Abstract
Dynamic regulation of gene expression is vital for proper cellular development and maintenance of differentiated states. Over the past 20 years, chromatin remodeling and epigenetic modifications of histones have emerged as key controllers of rapid reversible changes in gene expression. Mutations in genes encoding enzymes that modify chromatin have also been identified in a variety of human neurodevelopmental disorders, ranging from isolated intellectual disability and autism spectrum disorder to multiple congenital anomaly conditions that affect major organ systems and cause severe morbidity and mortality. In this study, we review recent evidence that chromodomain helicase DNA-binding (CHD) proteins regulate stem cell proliferation, fate, and differentiation in a wide variety of tissues and organs. We also highlight known roles of CHD proteins in human developmental diseases and present current unanswered questions about the pleiotropic effects of CHD protein complexes, their genetic targets, nucleosome sliding functions, and enzymatic effects in cells and tissues.
Introduction
Epigenetic modifier proteins are commonly divided into three classes: chromatin writers (eg, histone methyltransferases and acetylases), erasers (eg, histone demethylases and deacetylases), and readers (eg, chromodomain and tudor domain remodeling proteins). In this review, we focus on a set of chromatin readers and an important family of ATP-dependent helicase-containing DNA-binding proteins called chromodomain helicase DNA-binding (CHD) proteins. We review their structures, functions, and recently discovered roles in stem cells and human diseases. Interestingly, CHD proteins have been identified as critical regulators of cellular processes such as stem cell quiescence, proliferation, and cell fate determination. In addition, they are implicated in a wide variety of human disease processes, including autism, multiple organ system development, and cancer. Finally, we synthesize recent literature indicating that CHD proteins act at enhancer and promoter regions of genes that regulate key developmental processes, suggesting they orchestrate major cellular proliferation and fate decisions. For reference, a summary of CHD proteins, associated mouse and human phenotypes, stem cells, interacting proteins, and target binding sites is provided in Table 1.
Table 1.
Chromodomain Proteins, Associated Mouse and Human Phenotypes, Stem Cells, Interacting Proteins, and Target Binding Sites
| Protein | Mouse phenotypes | Human phenotypes | Stem cell type | Interacting proteins | Binding sites | Reference(s) |
|---|---|---|---|---|---|---|
| CHD1 | Prostate cancer | ES cells | Mediator complex | H3K4me3 | [11,22,30,31,84] | |
| AT-rich | ||||||
| iPS cells | NCoR, HDAC1/2 | sequences | ||||
| CHD2 | −/−lethal | Epilepsy | Mesenchymal stem cells | H3.3 | [39,69,70,77,96,97] | |
| CHD3 | NuRD, HDAC1/2, ATR, TRIM27 | [69,70,77,98] | ||||
| CHD4/Mi-2b | Epilepsy | Hematopoietic stem cells | NuRD | H3K4 | [26–28,44,46,86,98] | |
| Uterine cancer | Neural stem cells | Polycomb | H3K9me3 | |||
| Dermatomyositis | ||||||
| CHD5 | −/−viable; males infertile | 1p36 deletion | Neural stem cells | NuRD | H3K4 | [12,87,89,99,100] |
| Tumor suppressor | H3K27me3 | |||||
| CHD6 | ID | RNA Pol II | [73] | |||
| CHD7 | −/−lethal | CHARGE | ES cells | P300, others | Sox2, Oct4, Nanog Neurod, Rxrg, Rarb, Twist, Sox9 | [12,32,51,52,64,101] |
| Neural stem cells | SOX2, PBAF | |||||
| +/−CHARGE-like | Autism, ID | Neural crest cells | ||||
| CHD8 (Duplin) | −/−lethal | Autism | β-catenin | [82] | ||
| ID | CTCF, CHD7 | |||||
| CHD9 (CReMM) | Mesenchymal stem cells | GR, PPAR-α | Osteocalcin | [40,41] |
CHD, chromodomain helicase DNA-binding; ES, embryonic stem; iPS, induced pluripotent stem; CHARGE, Coloboma, Heart defects, Atresia of the choanae, Retardation of growth and development, Genital hypoplasia, and Ear abnormalities, including deafness and vestibular disorders; CReMM, chromatin-related mesenchymal modulator; ID, intellectual disability; NuRD, Nucleosome-Remodeling Deacetylase complex.
Structure and Function of the CHD Superfamily
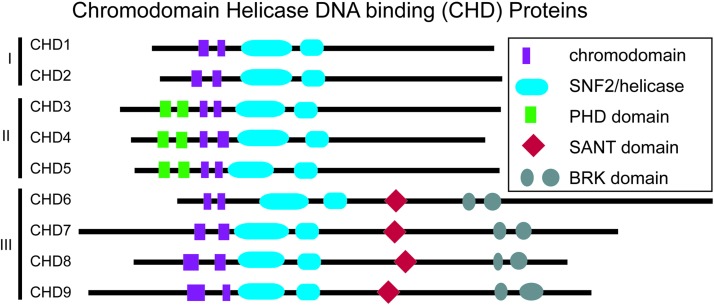
There are three major superfamilies of ATP-dependent chromatin remodeling enzymes in eukaryotic organisms: SWItch/Sucrose NonFermentable (SWI/SNF), Imitation SWI, and CHD, each of which has a characteristic histone interaction domain [1]. These chromatin remodeling enzymes interpret or read histone modifications through specialized protein domains that vary both between and among the protein families. Upon reading the chromatin state, these enzymes disrupt DNA–histone interactions by sliding nucleosomes either along the DNA strand or by translocating the nucleosome core particle to another DNA strand [2]. Ultimately, this chromatin remodeling function allows for improved or reduced access to DNA by transcription factors and other DNA-binding proteins that influence gene expression. The CHD family of ATP-dependent chromatin remodeling enzymes comprises nine proteins divided into three subfamilies based on domain homology (Fig. 1). All CHD proteins contain two tandem chromatin organization modifier (chromo) domains and two Sucrose NonFermentable2 (SNF2)-like ATP-dependent helicase domains [3,4]. The organization of these domains and how they differ between CHD proteins were recently reviewed [5]. In this study, we review highly important functions of specific CHD proteins and protein domains and focus on the roles of CHD proteins in stem cells and human developmental disorders.
FIG. 1.
Cartoon of chromodomain helicase DNA-binding (CHD) proteins and subfamilies. Shown are protein domains with relative positions to the amino (left) and carboxy (right) termini, not to scale. Adapted with permission from Layman et al. [4].
Chromodomains were originally identified in Drosophila heterochromatin protein 1 (HP1). HP1 has a single chromodomain that binds nucleosomes to promote closed chromatin states (heterochromatin) and downregulate homeotic genes during development [6–8]. Specifically, the HP1 chromodomain facilitates protein–protein interactions with the repressive histone modification H3K9me3, leading to the formation of heterochromatin [6,9,10]. It is now understood that the primary common function of chromodomains is binding to methylated histone residues. Indeed, CHD proteins contain a unique variant of the chromodomain containing a methyl-binding cage that facilitates interactions with lysine residue 4 of histone H3 (H3K4) [10,11]. CHD1 chromodomains (Fig. 1) interact with lysine 4 of methylated histone H3 (H3K4me), and CHD5 chromodomains bind to and maintain lysine 27 of trimethylated histone H3 (H3K27me3) [11,12]. Thus, specific CHD chromatin remodeling proteins exhibit unique functions and preferences for repressive or active histone marks, and the methyl-histone-binding chromodomains are essential for maintaining dynamic chromatin structures and proper gene expression.
The helicase–ATPase domains of CHD proteins are highly similar to those observed in the SWItch2/SNF2 superfamily of ATP-dependent chromatin remodeling enzymes [3,13]. The helicase–ATPase domain functions as a bilobed motor, which provides chemical energy and promotes mechanical disruption of DNA–histone contacts. This histone–DNA disruption leads to sliding of core histones along the DNA template or core histone evacuation and deposition onto another DNA strand [14–17]. Additionally, the CHD subfamilies are delineated by the presence of subfamily-specific protein domains (Fig. 1). CHD1 and CHD2 contain DNA-binding domains, which have been shown to be similar in function to SWI3, ADA2, N-COR, and TFIIB (SANT) domains present in CHD6-9 [18]. The SANT domain confers nonspecific DNA binding, particularly to linker DNA between nucleosomes [19–21].
Chromodomains in CHD1 exhibit preferential binding to AT-rich sequences [22]. Compared with wild-type, recombinant CHD1 and CHD2 lacking the DNA-binding domain lose the ability to bind both to DNA and to nucleosomes, demonstrating the critical roles of CHD1 in nucleosome targeting to DNA [18,23]. CHD3, CHD4, and CHD5 lack DNA-binding domains, yet contain two tandem plant homeodomains (PHDs) [24]. PHD domains are zinc finger motifs that facilitate interactions with methylated histone residues and protein cofactors, for example, between CHD3/4 and histone deacetylase 1, which is part of the potent negative transcriptional regulation Nucleosome-Remodeling Deacetylase (NuRD) complex [25]. PHD domains confer specificity to the target histone residues. For example, binding of the two tandem PHD domains in CHD4 to H3K4 (unmodified) and H3K9me3 (but not to H3K4me3) mediates the transcriptional repressive activity of CHD4/NuRD [26–28]. In addition to the chromodomain and helicase domains, CHD6–9 proteins also contain tandem BRahma Kismet (BRK) domains that are also present in Drosophila Brahma (brm), but the functions of these BRK domains are not understood [29].
CHD Proteins and Embryonic Stem Cells
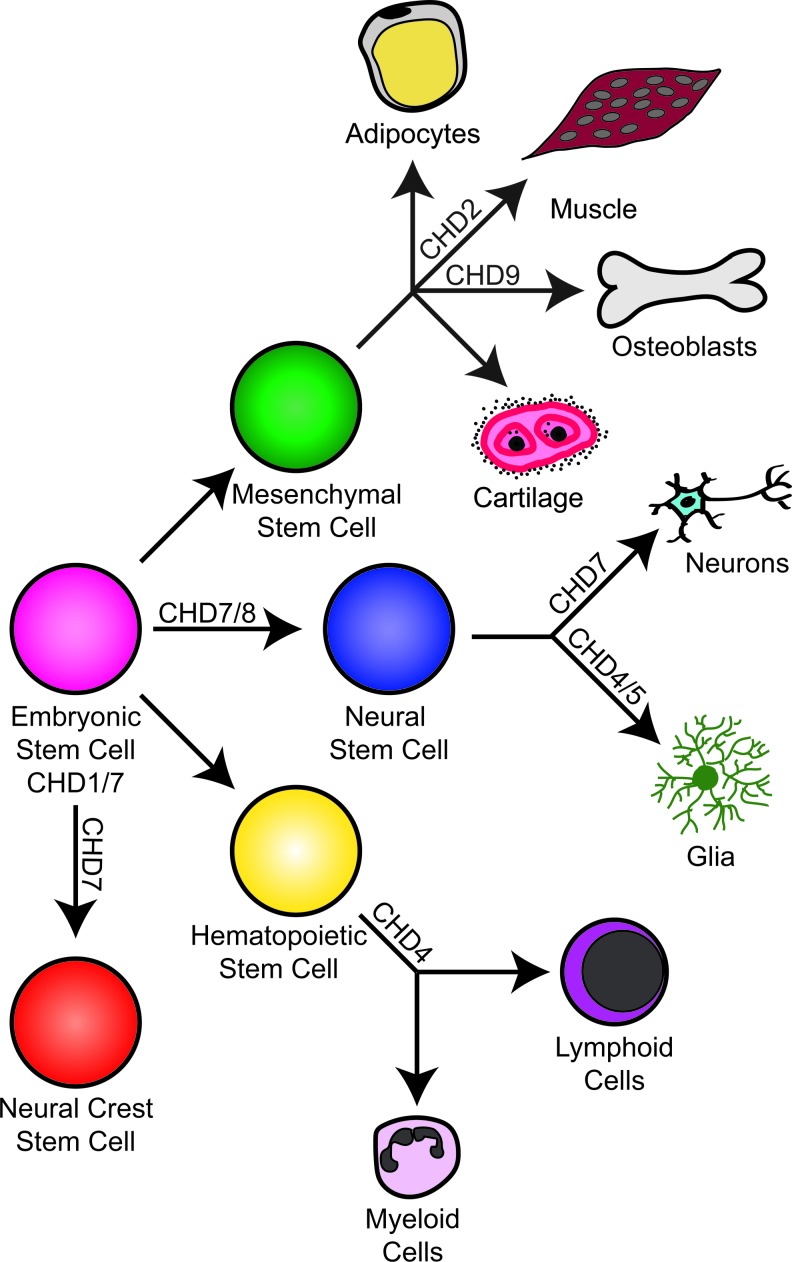
Regardless of the overall protein domain structure, the function of CHD superfamily proteins is intimately tied to regulation of gene expression through remodeling of nucleosomes. Importantly, emerging trends across the entire CHD family include roles in the maintenance, survival, or proliferation of stem cell populations and in directing cell fate decisions of their progeny (Fig. 2). Embryonic stem (ES) cells comprise a self-renewing and pluripotent cell population from which the majority of mammalian tissues are derived. ES cells can be viewed as a blank slate with an open chromatin environment, becoming progressively more differentiated toward neural, hematopoietic, mesenchymal, and other lineage-specific cells through activation or repression of various genetic pathways. CHD1, the first CHD protein implicated in stem cell function, was shown to participate in Mediator complex regulation of ES cells by maintaining open chromatin [30]. The Mediator complex is a multiprotein complex responsible for preinitiation of gene transcription that binds to CHD1 and recruits it to actively expressed genes [31]. Specifically, CHD1 binds to tracts of the active mark trimethylated histone H3 at the residue lysine 4 (H3K4me3) and excludes the repressive mark H3K27me3 [30]. When CHD1 is deleted in ES cells, chromatin condenses to form heterochromatin and pluripotent differentiation is impaired potentially due to promotion of ectodermal lineage gene expression at the expense of endodermal lineage gene expression [30]. In addition to its role in ES cells, CHD1 influences pluripotency since induction of CHD1 expression is required for efficient reprograming of mature fibroblasts into induced pluripotent stem (iPS) cells [30]. Together, these studies show that CHD1 is vital for maintaining pluripotency in ES and other stem cells. By extension, CHD1 may also play an important role in establishment of the pluripotent state, but this has yet to be formally tested.
FIG. 2.
CHD proteins function in a variety of stem cell types. Shown, in colored circles, are the various stem cell types and their associated CHD proteins, along with differentiated cell lineages to which they contribute. Specific functions (activation, inhibition) are omitted for simplicity or, in some cases, because this information is not yet available.
Like CHD1, CHD7 has been implicated in several stem cell populations and appears to be critical for regulating cell fate decisions through modulation of signaling and epigenetic pathways. The mouse Chd7 gene is highly expressed in ES cells, where it appears to function similarly to CHD1 by associating with signals of active gene expression and open chromatin at enhancer elements of critical stem cell pluripotency genes, including Sox2, Oct4, and Nanog [32]. While it is common for genes to be completely active or inactive in fully differentiated cells, the euchromatic chromatin environment in stem and progenitor cells is poised for either repression or activation [33,34]. Unlike their active and inactive counterparts, poised enhancers and promoters typically display both active and repressive marks [35]. CHD7 preferentially binds active (H3K4me1+, H3K27ac+) and poised (H3K4me1+, H3K27me3+) enhancer elements at ectodermal lineage genes in ES cells [32]. Thus, colocalization of CHD7 with poised genetic elements presents a paradigm whereby CHD7 may play a role in recruiting transcription factors, histone modifiers, and/or other chromatin remodeling proteins in a cell type-specific manner to promote activation or repression of certain classes of genes through resolution of the poised chromatin state.
CHD Proteins and Mesenchymal Stem Cells
Recent studies have provided evidence that CHD proteins also regulate mesenchymal development (Fig. 2). Mesenchymal stem cells are multipotent mesoderm-derived cells that give rise to myoblasts (muscle), adipocytes (fat), osteoblasts (bone), and chondrocytes (cartilage) [36–38]. The differentiation of mesenchymal stem cells into four distinct lineages is regulated by several different CHD proteins. Induction of myogenic cell fates requires the recruitment by CHD2 of a chromatin destabilizing histone variant, histone H3.3, to muscle differentiation genes, which then facilitates binding and gene activation by the transcription factor MyoD [39]. Interestingly, CHD9, also known as chromatin-related mesenchymal modulator, binds to and promotes the expression of osteocalcin (bone gamma-carboxyglutamate), one of the major genes responsible for promoting bone development [40,41]. Collectively, these observations emphasize the importance of CHD family proteins in mesenchymal cell fate decisions and highlight the basic principle that these proteins can have diverse and potentially nonredundant functions within the same stem cell type.
CHD Proteins and Hematopoietic Stem Cells
In addition to their roles in promoting development of mesenchymal derivatives, one CHD protein (CHD4) has been implicated in hematopoietic lineages (Fig. 2). The diverse cell types present in blood are derived from common hematopoietic stem cell progenitors that reside in the bone marrow and adopt one of two potential lineages: myeloid and lymphoid [42,43]. Myeloid lineage cells give rise to red blood cells, platelet-producing megakaryocytes, and granulocyte immune cells [42,43]. Lymphoid lineage cells give rise to the agranulocytic natural killer, T-, and B-cells that also contribute to the innate and adaptive immune responses [42,43]. CHD4, also known as Mi-2β, is an NuRD complex component that has been shown to promote self-renewal and multilineage differentiation of hematopoietic stem cells [44]. Importantly, loss of the Chd4 gene in mouse bone marrow leads to an abundance of erythroid progenitors and red blood cells and to loss of lymphoid and remaining myeloid lineage cell types [44]. Further examination of CHD4 activity in the regulation of hematopoietic stem cells may provide novel insights into mechanisms of human diseases such as cancers affecting lymphoid lineage cells, including leukemia and lymphoma. Functions for other CHD proteins in hematopoietic cell types have not yet been reported.
CHD Proteins and Neural Stem Cells
Neural stem cells play a vital role in the development and maintenance of the central nervous system and in sensory organs by producing neurons and supporting cells such as glia and oligodendrocytes. CHD4, CHD5, and CHD7 play pivotal roles in the function and differentiation of neural stem cell niches in the subventricular zone (SVZ) of the forebrain and dentate gyrus of the hippocampus through cooperation with major epigenetic modifiers, transcription factors, and signaling pathways. During cortical neurogenesis, CHD4 is expressed in murine SVZ neural progenitor cells and interacts with the Polycomb Repressive Complex 2 (PRC2), specifically with the H3K27 methyltransferase enzyme, Enhancer of Zeste 2 [45,46]. The CHD4/PRC2 complex directly binds to the promoter of the glial fibrillary acidic protein (GFAP) gene (Gfap), represses its expression, and prevents glial differentiation [46]. Through inhibition of the Gfap locus, CHD4 and PRC2 promote neuronal differentiation during the neurogenic period (between embryonic days 11 and 18 in mice) [46]. In addition to these roles for CHD4 in neural stem cells, a recent study demonstrated that CHD4 and other NuRD complex proteins actively repress genes that inhibit neuronal differentiation in the cerebellum [47]. Thus, individual CHD proteins have important roles in stem cell and differentiated cell populations, guiding decision making and developmental progression at multiple stages.
Similar to CHD4, CHD5 also interacts with the PRC2 and specifically associates with the repressive histone modification H3K27me3 in neural stem cells [12]. CHD5 is highly expressed in neural progenitor and neuroblast cells of the SVZ and subgranular zone (SGZ) of the hippocampus, which produces neurons that are important for learning and memory [12]. Reduced Chd5 expression in the developing cortex also leads to a significant loss of migratory neuroblasts [12], and Chd5-deficient neural stem cells exhibit downregulation of genes responsible for neuronal migration and maturation [12]. Taken together, these studies demonstrate that CHD4 and CHD5 are critical for the inhibition of glial differentiation during key neurogenic phases of mammalian brain development and suggest that chromatin remodeling is vital and dynamic during brain development.
CHD7 is another CHD protein that is enriched in and critical for proper function of neural stem cells. Chd7 is highly expressed in the SGZ of the hippocampus and the SVZ of adult mice, where it colocalizes with markers of neural stem cells (GFAP), neural progenitor cells (marked by the transcription factor ASCL1), and neuroblasts (marked by Doublecortin or DCX) [48,49]. Longitudinal studies in mice with temporally induced conditional deletion of Chd7 in adult SVZ neural stem cells show that Chd7 deficiency leads to a reduction in mature dopaminergic and GABAergic olfactory bulb interneurons and reduced expression of the proneural genes, Sox4 and Sox11 [49]. In the SGZ of the hippocampus, conditional knockout of Chd7 also leads to a reduction in neurogenesis, which can be rescued through exercise [49]. In a recent study, Chd7 was also shown to promote quiescence and maintenance of adult hippocampal neural stem cell populations [50]. Considering that Chd7 also promotes neural stem cell progenitor proliferation in the developing olfactory and otic placodes [51,52], these studies provide convincing evidence that CHD7 is essential for stem cell function in a variety of tissues. However, the precise mechanisms by which CHD7 regulates stem cell proliferation, quiescence, fate, or differentiation, which binding partners and genomic targets it associates with, and whether these mechanisms vary between developmental and postnatal stages remain to be determined. It is also not clear whether differences in CHD7-mediated mechanisms of neural development differ across the various neural stem cell populations in the SVZ, SGZ, and sensory organs.
CHD Proteins in Human Disease
CHD7 and CHARGE syndrome
CHARGE syndrome (Coloboma, Heart defects, Atresia of the choanae, Retardation of growth and development, Genital hypoplasia, and Ear abnormalities, including deafness and vestibular disorders) is a multiple congenital anomaly condition that occurs in approximately 1 in 10,000 live births [53]. Heterozygous mutations in CHD7 are found in over 90% of individuals with CHARGE syndrome, suggesting that it is a monogenic disorder with variable expressivity [54–56]. To date, there are no definitive genotype/phenotype correlations, likely due to the tremendous diversity of CHD7 nonsense, missense, deletion, and truncation mutations and the highly variable expressivity of CHARGE features, even between members of the same family who have the same mutation [56]. In vitro biochemical assays with recombinant CHD7 protein have confirmed that CHD7 is an ATP-dependent chromatin remodeling enzyme capable of modulating DNA–histone interactions [57]. Interestingly, when point mutations in the chromodomains from individuals with CHARGE are introduced into recombinant CHD7, its ATP-dependent chromatin remodeling enzymatic activity is reduced in a mutation-specific manner [57]. These studies provide additional evidence for haploinsufficiency as the major mechanism of CHARGE features. However, additional functions of other CHD7 protein domains may still be discovered and these new cellular functions, if they exist, may help explain the wide variability in clinical features in individuals with CHD7 mutations.
In individuals with CHARGE syndrome, CHD7 haploinsufficiency causes dysfunction in sensory processes and impaired hearing, vision, balance, and olfaction. To more fully understand the role of CHD7 in development and its impact on sensory processes, mouse models have been created and carefully analyzed. In our laboratory, a Chd7 gene trap null allele was generated through insertion of the lacZ reporter into the Chd7 allele, creating a functionally null allele with β-galactosidase reporter activity [58]. Interestingly, Chd7 null mouse embryos do not survive past embryonic day 10.5, presumably from respiratory and cardiovascular defects, while Chd7 heterozygous mice display many of the same defects observed in CHARGE [58]. Similarly, there have been no humans identified with homozygous mutations in CHD7, suggesting that complete loss of CHD7 (and perhaps other CHD genes) is embryonically lethal.
Chd7 is widely expressed in mammalian tissues, most notably in those affected in CHARGE syndrome, including the heart, inner ear, eye, olfactory epithelium, and brain [4,58]. In each organ thus far analyzed, Chd7 expression has been predictive of tissue or cellular defects in mutant mice. In the inner ear, Chd7 heterozygous mice display hypoplasia or aplasia of the lateral and posterior semicircular canals and innervation defects of the vestibular sensory epithelium [52,59]. Conditional knockout of Chd7 causes complete aplasia of vestibular and cochlear structures, reductions in fibroblast growth factor signaling, and decreased proliferation with reduced otic neural progenitors and reduced expression of proneural genes such as Ngn1 and Neurod1 [52]. These data indicate that CHD7 may function in some sensory tissues similarly as it does in brain neural stem cells, through regulation of critical molecular pathways that activate neurogenesis and inhibit gliogenesis.
Hyposmia and anosmia, reduction or loss of the sense of smell, are two of the most highly penetrant phenotypes observed in individuals with CHARGE. Olfactory deficits are commonly accompanied by hypoplasia or aplasia of the olfactory bulbs in the brain [55,60–62]. It was discovered, through electrophysiological and behavioral assays, that Chd7 heterozygous mice display complete anosmia, lack of odor discrimination, and olfactory bulb hypoplasia [61,63]. Chd7 is highly expressed in olfactory epithelial neural stem and progenitor cells, as demonstrated by colocalization with Ascl1 and Neurod1 [61]. In mice, loss of Chd7 correlates with a marked decrease in olfactory epithelial neural stem cell proliferation, a subsequent reduction in olfactory sensory neurons, and impaired recovery from damage [61]. Interestingly, Chd7 heterozygous mutant mice also show decreased dopaminergic tyrosine hydroxylase-positive interneurons in the olfactory bulb, which could be due to impaired efferent signals from the olfactory epithelium or a defect in olfactory bulb neurogenesis from the SVZ neural stem cell niche [61]. Thus, olfactory processing depends on CHD7 function not only in the peripheral olfactory epithelium but also in central nervous system-derived neurogenic niches.
Coimmunoprecipitation and chromatin immunoprecipitation studies have also shown that in neural stem cells, CHD7 physically interacts with SOX2 at genes that are associated with human diseases, including Alagille syndrome (caused by mutations in JAG1, a Notch signaling ligand), Feingold syndrome (caused by mutations in MYCN, a bHLH transcription factor), and Pallister–Hall syndrome (caused by mutations in GLI3, a mediator of Sonic hedgehog signaling) [64]. Several phenotypes observed in these syndromes are also common in CHARGE syndrome, including genital abnormalities (Pallister–Hall and SOX2 deficiency), tracheoesophageal defects (Alagille, Feingold, and SOX2 deficiency), pituitary and endocrine dysfunction (Pallister–Hall and SOX2 deficiency), and semicircular canal hypoplasia (Alagille). CHD7 binding to SOX2 is intriguing due to the extensive overlap in expression and function of these two proteins and their contributions to development of ectodermal lineages affected in CHARGE, including the brain, retina, and neural crest-derived cells, that populate the heart and craniofacial tissues [65]. Together, these observations suggest that CHD7 activity and protein–protein interactions may be important for stem cell differentiation and cell fate decisions in a wide variety of tissues and cell types.
CHD proteins in autism spectrum disorder, intellectual disability, and epilepsy
Application of cytogenetic and next-generation sequencing technologies to large cohorts of individuals with autism spectrum disorder (ASD), intellectual disability (ID), and/or epilepsy has uncovered de novo and inherited heterozygous frameshift, nonsense, or copy dosage mutations in several CHD genes, including CHD2, CHD6, CHD7, and CHD8 [66–74]. For CHD2, CHD6, and CHD7, mutations identified thus far are nonrecurrent (present in only individual cases), private mutations that account for a small fraction of ASD/ID/epilepsy cases. In contrast to the rare isolated mutations in these three CHD genes, 13 different recurrent alleles of CHD8 have been observed in individuals with ASD/ID in association with macrocephaly and gastrointestinal disturbance [66,70,75–78]. In cultured cells, CHD8 has been shown to bind CHD7 and to both bind and regulate p53 and inhibit its proapoptotic effects during development; thus, it is surprising that loss of CHD8 is not associated with broader phenotypic effects in humans [79–82]. Knockdown of Chd8 by shRNA does not alter the morphology or neural ectodermal markers of neural progenitors derived from human iPS cells, but does significantly impair their gene expression [74]. A very recent study also showed that CHD7 binds to and represses p53, suggesting some CHD proteins (at least those in the third subclass) may share common downstream mechanisms, interacting factors, and target genes [83]. Thus, CHD7 and CHD8 not only share common mechanistic pathways but also make unique contributions to developmental events in a wide variety of tissues and cell types.
Similar to CHD7 and CHD8 in ASD, CHD2 mutations have been observed in individuals with epilepsy [66–68]. This raises the likelihood that the spectrum of human mutations in CHD genes could be much broader than previously suspected. Given the complexities of CHD target genes and interacting partners, a major research goal is to clarify the exact mechanisms by which loss of CHD protein function disrupts neural stem cell and/or neuronal/glial development, and to determine exactly how this disruption results in the complex developmental abnormalities in CHARGE syndrome and the cognitive and behavioral profiles observed in ASD/ID and epilepsy. Such research could uncover common underlying mechanisms that may also potentially be targeted by therapies that directly or indirectly modulate chromatin. Treatments might then be directed toward altering the structure and/or function of synapses, dendrites, axons, and signaling pathways that are critical for proper progenitor, neuronal, and glial development.
CHD proteins in cancer and other disease processes
In addition to the varied developmental roles for CHD proteins in human biology, there are several other conditions where CHD proteins have been implicated either through genetic screens or analysis of tissue transcriptomic or biochemical data. CHD1 was identified as frequently deleted in the homozygous form in prostate cancer [84] and likely contributes to cellular invasiveness [85]. CHD4/Mi-2β has been found to be deleted in a high percentage (17%) of endometrial cancers and is implicated as an autoantigen in the inflammatory disorder dermatomyositis [86]. CHD5 is unique among CHD proteins, in that it is highly enriched in the nervous system and testis, and has been found to act as a tumor suppressor in a wide variety of cancers, including neuroblastoma, breast, ovarian, prostate, colon, and lung cancer, and has been shown to regulate spermatogenesis [87–89]. Notably, CHD6 mutations have been identified in bladder [90] and colon cancers [91], consistent with roles for CHD proteins in DNA damage repair and response pathways [92]. Analysis of gastric and colorectal cancers has revealed mutations or altered expression of several CHD proteins—CHD1, CHD2, CHD3, CHD4, CHD7, CHD8, and CHD9—providing further evidence of the broad roles for chromatin remodeling in cancer pathogenesis [93]. Somewhat surprisingly, scoliosis is also linked to disruptions or sequence variants in CHD proteins, including CHD2 [94] and CHD7 [95]. Together, these disease associations highlight the wide variety of cells, tissues, and pathophysiological processes that depend upon proper ATP-dependent chromatin dynamics for normal function.
Conclusions
In this study, we have summarized CHD proteins and recent evidence supporting their involvement in a variety of stem cell types. Studies aimed at identifying CHD protein-binding partners, target sites in the genome, and associated disease mechanisms are active areas of research. CHD proteins are emerging as critical contributors to health and disease, with major functions during development of the brain, eye, ear, heart, and skeletal systems. Future work dedicated to uncovering the mechanisms whereby CHD proteins mediate these effects are likely to reveal even more interesting clues about this complex and intriguing class of ATP-dependent chromatin remodelers.
Acknowledgments
J.A.M. was supported by the NIH Hearing, Balance, and Chemical Senses Training Grant (T32-DC000011) and an Individual NRSA (F31-DC013227) and is currently supported by T32-HL007439. E.D.S. is a fellow of the University of Michigan Medical Scientist Training Program (NIH T32-GM007863) and is supported by T32-DC000011. D.M.M. is supported by NIH R01-DC009410 and the University of Michigan Donita B. Sullivan, MD Research Professorship Funds.
Author Disclosure Statement
D.M.M. serves as Chair of the Scientific Advisory Board for the CHARGE Syndrome Foundation. There are no other disclosures.
References
- 1.de la Serna IL, Ohkawa Y. and Imbalzano AN. (2006). Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet 7:461–473 [DOI] [PubMed] [Google Scholar]
- 2.Smith CL. and Peterson CL. (2005). ATP-dependent chromatin remodeling. Curr Top Dev Biol 65:115–148 [DOI] [PubMed] [Google Scholar]
- 3.Hall JA. and Georgel PT. (2007). CHD proteins: a diverse family with strong ties. Biochem Cell Biol 85:463–476 [DOI] [PubMed] [Google Scholar]
- 4.Layman WS, Hurd EA. and Martin DM. (2010). Chromodomain proteins in development: lessons from CHARGE syndrome. Clin Genet 78:11–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W. and Mills AA. (2014). Architects of the genome: CHD dysfunction in cancer, developmental disorders and neurological syndromes. Epigenomics 6:381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball LJ, Murzina NV, Broadhurst RW, Raine AR, Archer SJ, Stott FJ, Murzin AG, Singh PB, Domaille PJ. and Laue ED. (1997). Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J 16:2473–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearce JJ, Singh PB. and Gaunt SJ. (1992). The mouse has a Polycomb-like chromobox gene. Development 114:921–929 [DOI] [PubMed] [Google Scholar]
- 8.Wreggett KA, Hill F, James PS, Hutchings A, Butcher GW. and Singh PB. (1994). A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet Cell Genet 66:99–103 [DOI] [PubMed] [Google Scholar]
- 9.Brehm A, Nielsen SJ, Miska EA, McCance DJ, Reid JL, Bannister AJ. and Kouzarides T. (1999). The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J 18:2449–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR., 3rd and Grant PA. (2005). Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433:434–438 [DOI] [PubMed] [Google Scholar]
- 11.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F. and Khorasanizadeh S. (2005). Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438:1181–1185 [DOI] [PubMed] [Google Scholar]
- 12.Egan CM, Nyman U, Skotte J, Streubel G, Turner S, O'Connell DJ, Rraklli V, Dolan MJ, Chadderton N, et al. (2013). CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev Cell 26:223–236 [DOI] [PubMed] [Google Scholar]
- 13.Hirschhorn JN, Brown SA, Clark CD. and Winston F. (1992). Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev 6:2288–2298 [DOI] [PubMed] [Google Scholar]
- 14.Winston F. and Carlson M. (1992). Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet 8:387–391 [DOI] [PubMed] [Google Scholar]
- 15.Peterson CL, Dingwall A. and Scott MP. (1994). Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci U S A 91:2905–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pazin MJ. and Kadonaga JT. (1997). SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions?. Cell 88:737–740 [DOI] [PubMed] [Google Scholar]
- 17.Becker PB. and Horz W. (2002). ATP-dependent nucleosome remodeling. Annu Rev Biochem 71:247–273 [DOI] [PubMed] [Google Scholar]
- 18.Ryan DP, Sundaramoorthy R, Martin D, Singh V. and Owen-Hughes T. (2011). The DNA-binding domain of the Chd1 chromatin-remodelling enzyme contains SANT and SLIDE domains. EMBO J 30:2596–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuster EF. and Stoger R. (2002). CHD5 defines a new subfamily of chromodomain-SWI2/SNF2-like helicases. Mamm Genome 13:117–119 [DOI] [PubMed] [Google Scholar]
- 20.Aasland R, Stewart AF. and Gibson T. (1996). The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci 21:87–88 [PubMed] [Google Scholar]
- 21.Boyer LA, Latek RR. and Peterson CL. (2004). The SANT domain: a unique histone-tail-binding module?. Nat Rev Mol Cell Biol 5:158–163 [DOI] [PubMed] [Google Scholar]
- 22.Stokes DG. and Perry RP. (1995). DNA-binding and chromatin localization properties of CHD1. Mol Cell Biol 15:2745–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu JC, Gerreira CG. and Yusufzai T. (2015). Human CHD2 is a chromatin assembly ATPase regulated by its chromo- and DNA-binding domains. J Biochem 290:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bienz M. (2006). The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 31:35–40 [DOI] [PubMed] [Google Scholar]
- 25.Xue Y, Wong J, Moreno GT, Young MK, Cote J. and Wang W. (1998). NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 2:851–861 [DOI] [PubMed] [Google Scholar]
- 26.Musselman CA, Ramirez J, Sims JK, Mansfield RE, Oliver SS, Denu JM, Mackay JP, Wade PA, Hagman J. and Kutateladze TG. (2012). Bivalent recognition of nucleosomes by the tandem PHD fingers of the CHD4 ATPase is required for CHD4-mediated repression. Proc Natl Acad Sci U S A 109:787–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansfield RE, Musselman CA, Kwan AH, Oliver SS, Garske AL, Davrazou F, Denu JM, Kutateladze TG. and Mackay JP. (2011). Plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J Biol Chem 286:11779–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Musselman CA, Mansfield RE, Garske AL, Davrazou F, Kwan AH, Oliver SS, O'Leary H, Denu JM, Mackay JP. and Kutateladze TG. (2009). Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem J 423:179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daubresse G, Deuring R, Moore L, Papoulas O, Zakrajsek I, Waldrip WR, Scott MP, Kennison JA. and Tamkun JW. (1999). The Drosophila kismet gene is related to chromatin-remodeling factors and is required for both segmentation and segment identity. Development 126:1175–1187 [DOI] [PubMed] [Google Scholar]
- 30.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E. and Ramalho-Santos M. (2009). Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460:863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin JJ, Lehmann LW, Bonora G, Sridharan R, Vashisht AA, Tran N, Plath K, Wohlschlegel JA. and Carey M. (2011). Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev 25:2198–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zentner GE, Tesar PJ. and Scacheri PC. (2011). Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21:1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M. and Fisher AG. (2006). Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8:532–538 [DOI] [PubMed] [Google Scholar]
- 34.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326 [DOI] [PubMed] [Google Scholar]
- 35.Zhou VW, Goren A. and Bernstein BE. (2011). Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12:7–18 [DOI] [PubMed] [Google Scholar]
- 36.Ralston SH. and de Crombrugghe B. (2006). Genetic regulation of bone mass and susceptibility to osteoporosis. Genes Dev 20:2492–2506 [DOI] [PubMed] [Google Scholar]
- 37.Uccelli A, Moretta L. and Pistoia V. (2008). Mesenchymal stem cells in health and disease. Nature reviews. Immunology 8:726–736 [DOI] [PubMed] [Google Scholar]
- 38.Chamberlain G, Fox J, Ashton B. and Middleton J. (2007). Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells 25:2739–2749 [DOI] [PubMed] [Google Scholar]
- 39.Harada A, Okada S, Konno D, Odawara J, Yoshimi T, Yoshimura S, Kumamaru H, Saiwai H, Tsubota T, et al. (2012). Chd2 interacts with H3.3 to determine myogenic cell fate. EMBO J 31:2994–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shur I. and Benayahu D. (2005). Characterization and functional analysis of CReMM, a novel chromodomain helicase DNA-binding protein. J Mol Biol 352:646–655 [DOI] [PubMed] [Google Scholar]
- 41.Shur I, Socher R. and Benayahu D. (2006). In vivo association of CReMM/CHD9 with promoters in osteogenic cells. J Cell Physiol 207:374–378 [DOI] [PubMed] [Google Scholar]
- 42.Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA. and Weissman IL. (2003). Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol 21:759–806 [DOI] [PubMed] [Google Scholar]
- 43.Ugarte F. and Forsberg EC. (2013). Haematopoietic stem cell niches: new insights inspire new questions. EMBO J 32:2535–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, Snippert HJ, Heller EJ, Qi X, Lawton LN, Williams CJ. and Georgopoulos K. (2008). The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev 22:1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS. and Zhang Y. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039–1043 [DOI] [PubMed] [Google Scholar]
- 46.Sparmann A, Xie Y, Verhoeven E, Vermeulen M, Lancini C, Gargiulo G, Hulsman D, Mann M, Knoblich JA. and van Lohuizen M. (2013). The chromodomain helicase Chd4 is required for Polycomb-mediated inhibition of astroglial differentiation. EMBO J 32:1598–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada T, Yang Y, Hemberg M, Yoshida T, Cho HY, Murphy JP, Fioravante D, Regehr WG, Gygi SP, Georgopoulos K. and Bonni A. (2014). Promoter decommissioning by the NuRD chromatin remodeling complex triggers synaptic connectivity in the mammalian brain. Neuron 83:122–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Micucci JA, Layman WS, Hurd EA, Sperry ED, Frank SF, Durham MA, Swiderski DL, Skidmore JM, Scacheri PC, Raphael Y. and Martin DM. (2014). CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum Mol Genet 23:434–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng W, Khan MA, Bellvis P, Zhu Z, Bernhardt O, Herold-Mende C. and Liu HK. (2013). The chromatin remodeler CHD7 regulates adult neurogenesis via activation of soxc transcription factors. Cell Stem Cell 13:62–72 [DOI] [PubMed] [Google Scholar]
- 50.Jones KM, Saric N, Russell JP, Andoniadou CL, Scambler PJ. and Basson MA. (2015). CHD7 maintains neural stem cell quiescence and prevents premature stem cell depletion in the adult hippocampus. Stem Cells 33:196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Layman WS, Hurd EA. and Martin DM. (2011). Reproductive dysfunction and decreased GnRH neurogenesis in a mouse model of CHARGE syndrome. Hum Mol Genet 20:3138–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hurd EA, Poucher HK, Cheng K, Raphael Y. and Martin DM. (2010). The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development 137:3139–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Issekutz KA, Graham JM, Jr., Prasad C, Smith IM. and Blake KD. (2005). An epidemiological analysis of CHARGE syndrome: preliminary results from a Canadian study. Am J Med Genet A 133:309–317 [DOI] [PubMed] [Google Scholar]
- 54.Vissers LE, van Ravenswaaij CM, Admiraal R, Hurst JA, de Vries BB, Janssen IM, van der Vliet WA, Huys EH, de Jong PJ, et al. (2004). Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet 36:955–957 [DOI] [PubMed] [Google Scholar]
- 55.Bergman JE, Janssen N, Hoefsloot LH, Jongmans MC, Hofstra RM. and van Ravenswaaij-Arts CM. (2011). CHD7 mutations and CHARGE syndrome: the clinical implications of an expanding phenotype. J Med Genet 48:334–342 [DOI] [PubMed] [Google Scholar]
- 56.Janssen N, Bergman JE, Swertz MA, Tranebjaerg L, Lodahl M, Schoots J, Hofstra RM, van Ravenswaaij-Arts CM. and Hoefsloot LH. (2012). Mutation update on the CHD7 gene involved in CHARGE syndrome. Hum Mutat 33:1149–1160 [DOI] [PubMed] [Google Scholar]
- 57.Bouazoune K. and Kingston RE. (2012). Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. Proc Natl Acad Sci U S A 109:19238–19243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hurd EA, Capers PL, Blauwkamp MN, Adams ME, Raphael Y, Poucher HK. and Martin DM. (2007). Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome 18:94–104 [DOI] [PubMed] [Google Scholar]
- 59.Adams ME, Hurd EA, Beyer LA, Swiderski DL, Raphael Y. and Martin DM. (2007). Defects in vestibular sensory epithelia and innervation in mice with loss of Chd7 function: implications for human CHARGE syndrome. J Comp Neurol 504:519–532 [DOI] [PubMed] [Google Scholar]
- 60.Pinto G, Abadie V, Mesnage R, Blustajn J, Cabrol S, Amiel J, Hertz-Pannier L, Bertrand AM, Lyonnet S, Rappaport R. and Netchine I. (2005). CHARGE syndrome includes hypogonadotropic hypogonadism and abnormal olfactory bulb development. J Clin Endocrinol Metab 90:5621–5626 [DOI] [PubMed] [Google Scholar]
- 61.Layman WS, McEwen DP, Beyer LA, Lalani SR, Fernbach SD, Oh E, Swaroop A, Hegg CC, Raphael Y, Martens JR. and Martin DM. (2009). Defects in neural stem cell proliferation and olfaction in Chd7 deficient mice indicate a mechanism for hyposmia in human CHARGE syndrome. Hum Mol Genet 18:1909–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Legendre M, Gonzales M, Goudefroye G, Bilan F, Parisot P, Perez MJ, Bonniere M, Bessieres B, Martinovic J, et al. (2012). Antenatal spectrum of CHARGE syndrome in 40 fetuses with CHD7 mutations. J Med Genet 49:698–707 [DOI] [PubMed] [Google Scholar]
- 63.Bergman JE, Bosman EA, van Ravenswaaij-Arts CM. and Steel KP. (2010). Study of smell and reproductive organs in a mouse model for CHARGE syndrome. Eur J Hum Genet 18:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engelen E, Akinci U, Bryne JC, Hou J, Gontan C, Moen M, Szumska D, Kockx C, van Ijcken W, et al. (2011). Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet 43:607–611 [DOI] [PubMed] [Google Scholar]
- 65.Pagon RA, Graham JM, Jr., Zonana J. and Yong SL. (1981). Coloboma, congenital heart disease, and choanal atresia with multiple anomalies: CHARGE association. J Pediatr 99:223–227 [DOI] [PubMed] [Google Scholar]
- 66.Chenier S, Yoon G, Argiropoulos B, Lauzon J, Laframboise R, Ahn JW, Ogilvie CM, Lionel AC, Marshall CR, et al. (2014). CHD2 haploinsufficiency is associated with developmental delay, intellectual disability, epilepsy and neurobehavioural problems. J Neurodev Disord 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Epi KC, Epilepsy Phenome/Genome Project, Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D, Eichler EE, Epstein MP, et al. (2013). De novo mutations in epileptic encephalopathies. Nature 501:217–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carvill GL, Heavin SB, Yendle SC, McMahon JM, O'Roak BJ, Cook J, Khan A, Dorschner MO, Weaver M, et al. (2013). Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet 45:825–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, et al. (2012). Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485:242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, et al. (2012). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485:246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tucker T, Zahir FR, Griffith M, Delaney A, Chai D, Tsang E, Lemyre E, Dobrzeniecka S, Marra M, et al. (2014). Single exon-resolution targeted chromosomal microarray analysis of known and candidate intellectual disability genes. Eur J Hum Genet 22:792–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang YH, Yuen RK, Jin X, Wang M, Chen N, Wu X, Ju J, Mei J, Shi Y, et al. (2013). Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet 93:249–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamada K, Fukushi D, Ono T, Kondo Y, Kimura R, Nomura N, Kosaki KJ, Yamada Y, Mizuno S. and Wakamatsu N. (2010). Characterization of a de novo balanced t(4;20)(q33;q12) translocation in a patient with mental retardation. Am J Med Genet A 152A:3057–3067 [DOI] [PubMed] [Google Scholar]
- 74.Sugathan A, Biagioli M, Golzio C, Erdin S, Blumenthal I, Manavalan P, Ragavendran A, Brand H, Lucente D, et al. (2014). CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci U S A 111:E4468–E4477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bernier R, Golzio C, Xiong B, Stessman HA, Coe BP, Penn O, Witherspoon K, Gerdts J, Baker C, et al. (2014). Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158:263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krumm N, O'Roak BJ, Shendure J. and Eichler EE. (2014). A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci 37:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, et al. (2012). Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338:1619–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suls A, Jaehn JA, Kecskes A, Weber Y, Weckhuysen S, Craiu DC, Siekierska A, Djemie T, Afrikanova T, et al. (2013). De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am J Hum Genet 93:967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nishiyama M, Oshikawa K, Tsukada Y, Nakagawa T, Iemura S, Natsume T, Fan Y, Kikuchi A, Skoultchi AI. and Nakayama KI. (2009). CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol 11:172–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sakamoto I, Kishida S, Fukui A, Kishida M, Yamamoto H, Hino S, Michiue T, Takada S, Asashima M. and Kikuchi A. (2000). A novel beta-catenin-binding protein inhibits beta-catenin-dependent Tcf activation and axis formation. J Biol Chem 275:32871–32878 [DOI] [PubMed] [Google Scholar]
- 81.Nishiyama M, Nakayama K, Tsunematsu R, Tsukiyama T, Kikuchi A. and Nakayama KI. (2004). Early embryonic death in mice lacking the beta-catenin-binding protein Duplin. Mol Cell Biol 24:8386–8394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Batsukh T, Pieper L, Koszucka AM, von Velsen N, Hoyer-Fender S, Elbracht M, Bergman JE, Hoefsloot LH. and Pauli S. (2010). CHD8 interacts with CHD7, a protein which is mutated in CHARGE syndrome. Hum Mol Genet 19:2858–2866 [DOI] [PubMed] [Google Scholar]
- 83.Van Nostrand JL, Brady CA, Jung H, Fuentes DR, Kozak MM, Johnson TM, Lin CY, Lin CJ, Swiderski DL, et al. (2014). Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature 514:228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu W, Lindberg J, Sui G, Luo J, Egevad L, Li T, Xie C, Wan M, Kim ST, et al. (2012). Identification of novel CHD1-associated collaborative alterations of genomic structure and functional assessment of CHD1 in prostate cancer. Oncogene 31:3939–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang S, Gulzar ZG, Salari K, Lapointe J, Brooks JD. and Pollack JR. (2012). Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene 31:4164–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Le Gallo M, O'Hara AJ, Rudd ML, Urick ME, Hansen NF, O'Neil NJ, Price JC, Zhang S, England BM, et al. (2012). Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet 44:1310–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhuang T, Hess RA, Kolla V, Higashi M, Raabe TD. and Brodeur GM. (2014). CHD5 is required for spermiogenesis and chromatin condensation. Mech Dev 131:35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kolla V, Zhuang T, Higashi M, Naraparaju K. and Brodeur GM. (2014). Role of CHD5 in human cancers: 10 years later. Cancer Res 74:652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thompson PM, Gotoh T, Kok M, White PS. and Brodeur GM. (2003). CHD5, a new member of the chromodomain gene family, is preferentially expressed in the nervous system. Oncogene 22:1002–1011 [DOI] [PubMed] [Google Scholar]
- 90.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, et al. (2011). Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 43:875–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, Arango D, Strausberg RL, Buchanan D, et al. (2014). Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res 74:3238–3247 [DOI] [PubMed] [Google Scholar]
- 92.Stanley FK, Moore S. and Goodarzi AA. (2013). CHD chromatin remodelling enzymes and the DNA damage response. Mutat Res 750:31–44 [DOI] [PubMed] [Google Scholar]
- 93.Kim MS, Chung NG, Kang MR, Yoo NJ. and Lee SH. (2011). Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology 58:660–668 [DOI] [PubMed] [Google Scholar]
- 94.Kulkarni S, Nagarajan P, Wall J, Donovan DJ, Donell RL, Ligon AH, Venkatachalam S. and Quade BJ. (2008). Disruption of chromodomain helicase DNA binding protein 2 (CHD2) causes scoliosis. Am J Med Genet A 146A:1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao X, Gordon D, Zhang D, Browne R, Helms C, Gillum J, Weber S, Devroy S, Swaney S, et al. (2007). CHD7 gene polymorphisms are associated with susceptibility to idiopathic scoliosis. Am J Hum Genet 80:957–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marfella CG, Ohkawa Y, Coles AH, Garlick DS, Jones SN. and Imbalzano AN. (2006). Mutation of the SNF2 family member Chd2 affects mouse development and survival. J Cell Physiol 209:162–171 [DOI] [PubMed] [Google Scholar]
- 97.Nagarajan P, Onami TM, Rajagopalan S, Kania S, Donnell R. and Venkatachalam S. (2009). Role of chromodomain helicase DNA-binding protein 2 in DNA damage response signaling and tumorigenesis. Oncogene 28:1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ge Q, Nilasena DS, O'Brien CA, Frank MB. and Targoff IN. (1995). Molecular analysis of a major antigenic region of the 240-kD protein of Mi-2 autoantigen. J Clin Invest 96:1730–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, Bredel M, Vogel H. and Mills AA. (2007). CHD5 is a tumor suppressor at human 1p36. Cell 128:459–475 [DOI] [PubMed] [Google Scholar]
- 100.Quan J. and Yusufzai T. (2014). The tumor suppressor chromodomain helicase dna-binding protein 5 (CHD5) remodels nucleosomes by unwrapping. J Biol Chem 289:20717–20726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T. and Wysocka J. (2010). CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463:958–962 [DOI] [PMC free article] [PubMed] [Google Scholar]