Abstract
Clinical specimens are each inherently unique, limited and non-renewable. As such, small samples such as tissue biopsies are often completely consumed after a limited number of analyses. Here we present a method that enables fast and reproducible conversion of a small amount of tissue (approximating the quantity obtained by a biopsy) into a single, permanent digital file representing the mass spectrometry-measurable proteome of the sample. The method combines pressure cycling technology (PCT) and SWATH mass spectrometry (MS), and the resulting proteome maps can be analyzed, re-analyzed, compared and mined in silico to detect and quantify specific proteins across multiple samples. We used this method to process and convert 18 biopsy samples from 9 renal cell carcinoma patients into SWATH-MS fragment ion maps. From these proteome maps we detected and quantified more than 2,000 proteins with a high degree of reproducibility across all samples. The identified proteins clearly separated tumorous kidney tissues from healthy tissue, and differentiated distinct histomorphological kidney cancer subtypes.
Introduction
For many diseases, especially cancers, examination of disease-affected tissues is essential for clinical diagnosis, prognosis and selecting therapy. Traditionally, tissues are fixed in paraffin and examined empirically by highly trained pathologists (e.g. by light microscopy); the detected histomorphological features are then used to diagnose and classify disease.
More recently, the use of molecular patterns to support histomorphological classification of tissues has attracted interest 1. mRNA transcript profiling has been used to group breast cancer tissue into classes that predict clinical course 2,3; analogous strategies have been applied to most other common cancer types 4. Conceptually similar efforts based on genotypic variants 5,6, microRNA 7 and metabolites 8 have also been described.
Proteins are arguably the most informative class of biomolecules for tissue classification, not only because they most directly indicate the functional state of disease-relevant molecular systems such as signaling or metabolic pathways, but also because they are themselves the major drug targets in many diseases, particularly cancers. Antibody-based assays have been widely used for protein measurement in medical research. For tissue classification, elaborate immunohistochemical staining protocols have been developed and are widely applied. However, they are restricted by the paucity and high cost of validated reagents, difficult batch-to-batch quality control and cross-reproducibility. Moreover, the limited ability to multiplex of these analyses precludes the measurement of multi-protein panels in biopsy tissue samples. A range of array-based methods have been developed to alleviate some of these limitations. These include tissue microarrays 9 where a tissue sample is cut into multiple sections, each of which is tested by a specific immunoassay, and reverse arrays where specific proteins are detected and quantified via arrays of available antibodies in extracts from tissue specimens 10. While these methods increase the degree of multiplexing and thus support, in principle, the analysis of protein patterns, they remain limited by the repertoire of available reagents, their quantitative accuracy and dynamic range.
Mass spectrometry has evolved as the method of choice for versatile, multiplexed and quantitative analysis of proteins and proteomes. So far, two main mass spectrometric strategies have been used for protein analysis, each of which is represented by a multiplicity of methods. Both strategies depend on the efficient and reproducible extraction of proteins from the biological source, and the transformation of the protein sample into a complex peptide sample by proteolysis. In both strategies, the peptide sample is then analyzed by mass spectrometry. The first strategy, referred to as discovery or shotgun proteomics, is characterized by data-dependent acquisition (DDA). It uses a simple heuristic to instruct the mass spectrometer to sequentially select peptide ions from the multitude of precursor ions detected in a survey scan, to fragment the isolated ions, and to record their fragment ion spectra. This strategy is capable of identifying thousands of peptides per sample. However, if the number of peptides in the sample significantly exceeds the number of sequencing cycles available during data acquisition—as is the case for unfractionated tissue extracts—the population of correctly identified peptides across multiple samples becomes poorly reproducible. The second strategy, referred to as targeted proteomics, is exemplified by selected/multiple reaction monitoring (S/MRM). In targeted proteomics, predetermined sets of peptides are selected from the available sample peptides and identified and quantified in a sample by means of specific mass spectrometric assays that have to be prepared a priori for each targeted peptide. S/MRM generates highly reproducible, quantitatively accurate and sensitive data sets but its capacity to multiplex is limited to approximately 100 proteins per analysis. Multiplexing up to a few hundreds proteins is possible, albeit generally at the cost of diminished quantification accuracy or limit of detection 11.
We recently introduced a mass spectrometric strategy that allows reproducible detection and accurate quantification of thousands of proteins from complex protein samples with performance characteristics that approach those of S/MRM. The method, termed sequential window acquisition of all theoretical fragment-ion (SWATH) 12, consists of data-independent acquisition (DIA) which generates fragment ion spectra from all MS-measurable peptides of a proteome within a user-defined retention time vs. mass-to-charge ratio space, and a post-acquisition targeted data analysis strategy that associates signature fragment ion patterns for targeted peptides with feature groups in the SWATH-MS signal map, thus confirming the presence and determining the relative quantity of query peptides among samples. As the SWATH-MS data sets represent a single, permanent digital file representing the gap-free mass spectrometry-measurable proteome of the sample, once acquired, they can be perpetually analyzed in silico to test new hypotheses (e.g. to quantify newly postulated measurable biomarkers across suitable sample cohorts). At a sufficient degree of pattern reproducibility, the method also supports cross study comparisons that are not anticipated at the time of data acquisition. To date, SWATH-MS has been applied to study the relatively uncomplicated yeast proteome 12, the subproteome of glycosylated proteins 13 and purified protein complexes 14, but not to a proteome as complex as that of a human tissue.
Regardless of the MS strategy used for tissue proteome analysis, the success of the measurements critically depends on the quality of the peptide sample analyzed. Numerous published protocols notwithstanding, the reproducible generation of peptide samples at high yields from small amounts of tissue, such as those in biopsies, remains challenging. Pressure cycling technology (PCT) recently emerged as a technology that can be used to perform various steps in proteomic sample preparation 15. Programmed cycles oscillating between ultra-high and ambient pressure induce dissolution of matrix and proteins, and facilitate and accelerate proteolysis16. For clinical samples, PCT also inactivates potential contaminant infectious microorganisms 15.
In this report, we describe a PCT-based protocol for MS-ready peptide preparation from biopsy-level tissues that integrates tissue lysis, protein extraction/digestion, and combine it with the SWATH-MS method to rapidly generate highly reproducible fragment ion maps from human tissue biopsies. This method may enable many applications in basic and clinical research, as well as digital biobanking.
Results
Establishment of PCT-SWATH workflow
First, we optimized a PCT-based protocol that integrates tissue lysis, protein extraction and digestion. The procedure is schematically illustrated in Fig. 1. Briefly, a small piece of wet biopsy tissue, weighing about 1 mg, is placed in a pressure-resistant MicroTube, protein extraction and digestion reagents are added, and the mixture is subjected to pressure cycling. Our method is different from existing preparation of tissue samples for proteomic analysis in that the entire procedure, including tissue lysis, protein extraction and digestion, are integrated as a seamless protocol with minimal sample transfer, permitting analysis of minimal sample amount, and that the reaction solvent volume and incubation time for each step are precisely controlled by the barocycler minimizing technical variation. The resulting peptides are cleaned on reversed-phase columns and are then ready for SWATH-MS.
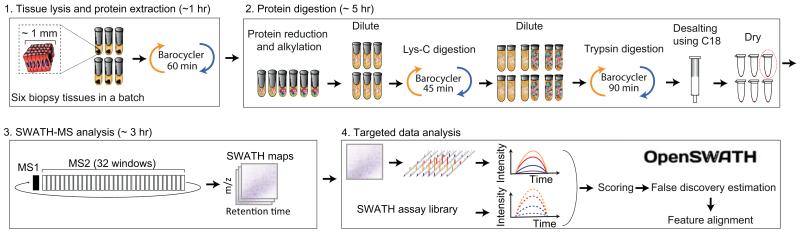
Figure 1. PCT-SWATH method flow chart.
A batch of 6 biopsy-scale tissues are placed in MicroTubes, and lysed in the barocycler for 60 min. Extracted proteins are reduced and alkylated. The urea concentration is then diluted prior to Lys-c digestion in the barocycler for 45 min. The MicroTubes are then taken out of the barocycler, and the urea concentration is further diluted before trypsin digestion for 90 min under barocycling. The resultant peptides are desalted using C18 cartridges and dried under vacuum. This process can be completed in about 6 hr. The peptide samples are then analyzed using SWATH-MS. The data are analyzed using OpenSWATH and a SWATH assay library.
In more detail, protein extraction and digestion were optimized with respect to yield, reproducibility and compatibility of the samples with MS. Among the many tissue lysis and protein extraction conditions tested, a mixture of 8 M urea and 100 mM ammonium bicarbonate supplemented with a protease inhibitor cocktail, in combination with a cycling program of 50 seconds of ultrahigh pressure (45,000 p.s.i.) and 10 seconds of ambient pressure, was found most efficient. Sixty pressure cycles (corresponding to 60 min runtime) were sufficient to extract proteins from different sources, including human kidney tissue, the osteosarcoma cell line U2OS and mouse liver.
Protein digestion was also accelerated by PCT at 33 °C, conditions where the reducing and alkylation agents tris(2-carboxyethyl)phosphine and iodoacetamide were stable in the dark, allowing protein reduction and alkylation in a single step at yields that were equivalent to conventional methods using glass homogenizor for tissue lysis / protein extraction, and overnight in-solution protein digestion (data not shown). Sequential Lys-C and trypsin proteolysis steps were used to efficiently digest proteins into peptides 17. Digestion was accelerated using PCT with a cycling scheme of 50 seconds at 20,000 p.s.i. and 10 seconds at ambient pressure. Lys-C digestion was carried out in 6 M urea for 45 cycles, whereas trypsin digestion was performed in 1.6 M urea for 90 cycles. The entire process from tissue to proteolyzed, injection-ready peptide samples took 6-8 hours. Six samples were processed concurrently in a single batch using this protocol. A higher number of samples can be processed with newer models of the barocycler.
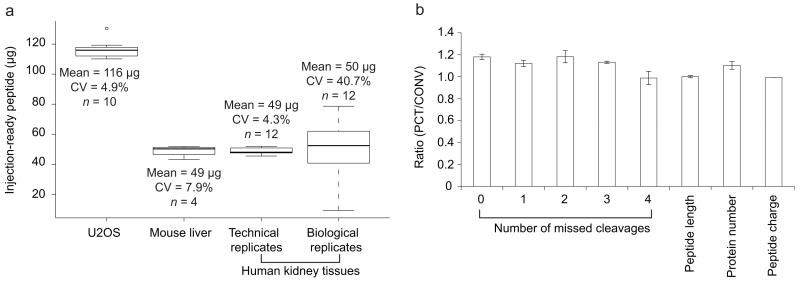
To assess the yield and reproducibility of the optimized PCT method, we determined the total peptide amount generated from a human cell line and two types of tissues. In every case, the total amount of processed, MS injection-ready peptide was determined based on its absorbance at 280 nm using NanoDrop. From the human osteosarcoma cell line U2OS, approximately 116 μg of total peptide mass was generated per million cells (Fig. 2a). From aliquots of 1-3 milligram samples of wet mouse liver and human kidney tissue, the method produced about 50 μg of total peptide mass per milligram of tissue with a high degree of reproducibility for technical replicates (coefficient of variation, CV < 8%). The CV of peptide yield increased to 41% for 12 human kidney tissues from different patients, probably due to biological variation.
Figure 2. Yield and efficiency of PCT-assisted tissue lysis and protein digestion.
(a) Ten aliquots of U2OS cells, four aliquots of mouse liver tissue, and twelve aliquots of human kidney tissue were processed with the PCT method. The final peptide yield was normalized to micrograms of peptides per million cells or milligram of tissue, respectively. Yields are shown in box-and-whiskers plots. The boundaries of the box denote 25% and 75% quantiles, whereas the horizontal line in the middle of the box indicates the median. Whiskers indicate the quantile boundaries for values beyond 1.5 times the interquartile range. Small circle beyond whiskers indicates outlier. (b) Mouse liver tissue was digested with duplicates using the PCT and a conventional (CONV) method. The produced peptides were analyzed by DDA mass spectrometry. Ratios of PCT and CONV in terms of a few parameters are displayed.
It was difficult to compare the yield of the PCT-assisted lysis and digestion method with that achieved by conventional methods at the same scale because we found it impractical to lyse and digest such small tissue samples using conventional glass homogenization or bead mill-based methods. Therefore, to compare the yield of conventional and PCT assisted methods, we lysed about 100 milligrams of mouse liver using a glass homogenizer. From the resulting lysate, we subjected 1% to conventional overnight in-solution digestion using Lys-C and trypsin sequentially. The yield of the conventional method was about 30 μg per milligram tissue, 40% lower than the median yield of PCT-based method carried out entirely on the 1 mg total tissue wet weight level. The respective peptide samples were analyzed by the widely used DDA method in an ion trap mass spectrometer and peptides were identified by sequence database searching. The PCT method produced peptides with a similar length and charge state distribution compared with the conventional method, and the number of peptides identified was 10-20% higher for PCT (Fig. 2b).
Generation of SWATH-MS maps
We next used SWATH-MS to acquire fragment ion spectra of the peptide samples generated by PCT. The peptides were separated by an optimized reversed-phase gradient of 135 min, ionized by electrospray ionization and injected into a TripleTOF 5600 MS operated in SWATH acquisition mode. Precursors in the range of 400-1200 Th were divided into 32 SWATH windows12. All ionized peptide precursors in this mass range were fragmented, generating comprehensive fragment ion maps. Identification of peptides from the SWATH map requires a priori information in the form of a SWATH assay library containing characteristic peptide fragment ion patterns and retention time information for all targeted peptides. Targeted data analysis was conducted using OpenSWATH 18.
We evaluated the reproducibility of the PCT-SWATH workflow by digesting 4 aliquots of a human kidney tissue sample and producing SWATH maps in triplicate. The 12 SWATH-MS maps were analyzed using the OpenSWATH software tool and a SWATH assay library containing 49,959 reference spectra for 41,542 proteotypic peptides from 4,624 reviewed Swissprot proteins, compiled using DDA analyses of the kidney tissues in a TripleTOF 5600 MS. Only one protein product from each gene was included in this analysis, therefore protein isoforms were excluded. The signals for peptides identified in different samples were aligned along the chromatography prior to label-free protein-level quantification using the aLFQ R-package 19. At an estimated false discovery rate of 0.1% at precursor (i.e. ionized peptide) level, we quantified 1,632 unique proteins (Supplementary Table 1). The overall sparsity of the quantified matrix across the 1,632 proteins and 12 samples was 12%, as visualized in a heatmap, indicating the high degree of data completeness for the method (Supplementary Fig. 1a).
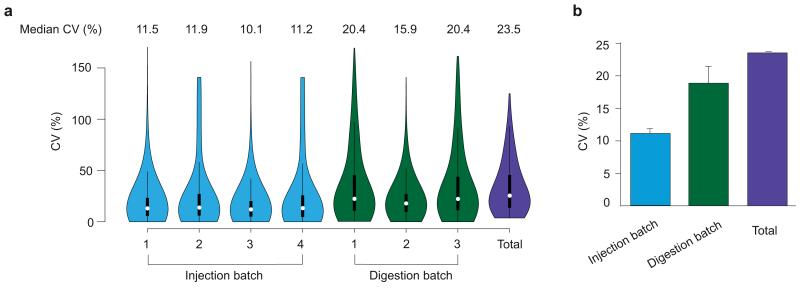
Next, we assessed variation among technical triplicates for each of the 4 digests. We determined the CV of triplicate injections of the 4 digests in three batches and plotted the CV distribution as violin plots (Fig. 3a). The 12% values missing in at least one map were not considered in this analysis. The median CV for repeated MS injection batches was between 10% and 12%. The variation of PCT-assisted digestion across the samples was slightly higher, with the median rising to 16-20%. Median CV of all 12 replicates (Total) was 24% (Fig. 3b). This is the overall median technical variation expected for the same tissue sample measured using the entire PCT-SWATH workflow. This value is comparable to that achieved in our laboratory by S/MRM in a label-free quantification mode if applied at high throughput to the model organism Saccharomyces cerevisiae 20. It is worth noting that excluding the 12% missing values may artificially influence the CV values, particularly among replicates. However, the abundance of proteins did not have significant influence on the CV (Supplementary Fig. 2).
Figure 3. Reproducibility of PCT-SWATH.
(a) Four aliquots of a human kidney tissue sample were separately digested using PCT, and each analyzed by SWATH-MS in duplicates. The distribution of CV values of protein quantification for each digestion batch, injection batch, and total variation is shown as violin plots where the kernel density curves shape the violin, the white node in the center denotes median value, and the black box inside the violin shows the same interquantile range as the box-and-whiskers plot. (b) Bar plot of average median CV of injection, digestion and the entire workflow (Total).
Dynamic range of proteins identified in kidney tissues
We applied the PCT-SWATH method to study a panel of 18 kidney tissue samples from 9 patients with renal cell carcinoma (RCC). From each individual, 2 biopsies were collected, one tumorous and the other non-tumorous as determined by histomorphological examination. Among the 9 patients, 6 had clear cell RCC (ccRCC), 2 were classified as having papillary RCC (pRCC), and 1 as chromophobe RCC (chRCC). Detailed information about these tissues is provided in Supplementary Table 2. Each sample was measured in technical duplicates. The resulting SWATH-MS maps were searched against a kidney tissue SWATH assay library. In total, 2375 proteins were quantified across all samples with a precursor FDR below 0.1% and a sparsity of 32% (Supplementary Table 3). The data are visualized in Supplementary Fig. 1b. The sparsity of the data matrix is higher than the previously tested 12 replicates, probably due to increased biological heterogeneity between individuals and tumor/non-tumorous tissues.
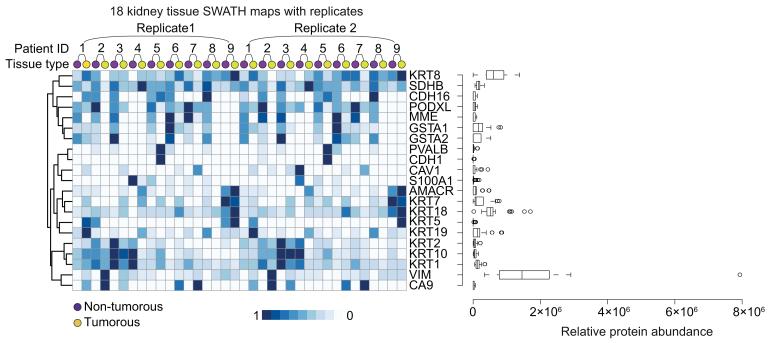
Next, we searched the literature and identified 53 Swiss-Prot proteins presently used as diagnostic or prognostic biomarkers for renal cancers 21,22, of which 21 proteins, including the widely used biomarkers VIM and AMACR, were quantified in our data set (Fig. 4). As we used exclusively proteotypic peptides, i.e. peptides that are unique to a specific protein 23, we were able to independently quantify even highly concordant protein isoforms. This is exemplified by cytokeratin isoforms, some of which showed idiotypic quantitative patterns across samples. For example, after an unsupervised clustering, while most cytokeratins clustered together, while cytokeratin 8 displayed a unique pattern.
Figure 4. Protein markers for RCC quantified in SWATH-MS maps.
Heat map shows relative abundance of 21 proteotypic proteins across 18 kidney tissue SWATH maps. Abundance values of each protein were normalized to a scale of 0 to 1, followed by unsupervised clustering of protein expression pattern. Box-and-whisker plots on the right side shows distribution of relative abundance for each protein. AMACR, alpha-methylacyl-CoA racemase; CA9, carbonate dehydratase IX; CAV1, caveolin 1; CDH1, E-cadherin; CDH16, cadherin 16; GSTA1/2, glutathione S-transferase alpha 1/2; KRTs, keratins; MME, membrane metallo-endopeptidase; PODXL, podocalyxin-like protein 1; PVALB, parvalbumin alpha; S100A1, S100 calcium binding protein A1; SDHB, succinate dehydrogenase; VIM, vimentin.
Next, we examined the dynamic range of proteins quantified in kidney tissues based on their signal intensity and compared the data to a few published data sets that contain estimates of absolute protein abundance in human cell lines 24,25. As shown in Supplementary Fig.3a, the dynamic range is about 4 orders of magnitude, in line with the dynamic range estimations determined on standardized samples with the SWATH method 12. DDA analysis of extensively fractionated cell line or tissue lysates have reached a dynamic range of 4.6-5.8 orders of magnitude.
To understand the subcellular localization of kidney proteins analyzed by PCT-SWATH, we compared the Gene Ontology annotations of the proteins identified in our data sets, the reference data sets, as well as the entire human proteome. The data in Supplementary Fig. 3b demonstrate that a higher number of mitochondrial proteins and fewer (P value = 0.011) nuclear proteins were present in our SWATH data compared to the reference populations, indicating that low abundance proteins are under-represented. We also observed that the percentage of surface proteins in our SWATH data was slightly higher (P value = 0.008) than the DDA data from cell lines and tissues, consistent with the study showing that PCT-assisted lysis and digestion facilitated detection of membrane proteins 15. It is not surprising that proteins from extracellular regions were high in both DDA and SWATH data of tissues, probably because secreted proteins were retained within tissues; proteins originating from blood contamination were also identified.
Variation analysis of tissue proteome PCT-SWATH maps
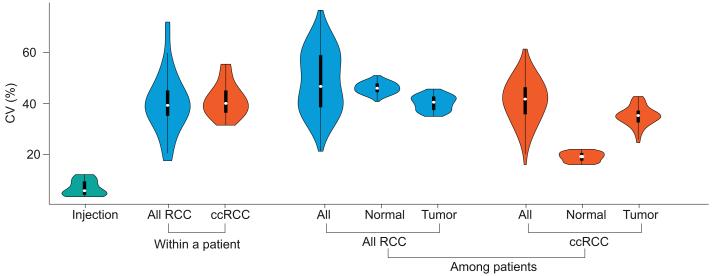
The availability of PCT-SWATH maps of the 9 paired kidney biopsies allowed us to again determine the technical and biological variation of PCT-SWATH analysis. The median CV of the injection batch variation was below 6%, consistent with the 12 kidney tissue test replicates data discussed above (Fig. 5). To estimate biological variation within an individual, we calculated the median CV of all proteins in the SWATH data quantified in every pair of tissue biopsies, either non-tumorous or tumorous, from each of the 9 RCC patients. The median of median CV values indicating within-patient variation rose to close to 40% (Fig.5). Inter-patient CV values in paired kidney biopsies were slightly higher than intra-patient CV. Interestingly, the median CV value of non-tumorous tissues from patients with ccRCC was 19%, significantly lower than the CV observed in all other variations, except injection batch variation. The tumor tissues among patients, even in the ccRCC subtype, displayed relatively high variation (Fig. 5).
Figure 5. Variation analysis of human kidney tissue proteomes by SWATH-MS.
The violin plots show the distribution of CV values from injection replicates, within-patient proteomic variation based on all RCC and ccRCC tissues, and among-patient variation based on all RCC and ccRCC tissues.
Classification of kidney tumor biopsies using PCT-SWATH
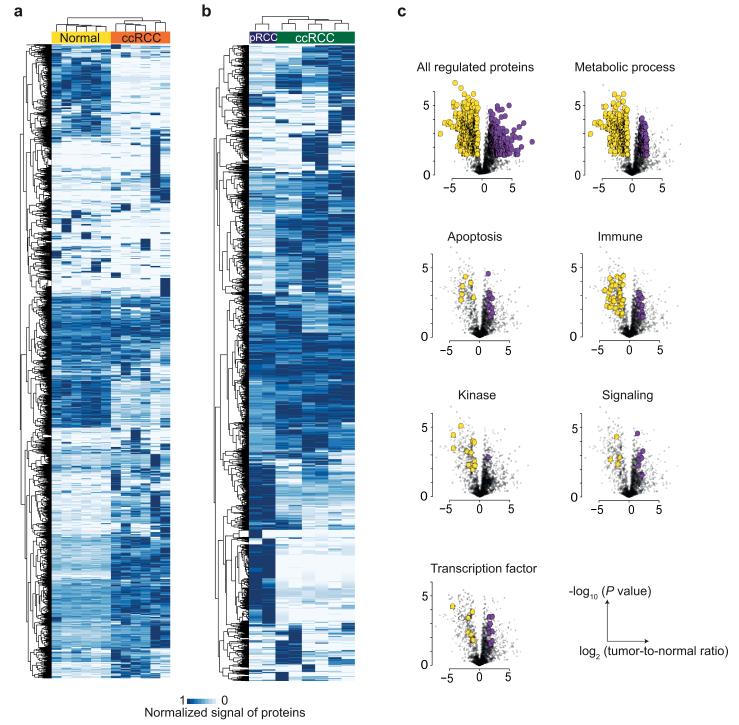
Reproducible quantification of clinically relevant proteins as outlined above suggests a high potential for using this method for tumor classification based on proteome profiling. We found that the proteomic profile is useful to categorize the tissue samples according to their degree of malignancy and histomorphological subtypes. Unsupervised clustering of more than 2,000 consistently quantified proteins resulted in the separation of tumor biopsies from neighboring non-tumorous biopsies from ccRCC patients as shown in Fig.6a. In total, 317 proteins were down-regulated, and 296 proteins were upregulated in tumor compared to non-tumorous biopsies (Supplementary Table 4). These included protein kinases and transcription factors, in addition to other proteins involved in the regulation of biological processes or pathways including apoptosis, metabolic pathways, signaling, and immune response (Fig. 6c).
Figure 6. Comparative proteomic analysis of clear cell RCC tissues.
(a) Unsupervised clustering of 2,317 proteins quantified in ccRCC as measured by PCT-SWATH in tumorous and non-tumorous tissues. (b) Unsupervised clustering of 2276 proteins quantified in pRCC and ccRCC tumorous tissues. (c) Volcano plots of all regulated proteins and proteins belonging to specific protein classes, biological processes, and pathways. P values were calculated using paired two-tailed t-test. Regulated proteins were defined as fold change higher than 2 with P value lower than 0.05. Up-regulated and down-regulated proteins are shown in purple and yellow, respectively.
Unsupervised clustering also correctly separated ccRCC tumors from pRCC tumors (Fig.6b). AMACR is used in clinic as a protein marker to distinguish pRCC (positive by immunohistochemistry) and ccRCC (negative by immunohistochemistry) 21. Here, the SWATH signal of AMACR is more than 13 times higher in pRCC than ccRCC with a P value of lower than 0.000001. GSTA1 and VIM are known to be highly expressed proteins in ccRCC, compared to other subtypes 21, consistent with our data. All the significantly regulated proteins are shown in Supplementary Table 5.
Discussion
Here, we introduce a fairly simple and fast methodology to perform high throughput, quantitative and highly reproducible proteomic analyses of clinical tumor biopsies. At present, PCT-SWATH is the fastest proteomic pipeline, requiring less than 12 hours for relatively deep proteomic analysis of human biopsy samples, with a level of technical reproducibility that is comparable to S/MRM, the most highly reproducible MS-based method at present 20,26 , and next generation sequencing techniques 27. However, in contrast to S/MRM, which is limited to the quantification of maximally a few 100s of proteins per sample injection, 1,000s of proteins could be quantified by SWATH-MS. Moreover, it is worth noting that the technical reproducibility we measured here for PCT-SWATH regards total variation coming from both sample preparation and mass spectrometric analysis.
The unprecedented speed and precision of this proteomic analysis was achieved by seamless integration of the PCT technique with different steps of protein sample preparation, and the conjugation with SWATH-MS. The method minimizes sample handling and chromatography, making it easily transferable to other laboratories given established standard operating procedures. This also contributes to minimizing technical variation and achieving a high level of reproducibility of the MS data, with a median CV of technical replicates below 20%. Further, the method also minimizes sample loss, thus maximizing sample yields, even from biopsy-level tissue samples. Moreover, the simple and fast protocol makes it possible to process a large number of samples with unprecedented reproducibility, an essential factor for clinical research as it provides timely data for clinical decision making.
The resultant SWATH-MS maps can be queried with targeted methods in analogy to “Big Data” datasets since they contain highly reproducible signature fragment ion series for all mass spectrometer-measurable peptides of a proteome within a user-defined retention time vs. mass-to-charge ratio space. Unlike DDA mass spectrometric data which are typically searched in an unbiased manner against a protein sequence database, the analysis of SWATH-MS data is dependent on a priori knowledge of peptide fragmentation and chromatographic behavior of the targeted peptides. Therefore, it is not possible to determine the exact number of proteins and peptides that are present in a SWATH map, because noise, interfering signals obscuring some of the peptide fragment ion groups, signals below the noise level, and absence of some peptides in the SWATH assay library place the number of identified peptides below the maximal number of identifiable peptides. This issue cannot be fully investigated until a complete SWATH assay library for the respective sample type has been generated. However, the PCT-SWATH technology generates a comprehensive and permanent digital record of a sample from which even low signal intensity peptides can be reproducibly quantified and where the absence of a peptide can be verified by the absence of a signal above noise level.
Typically, we processed 1 mg of wet tissue mass. The as such generated SWATH-MS maps offer unique opportunities for tissue biopsy research because maps generated from the same individual over time, from different tissue samples from the same person, or from tissue samples from patient cohorts can be perpetually analyzed and compared in silico without further wet lab experimentation. This feature is particularly unique and advantageous when considering most biopsy samples and early-stage tumor tissues could be analyzed for limited times. Over time, the systematic conversion of tissue samples (e.g. those contained in biobanks) into SWATH-MS maps could complement traditional biobanking efforts and make precious clinical samples accessible to a wide range of evaluations.
At present, there is no software tool to deconvolute all signals in a SWATH-MS map. We therefore used the previously described targeted data analysis strategy12 to compare reference fragment ion spectra of proteotypic peptides generated as prior information with signal groups contained in the SWATH-MS maps. The library used here was generated by DDA analysis in a model 5600 mass spectrometer of 33 samples and contained 49,959 reference spectra for 41,542 peptides from 4,624 reviewed SwissProt proteins. This assay library applied to the OpenSWATH software 18 quantified 2,375 proteins across the 9 paired kidney biopsies which classified the tissues into their correct histological and malignancy groups. The set of quantified proteins also contained common protein markers used in clinical and research laboratories (Fig. 4), suggesting that SWATH-MS maps, once acquired, could be used for clinical assessment. The number of proteins identified in this study is lower than the number of proteins that can be identified in studies using the most advanced DDA methods with extensive fractionation of the sample 25. This is in part due to the fact that in this study we focused on proteotypic peptides only, used the low redundancy SwissProt database as the source of protein identities and used a SWATH assay library of limited complexity and depth. It can be expected that from the same SWATH-MS maps additional proteins and protein variants will be identified and quantified in the future.
The present PCT-SWATH protocol relies on in-solution digestion. Therefore, the tissue being processed should not contain contaminants that interfere with enzymatic digestion. It is common that fresh biopsy tissues are embedded in Tissue-Tek O.C.T. compound, a formulation of water-soluble glycols and resins, as the specimen matrix for cryostat sectioning. The O.C.T. must be washed away prior to PCT-SWATH analysis.
PCT-SWATH is a generic method for proteomic analysis that can be extended beyond biopsy tissues. PCT can be used to prepare mass spectrometry-ready peptides from various types of samples, including bacteria, yeast and mammalian tissues. SWATH-MS can be used to digitize the resultant proteome. With this method, comparative quantitative analysis of large number of proteomes is becoming possible and feasible. In the present form, PCT-SWATH is not ready to be used as a clinical test, but we expect wide applications of PCT-SWATH in personalized medicine and translational studies in general, as large scale quantitative information on proteins is highly desirable for systems modeling and biomarker discovery. Also, if applied with the addition of carefully selected and calibrated isotope labeled reference peptides, PCT-SWATH may reach the quantitative accuracy and reproducibility required for clinical adoption.
Online Methods
Cells and tissues
U2OS cells were obtained from the American Type Culture Collection, grown in DMEM medium supplemented with 10% bovine serum, washed three times using PBS. Mouse liver tissues were provided by Dr. Nora Roesch in the group of Prof. Markus Stoffel, ETH Zurich, complying with official institutional ethical guidelines. The protocol for mouse tissue was approved by the institutional ethics committee. Tumor specimens were collected at the time of surgery. Nine patients with renal cell carcinoma were included in the study. Tissue samples (tumor and matched kidney tissues) were collected from nephrectomy specimens within 1 hr after surgical removal, snap-frozen and stored at −80 °C. Prior to tissue lysis, histological analysis was performed using haematoxylin-eosin stained frozen sections. Tumor cell content was evaluated by a trained pathologist. All tissue samples were collected at the Institute of Pathology, Kantonsspital St.Gallen. The study protocol was approved by the local ethics committee. Written informed consent had been obtained from each patient prior to tissue collection. Tissues embedded in OCT compounds must be washed prior to PCT-SWATH analysis. The OCT washing can be done following the protocol prepared by Misti Martinez and Kent Shaddox, Vanderbilt-Ingram Cancer Center, which is available on the website of Clinical Proteomic Tumor Analysis Consortium (CPTAC). The url for the protocol is https://cptac-data-portal.georgetown.edu/cptac/dataPublic/list?currentPath=%2FPhase_II_Data%2FOCT%2FOCT_Embedded_Tumor_Tissue_VU&nonav=true. Briefly, tissue is placed in a 1.5 mL microtube and vortexed in the following buffers in sequence: 1) 1 mL 70% ethanol / 30% water for 30 seconds; 2) 1 mL 100% water for 30 seconds; 3) 1 mL 70% ethanol / 30% water for 5 minutes, twice; 4) 1 mL 85% ethanol / 15% water for 5 minutes, twice; 5) 1 mL 100% ethanol for 5 minutes, twice.
PCT-based lysis and digestion
U2OS cell, mouse liver, and human kidney tissues were digested using a Barocycler NEP2320-45k (PressureBioSciences, Inc, South Easton, MA). Cell pellets (cell count range 300,000-900,000) and mechanistically disrupted tissue pieces (weight range: 1-3 mg) were placed in Microtubes (Pressure BioSciences) with lysis buffer consisting of 8 M urea, 0.1 M ammonium bicarbonate, COMPLETE protease inhibitor cocktail (Roche), and PhosSTOP phosphatase inhibitor cocktail (Roche). The solution was sonicated for 20 seconds on ice and placed in the barocycler for tissue lysis and protein extraction at 33 °C. The barocyling process was programmed to contain 60 cycles, each consisting of 50 seconds at 45,000 p.s.i. high and 10 sec at ambient pressure. The protein solution was transferred to new Microtubes for subsequent processes. Tris ( 2 - carboxyethyl ) phosphine and iodoacetamide were simultaneously added to the solution at a final concentration of 10 mM and 40 mM, respectively, followed by a 30-min incubation in the dark with gentle vortexing. The 8 M urea was diluted to 6 M using 0.1 M ammonium bicarbonate. Proteins were first digested using Lys-C (Wako, enzyme-to-substrate ratio 1:40) in the barocycler at 33 °C. The barocycling process was programmed to contain 45 cycles, each consisting of 50 sec at 20,000 p.s.i. and 10 seconds at ambient pressure. Tryptic digestion was then performed after diluting the urea concentration to 1.6 M using 0.1 M ammonium bicarbonate. Trypsin (Promega) was used with an enzyme-to-substrate ratio of 1:20. Tryptic digestion was performed in the barocycler at 33 °C using 90 cycles of 50 sec at 20,000 p.s.i. and 10 sec at ambient pressure. Subsequently, trifluoroacetic acid was added to the solution at a final concentration of 0.4% to stop digestion. The peptides were cleaned using SEP-PAK C18 cartridges (Waters Corp., Milford, MA), dried under vacuum, and stored in −80°C before MS analysis. Peptides were dissolved in HPLC grade water containing 0.1% formic acid and 2% acetonitrile. To facilitate peptide solubilization, peptide solution was sonicated in an icy water bath for 5 min. Peptide concentration was measured using NanoDrop 1000 spectrophotometer with the option of Protein A280 (1Ab = 1 mg mL−1). Peptide concentration was adjusted to 0.3 μg μl−1 using 0.1% formic acid / 2% acetonitrile in HPLC water. One microliter of iRT peptide cocktail 28 (Biognosis, Zurich, Switzerland) was spiked into every nine microliter of peptide sample prior to LC-MS analysis.
Orbitrap MS analysis
The peptide samples were analyzed on an LTQ Orbitrap XL mass spectrometer (Thermo Scientific) using Xcalibur version 2.0.7 coupled to an Eksigent NanoLC-2D HPLC nanoflow system (dual pump system with one analytical column; Eksigent) via a nanoelectrospray ion source using a liquid junction (Thermo Scientific), as described previously 29. The raw data were converted to mzXML format and searched using Sorcerer against a target-decoy Swissprot mouse protein database. False discovery rate was controlled below 1% using Trans Proteomic Pipeline (TPP, v 4.6.1).
Triple-TOF MS analysis in DDA and SWATH mode
A 5600 TripleTOF mass spectrometer (AB Sciex, Concord, Canada) and an Eksigent 1D+ Nano LC systems (Eksigent, Dublin, CA) were employed for SWATH-MS analysis. The LC gradient was formulated with a buffer A (2% acetonitrile and 0.1% formic acid in HPLC water) and buffer B (2% water and 0.1% formic acid in acetonitrile). The analytical column (75 μm × 20 cm) was home-packed directly in a fused silica PicoTip emitter (New Objective, Woburn, MA, USA) with 3 μm 200 Å Magic C18 AQ resin (Michrom BioResources, Auburn, CA, USA). The peptide samples were prepared at 0.3 μg μL−1 spiked-in with 1:20 (v:v) iRT peptides. Three microliters of each sample were injected and separated with a linear gradient of 2% buffer B to 35% buffer B over 120 min at a flow rate of 0.3 μl min−1. The column was then flushed with 90% buffer B for 5 min, and re-equilibrated with 2% buffer B for 10 min. For shotgun experiments the TripleTOF mass spectrometer was operated with a “top20” method. From the 500 milliseconds survey scan (TOF-MS), the top 20 most intense precursor ions were selected for subsequent automated MS/MS in measurements where each MS/MS event consisted of a 150 milliseconds scan. The selection criteria for parent ions included intensity, where ions had to be greater than 200 counts per second, a charge state greater than 2+. The precursors that were selected for fragmentation were added to a dynamic exclusion list for 20 s. Ions were isolated using a quadrupole resolution of 0.7 amu and fragmented in the collision cell using the collision energy equation (0.0625 × m/z - 3.5) with an additional collision energy spread of 15 eV within the 150 milliseconds accumulation time to mimic SWATH fragmentation conditions. In the instances where there were ≤ 20 parent ions per survey scan which met the selection criteria, those ions were subjected to extended MS/MS accumulation times to maintain a constant total cycle time of 3.5 s. For SWATH-MS based experiments the mass spectrometer was operated in a looped product ion mode. In this mode the instrument was specifically tuned to allow a quadrupole resolution of 25 amu per mass selection. The stability of the mass selection was maintained by the operation of the RF and DC voltages on the isolation quadrupole in an independent manner as described earlier12. Using an isolation width of 25 amu, a set of 32 overlapping windows (1 amu overlap) was constructed covering the mass range 400 to 1,200 amu. The collision energy for each window was determined based on the collision energy for a 2+ ion centered in the respective window (equation: 0.0625 × m/z - 3.5) with a collision energy spread of 15 eV. An accumulation time of 100 milliseconds was used for each fragment ion scan and for the survey scans acquired at the beginning of each cycle, resulting in a total cycle time of 3.3 sec. The sequential precursor isolation window set-up was as follows: 500-425, 424-450, 449-475, … ,1174-1200, with an effective (100%) transmission of ~ 25 Da and ~ 0.3 Da attrition on either side of the isolation window.
Kidney tissue SWATH assay library generation
A pool of the 18 kidney tissue samples used for SWATH analysis was resolved into 15 gel fractions using SDS-PAGE and digested individually in-gel. The peptides extracted from each gel band were subjected to DDA analysis in TripleTOF MS. The individual digests of whole proteome of the 18 kidney tissues were also analyzed in TripleTOF in DDA mode. iRT peptides28 were spiked in to these samples at a concentration of 10% prior to LC-MS analysis.
All 33 DDA data files were searched for peptide identification using X!Tandem 30 and OMSSA 31 against a target-decoy non-redundant human UniProtKB/SwissProt protein database (Oct 1, 2013) containing 20265 protein sequences and the iRT peptide sequences. The decoy sequences were generated by reversing each of the target sequences. Fully tryptic peptides with maximal 2 missed cleavages were allowed. The mass tolerance of parent mass was set to 50 ppm. Fragment mass tolerance was set at 0.1 Da. Carbamidomethyl at cysteine was set as static modification, and oxidation at methionine was set as variable modification. Search results from X!Tandem and OMSSA were further analyzed through Trans-Proteomic Pipeline (TPP, v4.6.0) 32 using PeptideProphet and iProphet. The cutoff of probability for peptides was 0.9. The resultant pep.xml files were combined into a single file named as interact.pep.xml.
SpectraST (version 4.0) in TPP and a previously described workflow 18 were used to process the resulting interact.pep.xml file, to normalize the retention time, to generate consensus spectra and to produce the TraML library file following the commands below:
spectrast -c_BIN! -cf’Protein!~DECOY’ -cP0.9 -cICID-QTOF -cNstep1 interact.pep.xml
spectrast2spectrast_irt.py -i step1.splib -o step2.splib
spectrast -cAC -c_BIN! -cf’Protein!~iRT’ -cNstep3 step2.splib
spectrast2tsv.py -l 400,2000 -s b,y -x 1,2,3,4 -o 6 -n 6 -p 0.05 -e -w swaths.txt -k openswath -a step4.tsv step3.splib
mv step4.tsv step4.csv (move step4.tsv step4.csv in windows)
ConvertTSVTTraML -in step4.csv -out step5.TraML
OpenSwathDecoyGenerator -in step5.TraML -out final.TraML-min_transitions 3 -max_transitions 6 -method shuffle -append -exclude_similar
All the tools used here are available under the modified BSD license at http://www.openswath.org/ and https://code.google.com/p/msproteomicstools. Documentation for OpenSWATH and a detailed protocol for SWATH assay library generation is available 18. In our laboratory, the entire pipeline was streamlined as an iPortal workflow 33.
SWATH data analysis
SWATH wiff files were converted into mzXML files using ProteoWizard msconvert 34 v.3.0.3316. A Python script (fix_swath_windows.py) was used to correct precursor isolation-window mass error introduced by msconvert as described previously. The mzXML file was converted to mzML file using OpenMS tool FileConverter 35. OpenSWATH was performed using the tool OpenSWATHWorkflow with input files including the mzXML file, the TraML library file, and TraML file for iRT peptides 18. The assays were identified with a false discovery rate below 0.1%. High confidence peptide features from different samples were aligned using the algorithm TRansition of Identification Confidence (TRIC) (version r238) (Roest, HL, et al, in preparation), which is available from https://pypi.python.org/pypi/msproteomicstools or https://code.google.com/p/msproteomicstools. The following parameters for the feature_alignment.py follows: max_rt_diff=30, method=global_best_overall, nr_high_conf_exp=2, target_fdr=0.001, use_score_filter=1.
Label-free protein-level quantification was performed using the function ProteinInference from the R package aLFQ (version 1.3.1) 19 on output from OpenSWATH. The parameters differ from the default for ProteinInference were:
peptide_method=“top”,peptide_topx=3,transition_topx=5,consensus_proteins=FALSE, consensus_peptides=FALSE, consensus_transitions=FALSE.
The mass spectrometry proteomics data and SWATH assay library have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository 36 with the dataset identifier PXD000672.
Statistical analysis
The protein expression values were subjected to the arcsinh variance stabilizing transformation37 and the effect was assessed by the Bland Altman diagnostic plot. We corrected for systematic differences in expression levels between the samples using the quantile normalization 38. Missing values were excluded from calculation of CV values, but considered as 0 in heatmap generation and t-test. Variation analysis of the RCC SWATH data was performed by comparing quantified proteins between each pair of SWATH data sets taking into account all combinations. The median CV of all proteins was plotted using R package vioplot. The MS injection variation analysis considered 18 pairs of data sets. The within-patient variation analysis of all RCC and ccRCC considered 36 and 24 pairs of data sets, respectively. Regarding inter-patient variation analysis in all RCC patients, one data set was sampled from each of the 9 patients and pooled for variation analysis. For all RCC tissues, altogether 262,144 combinations were analyzed. 512 combinations were included for non-tumorous and tumorous tissues from all RCC patients. Variation analysis of ccRCC was based on 4,096 combinations of ccRCC data sets, and 64 combinations of non-tumorous or tumorous ccRCC data sets. Heatmaps and violin plots were plotted using R. P values in the volcano plots were calculated using paired or un-paired two-tailed t-test as indicated in the respective legend.
Supplementary Material
Acknowledgements
We thank Prof. H.P. Schmid and Dr. Daniel Engeler for their help with tissue collection. We thank Dr. Alexander Leitner and Karel Novy for help in Orbitrap analysis. We also thank Dr. Alexander Leitner for assistance in setting up the barocycler and the ETH Brutus team for computational support, and O.L.Kon for critical reading of the manuscript. The work was supported by SystemsX.ch project PhosphoNetX. The project was further supported in part by the SNSF (Grant# 3100A0-688 107679), the European Research Council (Grant# ERC-2008-AdG 233226). P.K. was supported by Finnish Cultural Foundation. We thank the PRIDE team for support in mass spectrometry data deposition.
Footnotes
Competing financial interests R.A. holds shares of Biognosys AG, which operates in the field covered by the article. The research group of R.A. is supported by AB SCIEX by providing access to prototype instrumentation, and Pressure Biosciences by providing access to advanced sample preparation instrumentation.
Reference for main text
- 1.Liotta L, Petricoin E. Molecular profiling of human cancer. Nature reviews. Genetics. 2000;1:48–56. doi: 10.1038/35049567. [DOI] [PubMed] [Google Scholar]
- 2.van ’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. The New England journal of medicine. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 4.Barrett T, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic acids research. 2009;37:D885–890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic acids research. 2014;43:D805–811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research, N. et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature genetics. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 8.Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 9.Kononen J, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nature medicine. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 10.Haab BB. Antibody arrays in cancer research. Molecular & cellular proteomics : MCP. 2005;4:377–383. doi: 10.1074/mcp.M500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nature biotechnology. 2010;28:710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 12.Gillet LC, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Molecular & cellular proteomics : MCP. 2012;11:O111, 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, et al. Quantitative measurements of N-linked glycoproteins in human plasma by SWATH-MS. Proteomics. 2013;13:1247–1256. doi: 10.1002/pmic.201200417. [DOI] [PubMed] [Google Scholar]
- 14.Collins BC, et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nature methods. 2013;10:1246–1253. doi: 10.1038/nmeth.2703. [DOI] [PubMed] [Google Scholar]
- 15.Powell BS, Lazarev AV, Carlson G, Ivanov AR, Rozak DA. Pressure cycling technology in systems biology. Methods Mol Biol. 2012;881:27–62. doi: 10.1007/978-1-61779-827-6_2. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Ferrer D, et al. Application of pressurized solvents for ultrafast trypsin hydrolysis in proteomics: proteomics on the fly. Journal of proteome research. 2008;7:3276–3281. doi: 10.1021/pr7008077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glatter T, et al. Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion. Journal of proteome research. 2012;11:5145–5156. doi: 10.1021/pr300273g. [DOI] [PubMed] [Google Scholar]
- 18.Rost H, et al. OpenSWATH: Automated, targeted analysis of mass spectrometric data generated by data-independent acquisition. Nature biotechnology. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 19.Rosenberger G, Ludwig C, Rost HL, Aebersold R, Malmstrom L. aLFQ: An R-package for estimating absolute protein quantities from label-free LC-MS/MS proteomics data. Bioinformatics. 2014;30:2511–2513. doi: 10.1093/bioinformatics/btu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full Dynamic Range Proteome Analysis of S. cerevisiae by Targeted Proteomics. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Algaba F, et al. Current pathology keys of renal cell carcinoma. European urology. 2011;60:634–643. doi: 10.1016/j.eururo.2011.06.047. [DOI] [PubMed] [Google Scholar]
- 22.Tan PH, et al. Renal tumors: diagnostic and prognostic biomarkers. The American journal of surgical pathology. 2013;37:1518–1531. doi: 10.1097/PAS.0b013e318299f12e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuster B, Schirle M, Mallick P, Aebersold R. Scoring proteomes with proteotypic peptide probes. Nature reviews. Molecular cell biology. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- 24.Beck M, et al. The quantitative proteome of a human cell line. Molecular systems biology. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wisniewski JR, et al. Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Molecular systems biology. 2012;8:611. doi: 10.1038/msb.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Addona TA, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nature biotechnology. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.t Hoen PA, et al. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nature biotechnology. 2013;31:1015–1022. doi: 10.1038/nbt.2702. [DOI] [PubMed] [Google Scholar]
References for Online Methods
- 28.Escher C, et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12:1111–1121. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leitner A, et al. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Molecular & cellular proteomics. 2012;11:M111 014126. doi: 10.1074/mcp.M111.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacLean B, Eng JK, Beavis RC, McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics. 2006;22:2830–2832. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- 31.Geer LY, et al. Open mass spectrometry search algorithm. Journal of proteome research. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 32.Deutsch EW, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunszt P, et al. Improving the Swiss Grid Proteomics Portal: Requirements and new Features based on Experience and Usability Considerations. Proceedings of the 5th International Workshop on Science Gateways. 2013;993 [Google Scholar]
- 34.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sturm M, et al. OpenMS - an open-source software framework for mass spectrometry. BMC bioinformatics. 2008;9:163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vizcaino JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic acids research. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 38.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.