investigated the genomics of plumage color, a behaviorally important trait and longstanding example of sexual selection (86). Male birds have frequently evolved extravagant plumage color in response to both male-male competition and female choice (87, 88), resulting in remarkable sexual dichromatism. Analysis of 15 genes implicated in avian plumage coloration demonstrated rapid evolutionary rates over the genomic average in 8 of 46 lineages (table S51). This pattern suggests that these genes are evolving under adaptive evolution.
Carotenoids, which are responsible for the bright yellow and red pigments that underlie some of the most conspicuous coloration patterns in vertebrates, unlike melanins can be only acquired through diet and represent trade-offs between coloration and other physiological conditions. We identified a negative correlation between color discriminability and dN/dS across birds for the gene GSTA2, which is involved in the binding and deposition of carotenoids and in plumage dichromatism (R2 = 0.24, P = 0.045, Pearson’s test with phylogenetically independent contrasts) (Fig. 6D and fig. S41), and similarly for SLC24A4, which is associated with hair color in humans (R2 = 0.21, P = 0.056, Pearson’s test with phylogenetically independent contrasts) (Fig. 6D and fig. S42), suggesting that either diver-sifying and stabilizing selection or the effect of different population sizes is driving the evolution of plumage color genes.
Discussion and conclusions
The small genome size of birds with fragmented microchromosomes and reduced repeat transposon activity, in contrast to other vertebrates, has been a static feature in the avian clade for >100 million years. Avian genomes consistently contain fewer genes, ~70% of the number of the human genome, and with one detected exception (downy woodpecker), an extremely reduced fraction of repeat elements. Thus, the ancestral avian lineage has distinctly lost a large number of genes by means of large segmental deletions after their divergence from other extant reptiles. These large genomic sequence deletions appear to be linked to a second defining feature of avian genomes: the putatively ancestral fission of macrochromosomes into a relatively large number of microchromosomes.
Genome conservation in birds—along with regard to sequence, synteny, and chromosomal structure—is remarkable in light of their rapid historical radiation. This is considerably different from the evolution of mammalian genomes, which although are experiencing a rapid radiation at a similar time, today display richer genome shuffling and variation (89). By comparing the genomes of 48 birds that are constrained within a largely resolved phylogeny, we discovered millions of highly constrained elements comprising 7.5% of avian genomes. This evolutionary profiling of genomes across >100 million years (5) enables their interpretation in a functional genomic context not possible in previous genomic studies restricted to fewer taxa.
The analyses of genome sequences for taxa distributed across the avian phylogeny also explains the rich biodiversity of the avian clade because we identified selective constraints on certain categories of genes in different avian lineages. Convergent evolution also appears to be shaping the evolution of protein-coding genes and their regulatory elements, establishing similar morphological or behavioral features in distantly related bird species, as well as variation in specific gene families that correspond to avian traits and environmental adaptation. We believe that the data and analyses presented here open a new window into the evolution, diversification, and ecological adaptation of tetrapod vertebrates and offers a phylogenomic perspective that helps bridge the chasm between micro- and macroevolution.
ACKNOWLEDGMENTS
Genome assemblies and annotations of avian genomes in this study are available on the avian phylogenomics website (http://phybirds.genomics.org.cn), GigaDB (http://dx.doi.org/10.5524/101000), National Center for Biotechnology Information (NCBI), and ENSEMBL (NCBI and Ensembl accession numbers are provided in table S2). The majority of this study was supported by an internal funding from BGI. In addition, G.Z. was supported by a Marie Curie International Incoming Fellowship grant (300837); M.T.P.G. was supported by a Danish National Research Foundation grant (DNRF94) and a Lundbeck Foundation grant (R52-A5062); C.L. and Q.L. were partially supported by a Danish Council for Independent Research Grant (10-081390); and E.D.J. was supported by the Howard Hughes Medical Institute and NIH Directors Pioneer Award DP1OD000448.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/346/6215/1311/suppl/DC1
Supplementary Text
Figs. S1 to S42
Tables S1 to S51
References (92–192)
REFERENCES AND NOTES
- 1.Gill F, Donsker D. IOC World Bird List (version 3.5) 2013 [Google Scholar]
- 2.Chiappe LM, Witmer LM. Mesozoic Birds: Above the Heads of Dinosaurs. Berkeley, CA: Univ. California Press; 2002. [Google Scholar]
- 3.Dyke G, Kaiser GW. Living Dinosaurs the Evolutionary History of Modern Birds. Hoboken, NJ: Wiley-Blackwell; 2011. [Google Scholar]
- 4.Feduccia A. Trends Ecol. Evol. 2003;18:172–176. [Google Scholar]
- 5.Jarvis ED, et al. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holt BG, et al. Science. 2013;339:74–78. doi: 10.1126/science.1228282. [DOI] [PubMed] [Google Scholar]
- 7.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. Nature. 2012;491:444–448. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 8.Zeigler H, Marler PE. Behavioral Neurobiology of Birdsong. New York, NY, US: Hunter College, City University of New York; 2002. Dec, this volume is the result of the aforementioned conference which was one of an annual symposium series sponsored by the Hunter College Gene Center. (2004). [Google Scholar]
- 9.Stattersfield AJ, Crosby MJ, Long AJ, Wege DC. Endemic Bird Areas of the World: Priorities for Conservation. BirdLife Conservation. Cambridge, UK: BirdLife International; 1998. [Google Scholar]
- 10.Alexander DJ. Vet. Microbiol. 2000;74:3–13. doi: 10.1016/s0378-1135(00)00160-7. [DOI] [PubMed] [Google Scholar]
- 11.Hillier LW, et al. Nature. 2004;432:695–716. [Google Scholar]
- 12.Dalloul RA, et al. PLOS Biol. 2010;8:e1000475. [Google Scholar]
- 13.Warren WC, et al. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y, et al. Nat. Genet. 2013;45:776–783. doi: 10.1038/ng.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhan X, et al. Nat. Genet. 2013;45:563–566. doi: 10.1038/ng.2588. [DOI] [PubMed] [Google Scholar]
- 16.Shapiro MD, et al. Science. 2013;339:1063–1067. doi: 10.1126/science.1230422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickinson E, Remsen J. The Howard and Moore Complete Checklist of the Birds of the World. Eastbourne, UK: Aves Press; 2013. [Google Scholar]
- 18.Materials and methods are available as supplementary materials on Science Online.
- 19.Zhang G, et al. GigaScience. 2014;3:26. doi: 10.1186/2047-217X-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganapathy G, et al. GigaScience. 2013;3:11. [Google Scholar]
- 21.Gregory TR. The Animal Genome Size Database. 2005 available at www.genomesize.com. [Google Scholar]
- 22.Zhou Q, et al. Science. 2014;346:1246338. doi: 10.1126/science.1246338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lander ES, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 24.Green RE, et al. Science. 2014;346:1254449. doi: 10.1126/science.1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes AL, Hughes MK. Nature. 1995;377:391. doi: 10.1038/377391a0. [DOI] [PubMed] [Google Scholar]
- 26.Rho M, et al. Genome Biol. Evol. 2009;1:2–12. doi: 10.1093/gbe/evp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morand S, Ricklefs RE. Trends Genet. 2001;17:567–568. doi: 10.1016/s0168-9525(01)02414-3. [DOI] [PubMed] [Google Scholar]
- 28.Waltari E, Edwards SV. Am. Nat. 2002;160:539–552. doi: 10.1086/342079. [DOI] [PubMed] [Google Scholar]
- 29.Kidwell MG. Genetica. 2002;115:49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- 30.Feschotte C, Pritham EJ. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch M, Conery JS. J. Struct. Funct. Genomics. 2003;3:35–44. [PubMed] [Google Scholar]
- 32.Sela N, Kim E, Ast G. Genome Biol. 2010;11:R59. doi: 10.1186/gb-2010-11-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Böhne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN. Chromosome Res. 2008;16:203–215. doi: 10.1007/s10577-007-1202-6. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Q, Edwards SV. Genome Biol. Evol. 2012;4:1033–1043. doi: 10.1093/gbe/evs070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Organ CL, Shedlock AM, Meade A, Pagel M, Edwards SV. Nature. 2007;446:180–184. doi: 10.1038/nature05621. [DOI] [PubMed] [Google Scholar]
- 36.Hughes AL, Friedman R. Mol. Biol. Evol. 2008;25:2681–2688. doi: 10.1093/molbev/msn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffin DK, Robertson LBW, Tempest HG, Skinner BM. Cytogenet. Genome Res. 2007;117:64–77. doi: 10.1159/000103166. [DOI] [PubMed] [Google Scholar]
- 38.Bradnam KR, et al. GigaScience. 2013;2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J, et al. Proc. Natl. Acad. Sci. U.S.A. 2013;110:1785–1790. doi: 10.1073/pnas.1220349110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmann FG, Opazo JC, Storz JF. Mol. Biol. Evol. 2012;29:303–312. doi: 10.1093/molbev/msr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoffmann FG, Opazo JC, Storz JF. Mol. Biol. Evol. 2008;25:591–602. doi: 10.1093/molbev/msn004. [DOI] [PubMed] [Google Scholar]
- 42.Grispo MT, et al. J. Biol. Chem. 2012;287:37647–37658. doi: 10.1074/jbc.M112.375600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baer CF, Miyamoto MM, Denver DR. Nat. Rev. Genet. 2007;8:619–631. doi: 10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- 44.Venditti C, Pagel M. Trends Ecol. Evol. 2010;25:14–20. doi: 10.1016/j.tree.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Siepel A, et al. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindblad-Toh K, et al. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nabholz B, Glémin S, Galtier N. BMC Evol. Biol. 2009;9:54. doi: 10.1186/1471-2148-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lathrop WF, Carmichael EP, Myles DG, Primakoff P. J. Cell Biol. 1990;111:2939–2949. doi: 10.1083/jcb.111.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson WJ, Vacquier VD. Nat. Rev. Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- 50.Pennacchio LA, et al. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 51.ENCODE Project Consortium. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mank JE, Axelsson E, Ellegren H. Genome Res. 2007;17:618–624. doi: 10.1101/gr.6031907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Axelsson E, Webster MT, Smith NGC, Burt DW, Ellegren H. Genome Res. 2005;15:120–125. doi: 10.1101/gr.3021305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suh A, et al. Nat. Commun. 2011;2:443. doi: 10.1038/ncomms1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jarvis ED. Ann. N. Y. Acad. Sci. 2004;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitney O, et al. Science. 2014;346:1256780. doi: 10.1126/science.1256780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfenning AR, et al. Science. 2014;346:1256846. doi: 10.1126/science.1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karten HJ, et al. J. Comp. Neurol. 2013;521:3702–3715. doi: 10.1002/cne.23443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hubisz MJ, Pollard KS, Siepel A. Brief. Bioinform. 2011;12:41–51. doi: 10.1093/bib/bbq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dumont ER. Proc. Biol. Sci. 2010;277:2193–2198. doi: 10.1098/rspb.2010.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gutzwiller SC, Su A, O’Connor PM. Anat. Rec. 2013;296:867–876. doi: 10.1002/ar.22691. [DOI] [PubMed] [Google Scholar]
- 62.Duncker HR. Respir. Physiol. Neurobiol. 2004;144:111–124. doi: 10.1016/j.resp.2004.07.020. [DOI] [PubMed] [Google Scholar]
- 63.West JB, Watson RR, Fu Z. Respir. Physiol. Neurobiol. 2007;157:382–390. doi: 10.1016/j.resp.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gill F. Ornithology. New York: W.H. Freeman and Company; 1995. [Google Scholar]
- 65.Haake AR, König G, Sawyer RH. Dev. Biol. 1984;106:406–413. doi: 10.1016/0012-1606(84)90240-9. [DOI] [PubMed] [Google Scholar]
- 66.Ross-Ibarra J. Am. Nat. 2004;163:105–112. doi: 10.1086/380606. [DOI] [PubMed] [Google Scholar]
- 67.Blirt A, Bell G. Nature. 1987;326:803–805. doi: 10.1038/326803a0. [DOI] [PubMed] [Google Scholar]
- 68.Louchart A, Viriot L. Trends Ecol. Evol. 2011;26:663–673. doi: 10.1016/j.tree.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 69.Livezey BC, Zusi RL. Zool. J. Linn. Soc. 2007;149:1–95. doi: 10.1111/j.1096-3642.2006.00293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cracraft J. Syst. Biol. 1982;31:35–56. [Google Scholar]
- 71.Meredith RW, Zhang G, Gilbert MTP, Jarvis ED, Springer MS. Science. 2014;346:1254390. doi: 10.1126/science.1254390. [DOI] [PubMed] [Google Scholar]
- 72.Birdsey GM, Lewin J, Cunningham AA, Bruford MW, Danpure CJ. Mol. Biol. Evol. 2004;21:632–646. doi: 10.1093/molbev/msh054. [DOI] [PubMed] [Google Scholar]
- 73.Chatterjee IB. Science. 1973;182:1271–1272. doi: 10.1126/science.182.4118.1271. [DOI] [PubMed] [Google Scholar]
- 74.Cui J, Yuan X, Wang L, Jones G, Zhang S. PLOS ONE. 2011;6:e27114. doi: 10.1371/journal.pone.0027114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cui J, Pan Y-H, Zhang Y, Jones G, Zhang S. Mol. Biol. Evol. 2011;28:1025–1031. doi: 10.1093/molbev/msq286. [DOI] [PubMed] [Google Scholar]
- 76.Davies WIL, Collin SP, Hunt DM. Mol. Ecol. 2012;21:3121–3158. doi: 10.1111/j.1365-294X.2012.05617.x. [DOI] [PubMed] [Google Scholar]
- 77.Pires SS, et al. Proc. Biol. Sci. 2007;274:2791–2799. doi: 10.1098/rspb.2007.0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bowmaker JK, Martin GR. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 1985;156:71–77. [Google Scholar]
- 79.Ödeen A, Håstad O, Alström P. BMC Evol. Biol. 2011;11:313. doi: 10.1186/1471-2148-11-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chittka L. Naturwissenschaften. 1996;83:136–138. [Google Scholar]
- 81.Guraya SS. Ovarian Follicles in Reptiles and Birds. Berlin, Germany: Springer-Verlag; 1989. [Google Scholar]
- 82.Zheng X, et al. Nature. 2013;495:507–511. doi: 10.1038/nature11985. [DOI] [PubMed] [Google Scholar]
- 83.Jo M, Curry TE., Jr Biol. Reprod. 2004;71:1796–1806. doi: 10.1095/biolreprod.104.031823. [DOI] [PubMed] [Google Scholar]
- 84.Goodarzi MO, Antoine HJ, Azziz R. J. Clin. Endocrinol. Metab. 2007;92:2659–2664. doi: 10.1210/jc.2006-2600. [DOI] [PubMed] [Google Scholar]
- 85.Birkhead TR, Møller AP. Sperm competition and sexual selection. Academic Press; 1998. [Google Scholar]
- 86.Darwin C. The Descent of Man, and Selection in Relation to Sex. London: J. Murray; 1871. [Google Scholar]
- 87.Zuk M, Ligon JD, Thornhill R. Anim. Behav. 1992;44:999–1006. [Google Scholar]
- 88.Mateos C, Carranza J. Anim. Behav. 1997;54:1205–1214. doi: 10.1006/anbe.1997.0516. [DOI] [PubMed] [Google Scholar]
- 89.Quinlan AR, Hall IM. Trends Genet. 2012;28:43–53. doi: 10.1016/j.tig.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Storz JF, Opazo JC, Hoffmann FG. Mol. Phylogenet. Evol. 2013;66:469–478. doi: 10.1016/j.ympev.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hoffmann FG, Storz JF, Gorr TA, Opazo JC. Mol. Biol. Evol. 2010;27:1126–1138. doi: 10.1093/molbev/msp325. [DOI] [PMC free article] [PubMed] [Google Scholar]