Abstract
Nowadays molecular species delimitation methods promote the identification of species boundaries within complex taxonomic groups by adopting innovative species concepts and theories (e.g. branching patterns, coalescence). As some of them can efficiently deal with large single-locus datasets, they could speed up the process of species discovery compared to more time consuming molecular methods, and benefit from the existence of large public datasets; these methods can also particularly favour scientific research and actions dealing with threatened or economically important taxa. In this study we aim to investigate and clarify the status of economically important moths species belonging to the genus Spodoptera (Lepidoptera, Noctuidae), a complex group in which previous phylogenetic analyses and integrative approaches already suggested the possible occurrence of cryptic species and taxonomic ambiguities. In this work, the effectiveness of innovative (and faster) species delimitation approaches to infer putative species boundaries has been successfully tested in Spodoptera, by processing the most comprehensive dataset (in terms of number of species and specimens) ever achieved; results are congruent and reliable, irrespective of the set of parameters and phylogenetic models applied. Our analyses confirm the existence of three potential new species clusters (for S. exigua (Hübner, 1808), S. frugiperda (J.E. Smith, 1797) and S. mauritia (Boisduval, 1833)) and support the synonymy of S. marima (Schaus, 1904) with S. ornithogalli (Guenée, 1852). They also highlight the ambiguity of the status of S. cosmiodes (Walker, 1858) and S. descoinsi Lalanne-Cassou & Silvain, 1994. This case study highlights the interest of molecular species delimitation methods as valuable tools for species discovery and to emphasize taxonomic ambiguities.
Introduction
Though millions of species remain to be discovered and described [1–4], the discipline of finding, describing and naming species—Taxonomy—currently faces an important deficit in term of funding and manpower [5]. This ‘taxonomy crisis’ is a major issue because the speed at which species are being lost is much faster than the required time to find, describe and potentially protect them [6,7]. The problem is also aggravated by the fact that traditional morphological studies are usually extremely time consuming, especially when dealing with hyper diverse groups such as insects or plants, or when assessing species complexes with superficially indistinguishable morphological attributes [8–13]. Our lack of knowledge is even more problematic when facing species complexes that threaten public health [14,15] or target important crops [16–19]. In this context, some hopes came from the use of molecular data in studies on taxonomy and systematics. Thanks to the increased availability of molecular data over the last decades, DNA-based taxonomy has been proposed as an elegant and effective solution to speed up species identifications and discoveries [20–27]. However, this initiative was quickly criticized on both practical [28–32] and theoretical [33–39] grounds, with numerous studies underlining the fact that molecular data alone (especially molecular barcodes) cannot solve the current taxonomic crisis [40–43]. The general consensus that emerged from these debates was that faster and more accurate species identifications or delimitations can be achieved through the use of multiple complementary sources of information (e.g. behavioural, ecological, molecular, morphological) [17,44,45], hence underlining the need for a more ‘integrative’ [46,47] or ‘iterative’ taxonomy [48].
Because of their ever-increasing accessibility and decreasing cost, it is no wonder that molecular data are now routinely incorporated into taxonomy studies [7,17,45,49–52]. Of particular interest is the fact that multiple analytic procedures have been explicitly developed to delimit species boundaries using molecular data [53–57]. A first class of approaches—referred to as species discovery procedures—assigns samples to putative species clusters without a priori information [58]. A second class of approaches—referred to as species validation procedures—requires an a priori assignation of samples to putative species clusters, which can be later validated [58]. Both species discovery and species validation approaches can rely on molecular phylogenies, which are increasingly used to help identifying or delimiting species [17,54–56,59–60]. All tree-based species delimitation methods usually rely on the ‘phylogenetic species concept’ (sensu De Queiroz [61]), within a coalescent framework [55,62–63], whose central aim is to identify independently evolving lineages without gene flow, in which selection and drift operate, each representing a putative species [64]. Tree-based species delimitation methods can operate with single or multi locus data; the use of multi locus data is mandatory for species validation procedures and optional for species discovery approaches [58]. For species discovery procedures, the use of multi locus data is usually advocated because of its superior power to resolve complex evolutionary histories resulting from recent episodes of radiation, incomplete lineage sorting, gene introgression or species hybridization [58,65,66]. All these processes usually generate discordance between gene trees and species trees [67], which can biases the inference of putative species clusters. Nonetheless, single locus data have been proved useful to provide robust inferences of evolutionary histories [68] and quick assessments of species boundaries and diversity [26,69–72]. More recently they have been also associated with concise morphological descriptions to produce rapid descriptions of new species, an approach referred to as turbo-taxonomy [73–75]. Finally the use of single locus data also benefits from the fact that several genes (e.g. 16S ribosomal RNA) are well represented in large public databases such as GenBank (http://www.ncbi.nlm.nih.gov/genbank/) or the Barcode of Life Database (BOLD; http://www.boldsystems.org/; for the mitochondrial cytochrome c oxidase subunit I gene (COI)).
In this study, we explore the potential interest and efficiency of species discovery approaches to assess species status in moths belonging to the genus Spodoptera (Lepidoptera, Noctuidae). This genus encompasses 30 species [76], and includes several species whose status has been questioned in the past based on biological [77–79], morphological [80,81] and molecular evidence [82–85]. Clarifying Spodoptera species boundaries is of prime importance because half of known Spodoptera species are pests of economic importance that target major crops such as beans, cotton, maize, millet, rice, sorghum, soybean or tomatoes [76,86,87]. A particular focus will be made on the fall armyworm (FAW) Spodoptera frugiperda, whose identity is particularly debated [88–91]. The fall armyworm is a widespread and well-known agricultural pest in the Western hemisphere [76,87]. It is an extremely polyphagous species, which is known to develop on about 100 different plants species from 27 plant families [76]. Several features of the biology and the ecology of fall armyworm indicate that this species is composed of two morphologically undistinguishable ecological races (‘corn form’ and ‘rice form’ sensu Juarez et al., [91]), with distinct host-preferences [92–94]. The use of diagnostic molecular markers [95–99] have indicated that both strains are generally found in sympatry [92,100–101], hence leading some authors to argue that they could constitute a potential example of ongoing or sympatric speciation [78,88]. Both strains also exhibit a high level of genetic differentiation [85]. In addition to the case of S. frugiperda, we will also pay attention to five other Spodoptera species, whose species status is still disputed [81]. It is the case of S. marima and S. ornithogalli, which are morphologically related and possibly correspond to geographic races, as they have allopatric distributions and only differ by the absence of sexual dimorphism in S. marima males [76,81]. These two species were not found reciprocally monophyletic in a recent multilocus phylogenetic study [85], so we expect additional clarifications with the molecular species delimitation procedures. Another species pair (S. cosmiodes and S. descoinsi) was also recovered paraphyletic with a strong support in the study of Kergoat et al., [85], but similarly to the case of S. frugiperda, additional evidences (differences in pheromone blends, allochronism of females) led to think that these two entities could possibly constitute valid species [76,77]. Finally, the last case that remains to be investigated is the status of Western populations (from East Africa, Mauritius, the Malagasy Republic, and the Reunion Island) and Eastern populations (from the Indomalayan and Australasian regions) of S. mauritia, which may constitute valid species of their own [80]. Here we will explore Spodoptera species boundaries by using two different tree-based approaches that have drawn much attention in recent years [102–110]: the general mixed Yule-coalescent (GMYC) model [54,108] and the Poisson-Tree-Processes (PTP) [60]. The GMYC model uses the distinct branching patterns between a Yule model (modelling interspecific speciation events) and a coalescent model (modelling expected coalescent times of genes at an intraspecific level) to distinguish between species and population processes. This model is fitted to an ultrametric tree (a tree where all the path-lengths from the root to the tips are equal) and optimized under maximum likelihood to find the threshold time (T), which corresponds to the transition between the two distinct branching patterns. It is possible to visualise this transition with a lineage-through-time plot, in which a sharp increase in branching rates is expected at T. Potential species clusters are then determined by identifying the clades (or single lineages) that originate after this putative threshold. The PTP is a recently developed method, which has the advantage of not requiring an ultrametric tree (inferring an ultrametric tree is usually a time-consuming and potentially error-prone process [103,110]); instead the PTP model uses branch lengths to estimate the mean expected number of substitutions per site between two branching events. The model assumes that each substitution has a small probability of generating a speciation event; hence the number of substitutions between species is expected to be significantly higher than those within species [60]. The model then implements two independent classes of Poisson processes (one describing speciation and the other describing within species branching events) and searches for transition points between interspecific and intraspecific branching patterns. Potential species clusters are then determined by identifying the clades (or single lineages) that originate after these transition points. Because both methods efficiently deal with large single-locus datasets [110], they can readily benefit from the mass of sequence data generated through the Barcode of Life (BoL) initiative or deposited on public database such as GenBank. Here we combine Spodoptera sequences generated from previous studies that are available on GenBank or BOLD with newly generated sequences for the COI gene. The resulting dataset will be used to: (i) infer putative species boundaries in the genus Spodoptera by applying species discovery methods particularly suitable to deal with large single-locus datasets; (ii) to test the reliability and the usefulness of these species discovery methods by comparing our results with other sources of information.
Material and Methods
Sampling design
The sampling for this study covers 27 of the 30 known Spodoptera species [76]. We were unable to acquire sequence data for three species: S. compta (Walker, 1869) (from Peru), S. malagasy Viette, 1967 (from Madagascar) and S. roseae (Schaus, 1923) (from the Galapagos Islands). Both S. compta and S. roseae are extremely rare in collections (especially S. compta which is only known from three specimens collected in the 19th century). As for S. malagasy, previous sequencing attempts on several old museum specimens have been unsuccessful [85]. For this study we generated 86 new sequences for specimens obtained from the Muséum National d’Histoire Naturelle in Paris (MNHN). In addition we used 118 sequences from the study of Kergoat et al., [85], 463 additional sequences from GenBank and five sequences from BOLD. The resulting dataset encompasses 672 Spodoptera specimens, with a mean number of 24 individuals per species (from two individuals for S. apertura (Walker, 1865), S. pecten Guenée, 1852, S. pulchella (Herrich-Schäffer, 1868) and S. triturata (Walker, 1857) to 101 in S. frugiperda; S1 Table). All specimens, their geographic origin and the corresponding sequence accession number are listed in a supplementary table (S1 Table). For outgroup choice we used the results of the study of Mitchell et al., [111] to select the following seven noctuid taxa: Agrotis ipsilon (Hufnagel, 1766) (Noctuinae), Helicoverpa armigera (Hübner, 1805) (Heliothinae), Heliothis virescens (Fabricius, 1777) (Heliothinae), Mythimna unipuncta (Haworth, 1809) (Hadeninae), Noctua atlantica (Warren, 1905) (Noctuinae), Psychomorpha epimenis (Drury, 1782) (Agaristinae), Sesamia inferens (Walker, 1856) (Xyleninae). Based on Mitchell et al., [111] all trees were rooted with Psychomorpha epimenis, which belongs to a clade that is sister to the clade including both Spodoptera spp. and the subfamilies Hadeninae, Heliothinae, Noctuinae and Xyleninae.
DNA extraction, Polymerase chain reactions and Sequencing
Total genomic DNA of new samples was extracted by grinding up hind legs using DNAeasy Blood and Tissue Kit (Qiagen). Partial fragments of the COI gene were amplified following Kergoat et al., [86]. The polymerase chain reaction (PCR) mix (25 μL) consisted of 1xPCR buffer, 1.5mM MgCl2, 0.1mM of each desoxynucleotide triphosphates, 1μM of each primer, 1 unit of Taq DNA polymerase (Qiagen) and 2 μL (diluted to tenth) of extracted DNA and nuclease-free water to 25 μL. PCR cycling conditions were the following: initial denaturation step at 95°C for 3 min followed by 35 cycles including a denaturation step at 92°C for 1 min, an annealing step at 48°C for 1 min, and an elongation step at 72°C for 1 min and a final elongation step at 72°C for 10 min. The resulting PCR products were processed by Eurofins MWG Synthesis GmbH (Ebersberg, Germany). Both strands were sequenced for all specimens to minimize PCR artifacts and ambiguities.
Sequences of complementary strands were edited and reconciled using Geneious v5.1 software (available at: www.geneious.com/). The alignment of all sequences was carried out using MAFFT 7 [112] with default option settings. No gap was present in the corresponding alignment. We then used Mesquite v3.0 [113] to check the coding frame for possible errors or stop codons. Additional sequences generated for the study were deposited to GenBank under numbers KF854152 to KF854234. The final dataset encompasses 679 individuals and 658 aligned characters.
Gene tree inference
Phylogenetic analyses were conducted using Maximum likelihood (ML) and Bayesian inference (BI). For both methods we carried out partitioned analyses to improve phylogenetic accuracy [114]. Partitions and substitution models were determined using PartitionFinder v1.1.1 [115]. The corrected Akaike information criterion (AICc; [116]) was used as a metric for ML analyses whereas the Bayesian information criterion (BIC) was used for BI analyses.
Maximum Likelihood analyses were performed using RAxML v8 [117]. Based on the AICc results (S2 Table) we used only one partition with a General time reversible (GTR) model. Instead of using a standard GTR+G+I model we used the GTR CAT approximation model that accommodate searches incorporating rate heterogeneity with faster inference times and better maximum likelihood values [118]. The best ML tree was obtained using a heuristic search implementing 100 random-addition replicates. Clade support was then assessed using a non-parametric bootstrap procedure (1,000 replicates were used). Nodes supported by bootstrap values (BV) ≥ 70% were considered as strongly supported following Hillis and Bull [119].
Bayesian inference analyses were carried out using MrBayes 3.2.2 [120] and BEAST 1.8 [121]. Based on the BIC results (S2 Table) we used three partitions. For MrBayes analyses, instead of using a specific model for each partition we used the mixed model option. The latter allows to sample across the substitution model space in the Bayesian Markov Chains Monte Carlo (MCMC) analysis itself, removing the need for a priori model testing [122]. We conducted two independent runs with four MCMC (one cold and three incrementally heated) that ran for 50 million generations, with trees sampled every 1,000 generations. A conservative burn-in of 25% was then applied after checking for stability on the log-likelihood curves and the split-frequencies of the runs under MrBayes (a threshold of 0.05 was used). For the BEAST analyses, we implemented the models selected by PartitionFinder (S2 Table). Following Tang et al., [110] we also relied on distinct sets of models to infer trees with branch lengths proportional to time. To limit the size of parameter space the majority-rule consensus topology from MrBayes analyses was used as a guide tree. We alternatively used a strict clock model or an uncorrelated lognormal (UCLN) relaxed clock as clock priors, and a bird-death (BD) model as tree prior. The corresponding analyses were carried out using two independent runs of 50 million generations, with trees sampled every 1,000 generations. A conservative burn-in of 25% was then applied after checking for the effective sampling sizes (ESS) of parameters (a threshold of 200 was used). Support of nodes for both MrBayes and BEAST analyses was provided by clade posterior probabilities (PP) as directly estimated from the majority-rule consensus topology. Nodes supported by PP ≥ 0.95 were considered as strongly supported following Erixon et al., [123].
Molecular species delimitations
We followed the recommendations of Tang et al., [110], who advocated the use of model-based gene trees (generated with programs such as MrBayes or RAxML) for PTP analyses and the use of ultrametric trees generated with BEAST for the GMYC analyses. Poisson-Tree-Processes molecular species delimitations were conducted on the Web server of the Exelixis Lab (http://species.h-its.org/ptp/), with default settings. The corresponding analyses were carried out using trees either the best tree from the ML analyses or the majority-rule consensus topology resulting from the BI analyses. To avoid biases that may arise if some of the outgroup taxa are too distantly related from ingroup taxa, outgroups were pruned before conducting the PTP analyses for both ML and BI trees. General mixed Yule-coalescent analyses were performed under R (R Development Core Team 2010) using the latest implementation of the GMYC procedure [108], which relies on the following packages: splits [124], APE [125] and apTreeshape [126]. All corresponding analyses were carried out using a single threshold with a confidence interval falling within 2 log-likelihood units of the ML solution [54].
Results
Gene tree inference
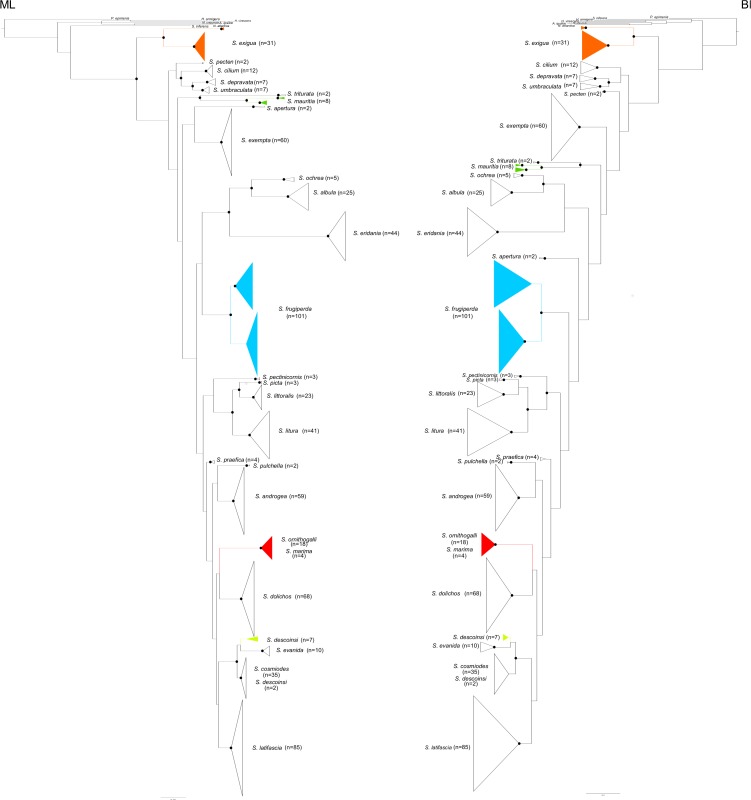
The phylogenetic analyses in ML and BI show similar results (Fig 1; see also S1 and S2 Figs for details), which only differ by the position of some outgroups and the placement of two Spodoptera species (S. apertura and S. pecten). In both ML and BI analyses, the genus Spodoptera is recovered monophyletic. Support for Spodoptera monophyly was higher for the ML tree (BV of 95%) in comparison to the BI tree (PP of 0.94). The phylogenies are overall not resolved, except for the terminal nodes. Support values are equally high for the nodes leading to morphospecies (BV ≥ 70% for 23 out of the 27 corresponding nodes under ML; PP ≥ 0.95 for 21 out of the 27 corresponding nodes under BI).
Fig 1. COI-based phylogenetic relationships of Spodoptera species.
The tree on the left summarizes the results of ML analyses while the tree on the right summarizes the results of BI analyses. Black dots indicate nodes supported by bootstrap values ≥ 70% (ML tree) or supported by posterior probability ≥ 0.95 (BI tree).
Four of the 27 Spodoptera morphospecies (S. cosmiodes, S. descoinsi, S. marima and S. ornithogalli) are consistently recovered paraphyletic in all analyses. Out of the nine sampled S. descoinsi individuals (all from French Guiana), seven of them constitute a well-supported clade, sister to S. evanida Schaus, 1914 (specimens of S. evanida were collected in French Guiana and Peru). The two remaining specimens identified as S. descoinsi (voucher codes B19 and LSU30) are grouped in a moderately to highly supported clade (BV of 88%, PP of 0.87) that includes all sampled S. cosmiodes individuals (from Brazil and French Guiana). Regarding S. marima and S. ornithogalli, the four sampled specimens of S. marima (from French Guiana and Venezuela) are mixed with the 18 sampled specimens of S. ornithogalli (from Canada, Dominican Republic, Guatemala, French Guiana and USA) in a well-supported clade (BV of 100%, PP of 1.0).
Two distinct and well-supported clades were inferred for S. frugiperda (BV of 100 and PP of 1.0). A first clade (58 specimens) includes all the specimens that have been assigned to the ‘rice form’ in the studies of Nagoshi et al., [127] and Kergoat et al., [85] whereas the other one (43 specimens) includes all the specimens previously assigned to the ‘corn form’. Two distinct and well-supported clades were also recovered for S. mauritia. A first clade (BV of 96%, PP of 1.0) groups three specimens collected in the Réunion Island that belong to the nominate form (S. m. mauritia) distributed in Madagascar and neighboring islands (Comoro, Mauritius and Réunion). A second clade (BV of 98%, PP of 1.0) groups five specimens collected in Australia (KF389305), Japan (AB733407, AB733408 and AB733409) and Papua New Guinea (voucher code B152) that belong to S. m. acronyctoides, which is distributed in the Oriental, Indo-Australian and Pacific regions.
Molecular species delimitations
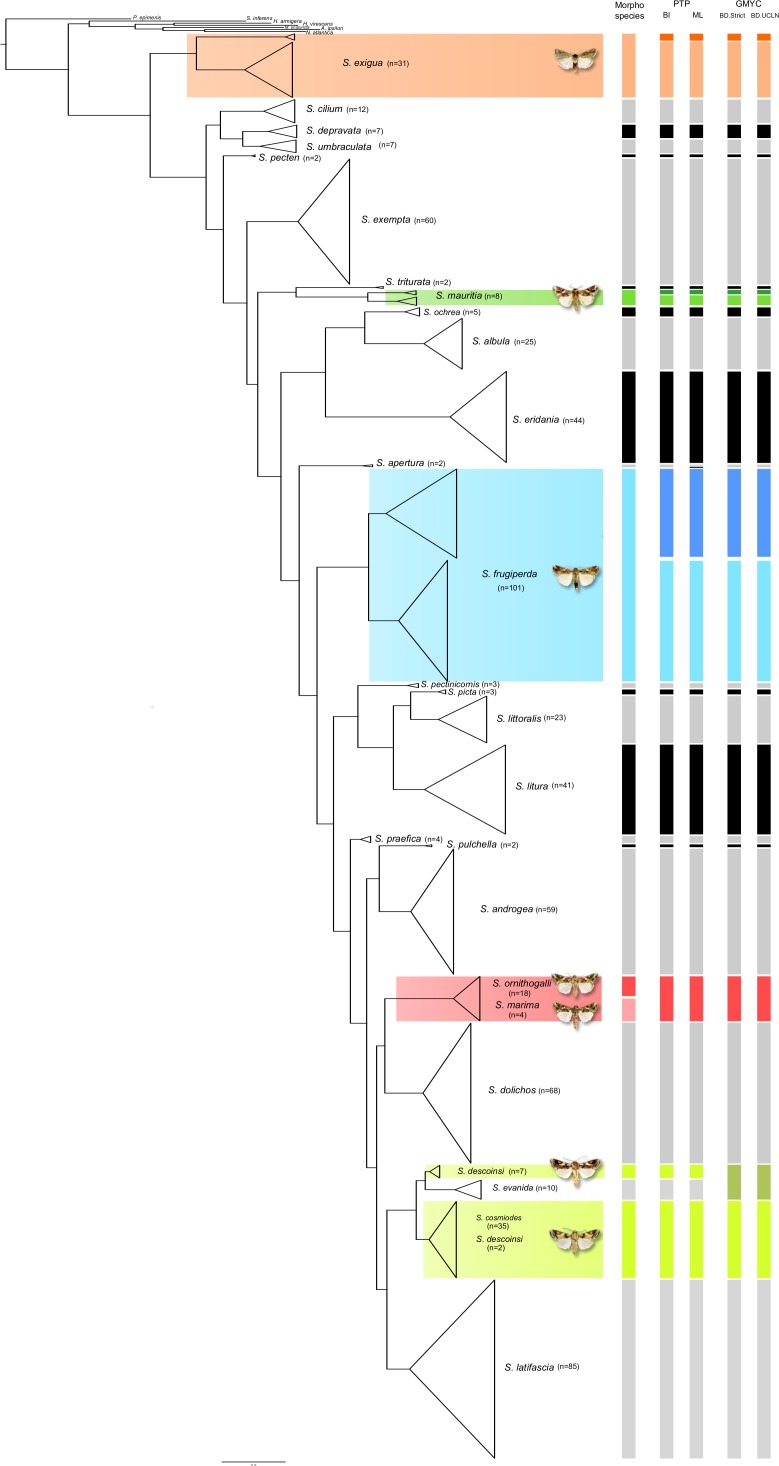
The results of phylogenetic molecular species delimitations were compared to morphological assignations that follow the checklist of species provided by Pogue [76].
Regarding the PTP analyses, at least 29 putative species clusters were recovered for the 67 specimens sampled from 27 Spodoptera morphospecies (Fig 2, S3 and S4 Figs and S3 Table). 30 putative species clusters were inferred for the analyses based on the best-fit ML tree (S3 Fig) while 29 putative species clusters were inferred for the analyses based on the BI majority-rule consensus topology (S4 Fig). Two distinct putative species clusters were consistently found for the following three species: S. exigua, S. frugiperda and S. mauritia. In addition the two individuals of S. apertura were also recovered as distinct putative species in the analyses based on the ML tree. Regarding S. exigua, a putative species cluster groups the four sampled Australian specimens (KF522648, KF522649, KF522650 and KF522651) whereas a second one groups the remaining 27 S. exigua specimens. For S. frugiperda the two putative species clusters consist of 43 and 58 specimens, respectively. The smaller FAW cluster groups all individuals that have been assigned to the ‘corn form’ by previous studies [85,127] whereas the bigger FAW cluster groups all individuals previously assigned to the ‘rice form’ [85,127]. For S. mauritia the two putative species clusters distinguish the specimens belonging to S. m. mauritia (three individuals collected in the Réunion Island) with the specimens belong to S. m. acronyctoides (five individuals collected in Australian, Japan an Papua New Guinea). All individuals belonging to S. marima and S. ornithogalli are lumped in one putative species cluster encompassing 22 specimens (18 for S. ornithogalli and four S. marima). Concerning S. cosmiodes and S. descoinsi, the PTP species delimitation recovered two putative species clusters (see S4 Fig). A first potential species cluster is made of seven S. descoinsi individuals, which are sister to the clade corresponding to S. evanida. A second cluster encompasses all S. cosmiodes individuals plus two S. descoinsi specimens.
Fig 2. Putative species clusters corresponding to: (i) morphological delineations (column 1); (ii) PTP analyses using either the majority-rule consensus topology resulting from the BI analyses (column 2) or the best tree from the ML analyses (column 3); (iii) GMYC analyses based either on a strict clock and a BD model (column 4) or on a UCLN clock and a BD model (column 5).
The reference tree used here corresponds to the tree resulting from the BI analyses carried out with MrBayes.
For the GMYC procedure, the same putative species clusters were inferred for the analyses based either on a strict clock and BD model or on a UCLN clock and BD model (see Fig 2, S5 and S6 Figs and S3 Table). 28 putative species clusters were recovered, with a confidence interval of 27–31 for the tree based on a strict clock and BD model and a confidence interval of 27–30 for the tree based on a UCLN clock and BD model. In comparison with the PTP analyses, the exact same putative species clusters were inferred with the exception of a larger cluster which groups a clade of seven S. descoinsi individuals with the clade corresponding to S. evanida (see Fig 2 and S3 Table). When examining the taxa for which changes of status are suggested we found out that most changes are statistically supported (see S5 and S6 Figs). It is the case of the grouping of S. marima with S. ornithogalli and of the splits of S. exigua, S. frugiperda and S. mauritia. The only exception is the case of the grouping of the seven S. descoinsi individuals with the clade corresponding to S. evanida, which is not statistically supported.
Discussion
A cautionary tale of consistency
The results of our phylogenetic species delimitations analyses are remarkably congruent, as they recover the same species clusters (except for the case of S. apertura in one of the PTP analyses and the case of S. descoinsi and S. evanida in one of the GMYC analyses) whatever the set of parameters or methods used. The number of species clusters inferred using PTP procedures also falls well within the confidence interval of the GMYC analyses (27–31 and 27–30 for the strict clock/BD model and UCLN clock/BD model associations, respectively). Although our study may give the impression that the results of species discovery procedures are expected to be always consistent whatever the models and sets of parameters used, we would like to insist on the fact that reliable and consistent results cannot be always achieved. Several studies have clearly highlighted the fact that most molecular species delimitation approaches (including tree-based species delimitation procedures) are highly susceptible to multiple biases or to the way the analyses are parameterized (see the review of Carstens et al., [58]). For instance, model performances are known to be extremely sensitive to biases such as insufficient or unbalanced sampling [31,34], heterogeneous rates of gene evolution [128], incomplete lineage sorting, or hybridization after introgression events [129]. Though rarely look upon, tree building procedures also matter, as different methods and sets of parameters can potentially yield very distinct topologies, which will likely generate contrasting putative species clusters. For example, a potential source of discrepancy is the estimation of branch lengths, as branch length estimates are known to be sensitive to the choice of substitution models [130–132] or partitioning strategies [133,134]. It is especially the case in phylogenetic analyses that use multiple partitions and/or complex models, for which over-parameterization can potentially bias branch length estimates [134,135]. In addition, it has been recently demonstrated [136] that branch length priors (within a Bayesian framework) can significantly impact branch length estimation, with shorter priors generating shorter branch lengths and longer priors generating longer branch lengths. Another potential source of discrepancy is the choice and parameterization of the ultrametrization procedures for methods (such as GMYC) that require ultrametric trees as input trees. In the study of Astrin et al., [103] on cryptorhynchine weevils (Coleoptera) the authors used 21 distinct sets of parameters and methods. They showed that the number of putative species clusters inferred using GMYC could vary drastically with numbers varying from 5 to 564 depending on the choice of methods and parameters. Similarly Tang et al., [110] highlighted the fact that the branch smoothing procedures are a potential source of biases for the approaches that require ultrametric trees.
In summary all these studies underline that molecular species delimitation approaches should not be used blindly. They nonetheless constitute powerful tools that can be effectively used in a concerted way to assess species boundaries (e.g. [137]). In our study, a high level of consistency was likely achieved because of the size of our dataset and because we carefully followed some of the recommendations of Tang et al., [110]. Because the focus of our study was to reassess Spodoptera species boundaries, and not to explore the impact of tree building procedures on molecular species delimitations, we feel that our analytical pipeline is appropriate and allows us to discuss with some confidence the ambiguous species status of several taxa.
Spodoptera species boundaries, a reappraisal
A clear pattern is evidenced for the species that is the main focus of our study, Spodoptera frugiperda. Individuals are separated into two clusters that likely correspond to the two known host-forms (rice and corn) of S. frugiperda. Both PTP and GMYC analyses indicate that the level of genetic differentiation exhibited by the two host-forms is compatible with a potential sister-species status, with a significant statistical support under the GMYC procedure. This result is in agreement with the results of several studies that have highlighted the existence of prezygotic and postzygotic isolation between the two host-forms (Table 1). Different prezygotic isolation mechanisms have been found, including differences in the composition of females' pheromones [78,138], allochronism in mating activity between the two strains [79,139] and oviposition preferences [94,140]. Regarding postzygotic isolation, the success of mating between the two strains remains highly controversial. On the one hand Pashley and Martin [141] and Dumas et al., [142] have observed unidirectional mating between corn females and rice males while the reciprocal cross gave viable offsprings. Furthermore, in backcrosses the hybrid females mated with low success with their brothers but not at all with males of either parental strain [140,141,143]. On the other hand, another study has shown that the two strains crossed successfully in both directions [144]. The latter is supported by the fact that a high proportion of hybrid has been evidenced is several surveys of natural populations [90,145]. Finally, a postzygotic isolation for several life-history traits in both host-forms has been also evidenced by Velasquez-Velez et al., [146] who identified a reduction of the number of hybrid females and a reduction in hybrid fertility in S. frugiperda. Taken altogether all these studies suggest that both host-forms are situated somewhere in between a ‘continuum’ of speciation [147]. In this context, the results of our study tend to favour the hypothesis that the two host-forms are well beyond the early steps of speciation as they present a level of genetic differentiation which is comparable with those observed between other distinct Spodoptera species.
Table 1. List of evidences arguing in favor of a high level of differentiation between the two FAW strains.
| Suitability | References | |
|---|---|---|
| Pre-zygotic barrier | ||
| Habitat isolation | + / - | Whitford et al., [140] Meagher et al., [94] Juárez et al., [91] |
| Temporal isolation | ++ | Schöfl et al., [79,139] |
| Behavioral isolation | ++ | Groot et al., [78] Lima and McNeil [138] |
| Unidirectional mating | +/- | Pashley and Martin [141] Dumas et al., [142] |
| Post-zygotic barrier | ||
| Reduction of hybrid fertility and reduction of hybrid female numbers | + | Velásquez-Vélez et al., [146] |
| Genetic differentiation | ||
| Phylogenetic analyses | ++ | Kergoat et al., [85] |
As indicated by the combination of pros and cons some results are more or less contradictory.
In the case of Spodoptera mauritia, the results of all phylogenetic species delimitation procedures suggested that Western (S. m. mauritia sensu Fletcher [148]) and Eastern (S. m. acronyctoides sensu Fletcher [148]) populations should be considered as distinct species. Though we cannot exclude the hypothesis of a possible analytical bias resulting from our limited sampling (only three individuals from La Reunion for S. m. mauritia and five individuals from Australia, Japan and Papua New Guinea), the suggested split is nonetheless statistically supported. From a morphological point of view, several authors (e.g. Fletcher [148]) noted differences in forewing coloration, where the conspicuous white areas of the forewing of Western populations are reduced in extent in Eastern populations. However the examination of long series of specimens from the two distribution areas by Brown and Dewhurst [80] did not reveal marked differences between putative subspecies. Additional studies are thus definitely required here to precise the status of both entities, such as comparisons of genitalia or pheromone compounds in female.
Regarding Spodoptera marima and Spodoptera ornithogalli, for which only few morphological (hindwing coloration and the lack of sexual dimorphism in S. marima [81]; differences exist, both analyses merge them into a single species cluster (with a significant statistical support under GMYC), hence providing more support to put these taxa into synonymy [85]. Because the two species occur in different geographic areas—S. marima is only found in the Western and Eastern Neotropics whereas S. ornithogalli is found in the Northern Neotropics and in the South, Western and Eastern Nearctic [76]—they could be considered as distinct subspecies of S. ornithogalli (due to priority, as S. ornithogalli was described before S. marima).
Interestingly our analyses suggest that Spodoptera exigua, a taxon with no ambiguous status, seems well differentiated, and may correspond to two distinct species. 27 of the 31 sampled specimens (including specimens from Canada, Egypt, Germany, Kenya, India, Japan, Thailand and United States of America) constitute a first putative species cluster whereas the four specimens from Australia constitute another one (note that no Australian samples were included in the study of Kergoat et al., [85]). This result is also statistically supported under GMYC and does not seem artefactual given the marked level of genetic differentiation between the two clades. It is also supported by additional researches on the Barcoding of life database (BOLD; available at http://www.boldsystems.org/); 81 S. exigua specimens from Australia are grouped into a distinct barcode cluster (BOLD:AAA6645) distinct from another barcode cluster (BOLD:AAA6644) grouping all remaining S. exigua individuals (n = 281). Obviously this result requires more investigation in order to determine whether these specimens correspond to a new species with marked morphological differences or to a case of cryptic species complex.
For Spodoptera apertura, only one of the four distinct sets of analyses (PTP using the ML tree; S3 Fig) recovers two distinct species clusters for the two sampled specimens of the species. We suspect that this result is artefactual; it possibly results from an artefact in the estimation of branch lengths under ML, where the two terminal branches leading to both individuals are really unbalanced (S3 Fig) whereas there are balanced under BI (S4 Fig). Insufficient sampling (only two specimens from Australia were sequenced for this species) could also have caused this likely error.
The situation is more complex in the case of Spodoptera cosmiodes and Spodoptera descoinsi, two taxa for which marked morphological and biological differences exist [76,77]. Both phylogenetic species delimitation procedures suggested the existence of two distinct species clusters. Analyses following the GMYC procedure also included representatives of S. evanida in one of the two clusters (the one with seven specimens S. descoinsi) but this grouping seems artefactual as the two species are morphologically not related [76]; in addition a closer examination of the GMYC output indicates that this grouping is not statistically supported. If we consider the remaining specimens, we can distinguish two species clusters, one that groups seven S. descoinsi and another that groups all (n = 35) S. cosmiodes individuals plus two representatives of S. descoinsi. To precise the status of these two individuals, additional studies are likely required; we advocate for the use of species validation procedures (with packages such as BPP [55] or SpedeSTEM [56]), using more specimens and a multi locus dataset. By doing so, it will be possible to better estimate the actual evolutionary lineages that compose the two species.
Conclusions
This study constitutes another case study that underlines the interest and effectiveness of fast species discovery procedures; the achieved results are congruent and reliable, irrespective of the set of parameters and phylogenetic models applied, even though we only relied on a single locus dataset. Our results unravel at least three potential new species clusters (for S. exigua, S. frugiperda and S. mauritia) and also support the synonymy of S. marima with S. ornithogalli. Though these redefined species boundaries have to be considered cautiously, they nonetheless constitute a new line of evidence that may fuel the debate on Spodoptera species boundaries and inspire future studies on species such as the beet armyworm S. exigua, for which a potential cryptic diversity was not expected. In contrast, the case of S. descoinsi is clearly more complex as illustrated by the inconsistencies between the results of PTP and GMYC procedures. Here further studies relying on additional markers and species validation procedures will be necessary to precise the status of this species.
Supporting Information
Support values are provided on major nodes (only BV ≥ 50% are shown).
(PDF)
Support values are provided on major nodes (only PP ≥ 0.5 are shown).
(PDF)
Putative species clusters are indicated using transitions between black-coloured to red-coloured branches.
(PDF)
Putative species clusters are indicated using transitions between black-coloured to red-coloured branches.
(PDF)
Putative species clusters are indicated using transitions between black-coloured to red-coloured branches. The inter- and intraspecific portions of the tree are divided with a dotted line (95% confidence intervals are figured using thinner dotted lines).
(PDF)
Putative species clusters are indicated using transitions between black-coloured to red-coloured branches. The inter- and intraspecific portions of the tree are divided with a dotted line (95% confidence intervals are figured using thinner dotted lines).
(PDF)
Sequences generated in a previous study of our research group are highlighted using ‘*’ whereas newly generated sequences are highlighted using ‘**’. The Democratic Republic of the Congo was abbreviated using DRC. When the country of origin is known we also provide the corresponding ISO 3166 alpha-3 code. We used the following abbreviations for the repository institutes and collections: ACG: Área de Conservación Guanacaste—Centro de Investigación y Estaciones Biológicas Programa de Educación Biológica, Costa Rica; ANIC: Australian National Insect Collection, Australia; BIO: Biodiversity Institute of Ontario, Canada; BSCZ: Bavarian State Collection of Zoology, Munich, Germany; CBGP: INRA—Centre de Biologie pour la Gestion des Populations, Montferrier/Lez, France; CLS: College of Life Science, Capital Normal University, Beijing, China; CIRN: CIRN, University of the Azores, Portugal; CTAG: Center for Theoretical and Applied Genetics, New Brunswick, USA; EMBRAPA: Embrapa National Soybean Research Center, Brazil; FERA: Fond and Environment Research Agency, York, UK; GSA: Graduate School of Agriculture, Kita-Ku, Japan; HUT: Hefei University of Technology, China; LEC: Lancaster Environment Centre, UK; MNHN: Muséum National d'Histoire Naturelle, Paris, France; NIAES: National Institute for Agro-Environmental Sciences, Tsukuba, Japan; NIBGE: National Institute for Biotechnology and Genetic Engineering, Pakistan; NIMBB: National Institute of Molecular Biology and Biotechnology, Los Banos, Philippines; NRIC: Natural Resources Inventory Center, Tsukuba, Japan; PAU: Punjab Agricultural University, India; PSCAS: Penn State College of Agricultural Sciences, USA; RCG: Research Collection of Theo Gruenewald, Germany; RCH: Research Collection of Alfred Haslberger, Munich, Germany; RCJH: Research Collection of D.H. Janzen & W. Hallwachs, USA; RCW: Research Collection of Jeremy deWaard, Guelph, Canada. SI: Smithsonian Institution, Washington, USA; TAU: Tamilnadu Agricultural University, India; UBC: University of British Columbia, Vancouver, Canada; UFP: Universidade Federal do Parana, Curitiba, Brazil; UM: University of Maryland, USA; UR: Université de Rouen, Rouen, France; USDA: USDA ARS, Gainesville, USA; YU: Yangzhou University, China; ZIQ: Zuhai Inspection and Quarantine bureau, China.
(DOCX)
Three partitions were specified (one per codon position). Selected models and partitions based on the AICc are figured on the left whereas selected models and partitions based on the BIC are figured on the right.
(DOCX)
The geographic origin and assignment to putative species clusters is provided for each Spodoptera specimen. Similarly to the sidebars presented on Fig 2, we have used one column per set of analyses (PTP with RAxML, PTP with MrBayes, GMYC with BEAST (strict clock) and GMYC with BEAST (UCLN clock). For more clarity, putative species clusters also are highlighted using a combination of numbers and distinct shades of grey.
(DOCX)
Acknowledgments
We would to thank Diego Fontaneto and two anonymous reviewers for their numerous constructive comments on a previous version of the manuscript. Some of the molecular data used in this work were produced through technical facilities of the "Laboratoire d'Excellence—Centre Méditerranéen de l'Environnement et de la Biodiversité" (LabEx—CeMEB), Montpellier, France. The authors also wish to thank A. Dehne Garcia for his help on the CBGP HPC computational platform.
Data Availability
All new sequences generated for the study are available from the GenBank database (accession numbers) KF854152 to KF854234.
Funding Statement
Part of the sequencing was supported by a Genoscope project @-Speed-Id (Accurate SPEciEs Delimitation and Identification of eukaryotic biodiversity using DNA markers) proposed by F-BoL, the French Barcode of life initiative and by the program ‘Bibliothèque du Vivant’ (Project SPODOBAR) supported by a joint "Centre national de la recherche scientifique", "Institut National de la Recherche Agronomique" and "Muséum national d'Histoire naturelle" consortium. The project was also supported by INRA (AAP SPE SPODOPTERA) and a grant from the French National Research Agency (ANR-12-BSV7-0004-01 ADA-SPODO).
References
- 1. Wilson EO (2004) Taxonomy as a fundamental discipline. Philos Trans R Soc Lond B Biol Sci 359: 739–739. 10.1098/rstb.2003.1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Joppa LN, Roberts DL, Myers N, Pimm SL (2011) Biodiversity hotspots house most undiscovered plant species. Proc Natl Acad Sci USA 108: 13171–13176. 10.1073/pnas.1109389108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mora C, Tittensor D, Adl S, Simpson A, Worm B (2011) How many species are there on Earth and in the ocean? PLoS Biol 9 10.1371/journal.pbio.1001127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scheffers BR, Joppa LN, Pimm SL, Laurance WF (2012) What we know and don’t know about Earth’s missing biodiversity. Trends Ecol Evol 27: 501–510. 10.1016/j.tree.2012.05.008 [DOI] [PubMed] [Google Scholar]
- 5. Godfray HCJ (2002) Challenges for taxonomy. Nature 417: 17–19. 10.1038/417017a [DOI] [PubMed] [Google Scholar]
- 6. Mace GM (2004) The role of taxonomy in species conservation. Philos Trans R Soc Lond B Biol Sci 359: 711–719. 10.1098/rstb.2003.1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Padial J, De la Riva I (2009) Integrative taxonomy reveals cryptic Amazonian species of Pristimantis (Anura: Strabomantidae). Zool J Linn Soc 155: 97–122. 10.1111/j.1096-3642.2008.00424.x [DOI] [Google Scholar]
- 8. Bond JE, Sierwald P (2002) Cryptic speciation in the Anadenobolus excisus millipede species complex on the island of Jamaica. Evolution 56: 1123–1135. 10.1111/j.0014-3820.2002.tb01426.x [DOI] [PubMed] [Google Scholar]
- 9. Collin R (2003) The utility of morphological characters in gastropod phylogenetics: an example from the Calyptraeidae. Biol J Linn Soc 78: 541–593. 10.1046/j.0024-4066.2002.00166.x [DOI] [Google Scholar]
- 10. Álvarez N, Hossaert-McKey M, Rasplus J-Y, McKey D, Mercier L, Soldati L, et al. (2005) Sibling species of bean bruchids: a morphological and phylogenetic study of Acanthoscelides obtectus Say and Acanthoscelides obvelatus Bridwell. J Zool Syst Evol Res 43: 29–37. 10.1111/j.1439-0469.2004.00286.x [DOI] [Google Scholar]
- 11. Huber BA, Rheims CA, Brescovit AD (2005) Speciation without changes in genital shape: a case study on Brazilian pholcid spiders (Araneae: Pholcidae). Zool Anz—J Comp Zool 243: 273–279. 10.1016/j.jcz.2004.12.001 23907846 [DOI] [Google Scholar]
- 12. Kodandaramaiah U, Weingartner E, Jank N, Leski M, Slove J, Warren A, et al. (2012) Investigating concordance among genetic data, subspecies circumscriptions and hostplant use in the Nymphalid butterfly Polygonia faunus . PLoS ONE 7: e41048 10.1371/journal.pone.0041058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kergoat GJ, Álvarez N (2008) Assessing the phylogenetic usefulness of a previously neglected morphological structure through elliptic Fourier analyses: a case study in Bruchus seed-beetles (Coleoptera: Chrysomelidae: Bruchinae). Syst Entomol 33: 289–300. 10.1111/j.1365-3113.2007.00405.x [DOI] [Google Scholar]
- 14. Gentile G, della Torre A, Maegga B, Powell JR, Caccone A (2002) Genetic differentiation in the African Malaria vector, Anopheles gambiae s.s., and the problem of taxonomic status. Genetics 161: 1561–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pringle A, Baker DM, Platt JL, Wares JP, Latgé JP, Taylor JW (2005) Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus . Evolution 59: 1886–1899. 10.1111/j.0014-3820.2005.tb01059.x [DOI] [PubMed] [Google Scholar]
- 16. Armstrong K, Ball S (2005) DNA barcodes for biosecurity: invasive species identification. Philos Trans R Soc B Biol Sci 360: 1813–1823. 10.1098/rstb.2005.1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roe AD, Sperling FAH (2007) Population structure and species boundary delimitation of cryptic Dioryctria moths: an integrative approach. Mol Ecol 16: 3617–3633. 10.1111/j.1365-294X.2007.03412.x [DOI] [PubMed] [Google Scholar]
- 18. Ross KG, Gotzek D, Ascunce MS, Shoemaker DD (2009) Species delimitation: a case study in a problematic ant taxon. Syst Biol. 59: 162–164. 10.1093/sysbio/syp089 [DOI] [PubMed] [Google Scholar]
- 19. Frey J, Guillen L, Frey B, Samietz J, Rull J, Aluja M (2013) Developing diagnostic SNP panels for the identification of true fruit flies (Diptera: Tephritidae) within the limits of COI-based species delimitation. BMC Evol Biol 13: 106 10.1186/1471-2148-13-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blaxter ML (2004) The promise of a DNA taxonomy. Philos Trans R Soc Lond B Biol Sci 359: 669–679. 10.1098/rstb.2003.1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dincă V, Zakharov EV, Hebert PDN, Vila R (2011) Complete DNA barcode reference library for a country’s butterfly fauna reveals high performance for temperate Europe. Proc R Soc B Biol Sci 278: 347–355. 10.1098/rspb.2010.1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hebert PDN, Gregory TR (2005) The promise of DNA barcoding for taxonomy. Syst Biol 54: 852–859. 10.1080/10635150500354886 [DOI] [PubMed] [Google Scholar]
- 23. Hebert PDN, Ratnasingham S, de Waard JR (2003) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B Biol Sci 270: S96– S99. 10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator . Proc Natl Acad Sci USA 101: 14812–14817. 10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM (2004) Identification of birds through DNA barcodes. PLoS Biol 2: e312 10.1371/journal.pbio.0020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Janzen DH, Hallwachs W, Blandin P, Burns JM, Cadiou J-M, Chacon I (2009) Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity. Mol Ecol Resour 9; 1–26. 10.1111/j.1755-0998.2009.02628.x [DOI] [PubMed] [Google Scholar]
- 27. Prado BR, Pozo C, Valdez-Moreno M, Hebert PDN (2011) Beyond the colours: discovering hidden diversity in the Nymphalidae of the Yucatan peninsula in Mexico through DNA barcoding. PLoS ONE 6: e27776 10.1371/journal.pone.0027776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Will KW, Mishler BD, Wheeler QD (2005) The perils of DNA barcoding and the need for integrative taxonomy. Syst Biol 54: 844–851. 10.1080/10635150500354878 [DOI] [PubMed] [Google Scholar]
- 29. Gompert Z, Nice CC, Fordyce JA, Forister ML, Shapiro AM (2006) Identifying units for conservation using molecular systematics: the cautionary tale of the Karner blue butterfly. Mol Ecol 15: 1759–1768. 10.1111/j.1365-294X.2006.02905.x [DOI] [PubMed] [Google Scholar]
- 30. Meier R, Shiyang K, Vaidya G, Ng PKL (2006) DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol 55: 715–728. 10.1080/10635150600969864 [DOI] [PubMed] [Google Scholar]
- 31. Wiemers M, Fiedler K (2007) Does the DNA barcoding gap exist?—a case study in blue butterflies (Lepidoptera: Lycaenidae). Front Zool 4: 8 10.1186/1742-9994-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Whitworth T, Dawson R, Magalon H, Baudry E (2007) DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae). Proc R Soc B Biol Sci 274: 1731–1739. 10.1098/rspb.2007.0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moritz C, Cicero C (2004) DNA barcoding: promise and pitfalls. PLoS Biol 2: e354 10.1371/journal.pbio.0020354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meyer CP, Paulay G (2005) DNA barcoding: error rates based on comprehensive sampling. PLoS Biol 3: e422 10.1371/journal.pbio.0030422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hickerson MJ, Meyer CP, Moritz C (2006) DNA barcoding will often fail to discover new animal species over broad parameter space. Syst Biol 55: 729–739. 10.1080/10635150600969898 [DOI] [PubMed] [Google Scholar]
- 36. Lohse K (2009) Can mtDNA barcodes be used to delimit species? A response to Pons et al. (2006). Syst Biol 58: 439–442. 10.1093/sysbio/syp039 [DOI] [PubMed] [Google Scholar]
- 37. Galtier N, Nabholz B, Glémin S, Hurst GD (2009) Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol 18: 4541–4550. 10.1111/j.1365-294X.2009.04380.x [DOI] [PubMed] [Google Scholar]
- 38. Hendrich L, Ribera I, Balke M (2010) Mitochondrial Cox1 sequence data reliably uncover patterns of insect diversity but suffer from high lineage-idiosyncratic error rates. PLoS ONE 5: e14448 10.1371/journal.pone.0014448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Velzen R, Weitschek E, Felici G, Bakker FT (2012) DNA barcoding of recently diverged species: relative performance of matching methods. PLoS ONE 7: e30490 10.1371/journal.pone.0030490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Will KW, Rubinoff D (2004) Myth of the molecule: DNA barcodes for species cannot replace morphology for identification and classification. Cladistics 20: 47–55. 10.1111/j.1096-0031.2003.00008.x [DOI] [PubMed] [Google Scholar]
- 41. Ebach MC, Holdrege C (2005) DNA barcoding is no substitute for taxonomy. Nature 434: 697–697. 10.1038/434697b [DOI] [PubMed] [Google Scholar]
- 42. Rubinoff D, Cameron S, Will K (2006) A genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. J Hered 97: 581–594. 10.1093/jhered/esl036 [DOI] [PubMed] [Google Scholar]
- 43. Dupuis JR, Roe AD, Sperling FAH (2012) Multi-locus species delimitation in closely related animals and fungi: one marker is not enough. Mol Ecol 21: 4422–4436. 10.1111/j.1365-294X.2012.05642.x [DOI] [PubMed] [Google Scholar]
- 44. Padial J, Miralles A, De la Riva I, Vences M (2010) The integrative future of taxonomy. Front Zool 7: 16 10.1186/1742-9994-7-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gebiola M, Gómez-Zurita J, Monti MM, Navone P, Bernardo U (2012) Integration of molecular, ecological, morphological and endosymbiont data for species delimitation within the Pnigalio soemius complex (Hymenoptera: Eulophidae). Mol Ecol 21: 1190–1208. 10.1111/j.1365-294X.2011.05428.x [DOI] [PubMed] [Google Scholar]
- 46. Dayrat B (2005) Towards integrative taxonomy. Biol J Linn Soc 85: 407–415. 10.1111/j.1095-8312.2005.00503.x [DOI] [Google Scholar]
- 47. Schlick-Steiner BC, Steiner FM, Seifert B, Stauffer C, Christian E, Crozier RH (2009) Integrative taxonomy: a multisource approach to exploring biodiversity. Annu Rev Entomol 55: 421–438. 10.1146/annurev-ento-112408-085432 [DOI] [PubMed] [Google Scholar]
- 48. Yeates DK, Seago A, Nelson L, Cameron SL, Joseph L, Trueman JWH (2011) Integrative taxonomy, or iterative taxonomy? Syst Entomol 36: 209–217. 10.1111/j.1365-3113.2010.00558.x [DOI] [Google Scholar]
- 49. Le Ru BP, Dulac CC, Toussaint E, Conlong D, Van den Berg J, Pallangyo B (2014) Integrative taxonomy of Acrapex stem borers (Lepidoptera, Noctuidae, Apameini): combining morphology and Poisson tree process analyses. Invertebr Syst. 28: 451–475. 10.1071/IS13062 [DOI] [Google Scholar]
- 50. Lumley LM, Sperling FAH (2010) Integrating morphology and mitochondrial DNA for species delimitation within the spruce budworm (Choristoneura fumiferana) cryptic species complex (Lepidoptera: Tortricidae). Syst Entomol 35: 416–428. 10.1111/j.1365-3113.2009.00514.x [DOI] [PubMed] [Google Scholar]
- 51. Melville J, Smith K, Hobson R, Shoo L (2014) The role of integrative taxonomy in the conservation management of cryptic species: the taxonomic status of endangered earless dragons (Agamidae: Tympanocryptis) in the grasslands of Queensland, Australia. PLoS ONE 9: e101847 10.1371/journal.pone.0101847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rougerie R, Naumann S, Nässig WA (2011) Morphology and molecules reveal unexpected cryptic diversity in the enigmatic genus Sinobirma Bryk, 1944 (Lepidoptera: Saturniidae). PLoS ONE 7: e43920 10.1371/journal.pone.0043920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. O’Meara BC, Ané C, Sanderson MJ, Wainwright PC (2006) Testing for different rates of continuous trait evolution using likelihood. Evolution 60: 922–933. 10.1111/j.0014-3820.2006.tb01171.x [DOI] [PubMed] [Google Scholar]
- 54. Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S (2006) Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol 55: 595–609. 10.1080/10635150600852011 [DOI] [PubMed] [Google Scholar]
- 55. Yang Z, Rannala B (2010) Bayesian species delimitation using multilocus sequence data. Proc Natl Acad Sci USA 107: 9264–9269. 10.1073/pnas.0913022107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ence DD, Carstens BC (2011) SpedeSTEM: a rapid and accurate method for species delimitation. Mol Ecol Resour 11: 473–480. 10.1111/j.1755-0998.2010.02947.x [DOI] [PubMed] [Google Scholar]
- 57. Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol 21: 1864–1877. 10.1111/j.1365-294X.2011.05239.x [DOI] [PubMed] [Google Scholar]
- 58. Carstens BC, Pelletier TA, Reid N, Satler JD (2013) How to fail at species delimitation. Mol Ecol 22:4369–4383. 10.1111/mec.12413 [DOI] [PubMed] [Google Scholar]
- 59. Hamback P, Weingartner E, Ericson L, Fors L, Cassel-Lundhagen A, Stenberg JA et al. (2013) Bayesian species delimitation reveals generalist and specialist parasitic wasps on Galerucella beetles (Chrysomelidae): sorting by herbivore or plant host. BMC Evol Biol 13: 92 10.1186/1471-2148-13-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang J, Kapli P, Pavlidis P, Stamatakis A (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29: 2869–2876. 10.1093/bioinformatics/btt499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. De Queiroz K (2007) Species concepts and species delimitation. Syst Biol 56: 879–886. 10.1080/10635150701701083 [DOI] [PubMed] [Google Scholar]
- 62. Rannala B, Yang Z (2003) Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci. Genetics 164: 1645–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Knowles LL, Carstens BC (2007) Delimiting species without monophyletic gene trees. Syst Biol 56: 887–895. 10.1080/10635150701701091 [DOI] [PubMed] [Google Scholar]
- 64. Fujita MK, Leaché AD, Burbrink FT, McGuire JA, Moritz C (2012) Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol Evol 27: 480–488. 10.1016/j.tree.2012.04.012 [DOI] [PubMed] [Google Scholar]
- 65. Dupuis JR, Roe AD, Sperling FAH (2012) Multi-locus species delimitation in closely related animals and fungi: one marker is not enough. Mol Ecol 21: 4422–4436. 10.1111/j.1365-294X.2012.05642.x [DOI] [PubMed] [Google Scholar]
- 66. Funk DJ, Omland EK (2003) Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu Rev Ecol Evol Syst 34: 397–423. 10.1146/annurev.ecolsys.34.011802.132421 [DOI] [Google Scholar]
- 67. Hailer F, Kutschera VE, Hallstrom BM, Klassert D, Fain SR, Leonard JA, et al. (2012) Nuclear genomic sequences reveal that polar bears are an old and distinct bear lineage. Science 336:344–347. 10.1126/science.1216424 [DOI] [PubMed] [Google Scholar]
- 68. Avise JC (2009) Phylogeography: retrospect and prospect. J. Biogeogr. 36: 3–15. 10.1111/j.1365-2699.2008.02032.x [DOI] [Google Scholar]
- 69. Barraclough TG, Hughes M, Ashford-Hodges N, Fujisawa T (2009) Inferring evolutionarily significant units of bacterial diversity from broad environmental surveys of single-locus data. Biol. Lett. 5: 425–428. 10.1098/rsbl.2009.0091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Avise JC (2009) Phylogeography: retrospect and prospect. J. Biogeogr. 36: 3–15. 10.1111/j.1365-2699.2008.02032.x [DOI] [Google Scholar]
- 71. Hebert PDN, deWaard JR, Landry JF (2010) DNA barcodes for 1/1000 of the animal kingdom. 6: 359–362. 10.1098/rsbl.2009.0848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Haussmann A, Haszprunar G, Hebert PDN (2011) DNA barcoding the geometrid fauna of Bavaria (Lepidoptera): successes, surprises, and questions. PLoS ONE 6: e17134 10.1371/journal.pone.0017134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Butcher BA, Smith MA, Sharkey MJ, Quicke DLJ (2012) A turbo-taxonomic study of Thai Aleiodes (Aleiodes) and Aleiodes (Arcaleiodes) (Hymenoptera: Braconidae: Rogadinae) based largely on COI barcoded specimens, with rapid descriptions of 179 new species. Zootaxa 3457: 1–232. [Google Scholar]
- 74. Riedel A, Sagata K, Surbakti S, Tänzler R, Balke M (2013. a) One hundred and one new species of Trigonopterus weevils from New Guinea. ZooKeys 280: 1–150. 10.3897/zookeys.280.3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Riedel A, Sagata K, Suhardjono YR, Tänzler R, Blake M (2013. b) Integrative taxonomy on the fast track—towards more sustainability in biodiversity research. Front. Zool. 10: 15 10.1186/1742-9994-10-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pogue M (2002) A world revision of the genus Spodoptera Guenée (Lepidoptera: Noctuidae) American Entomological Society; Philadelphia. 202 p. [Google Scholar]
- 77. Lalanne-Cassou B, Silvain JF, Monti L, Malosse C (1999) Mécanismes d'isolement reproducteur chez les espèces du complexe Neotropical Spodoptera latifascia—S. cosmioides—S. descoinsi (Lepidoptera: Noctuidae). Ann Soc Entomol Fr (NS) 35: S109–S116. [Google Scholar]
- 78. Groot A, Marr M, Schofl G, Lorenz S, Svatos A, Heckel DG (2008) Host strain specific sex pheromone variation in Spodoptera frugiperda . Front Zool 5: 20 10.1186/1742-9994-5-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Schöfl G, Heckel David G., Groot AT (2009) Time-shifted reproductive behaviours among fall armyworm (Noctuidae: Spodoptera frugiperda) host strains: evidence for differing modes of inheritance. J Evol Biol 22: 1447–1459. 10.1111/j.1420-9101.2009.01759.x [DOI] [PubMed] [Google Scholar]
- 80. Brown ES, Dewhurst CF (1975) The genus Spodoptera (Lepidoptera: Noctuidae) in Africa and the Near East. Bull Entomol Res 65: 221–262. 10.1017/S0007485300005939 [DOI] [Google Scholar]
- 81. Todd EL, Poole RW (1980) Keys and illustrations for the armyworm moths of the Noctuid genus Spodoptera Guenée from the western hemisphere. Ann Entomol Soc Am 73: 722–738. [Google Scholar]
- 82. Silvain J-F, Lalanne-Cassou B (1997) Distinction entre Spodoptera latifascia (Walker) et Spodoptera cosmioides (Walker), bona species (Lepidoptera, Noctuidae). Rev Fr Entomol 19: 95–97. [Google Scholar]
- 83. Martinelli S, Barata RM, Zucchi MI, Silva-Filho MDC, Omoto C (2006) Molecular variability of Spodoptera frugiperda (Lepidoptera: Noctuidae) populations associated to maize and cotton crops in Brazil. J Econ Entomol 99: 519–526. 10.1603/0022-0493-99.2.519 [DOI] [PubMed] [Google Scholar]
- 84. Salinas-Hernandez H, Saldamando-Benjumea CI (2011) Haplotype identification within Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae) corn and rice strains from Colombia. Neotrop Entomol 40: 421–430. 10.3958/059.037.0213 [DOI] [PubMed] [Google Scholar]
- 85. Kergoat GJ, Prowell DP, Le Ru BP, Mitchell A, Dumas P, Clamens A-L (2012) Disentangling dispersal, vicariance and adaptive radiation patterns: a case study using armyworms in the pest genus Spodoptera (Lepidoptera: Noctuidae). Mol Phylogenet Evol 65: 855–870. 10.1016/j.ympev.2012.08.006 [DOI] [PubMed] [Google Scholar]
- 86. Greenberg SM, Sappington TW, Legaspi BC, Liu T-X, Sétamou M (2001) Feeding and life history of Spodoptera exigua (Lepidoptera: Noctuidae) on different host plants. Ann Entomol Soc Am 94: 566–575. [Google Scholar]
- 87. Barros EM, Torres JB, Ruberson JR, Oliveira MD (2010) Development of Spodoptera frugiperda on different hosts and damage to reproductive structures in cotton. Entomol Exp Appl 137: 237–245. 10.1111/j.1570-7458.2010.01058.x [DOI] [Google Scholar]
- 88. Drès M, Mallet J (2002) Host races in plant-feeding insects and their importance in sympatric speciation. Philos Trans R Soc Lond B Biol Sci 357: 471–492. 10.1098/rstb.2002.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Prowell DP, McMichael M, Silvain J-F (2004) Multilocus genetic analysis of host use, introgression, and speciation in host strains of fall armyworm (Lepidoptera: Noctuidae). Ann Entomol Soc Am 97: 1034–1044. [Google Scholar]
- 90. Bolnick DI, Fitzpatrick BM (2007) Sympatric speciation: models and empirical evidence. Annu Rev Ecol Evol Syst 38: 459–487. 10.1146/annurev.ecolsys.38.091206.095804 [DOI] [Google Scholar]
- 91.Juárez ML, Schöfl G, Vera MT, Vilardi JC, Murúa MG, Willink E, et al. (2014) Population structure of Spodoptera frugiperda maize and rice host forms in South America: are they host strains? Entomol Exp Appl. 10.1111/eea.12215 [DOI]
- 92. Pashley DP (1986) Host-associated genetic differentiation in fall armyworm (Lepidoptera: Noctuidae): a sibling species complex? Entomol Soc Am 79: 989–904. [Google Scholar]
- 93. Prowell DP, McMichael M, Silvain J-F (2004) Multilocus genetic analysis of host use, introgression, and speciation in host strains of fall armyworm (Lepidoptera: Noctuidae). Ann Entomol Soc Am 97: 1034–1044. [Google Scholar]
- 94. Meagher R, Nagoshi R, Stuhl C (2011) Oviposition choice of two fall armyworm (Lepidoptera: Noctuidae) host strains. J Insect Behav 24: 337–347. 10.1007/s10905-011-9259-7 [DOI] [Google Scholar]
- 95. Lu Y, Adang MJ (1996) Distinguishing fall armyworm (Lepidoptera: Noctuidae) strains using a diagnostic mitochondrial DNA marker. Fla Entomol 79: 48–55. 10.2307/3495753 [DOI] [Google Scholar]
- 96. McMichael M, Prowell DP (1999) Differences in amplified fragment-length polymorphisms in fall armyworm (Lepidoptera: Noctuidae) host strains. Ann Entomol Soc Am 92: 175–181. [Google Scholar]
- 97. Levy HC, Garcia-Maruniak A, Maruniak JE (2002) Strain identification of Spodoptera frugiperda (Lepidoptera: Noctuidae) insects and cell line: pcr-RFLP of cytochrome oxidase C subunit I gene. Fla Entomol 85: 186–190. [Google Scholar]
- 98. Meagher RL, Gallo-Meagher M (2003) Identifying host strains of fall armyworm (Lepidoptera: Noctuidae) in Florida using mitochondrial markers. Fla Entomol 86: 450–455. [Google Scholar]
- 99. Nagoshi RN, Armstrong JS, Silvie P, Meagher RL (2008) Structure and distribution of a strain-biased tandem repeat element in fall armyworm (Lepidoptera: Noctuidae) populations in Florida, Texas, and Brazil. Ann Entomol Soc Am 101: 1112–1120. 10.1603/0013-8746-101.6.1112 [DOI] [Google Scholar]
- 100. Pair SD, Raulston JR, Sparks AN, Westbrook JK, Douce GK (1986) Fall armyworm distribution and population dynamics in the Southeastern states. Fla Entomol 69: 468–487. 10.2307/3495380 [DOI] [Google Scholar]
- 101. Machado V, Wunder M, Baldissera VD, Oliveira JV, Fiúza LM, Nagoshi RN (2008) Molecular characterization of host strains of Spodoptera frugiperda (Lepidoptera: Noctuidae) in Southern Brazil. Ann Entomol Soc Am 101: 619–626. [Google Scholar]
- 102. Fontaneto D, Herniou EA, Boschetti C, Caprioli M, Melone G, Ricci C, et al. (2007) Independently evolving species in asexual bdelloid rotifers. PLoS Biol 5: e87 10.1371/journal.pbio.0050087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Astrin JJ, Stüben PE, Misof B, Wägele JW, Gimnich F, Raupach MJ, et al. (2012) Exploring diversity in cryptorhynchine weevils (Coleoptera) using distance-, character- and tree-based species delineation. Mol Phylogenet Evol 63: 1–14. 10.1016/j.ympev.2011.11.018 [DOI] [PubMed] [Google Scholar]
- 104. Bergsten J, Bilton DT, Fujisawa T, Elliott M, Monaghan MT, Balke M, et al. (2012) The effect of geographical scale of sampling on DNA barcoding. Syst Biol. 61: 851–869. 10.1093/sysbio/sys037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Esselstyn JA, Evans BJ, Sedlock JL, Anwarali Khan FA, Heaney LR (2012) Single-locus species delimitation: a test of the mixed Yule-coalescent model, with an empirical application to Philippine round-leaf bats. Proc R Soc Lond B Biol Sci. 279: 3678–3686. 10.1098/rspb.2012.0705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Papadopoulou A, Monaghan MT, Barraclough TG, Vogler AP (2009) Sampling error does not invalidate the Yule-coalescent model for species delimitation. A response to Lohse (2009). Syst Biol. 58: 442–444. 10.1093/sysbio/syp038 [DOI] [Google Scholar]
- 107. Reid N, Carstens B (2012) Phylogenetic estimation error can decrease the accuracy of species delimitation: a bayesian implementation of the general mixed Yule-coalescent model. BMC Evol Biol 12: 196 10.1186/1471-2148-12-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Fujisawa T, Barraclough TG (2013) Delimiting species using single-locus data and the generalized mixed Yule coalescent (GMYC) approach: a revised method and evaluation on simulated datasets. Syst Biol. 62: 707–724. 10.1093/sysbio/syt033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Talavera G, Dinca V, Vila R (2013). Factors affecting species delimitations with the GMYC model: insights from a butterfly survey. Methods Ecol Evol 4: 1101–1110. 10.1111/2041-210X.12107 [DOI] [Google Scholar]
- 110. Tang CQ, Humphreys AM, Fontaneto D, Barraclough TG (2014) Effects of phylogenetic reconstruction method on the robustness of species delimitation using single-locus data. Methods Ecol Evol 5: 1086–1094. 10.1111/2041-210X.12246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Mitchell A, Mitter C, Regier JC (2006) Systematics and evolution of the cutworm moths (Lepidoptera: Noctuidae): evidence from two protein-coding nuclear genes. Syst Entomol 31: 21–46. 10.1111/j.1365-3113.2005.00306.x [DOI] [Google Scholar]
- 112. Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maddison WP (2010) Maddison. DR (2010). Mesquite: a modular system for evolutionary analysis. Version 3.
- 114. Nylander JAA, Ronquist F, Huelsenbeck JP, Nieves-Aldrey J (2004) Bayesian phylogenetic analysis of combined data. Syst Biol 53: 47–67. 10.1080/10635150490264699 [DOI] [PubMed] [Google Scholar]
- 115. Lanfear R, Calcott B, Ho SYW, Guindon S (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29: 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- 116. Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol 53: 793–808. 10.1080/10635150490522304 [DOI] [PubMed] [Google Scholar]
- 117. Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 119. Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42: 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- 120. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. (2012) MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Huelsenbeck JP, Larget B, Alfaro ME (2004) Bayesian phylogenetic model selection using reversible jump Markov chain monte carlo. Mol Biol Evol 21: 1123–1133. 10.1093/molbev/msh123 [DOI] [PubMed] [Google Scholar]
- 123. Erixon P, Svennblad B, Britton T, Oxelman B (2003) Reliability of bayesian posterior probabilities and bootstrap frequencies in phylogenetics. Syst Biol 52: 665–673. 10.1080/10635150390235485 [DOI] [PubMed] [Google Scholar]
- 124.Ezard T, Fujisawa T, Barraclough TG (2009) Splits: species’ limits by threshold statistics. R Package Version 1.
- 125. Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- 126. Bortolussi N, Durand E, Blum M, François O (2006) apTreeshape: statistical analysis of phylogenetic tree shape. Bioinformatics 22: 363–364. 10.1093/bioinformatics/bti798 [DOI] [PubMed] [Google Scholar]
- 127. Nagoshi RN, Meagher RL, Hay-Roe M (2012) Inferring the annual migration patterns of fall armyworm (Lepidoptera: Noctuidae) in the United States from mitochondrial haplotypes. Ecol Evol 2: 1458–1467. 10.1002/ece3.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Williams ST, Knowlton N (2001) Mitochondrial pseudogenes are pervasive and often insidious in the snapping shrimp genus Alpheus . Mol Biol Evol 18: 1484–1493. [DOI] [PubMed] [Google Scholar]
- 129. Petit RJ, Excoffier L (2009) Gene flow and species delimitation. Trends Ecol Evol 24: 386–393. 10.1016/j.tree.2009.02.011 [DOI] [PubMed] [Google Scholar]
- 130. Brown JM, Hedtke SM, Lemmon AR, Lemmon EM (2010) When trees grow too long: investigating the causes of highly inaccurate bayesian branch-length estimates. Syst Biol 59: 145–161. 10.1093/sysbio/syp081 [DOI] [PubMed] [Google Scholar]
- 131. Lemmon AR, Moriarty EC (2004) The importance of proper model assumption in bayesian phylogenetics. Syst Biol 53: 265–277. 10.1080/10635150490423520 [DOI] [PubMed] [Google Scholar]
- 132. Phillips MJ (2009) Branch-length estimation bias misleads molecular dating for a vertebrate mitochondrial phylogeny. Gene. 441: 132–140. 10.1016/j.gene.2008.08.017 [DOI] [PubMed] [Google Scholar]
- 133. Brandley MC, Schmitz A, Reeder TW (2005) Partitioned bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst Biol 54: 373–390. 10.1080/10635150590946808 [DOI] [PubMed] [Google Scholar]
- 134. Brown JM, Lemmon AR (2007) The importance of data partitioning and the utility of bayes factors in bayesian phylogenetics. Syst Biol 56: 643–655. 10.1080/10635150701546249 [DOI] [PubMed] [Google Scholar]
- 135. Marshall DC, Simon C, Buckley TR (2006) Accurate branch length estimation in partitioned bayesian analyses requires accommodation of among-partition rate variation and attention to branch length priors. Syst Biol 55: 993–1003. 10.1080/10635150601087641 [DOI] [PubMed] [Google Scholar]
- 136. Ekman S, Blaalid R (2011) The devil in the details: interactions between the branch-length prior and likelihood model affect node support and branch lengths in the phylogeny of the Psoraceae. Syst Biol 60: 541–561. 10.1093/sysbio/syr022 [DOI] [PubMed] [Google Scholar]
- 137. Hamilton CA, Hendrixson BE, Brewer MS, Bond JE (2014) An evaluation of sampling effects on multiple DNA barcoding methods leads to an integrative approach for delimiting species: a case study of the North American tarantula genus Aphonopelma (Araneae, Mygalomorphae, Theraphosidae). Mol Phylogenet Evol 71: 79–93. 10.1016/j.ympev.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 138. Lima E, McNeil J (2009) Female sex pheromones in the host races and hybrids of the fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae). Chemoecology 19: 29–36. 10.1007/s00049-009-0005-y [DOI] [Google Scholar]
- 139. Schöfl G, Dill A, Heckel David G, Groot AT (2011) Allochronic separation versus mate choice: nonrandom patterns of mating between fall armyworm host strains. Am Nat 177: 470–485. 10.1086/658904 [DOI] [PubMed] [Google Scholar]
- 140. Whitford F, Quisenberry SS, Riley TJ, Lee JW (1988) Oviposition preference, mating compatibility, and development of two fall armyworm strains. Fla Entomol 71: 234–243. 10.2307/3495426 [DOI] [Google Scholar]
- 141. Pashley DP, Martin JA (1987) Reproductive incompatibility between host strains of the fall armyworm (Lepidoptera: Noctuidae). Ann Entomol Soc Am 80: 731–733. [Google Scholar]
- 142.Dumas P, Legeai F, Lemaitre C, Scaon E, Orsucci M, Labadie K, et al. (2015) Spodoptera frugiperda (Lepidoptera: Noctuidae) host plant variants: two strains or two distinct species? Genetica in press. 10.1007/s10709-015-9829-2 [DOI] [PMC free article] [PubMed]
- 143. Groot AT, Marr M, Heckel David G, Schofl G (2010) The roles and interactions of reproductive isolation mechanisms in fall armyworm (Lepidoptera: Noctuidae) host strains. Ecol Entomol 35: 105–118. 10.1111/j.1365-2311.2009.01138.x [DOI] [Google Scholar]
- 144. Quisenberry SS (1991) Fall armyworm (Lepidoptera: Noctuidae) host strain reproductive compatibility. Fla Entomol 74: 194–199. 10.2307/3495297 [DOI] [Google Scholar]
- 145. Nagoshi RN, Fleischer S, Meagher RL (2009) Texas is the overwintering source of fall armyworm in central Pennsylvania: implications for migration into the Northeastern United States. Environ Entomol 38: 1546–1554. 10.1603/022.038.0605 [DOI] [PubMed] [Google Scholar]
- 146. Velásquez-Vélez MI, Saldamando-Benjumea CI, Ríos-Diez JD (2011) Reproductive isolation between two populations of Spodoptera frugiperda (Lepidoptera: Noctuidae) collected in corn and rice fields from Central Colombia. Ann Entomol Soc Am 104: 826–833. 10.1603/AN10164 [DOI] [Google Scholar]
- 147. Nosil P (2012) Ecological speciation Oxford University; Oxford. 304 p. [Google Scholar]
- 148. Fletcher DS (1956) Spodoptera mauritia (Boisduval) and S. triturata (Walker), two distinct species. Bull Entomol Res 47: 215–217. 10.1017/S0007485300046666 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Support values are provided on major nodes (only BV ≥ 50% are shown).
(PDF)
Support values are provided on major nodes (only PP ≥ 0.5 are shown).
(PDF)
Putative species clusters are indicated using transitions between black-coloured to red-coloured branches.
(PDF)
Putative species clusters are indicated using transitions between black-coloured to red-coloured branches.
(PDF)
Putative species clusters are indicated using transitions between black-coloured to red-coloured branches. The inter- and intraspecific portions of the tree are divided with a dotted line (95% confidence intervals are figured using thinner dotted lines).
(PDF)
Putative species clusters are indicated using transitions between black-coloured to red-coloured branches. The inter- and intraspecific portions of the tree are divided with a dotted line (95% confidence intervals are figured using thinner dotted lines).
(PDF)
Sequences generated in a previous study of our research group are highlighted using ‘*’ whereas newly generated sequences are highlighted using ‘**’. The Democratic Republic of the Congo was abbreviated using DRC. When the country of origin is known we also provide the corresponding ISO 3166 alpha-3 code. We used the following abbreviations for the repository institutes and collections: ACG: Área de Conservación Guanacaste—Centro de Investigación y Estaciones Biológicas Programa de Educación Biológica, Costa Rica; ANIC: Australian National Insect Collection, Australia; BIO: Biodiversity Institute of Ontario, Canada; BSCZ: Bavarian State Collection of Zoology, Munich, Germany; CBGP: INRA—Centre de Biologie pour la Gestion des Populations, Montferrier/Lez, France; CLS: College of Life Science, Capital Normal University, Beijing, China; CIRN: CIRN, University of the Azores, Portugal; CTAG: Center for Theoretical and Applied Genetics, New Brunswick, USA; EMBRAPA: Embrapa National Soybean Research Center, Brazil; FERA: Fond and Environment Research Agency, York, UK; GSA: Graduate School of Agriculture, Kita-Ku, Japan; HUT: Hefei University of Technology, China; LEC: Lancaster Environment Centre, UK; MNHN: Muséum National d'Histoire Naturelle, Paris, France; NIAES: National Institute for Agro-Environmental Sciences, Tsukuba, Japan; NIBGE: National Institute for Biotechnology and Genetic Engineering, Pakistan; NIMBB: National Institute of Molecular Biology and Biotechnology, Los Banos, Philippines; NRIC: Natural Resources Inventory Center, Tsukuba, Japan; PAU: Punjab Agricultural University, India; PSCAS: Penn State College of Agricultural Sciences, USA; RCG: Research Collection of Theo Gruenewald, Germany; RCH: Research Collection of Alfred Haslberger, Munich, Germany; RCJH: Research Collection of D.H. Janzen & W. Hallwachs, USA; RCW: Research Collection of Jeremy deWaard, Guelph, Canada. SI: Smithsonian Institution, Washington, USA; TAU: Tamilnadu Agricultural University, India; UBC: University of British Columbia, Vancouver, Canada; UFP: Universidade Federal do Parana, Curitiba, Brazil; UM: University of Maryland, USA; UR: Université de Rouen, Rouen, France; USDA: USDA ARS, Gainesville, USA; YU: Yangzhou University, China; ZIQ: Zuhai Inspection and Quarantine bureau, China.
(DOCX)
Three partitions were specified (one per codon position). Selected models and partitions based on the AICc are figured on the left whereas selected models and partitions based on the BIC are figured on the right.
(DOCX)
The geographic origin and assignment to putative species clusters is provided for each Spodoptera specimen. Similarly to the sidebars presented on Fig 2, we have used one column per set of analyses (PTP with RAxML, PTP with MrBayes, GMYC with BEAST (strict clock) and GMYC with BEAST (UCLN clock). For more clarity, putative species clusters also are highlighted using a combination of numbers and distinct shades of grey.
(DOCX)
Data Availability Statement
All new sequences generated for the study are available from the GenBank database (accession numbers) KF854152 to KF854234.