Abstract
Physically interacting proteins or parts of proteins are expected to evolve in a coordinated manner that preserves proper interactions. Such coevolution at the amino acid-sequence level is well documented and has been used to predict interacting proteins, domains, and amino acids. Interacting proteins are also often precisely coexpressed with one another, presumably to maintain proper stoichiometry among interacting components. Here, we show that the expression levels of physically interacting proteins coevolve. We estimate average expression levels of genes from four closely related fungi of the genus Saccharomyces using the codon adaptation index and show that expression levels of interacting proteins exhibit coordinated changes in these different species. We find that this coevolution of expression is a more powerful predictor of physical interaction than is coevolution of amino acid sequence. These results demonstrate that gene expression levels can coevolve, adding another dimension to the study of the coevolution of interacting proteins and underscoring the importance of maintaining coexpression of interacting proteins over evolutionary time. Our results also suggest that expression coevolution can be used for computational prediction of protein-protein interactions.
Coevolution is an evolutionary process in which a heritable change in one entity establishes selective pressure for a change in another entity. These entities can range from nucleotides to amino acids to proteins to entire organisms and perhaps even ecosystems. A relatively simple and well studied example of coevolution involves physically interacting proteins, in which precise, complementary structural conformations of interacting partners are usually needed to maintain a functional interaction. If the conformation of one protein is interrupted by mutation, a compensatory change may be selected for in its interacting partner. When such compensatory changes occur, the two proteins are said to coevolve.
Coevolution of interacting amino acids and proteins has been studied intensively for more than a decade (see refs. 1-8). The identification of coevolving pairs of genes is interesting and important for several reasons. First, it can aid in functional annotations: When an uncharacterized gene is found to coevolve with several different genes, all of which encode proteins of a single function, the unknown gene is likely to share that same function. Second, identification of likely physical interactions through detection of coevolution can contribute to our understanding of how proteins work together to execute their functions. Third, coevolution may be a critical process by which complex cellular components, such as multimolecule machines and metabolic pathways, undergo adaptive or constructive change without disruption of organismal integrity.
Various methods have been developed to detect coevolution of proteins, most based on a common principle: Evolutionary distances between all possible pairs of amino acid sites or proteins are estimated from multiple alignments of protein sequences, and the extent of coevolution for each pair is determined by measuring the correlation of their evolutionary rates across different lineages. Such methods have been successful in quantifying the extent of coevolution between proteins, protein domains, and amino acid residues known to interact physically (3-8). These methods also have been used to predict specific interactions between receptors and their substrates in large paralogous protein families (4, 8) and between proteins from the bacterium Escherichia coli (6, 7).
In previous applications of this approach to the study of protein coevolution, ≥11 sequences (and sometimes many more) have been used in each multiple alignment (3-8). Whereas such extensive taxonomic sampling is possible in studies of prokaryotes, for which >100 genome sequences are available, it remains difficult in studies of eukaryotes.
Here, we examine whether coevolution can be detected not only in protein sequences but also in their levels of expression. The expectation that expression levels should coevolve stems in part from the observation that the expression levels of genes encoding interacting proteins are strongly correlated over different experimental conditions in Saccharomyces cerevisiae (9-11). This observation is thought to reflect the requirement for interacting proteins to be present in the cell in similar amounts at the same time to properly form stoichiometric complexes and execute their function. When protein complex subunits are misexpressed, they tend to have more severe consequences on growth than proteins that do not participate in stable protein interactions (12). Thus, we predicted that natural selection would maintain precise coexpression of interacting proteins; if the expression of one gene changes, it would be expected to result in a selection pressure for a similar expression change in its interacting partners, analogous to the coevolution of amino acid sequence described above.
In this study, we use the genome sequences of four closely related yeasts (Saccharomyces cerevisiae, Saccharomyces paradoxus, Saccharomyces mikatae, and Saccharomyces bayanus) along with protein interaction data from S. cerevisiae to introduce a method to detect coevolution of gene expression based on coordinated changes in gene expression, as estimated by codon usage bias. We also examine protein sequence coevolution to evaluate whether sequence data from these four species alone allow the coevolution of interacting proteins to be detected on a genomic scale and to compare the strength of expression coevolution with the strength of sequence coevolution.
Materials and Methods
Sequence Data. For all analyses described in this work, we used the complete genome sequences of four closely related (<20 million years divergence, corresponding to an average of 2.2 synonymous substitutions per site after correcting for nonneutral synonymous sites) yeast species in the genus Saccharomyces: S. cerevisiae (13), S. paradoxus, S. mikatae, and S. bayanus (14). Rigorous assignments of orthology were made based on both high-sequence identity and synteny between species (14), and alignments were performed on protein sequences by using clustalw (15). Alignments were discarded if their maximum likelihood phylogeny (16) was not consistent with the known phylogeny of the species, or if they contained either real or spurious (because of sequencing errors) frameshift mutations because frameshifts result in unrealistic estimates of evolutionary rates. Frameshifts were detected by establishing a majority-rule consensus sequence from the four sequences; if any sequence failed to match the consensus for at least five consecutive positions, it was counted as having a frameshift and was discarded from the alignment.
Detection of Sequence Coevolution. Our test for protein sequence coevolution of interacting proteins is similar to methods that search for strong correlations between pair-wise sequence distances or similarity of phylogenetic trees (3-8). For each set of orthologous genes, we used paml (16) to estimate the evolutionary rate (nonsynonymous to synonymous substitution ratio) in each branch of the yeast phylogenetic tree. Five branch lengths were calculated for each set of orthologs (one for each species plus one internal branch). These five lengths were normalized by dividing each by the average length of that branch over all of the trees calculated to control for the fact that some branches tended to be longer than others. The normalized lengths could then be plotted against each other for any pair of genes, and the Pearson correlation coefficient (17) could be calculated as a measure of the degree of coevolution. To calculate the significance of the observed distribution of correlation coefficients among interacting pairs, we compared it with the distribution of all possible pairs, except for those in the list of interactors. The nonparametric Kolmogorov-Smirnov (KS) test (17) was used to estimate the probability that both were sampled from the same underlying distribution.
Detection of Expression Coevolution. Our method for detecting expression coevolution was quite similar to our method for detecting sequence coevolution. Codon bias values, as represented by the codon adaptation index (CAI) (18), were calculated for each of the four orthologous sequences by using the codon frequencies of the 20 most highly expressed genes in S. cerevisiae, as estimated by Arava et al. (19). Results were not affected by using species-specific codon usage tables (data not shown). The four values for each gene then were plotted against each other for each pair of genes, and the Pearson correlation coefficient was calculated for each pair. Details of significance testing by the KS test were as described above.
Protein-Protein Interaction Data. A list of 4,175 putative interactions involving 1,360 S. cerevisiae proteins was taken from a study by von Mering et al. (20). Only those interactions listed with “high confidence” (interactions found by multiple independent methods) or listed as previously annotated (by non-high-throughput methods) were used to minimize the effects of false positives. High-throughput methods used to identify interactions were yeast two-hybrid, MS, synthetic lethality, and synexpression; computational methods used were conserved gene neighborhood, gene fusion, and phylogenetic profiling (20). Exclusion of interactions whose membership in the high-confidence category depended on synexpression (correlated expression levels in S. cerevisiae microarray experiments), because of a possible circularity when measuring CAI coevolution of these putatively interacting proteins, did not appreciably affect the results. Any interactions involving a protein with itself were discarded because these would indicate perfect coevolution for a trivial reason.
Results
Coevolution of Protein Sequences. We began by examining metrics of coevolution for proteins that have been observed to interact in S. cerevisiae. From a set of 4,175 relatively high-confidence protein-protein interactions involving 1,360 proteins (20), we identified 1,377 interacting pairs involving 621 proteins in which both proteins had clear orthologs in all four Saccharomyces species and the alignments of the protein sequences were of high quality. We used the multiple alignments to estimate rates of evolution for each protein in each lineage. As a measure of their coevolution, for all pairs of proteins we computed the correlation coefficient between their rates of evolution in the different lineages (see Materials and Methods). For comparison to the set of interacting proteins, we generated a list of all 192,510 possible pairs (involving the same 621 proteins) that were not in our list of 1,377 interactions.
Because there was a wide range in the amount of variance in evolutionary rates for different pairs of proteins (Fig. 1A), we reasoned that pairs in which one or both proteins had very little variance in evolutionary rates would not be very informative for detecting coevolution because the small changes that are indicated by a small variance are more likely to reflect random fluctuations or noise instead of authentic changes in the evolutionary rates of a gene along different lineages. For this reason, we restricted our analysis to the 200 interacting pairs (of the 1,377 total) with the greatest variance in both proteins of the pair (i.e., only the variance in the less variable of the two proteins was used to represent the pair). This variance cutoff (Fig. 1 A, dashed line) was then applied to the complete list of 192,510 random pairs, resulting in a list of 26,796 pairs (200 known interactions and 26,596 others) with a variance in evolutionary rates above the cutoff for every protein in the list. In other words, a minimum variance cutoff was applied to all 621 proteins, and all possible pairs among those satisfying the cutoff were included for further analysis.
Fig. 1.
Coevolution of sequence. (A) A histogram of the base 10 logarithms of variance in evolutionary rates for all 192,510 possible pairs of proteins in this study. The variance for each protein in a pair was calculated, and the lower of the two was used to represent the pair. The dashed line indicates the variance cutoff described in the main text. Note that evolutionary rates were normalized by the mean rate for each branch of the phylogenetic tree (see Materials and Methods). (B) A histogram of the correlation coefficients indicating the strength of amino acid sequence coevolution for 200 pairs of interacting proteins (solid line) and 26,596 pairs of noninteracting proteins (dashed line). The two distributions are significantly different from one another (KS test, P = 0.0069). Bin labels are the upper bound for each bin (e.g., the label 0.9 indicates 0.8 < r ≤ 0.9).
If the amino acid sequences of our 200 interacting proteins were coevolving, we would expect to see the distribution of correlation coefficients (our metric of coevolution) to be greater in the 200 interacting pairs than in the 26,596 noninteracting pairs. To test this hypothesis, we separated the interacting and noninteracting pairs into 10 bins each, separating protein pairs by the strength of the correlation between their sets of evolutionary rates. This analysis confirmed that we could observe such coevolution at a genomic scale: For all bins of correlation coefficients greater than or equal to the 0.4 < r ≤ 0.5 bin, there was a greater fraction of interacting protein pairs than random pairs (Fig. 1B). These two distributions are significantly different from one another, as measured by the KS test (P = 0.0069). This difference also can be summarized by comparing the medians of these two distributions; as expected from Fig. 1B, the median correlation coefficient for interacting pairs (r = 0.088) was higher than that of random pairs (r = -0.050).
Although these results establish that we can detect coevolution of interacting protein sequences by using just four genome sequences, they do not quantify the fraction of our interacting proteins for which we have detected coevolution. Another way to pose this same question is: For what fraction of our interacting proteins do we find a correlation coefficient higher than that expected for protein pairs that are not known to interact? Because the distribution of correlation coefficients among noninteracting pairs (Fig. 1B, dashed line) represents what is expected by chance, the value we seek is the difference between the values that form this curve and those that form our distribution of interaction correlation coefficients (Fig. 1B, solid line) at high correlation coefficients (specifically, at all correlation coefficient bins greater than the largest correlation coefficient at which the distributions cross). In other words, we are simply subtracting an estimate of the fraction of false positives from the fraction of true positives to find the number of true positives not explainable by random chance. We calculated this value to be 0.113, indicating that we detected coevolution in the sequences of ≈23 (11.3%) of our 200 interacting pairs. Because this calculation assumes that our list of interactions is free of false positives and that our noninteractor list is free of false negatives, it should be interpreted as a lower bound for the amount of sequence coevolution that we can detect with four genome sequences.
Coevolution of Gene Expression. Although our finding coevolution for 11.3% of the interacting pairs is significant, it still represents only a small fraction of the interactions in our list. Thus, we wished to develop a method to extract more information about protein interactions than we could from the coevolution of protein sequence alone. Because it has been shown that genes coding for physically interacting proteins tend to be coexpressed (9-11), we reasoned that interacting proteins might have detectable coevolution of expression levels if such coexpression must be maintained even as expression patterns change over evolutionary time.
One method to test whether expression levels coevolve would be to use DNA microarrays to measure the expression levels in various species and conditions and then to search for cases in which expression patterns of mRNAs encoding a protein and its interacting partner have changed in a coordinated fashion. Although such experiments are feasible, they are labor-intensive and expensive, and we can expect the generation of expression data to lag behind genome sequencing for some time. Therefore, we asked instead whether we could detect coevolution of gene expression using sequence alone. Although we have no method to accurately infer patterns of expression from sequence, there does exist a very well characterized method to estimate a gene's average expression level from its sequence. Bias in the usage of synonymous codons, which was first noted more than 20 years ago (21), is a remarkably good predictor of average expression level. The strong association between codon bias and expression is thought to be because of selection for translational efficiency and accuracy of highly expressed genes (22). (Because the changes in gene-expression levels we are interested in occurred over the last several million years of evolution in our four Saccharomyces species, codon bias may reflect aspects of previous selection on gene expression that may not be apparent in microarray expression data because microarray data are measured in laboratory conditions that are undoubtedly quite different from those of a natural yeast habitat. Also for this reason, the strength of the correlation between codon bias values and microarray expression data from the laboratory cannot be taken as a precise indicator of how well codon bias reflects historical expression levels.) Because codon bias can be calculated easily for any gene sequence, we tested the hypothesis that genes encoding interacting proteins tend to coevolve in expression and thus indicate coordinated changes in codon bias in different species. In other words, if codon bias for gene X is greater in species A than in species B, then we might expect codon bias for some or all genes whose protein products interact with the protein encoded by X to be greater in species A than in species B as well.
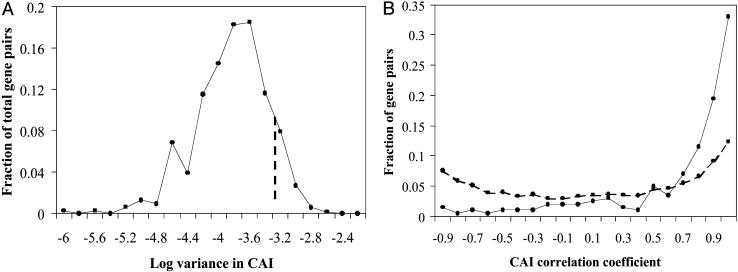
To test this hypothesis, we again began with our list of 1,377 interactions among 621 proteins. We used the codon usage from the 20 most highly expressed genes in S. cerevisiae (19) to parameterize the CAI (see Materials and Methods) for each species and used the CAI to estimate expression levels for each of the 621 genes in all four species. There was a wide range of variances in CAI for the 192,510 pairs (Fig. 2A), so, for the same reasons described above, we restricted our attention to the 200 interacting pairs with the highest variance in CAI for both members. Application of this cutoff (Fig. 2A, dashed line) to the list of all possible pairs yielded 11,781 pairs (of which 200 were known interactions and 11,581 were not).
Fig. 2.
Coevolution of expression. (A) A histogram of the base 10 logarithms of variance in CAI for all 192,510 possible pairs of the 621 proteins in this study. The variance for each protein in a pair was calculated, and the lower of the two was used to represent the pair. The dashed line indicates the variance cutoff described in the main text. (B) A histogram of the correlation coefficients indicating the strength of CAI coevolution for 200 pairs of interacting proteins (solid line) and 11,581 pairs of noninteracting proteins (dashed line). The two distributions are significantly different from one another (KS test, P < 10-26). Bin labels are the upper bound for each bin (e.g., the label 0.9 indicates 0.8 < r ≤ 0.9).
Comparison of the distribution of correlation coefficients for the 200 interacting pairs with the 11,581 noninteractors revealed a striking difference, with the interacting pair distribution sharply skewed toward high values (Fig. 2B, solid line). The median correlation coefficient for interacting pairs was 0.822, whereas that of noninteractors was only 0.1997. The KS test confirmed that the difference between the two distributions was quite significant (P < 10-26). Calculating the fraction of interacting pairs for which we could detect expression coevolution (as described above for protein sequence coevolution) resulted in a value of 37.3%, or ≈75 of our 200 interacting pairs, which again should be interpreted only as a lower bound. Thus, we were able to detect expression coevolution at a level above the random background for more than one-third of the interacting protein pairs.
Although our finding of strong correlations between expression levels of interacting proteins in different organisms is consistent with our hypothesis of coevolution occurring by sequential mutations, another possibility must also be considered. If the genes encoding interacting proteins are often regulated by the same transacting factor, then a single change affecting that factor could lead to up- or down-regulation of both interacting proteins in one species. Even though this scenario does lead to correlated changes in expression, it would not truly be coevolution. To distinguish between the true coevolution possibility and the single transacting mutation possibility, we used experimental genome-wide transcription factor binding data that are available for 113 transcription factors in yeast (23). We reasoned that, if single mutations in transcription factors account for some or all of our apparent expression coevolution, then genes encoding pairs of interacting proteins that are regulated by the same transcription factor should indicate stronger coevolution, on average, than those that are regulated by different transcription factors. Among our 1,377 interacting pairs, we found 59 that were coregulated (both genes being bound by one transcription factor with a confidence of P < 0.001). Surprisingly, these 59 had a median CAI correlation coefficient of 0.111, significantly lower than that of the rest of the interacting pairs (KS test, P = 0.047). Although we expect that we have missed many interacting pairs that are regulated by the same transcription factor (due to both false negatives in the binding data and our lack of binding data for all transcription factors), this shortcoming should only serve to weaken any bias we find. Our finding that interacting pairs regulated by the same transcription factor actually have weaker coevolution than others supports our interpretation of the correlations as evidence of coevolution by sequential mutations; however, we note that this analysis does not address whether those sequential mutations occurred in cis or in trans. We do not have an explanation for why interacting proteins whose genes are regulated by the same transcription factor indicate less expression coevolution than other interacting proteins.
Prediction of Protein Interactions. Considering that we have two metrics that are both indicative of physical interaction between proteins, we asked whether protein pairs with coevolving expression levels are more likely to indicate detectable protein sequence coevolution or whether instead the two metrics are largely independent. We found the latter to be the case, because the correlation between our two metrics of coevolution was extremely weak (Pearson r = 0.016). Because the metrics are independent, it is possible that they could be combined to yield more information than either in isolation.
To test the power of combining the two metrics, we generated predictions of previously uncharacterized protein interactions. We started with the list of random protein pairs that satisfied the variance cutoffs used above for both evolutionary rates and CAI (1,711 total pairs), and we applied cutoffs for both correlation coefficients. We began with the arbitrary cutoffs of r > 0.75 for protein sequence coevolution and r > 0.9 for CAI coevolution, which yielded a list of 21 predictions (Table 1) involving proteins of both high and low CAI (ranging from 0.197 to 0.85 in S. cerevisiae). Of these 21 pairs, four were interactions from our list of 1,377, which is 27-fold higher than expected by chance and is thus unlikely to occur randomly (P = 3 × 10-5). This enrichment can be interpreted as the approximate enrichment for interacting proteins for all pairs in the list that are not known to interact. In other words, each pair in Table 1 (aside from known interactors) is ≈27-fold more likely to interact than a random pair of yeast proteins. More or less stringent cutoffs also can be used to generate either more predictions with less confidence or fewer predictions with greater confidence. For example, use of a more stringent cutoff (evolutionary rate r > 0.9, CAI r > 0.95) on these same 1,711 pairs resulted in a list of 10 predictions (Table 1, first 10 rows), of which three were from our list of known interactions (42-fold enrichment, P = 4 × 10-5). These enrichments are stronger than those resulting from the application of either metric alone (data not shown), confirming our expectation that combining the two increases their power. Although we could undoubtedly have improved these enrichments for known interacting pairs by testing many different cutoffs to finely tune them, one must be careful not to overfit the data or to perform multiple tests without the appropriate statistical corrections; thus, we have chosen not to do this fine tuning.
Table 1. Predictions of protein interactions.
| ORF 1 | ORF 2 | Known interaction? | CAI r | Evol rate r |
|---|---|---|---|---|
| FUR1 | NOP7 | No | 1.000 | 0.999 |
| RLP24 | NOP7 | No | 0.962 | 0.995 |
| RLP24 | FUR1 | No | 0.953 | 0.991 |
| MRPL33 | RIB3 | No | 0.986 | 0.944 |
| PNO1 | TIF35 | No | 0.995 | 0.927 |
| NOG1 | RLP24 | Yes | 0.998 | 0.920 |
| SWP1 | ATP3 | No | 0.980 | 0.933 |
| NOG1 | FUR1 | No | 0.951 | 0.948 |
| NOG1 | NOP7 | Yes | 0.960 | 0.936 |
| TIF2 | YBR025C | Yes | 0.967 | 0.903 |
| TAF17 | QCR7 | No | 0.903 | 0.969 |
| VMA13 | MLC1 | No | 0.904 | 0.959 |
| TIF2 | RPL9A | No | 0.988 | 0.851 |
| WBP1 | YPT10 | No | 0.953 | 0.844 |
| RPL9A | YBR025C | No | 0.945 | 0.829 |
| NOP7 | UTP6 | No | 0.953 | 0.813 |
| FUR1 | UTP6 | No | 0.962 | 0.803 |
| RPL5 | YBR025C | No | 0.901 | 0.828 |
| RPP0 | TIF2 | No | 0.954 | 0.761 |
| RPL5 | RPP0 | Yes | 0.936 | 0.750 |
| NOB1 | APT1 | No | 0.912 | 0.765 |
A list of 21 protein-interaction predictions made by combining the sequence and expression coevolution metrics. The first 10 pairs satisfy the stringent cutoffs of evolutionary rate r > 0.9, CAI r > 0.95; all 21 satisfy the cutoffs of evolutionary rate r > 0.75, CAI r > 0.9.
It should be noted that several genes appear multiple times in the list of our predictions (Table 1), indicating that our method may prove useful at predicting small networks of interacting proteins. For example, our method predicted a fully connected network of four proteins (Nog1p, Rlp24p, Fur1p, and Nop7p), with all six interactions of that network among our top 10 predictions. Two of these interactions, namely Nog1p with Nop7p and Rlp24p, were previously known. Other predictions in this group, such as the interaction between Nop7p and Rlp24p, are quite plausible because they both interact with Nog1p and such clustering of interactions within small groups of proteins is common (24). Other proteins are also predicted to interact with at least one member of this group; for instance, Utp6p is predicted to interact with Nop7p, a hypothesis that is quite reasonable because both of these proteins are located in the nucleolus (25, 26). Whereas alternative methods for computational prediction of protein interactions and functional linkages have yielded more predictions than our method, we note that they have all used far more genome sequences as well (e.g., 57 genomes were used by Date and Marcotte in ref. 27). Thus, although we present very few predictions in this study, we anticipate that applying this method to more genomes will greatly enhance its power.
Discussion
We have shown that the expression levels of genes encoding interacting proteins tend to coevolve in yeast. This coevolution is of a nature fundamentally different from the only other type of coevolution that has thus far been studied in interacting proteins, namely the coevolution of amino acid sequence, and it may represent a widespread and important mode of evolutionary change. Both types of coevolution can be detected in scores of genes by using a large set of protein interactions in yeast, although >3-fold more interacting pairs showed detectable coevolution of expression than of protein sequence in this study.
What is perhaps most surprising is the extent of coevolution we were able to detect using only four genome sequences. We did not use partial genome sequences that are available for many more yeast species (28, 29), because including them dramatically reduced the number of genes for which alignments of orthologous genes in all species were available. However, because many more yeast species will soon have complete genome sequences available, we expect that the power of the tests introduced here will increase greatly. Furthermore, our use of four genome sequences provides a reasonable benchmark for future studies in other eukaryotes such as Drosophila melanogaster, Caenorhabditis elegans, and others because close relatives of these species (Drosophila pseudoobscura and Caenorhabditis briggsae) already have been fully sequenced and several close relatives soon will have sequenced genomes. Our method may not be as easily applicable in species with very little codon bias determined by gene expression levels, such as humans.
Aside from being useful for studying the evolution of gene regulation, our finding of expression coevolution has a practical application in predicting pairs of interacting proteins. Because these predictions are more accurate when the expression coevolution metric is combined with another method of interaction prediction based on amino acid sequence coevolution, we suggest that future studies in which protein interactions are predicted from genome sequences will be more comprehensive if expression coevolution is included. Because even our combined metric cannot detect most protein interactions when only four genome sequences are used, we have not yet attempted to make large-scale predictions of interacting proteins in yeast.
In addition to the metric of expression coevolution that we introduce here, several other purely sequence-based methods for predicting protein interactions exist, such as phylogenetic profiling (30), conservation of gene neighborhood (31), and gene fusions (32, 33). Because these methods are mostly independent, combining them might greatly increase the power to predict protein interactions based on genome sequences alone. The methods could be integrated in a Bayesian framework (34); for example, the extent of expression coevolution could serve as a prior probability of interaction, which can then be increased or decreased based on any other metric for interaction prediction. We note, however, that these other methods of protein interaction prediction would not have added any information in this study. Phylogenetic profiling depends on the absence of some genes from some genomes, but all genes we used were present in all four genomes; conservation of gene neighborhood requires shuffling of genes, but all genes we used had conserved synteny in the four genomes; and the method of gene fusions depends on relatively rare fusion events, which none of our genes have undergone in these four species.
Another unexplored application of both sequence and expression coevolution metrics is assessment of the quality of high-throughput protein-interaction data sets (e.g., ref. 20). One could use the degree of expression and sequence coevolution in a set of putative protein interactions to determine the accuracy of the data using a set of well established interactions to determine a baseline of the maximum amount of coevolution expected if all interactions in a list were correct.
It is interesting to speculate about the future direction of work investigating expression coevolution. Current research into the cis-regulatory gene expression “code” of yeast, Drosophila, and other organisms may soon make it possible to predict the approximate expression patterns of genes in different conditions on a genome-wide scale (35). If such prediction becomes possible, it will greatly increase the power to detect expression coevolution from sequence alone: Instead of a single number (mean gene expression level, estimated by codon bias), one could calculate a vector representing the expression over many conditions for each gene in each organism. With this more detailed picture of gene expression regulation across different species, expression coevolution could be studied in far greater detail.
Finally, it is possible that coevolution of both protein sequences and expression levels may also be a property of pairs or groups of genes that do not necessarily interact physically. Larger groups, or modules, of genes that work together to produce some output or trait (e.g., a single metabolic pathway) may show coordinated changes in expression levels and/or evolutionary rates because of increased or decreased utilization of those genes over evolutionary time. For example, if the genes specifically responsible for galactose transport and metabolism in yeast (the GAL genes) were used frequently in one species but seldom or never in another related yeast, we would expect to see an increase in the average expression (and thus codon bias) of those genes in the species that metabolized galactose more often. Changes in evolutionary rates also might be seen because the species that seldom use galactose for energy would have little selective pressure to maintain the amino acid sequences of those genes; they would drift more than their orthologous counterparts in the other species, and this lineage-specific drift may be reflected as coevolution of amino acid sequences. Such coevolution at the levels of both expression and sequence evolution may allow inference of functional relationships between groups of genes that do not necessarily physically interact; this evolutionary approach to prediction of genetic relationships and functions may prove to be quite useful as the amount of genome sequence data continues to increase.
Acknowledgments
We thank several anonymous referees for their comments. H.B.F. is a National Science Foundation predoctoral fellow, and M.B.E. is a Pew Scholar in the biomedical sciences.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CAI, codon adaptation index; KS, Kolmogorov-Smirnov.
References
- 1.Altschuh, D., Lesk, A. M., Bloomer, A. C. & Klug, A. (1987) J. Mol. Biol. 193, 693-707. [DOI] [PubMed] [Google Scholar]
- 2.Moyle, W. R., Campbell, R. K., Myers, R. V., Bernard, M. P., Han, Y. & Wang, X. (1994) Science 368, 251-255. [DOI] [PubMed] [Google Scholar]
- 3.Pazos, F., Helmer-Citterich, M., Ausiello, G. & Valencia, A. (1997) J. Mol. Biol. 271, 511-523. [DOI] [PubMed] [Google Scholar]
- 4.Goh, C. S., Bogan, A. A., Joachimiak, M., Walther, D. & Cohen, F. E. (2000) J. Mol. Biol. 299, 283-293. [DOI] [PubMed] [Google Scholar]
- 5.Ramani, A. K. & Marcotte, E. M. (2003) J. Mol. Biol. 327, 273-284. [DOI] [PubMed] [Google Scholar]
- 6.Pazos, F. & Valencia, A. (2001) Protein Eng. 14, 609-614. [DOI] [PubMed] [Google Scholar]
- 7.Pazos, F. & Valencia, A. (2002) Proteins 47, 219-227. [DOI] [PubMed] [Google Scholar]
- 8.Goh, C. S. & Cohen, F. E. (2002) J. Mol. Biol. 34, 177-192. [DOI] [PubMed] [Google Scholar]
- 9.Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. (1998) Proc. Natl. Acad. Sci. USA 95, 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grigoriev, A. (2001) Nucleic Acids Res. 29, 3513-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge, H., Liu, Z., Church, G. M. & Vidal, M. (2001) Nat. Genet. 29, 482-486. [DOI] [PubMed] [Google Scholar]
- 12.Papp, C., Pal, B. & Hurst, L. D. (2003) Nature 424, 194-197. [DOI] [PubMed] [Google Scholar]
- 13.Goffeau, A., Barrell, B. G., Bussey, H., Davis, R. W., Dujon, B., Feldmann, H., Galibert, F., Hoheisel, J. D., Jacq, C., Johnston, M., et al. (1996) Science 274, 563-567. [DOI] [PubMed] [Google Scholar]
- 14.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241-254. [DOI] [PubMed] [Google Scholar]
- 15.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555-556. [DOI] [PubMed] [Google Scholar]
- 17.Sokal, R. R. & Rohlf, F. J. (1995) Biometry (Freeman, New York).
- 18.Sharp, P. M. & Li, W. H. (1987) Nucleic Acids Res. 15, 1281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arava, Y., Wang, Y., Storey, J. D., Liu, C. L., Brown, P. O. & Herschlag, D. (2003) Proc. Natl. Acad. Sci. USA 100, 3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Mering, C., Krause, R., Snel, B., Cornell, M., Oliver, S. G., Fields, S. & Bork, P. (2002) Nature 417, 399-403. [DOI] [PubMed] [Google Scholar]
- 21.Ikemura, T. (1982) J. Mol. Biol. 158, 573-597. [DOI] [PubMed] [Google Scholar]
- 22.Akashi, H. (2003) Genetics 164, 1291-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799-804. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg, D. S. & Roth, F. P. (2003) Proc. Natl. Acad. Sci. USA 100, 4372-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams, C. C., Jakovljevic, J., Roman, J., Harnpicharnchai, P. & Woolford, J. L., Jr. (2002) RNA 8, 150-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dragon, F., Gallagher, J. E., Compagnone-Post, P. A., Mitchell, B. M., Porwancher, K. A., Wehner, K. A., Wormsley, S., Settlage, R. E., Shabanowitz, J., Osheim, Y., et al. (2002) Nature 417, 967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Date, S. V. & Marcotte, S. M. (2003) Nat. Biotechnol. 21, 1055-1062. [DOI] [PubMed] [Google Scholar]
- 28.Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B. A. & Johnston, M. (2003) Science 301, 71-76. [DOI] [PubMed] [Google Scholar]
- 29.Souciet, J. L., Aigle, M., Artiguenave, F., Glandin, G., Bolotin-Fukuhara, M., Bon, E., Brottier, P., Casaregola, S., de Montigny, J., Dujon, B., et al. (2000) FEBS Lett. 487, 3-12. [DOI] [PubMed] [Google Scholar]
- 30.Pellegrini, M., Marcotte, E. M., Thompson, M. J., Eisenberg, D. & Yeates, T. O. (1999) Proc. Natl. Acad. Sci. USA 96, 4285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dandekar, T., Snel, B., Huynen, M. & Bork, P. (1998) Trends Biochem. Sci. 23, 324-328. [DOI] [PubMed] [Google Scholar]
- 32.Marcotte, E. M., Pellegrini, M., Ho-Leung, N., Rice, D. W. & Yeates, T. O. (1999) Science 285, 751-753. [DOI] [PubMed] [Google Scholar]
- 33.Enright, A. J., Iliopoulos, I., Kyrpides, N. C. & Ouzounis, C. A. (1999) Nature 402, 86-90. [DOI] [PubMed] [Google Scholar]
- 34.Jansen, R., Yu, H., Greenbaum, D., Kluger, Y., Krogan, N. J., Chung, S., Emili, A., Snyder, M., Greenblatt, J. F. & Gerstein, M. (2003) Science 302, 449-453. [DOI] [PubMed] [Google Scholar]
- 35.Beer, M. A. & Tavazoie, S. (2004) Cell 117, 185-198. [DOI] [PubMed] [Google Scholar]