Abstract
mDia2 is an auto-inhibited Formin influencing actin dynamics upon conversion to the active conformation. mDia2 regulates actin-based protrusions and cell invasion, cell differentiation, vesicle trafficking, and cytokinesis. However, whether mDia2 has additional functions and how its action is functionally specified remain unknown. Here we draw the interactome of auto-inhibited and constitutively active mDia2 to address these issues. We embed mDia2 in protein networks accounting for its attributed functions and unexpectedly link it to the Ubiquitin Proteasome System. Taking FBXO3 as a test case, we show that mDia2 binds FBXO3 and p53, and regulates p53 transcriptional activity in an actin-nucleation-independent and conformation-insensitive manner. Increased mDia2 and FBXO3 levels elevate p53 activity and expression thereby sensitizing cells to p53-dependent apoptosis, whereas their decrease produces opposite effects. Thus, we discover a new role of mDia2 in p53 regulation suggesting that the closed conformation is biologically active and an FBXO3-based mechanism to functionally specify mDia2's activity.
mDia2 (mouse homolog of Diaphanous 2) and other nine proteins compose the Diaphanous-related Formin (Drf)1 family. Members of this family are defined by a tandem Formin Homology (FH) FH1 and FH2 domains, the so-called Formin signature, and a characteristic C-terminal Diaphanous Auto-regulatory Domain (DAD) (1). The homo-dimeric FH2 domain of mDia2 nucleates linear actin filaments and favors their elongation while remaining associated with the barbed ends. The FH1 domain harbors several Profilin-binding motifs and modulates the activity of the FH2 (2). Recently, the FH1-FH2 region of mDia2 has been shown to directly interact with and to stabilize microtubules (3).
Drfs display a common modular architecture also in their regulatory N-terminal part. The N terminus of mDia2 shows a Basic Domain (BD) followed by a GTPase-binding domain (GBD) to which activated Rho A-C, Rif, and Cdc42 bind (4). Next to it, there are a Diaphanous Inhibitory Domain (DID) and a Dimerization Domain (DD) mediating homotypic interactions (1).
An interaction between the C-terminal DAD and the N-terminal DID prevents actin nucleation by the FH2 both in vitro and in vivo, thus keeping Drfs in an auto-inhibited or “closed” state (5). Two amino acid stretches in the DAD are implicated in auto-inhibition: an amphipathic helix (MDXLLXL) and a nearby Lys- and Arg-rich sequence contribute to DID binding. Single point mutations, such as the M1041-to-A substitution in the amphipathic helix of mDia2, impair the DID-binding abilities of DAD and result in mDia2 remaining in an “open” biologically active state (6). Signals regulating the DID-DAD interaction control mDia2 activity: binding of activated Rho proteins to the GBD induces a conformational change in the adjacent DID that displaces the DAD and allows mDia2 to influence actin dynamics. Other Formin-binding proteins and post-translational modifications might cooperate with Rho GTPases to fully activate mDia2 (4).
mDia2 has been implicated in filopodium (7, 8), bleb (9, 10), and invadopodium formation (11), cell invasion (11, 12), erythropoiesis (13), vesicle trafficking (14), and cytokinesis (15). Besides contributing to the formation of actin-based protrusions, regulation of actin and microtubule dynamics by mDia2 has been shown to a play a role in cell invasiveness (2, 16). Yet, mechanistic insight into the other known functions of mDia2 is lacking. It is also unclear whether these seemingly diversified processes describe the mDia2 functional landscape in full and how mDia2 achieves functional specificity. Although mounting evidence suggests that the activity cycle of Drfs is controlled by protein–protein interactions, the protein networks harnessing mDia2 remain largely unknown.
Here, we have exploited affinity purification (AP) in combination with MS-based quantitative proteomics (17) and a computational analysis tool termed significance analysis of INTeractome (SAINT) (18) to draw the interactome of wild-type and constitutively active mDia2. Proteomic and bioinformatic analyses embed mDia2 in protein networks accounting for its attributed functions and unveil a link with the Ubiquitin Proteasome System (UPS). Taking the Ubiquitin ligase SCFFBXO3 as a test case, we first showed that mDia2 forms a complex with FBXO3 and p53. We then performed in-depth functional studies indicating that mDia2 stimulates p53′s transcriptional activity in an actin-independent and conformation-insensitive manner through FBXO3. Finally, complementary gain-of-function and loss-of-function approaches show that p53 regulation by mDia2 and FBXO3 contributes to p53-dependent apoptosis. In summary, we discover a novel role of mDia2 in p53 regulation, shed new light on its mode of action, and provide an open-access resource for dissecting the mDia2-dependent processes at the molecular level.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
High-glucose DMEM supplemented with pyruvate and GlutaMax® was from Invitrogen (Carlsbad, CA). Dual-Luciferase® Reporter Assay System was from Promega (Madison, WI). Colloidal Blue Staining Kit was from Invitrogen. Lactacystin was from Cayman Chemicals (Ann Arbor, MI), Protease inhibitor EDTA-free mixture was from Roche. If not otherwise specified, all other chemicals were from Sigma-Aldrich.
Antibodies
Antibodies were as follows: mouse monoclonal anti-Flag M2, anti-β-actin (AC-15) and anti-FBXO3 (Sigma-Aldrich), mouse monoclonal anti-mDia1 (BD Transduction Laboratories, Lexington, KY), rabbit polyclonal anti-CHMP5 (H-90), goat monoclonal anti-GRP78 (N-20), rabbit polyclonal anti-Profilin1/2 (FL-140) and anti-p300 (C-20), mouse monoclonal anti-p53 (DO-1), anti-HIPK2 (F-189) and anti-Cdk2 (A-1) (Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-Myc 9E10, anti-HA-11 (Covance), rat monoclonal anti-HA (3F10) (Roche), mouse monoclonal anti-tubulin (Cell Signaling Technology, Danvers, MA), rabbit polyclonal anti-mDia3 and anti-CacyBP (Bethyl Laboratories, Montgomery, TX), rabbit polyclonal anti-ERK1/2 (Cell Signaling), rabbit polyclonal anti-VTA1 (ProteinTech, Chicago, IL). Anti-GFP and anti-mDia2 sera were generated in house.
Expression Vectors
Flag-tagged mDia2, GST-mDia2, and deletion mutants thereof were previously described (7). mDia2 point mutants were generated with the Quick Change XL Site-directed mutagenesis kit (Stratagene, La Jolla, CA) and sequence verified. Primers are available on request. The vectors obtained from third parties are listed in the supplemental Experimental Procedures.
Cell Culture, Transfections, and Knockdowns
293T, U2OS, and HeLa cells were cultured in DMEM GlutaMax® (Invitrogen) supplemented with 10% FCS. 293T cells were transfected using a standard calcium phosphate protocol. U2OS cells were transfected with FuGENE6 or X-tremeGene 9 (Roche) according to the manufacturer's instructions. Transient mDia2 knockdown 293T cells were generated as previously described (7) and assessed 48 h post-transfection. Stable mDia2 knockdown cells were obtained by cloning previously described DIAPH3-targeting sequences (7) in the pLL3.7 lentiviral vector and infecting 293T cells (19) or using MISSION® TRC shRNA TRCN0000150903 and TRCN0000150850. Knockdown of FBXO3 was obtained using MISSION® TRC shRNA TRCN0000004327 and TRCN0000004328 (Sigma-Aldrich). Knockdown of FMN1 was obtained using MISSION® TRC shRNA TRCN0000242235 and TRCN000024223 (Sigma-Aldrich). Stable p53 knockdown in U2OS cells was obtained using MISSION® TRC shRNA TRCN0000010814.
Standard Biochemical Assays and Immunocytochemistry
The expression and purification of GST-mDia2 and its deletion mutants were previously described (7). Pull-down assays were performed as previously described (7): cleared cell lysates (500 μg) overexpressing the protein of interest were incubated for 2 h at 4°C with immobilized GST-fusion proteins (62.5 pmol). Beads were extensively washed and bound proteins eluted with Laemmli buffer. Correct loading in all pull-down experiments was confirmed by Ponceau staining. Co-immunoprecipitation and immunocytochemistry experiments were performed as previously described (20). Total cell lysates were obtained as previously described (20) and 30 μg were employed in all immunoblotting experiments. Luciferase activity assays and kinetic measurement of apoptosis are illustrated in the supplemental Experimental Procedures.
Generation of mDia2-Based Immunocomplexes and Mass Spectrometry
293T cells were transfected with Flag-tagged full-length mDia2 (either the wild type or the MA mutant) or empty vector. Cell lysates were prepared as previously described (7, 20). One and a half milligrams of cell lysates were immunoprecipitated using anti-Flag M2® Affinity gel (Sigma-Aldrich) for 2 h at 4°C. Beads were washed three times in NET buffer (50 mm Tris-HCl pH 7.6, 150 mm NaCl, 5 mm EDTA and 0.1% Triton X-100) supplemented with protease inhibitor mixture (Roche), 5 mm NaF, and 1 mm NaVO4. Proteins were eluted with Laemmli buffer and separated by SDS-PAGE (NuPage 4–12% Bis-Tris gradient gel (Invitrogen)). The gel was fixed and stained with Colloidal Blue according to manufacturer's instructions (Invitrogen).
Mass Spectrometry
Protein reduction and alkylation was performed in gel with dithiotreitol (56°C, 1 h) and 2-chloro-iodoacetamide (dark, RT, 30 min), respectively, after which digestion was performed with trypsin over night at 37°C. Peptides were extracted with 100% acetonitrile. The samples were analyzed on an LTQ Orbitrap or LTQ OrbitrapVelos instrument (Thermo Scientific, Bremen) connected to an Agilent 1200 HPLC system. The nanoLC was equipped with a 20 mm 100 μm i.d.Reprosil C18 trap column and a 400 mm 50 μm i.d.Reprosil C18 analytical column (Dr Maisch, Ammerbuch-Entringen, Germany) all packed in-house. Solvent A consisted of 0.1 m acetic acid (Merck) in deionized water (Milli-Q, Millipore), and solvent B consisted of 0.1 m acetic acid in 80% acetonitrile (Biosolve). Trapping was performed at a flow of 5 μl/min for 10 min and the fractions were eluted using a flow rate passively split to either 100 nl/min (60 min LC method) or 50 nl/min (90 min LC method). The gradient used was: 90 min LC method, 10 min solvent A; 13–28% solvent B in 45 min; 28–50% solvent B in 10 min; 50–100% solvent B in 3 min; 100% solvent B for 1 min; 20 min solvent A. The mass spectrometer was operated in positive ion mode and in data-dependent mode to automatically switch between MS and MS/MS. For the Orbitrap analysis the three most intense ions in the survey scan (350 to 1500 m/z, resolution 60,000, AGC target 5e5) were fragmented in the linear ion trap (AGC target 1e4), and for the OrbitrapVelos analysis the five most intense ions in the survey scan (350 to 1500 m/z, resolution 30,000, AGC target 5e5) were subjected to higher energy collision induced dissociation (HCD) fragmentation (resolution 7500, AGC target 3e4), with the normalized collision energy set to 35% for both collision induced dissociation (CID) and HCD. The signal threshold for triggering an MS/MS event was set to 500 counts. For internal mass calibration the 445.120025 ion was used as lock mass with a target lock mass abundance of 0%. The low mass cutoff for HCD was set to 180 m/z. Charge state screening was enabled, and precursors with unknown charge state or a charge state of 1 were excluded. Dynamic exclusion was enabled (exclusion size list 500, exclusion duration 25 s).
MS Data Analysis
Peak lists were generated from the raw data files using Proteome Discoverer version 1.3 (Thermo Scientific, Bremen). For each IP, one peak list was generated per entire gel lane. Peak lists were searched against concatenated reversed Uniprot Human (version from 08 - 2014) including a list of common contaminants using Mascot software version 2.3.02 (Matrix Science, UK). Trypsin was chosen as cleavage specificity with a single missed cleavage allowed. Carbamidomethylation (C) was set as a fixed modification and oxidation (M) was set as a full as variable modification. The searches were performed using a peptide tolerance of 50 ppm, which was sliced to the real mass accuracy of 7 ppm after the database search, and a product ion tolerance of 0.6 Da (ion trap CID) or 0.05 Da (HCD). Only PSMs with Mascot scores >20 were accepted to ensure that only high quality data is allowed for this study. The following result filters were applied for all data sets: peptide length 6–35; peptide score >20; maximum search engine rank 1; peptide mass deviation 7 ppm. Leucine and isoleucine were considered as equal for protein grouping. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (21) through the PRIDE partner repository with the data set identifier PXD000175 and are publicly available.
Confidence scores to protein–protein interactions were calculated using the SAINT computational tool, as described previously (17, 20), which uses label-free quantitative data to construct separate distributions for true and false interactions to derive the probability of a bona fide protein–protein interaction. The probability of Cdk2 (p = 0.24), the validated mDia2-binding protein having the poorest confidence score, was taken as cutoff to discriminate between true and false interactions.
Bioinformatic Analyses
Bioinformatic analyses were carried out using Ingenuity Pathways Analysis (IPA) (Ingenuity Systems, Redwood City, CA). The mDia2-binding proteins were submitted for Biological function and Canonical Pathway Analysis to identify the pathways that were most significantly associated with the query among those present in the IPA library. The significance of the association between the data set and the canonical pathway was measured with two parameters: (1) the ratio of the number of genes from the data set that are above the cutoff assigned to a given pathway divided by the total number of proteins belonging to the same canonical pathway, (2) the Fischer's exact test determining the probability (p value) that the association between the genes in the data set and the canonical pathway occurs by chance.
Protein Interaction Maps
Protein–protein interaction maps were built using the STRING protein–protein interaction database (21) and Cytoscape (22). The STRING software assembles functional protein networks based on compiled evidence. We then visualized the obtained interaction maps in Cytoscape with the STRING embedded layout. We used the STRING interaction confidence for the edge thickness, and the SAINT p value for the node size.
Total RNA Isolation and RT-qPCR Analyses
Total RNA from adherent cells was extracted using RNeasy Mini kit (Qiagen GmbH, Hilden, Germany) according to manufacturer's instructions. Complementary DNA synthesis was performed using 1–2 μg of mRNA with SuperScript-II® reverse transcriptase according to the manufacturer's instructions (Invitrogen). Real-time qPCR reactions were set up using 100 ng of cDNA as a template and gene specific primers (200 nm) in a StepOnePlusTM Real-Time PCR system (Applied Biosystems). All reactions produced single amplicons (100–200 bp), which allowed us to equate one threshold cycle difference. RT-qPCR analyses were performed using gene-specific primers and cyclophilin as a normalizing gene. Data are always presented as mean ± S.D. and technical triplicates were performed in all cases. The relative expression levels (Relative mRNA, arbitrary units (a. u.)) in the control cells were taken as a reference for normalization, unless otherwise specified. qPCR primers are listed in the supplemental Experimental Procedures. Note that Delphilin expression is not reported because all tested Delphilin-specific produced multiple amplicons (not shown).
Statistics
Student's paired t test, Fischer's Exact t test, One-way ANOVA (Bonferroni's Multiple Comparison Test) and Two-way ANOVA (Bonferroni's Multiple Comparison Test) were employed. p < 0.05 was considered statistically significant. Brackets mark Bonferroni's Multiple Comparison Test couples. In all cases: * = p < 0.05; ** = p < 0.01; *** = p < 0.001; **** = p < 0.0001.
RESULTS
Affinity Purification Coupled to Quantitative Mass Spectrometry Identifies the mDia2 Interactome
Wild-type mDia2 and its constitutively active M1041-to-A mutant (hereafter referred to as MA) were tagged with a Flag epitope (supplemental Fig. S1A). Transient expression of wild-type mDia2 in 293T cells resulted in a prominent cytosolic and a faint nuclear anti-Flag staining without noticeable alterations in the actin cytoskeleton (supplemental Fig. S1B). Importantly, localization and distribution of overexpressed and endogenous mDia2 were similar (supplemental Fig. S1B). The MA mutant showed a similar pattern but it also triggered the formation of slender actin-rich protrusions and readily accumulated at their distal tips (supplemental Fig. S1C). We observed similar phenotypes also in HeLa, U2OS, and MEF cells (data not shown). Therefore, wild-type and MA mDia2 can serve as models for the closed and open conformation, respectively.
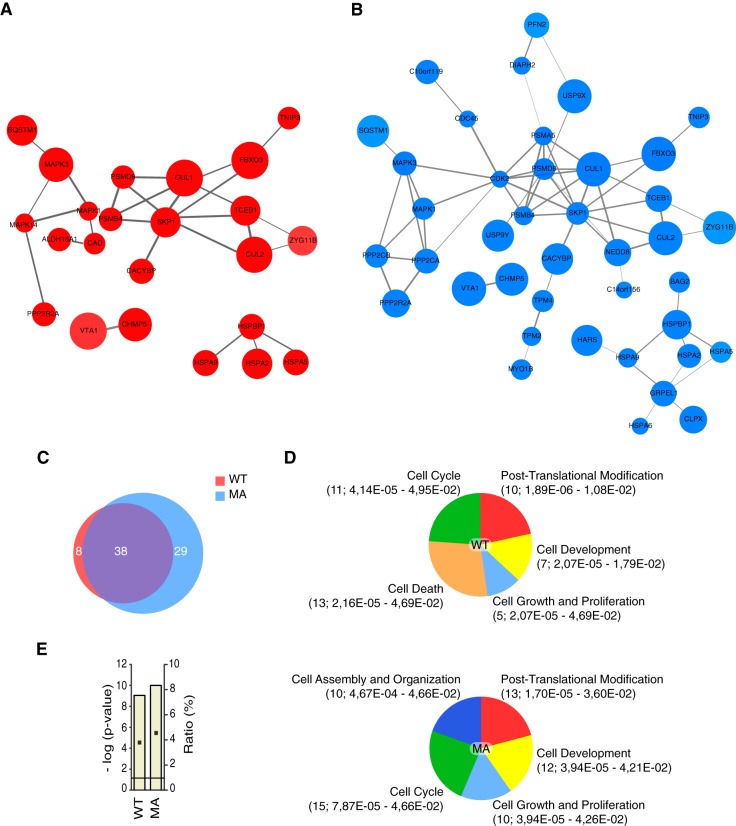
To gain insight into the protein interactions engaged by mDia2, we devised a strategy relying on affinity purification and mass spectrometry (AP-MS). 293T cells were transfected with either wild-type mDia2 or its constitutively active mutant, or with an empty vector as a control. mDia2-based complexes were affinity-purified by using anti-Flag antibodies, separated by one-dimensional SDS-PAGE, and then processed for MS (supplemental Fig. S1D). We analyzed three biological replicas and produced three independent datasets whose label-free quantitative information was exploited by SAINT to assign confidence values to individual protein–protein interactions (18). We ranked the interactions according to the computed confidence values and inventoried 46 and 67 proteins having an average probability higher than 0.24 for wild-type and constitutively active mDia2, respectively (supplemental Table S1). Among them were importin α (IMA8) and Profilin-2, two known mDia2-binding proteins (1, 2, 22). Interaction maps built using the STRING protein–protein interaction database (23) and Cytoscape (24) illustrated the connectivity of both interactomes (Fig. 1A and 1B). We obtained 75 nonredundant mDia2-binding proteins: 38 common to both wild-type and constitutively active mDia2, eight and 29 unique for the former and the latter, respectively (Fig. 1C). Many proteins could be clustered into the cell cycle, the cell growth and proliferation, the cell assembly and organization, and the cell development functional groups accounting for the known biological roles of mDia2 (Fig. 1D). Furthermore, the presence of the cell assembly and organization functional group exclusively in the interactome of the MA confirmed that the open conformation is associated with actin- and microtubule-based processes. Bioinformatics conveyed also new information: (1) the post-translational modification functional group unveiled the potential involvement of mDia2 in Ubiquitin biology, and (2) the presence of the cell death functional group only in the interactome of the wild type suggested a conformation-specific role of mDia2 in apoptosis (Fig. 1D). Noticeable was the enrichment for proteins implicated in ubiquitin biology: E3 ubiquitin ligases (FBXO3-Cullin 1-containing (25), Zhg-11-Cullin 2-containing (26), and CACYBP/SIP-containing (27) cullin-RING ubiquitin ligases), the ubiquitin-like protein and cullin-regulator NEDD8 (28), the de-ubiquitinating enzyme USP9X, several proteasome subunits and proteasome-associating proteins (NipSnap2, p62/SQSTM1 (29)) could be isolated as mDia2-binding proteins (supplemental Table S1). Consistently, the protein ubiquitination pathway was both significantly enriched and highly represented in the mDia2 interactomes (Fig. 1E).
Fig. 1.
Emergent properties of the interactome of wild-type and constitutively active mDia2. A, Interaction map of wild-type mDia2. Proteins interaction maps were obtained as described in the Experimental Procedures. The maps contain only those mDia2-binding proteins reported to entertain interactions in the STRING database according to the specified criteria. B, Interaction map of constitutively active mDia2. Proteins interaction maps were obtained as in A. C, The overlap between the interactome of wild-type and constitutively active mDia2 is 50.7%. Venn diagram showing the number proteins unique to either wild-type (WT; red) or constitutively active (MA; blue) mDia2, as well the number of the common ones (purple). Percentage of overlap was obtained as follows: common proteins (38)/total nonredundant proteins (75) × 100. D, mDia2-binding proteins cluster into common and conformation-specific functional groups. Proteins binding to wild-type (WT) and constitutively active (MA) mDia2 were assigned to pathways regulating biological processes that define a functional group in Ingenuity Pathway Analysis (IPA). Between brackets are: number of proteins specific for the indicated functional group; range of Fischer's Exact t test p values of the biological processes associated with the indicated functional group (supplemental Table S1, Sheet 4). E, mDia2 is linked to the protein ubiquitination pathway. The IPA protein ubiquitination pathway is the top canonical pathway in the interactome of both wild-type (WT) and constitutively active (MA) mDia2. Bar graphs show Fischer's Exact t test p values (as -log (p-value)), and ratios (% of the clustered proteins with respect to the total number of proteins belonging to the IPA protein ubiquitination pathway).
Validation of Candidate mDia2-interacting Proteins
We validated the mDia2 interactome assessing the binding of selected candidates to mDia2 through pull-down and co-immunoprecipitation experiments. The selection process aimed at covering as many functional groups as possible and relied on the availability of appropriate antibodies and expression vectors.
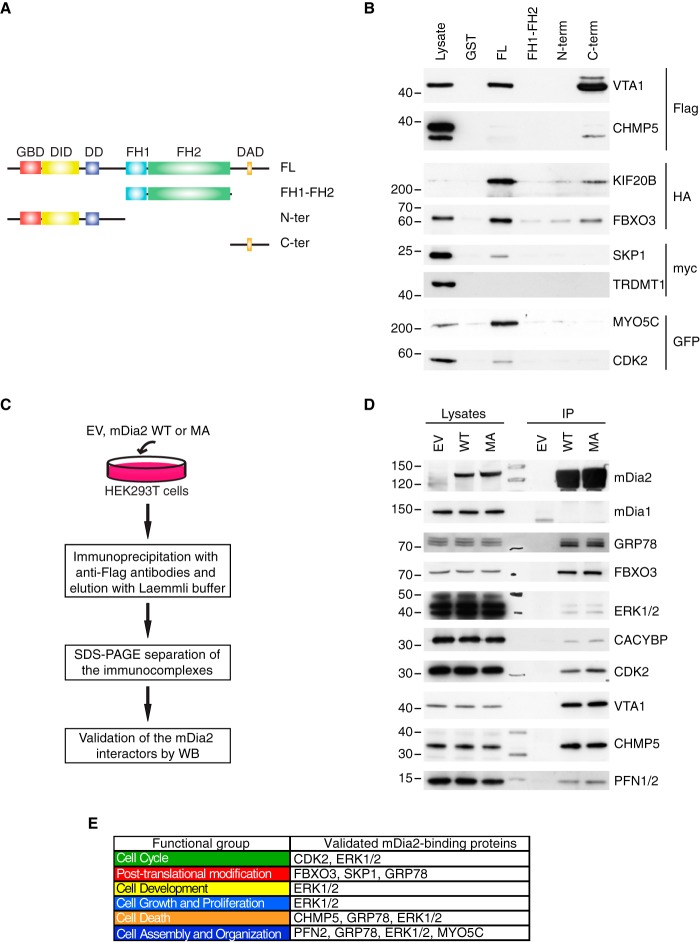
Immobilized full-length GST-mDia2 and deletion mutant thereof (7), or GST as a negative control, served as baits to fish out an epitope-tagged version of the candidate under scrutiny in all pull-down experiments (Fig. 2A). CHMP5 and VTA-1 are subunits of the ESCRT-III complex, which regulates membrane budding, endosomal sorting, and multivesicular body biogenesis, receptor signaling, cytokinesis, autophagy, cell polarity and migration, and ribonucleic acid biology (30). CHMP5 and VTA-1 bound to mDia2, the former less efficiently than the latter (Fig. 2B). The ESCRT-III complex may contribute to the role of mDia2 in both cell motility and cytokinesis. Interestingly, microtubule plus-end-directed kinesin KIF20B also acts during cytokinesis (31) and turned out to be a genuine partner of mDia2. Tropomyosin-2, Tropomyosin-4, and Myosin-Vc are three actin-regulatory proteins belonging to the cell assembly and organization functional group unique for constitutively active mDia2. The interplay between mDia2 and Tropomyosins in stress fiber formation (32) raised our interest in testing Myosin-Vc, a nonconventional myosin involved in exocytosis (33). The interaction between Myosin-Vc (MYO5C) and mDia2 (Fig. 2B) suggest that they may cooperate in regulating actin dynamics in vesicle trafficking. The cyclin-dependent kinase CDK2 controlling cell cycle progression and centrosome duplication (34) specifically bound to full-length mDia2 (Fig. 2B) and may link the microtubule-stabilizing action of mDia2 to a microtubule-dependent process. The SKP1-Cullin 1-F-box-complex subunits SKP1 and FBXO3 (35) interacted with mDia2, providing a first-line validation for the link between mDia2 and the UPS. Specificity of the above interactions was further corroborated by the fact that none of the employed baits pulled down TRDMT1, a control nonrelated protein (Fig. 2B). The seven hits that we confirmed to be genuine mDia2-binding proteins interact with either full-length mDia2 only or both the full length and specific mDia2-deletion mutants, thereby suggesting the existence of multiple binding surfaces (Fig. 2B).
Fig. 2.
Validation of selected mDia2-interacting proteins. A, Schematic of the mDia2-deletion mutants. GST-mDia2 proteins used in the pull-down assays: full-length mDia2 = FL; Formin homology domain 1 and 2 (aa: 530–1033) = FH1-FH2; N terminus (aa: 1–530) = N-ter; C terminus (aa: 1033–1172) = C-ter. B, Validations by pull-down. Lysates obtained from 293T cells transfected with an epitope-tagged version of the candidate of interest (on the right) were incubated with immobilized GST-tagged, full-length mDia2, the mDia2-deletion mutants depicted in A, or GST as a negative control (on the top). Lysates (2%) and affinity-precipitated material were probed as indicated. C, Overview of the co-immunoprecipitation approach. The flowchart is as in supplemental Fig. S1D with the exception of the two following changes: (1) immunoprecipitation (IP) was carried out starting from 1 mg of total cell lysate, (2) Western blots were performed. D, Validations by co-immunoprecipitation. Endogenous proteins in the lysates (2%) and in the bound material were detected with specific antibodies (on the right). The expression of mDia2 was confirmed using anti-Flag antibodies. E, Validated functional groups and proteins. Functional groups are color-coded as in Fig. 1D. Proteins are listed according to their respective functional group. The absence of some validated proteins is because of the IPA annotation.
To further the validation process, we looked at the coprecipitation of endogenous candidates with Flag-tagged mDia2 using the pipeline described in Fig. 2C. We found eight proteins forming a complex with mDia2, whose specificity was corroborated by mDia1 being both absent in our lists of mDia2-binding proteins and unable to interact with mDia2 (Fig. 2D). FBXO3, CHMP5, VTA-1, and CDK2, for which both plasmids and antibodies could be collected, proved the reliability of our MS datasets by means of both pull-down and co-immunoprecipitation. Additionally, both types of validations showed that proteins involved in Ubiquitin metabolism (FBXO3, SKP1 and CACYBP) bind to mDia2. We noted that overexpressed CHMP5 bound poorly to mDia2 in the pull downs whereas endogenous CHMP5 clearly coprecipitated with mDia2 (Fig. 2B and 2D). Interestingly, CHMP5 forms a complex with VTA-1 (36) and VTA-1 associates with mDia2 robustly (Fig. 2B and 2D). Thus, it is likely that the interaction between mDia2 and CHMP5 is mediated by VTA-1, which might become limiting upon CHMP5 overexpression. Most importantly, the validated proteins have biological roles overlapping with mDia2, are intertwined with protein networks accounting for all mDia2's attributed functions, and also substantiate the existence of a link between mDia2 and the UPS (Fig. 2E). Although a previous study suggested that mDia2 is ubiquitinated and degraded through a proteasome-dependent pathway (37), mDia2 levels did not significantly change upon inhibition of the proteasome in either 293T or HeLa cells (supplemental Fig. S2A and not shown, respectively). Thus, the emergent link between mDia2 and the UPS is likely to fulfil functions other than the mere regulation of mDia2 protein levels.
mDia2 Forms a Complex with FBXO3 and p53
We explored the link between mDia2 and the UPS by focusing on FBXO3, the top hit among all identified mDia2-binding proteins (supplemental Table S1). FBXO3 is an F-box-family protein hallmarked by a short F-box motif interacting with SKP1 (35). F-box proteins are subunits of a class of E3 ubiquitin protein ligases called SKP1-Cullin 1-F-box (SCF) complex, act as adaptors dictating the ubiquitination of specific substrates and confer the name on the SCF complex they belong (38).
In Fig. 2D, we showed that endogenous FBXO3 co-precipitated equally with both wild-type and constitutively active mDia2. We expressed HA-tagged FBXO3 at low levels to circumvent the lack of antibodies immunoprecipitating endogenous FBXO3 and surrogate the reciprocal experiment (supplemental Fig. S2B). The cotransfection of FBXO3 with either wild-type or MA mDia2, or a control empty vector, allowed us to confirm that the FBXO3-mDia2 interaction is independent of mDia2's conformation (supplemental Fig. S2C). Thus, we used wild-type mDia2 in all subsequent studies.
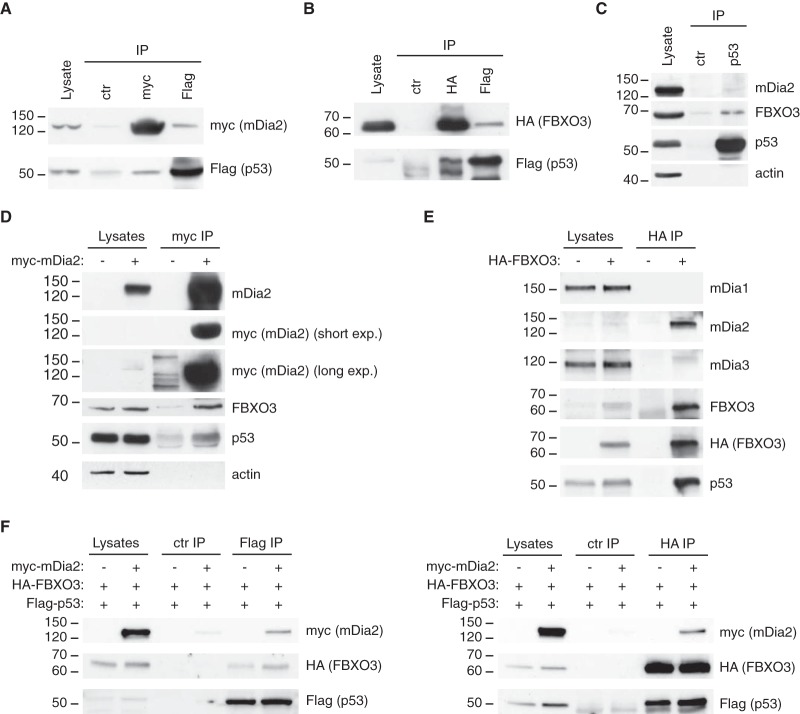
The fact that FBXO3 regulates p53 activity through a mechanism that does not involve inhibition of p300 and HIPK2 ubiquitination (25) raised the possibility that mDia2 binds to FBXO3 and p53. As all available antibodies did not allow us to selectively immunoprecipitate either endogenous mDia2 or endogenous FBXO3, we tested this hypothesis as explained below. First, we demonstrated that myc-tagged mDia2 and HA-tagged FBXO3 coprecipitate with Flag-tagged p53 and vice versa, mDia2 less robustly than FBXO3 (Fig. 3A and 3B, respectively). In keeping with this, endogenous FBXO3 coprecipitated with endogenous p53 more efficiently than endogenous mDia2 and the absence of actin from the p53-based immunocomplexes demonstrated the specificity of these interactions (Fig. 3C). Second, we proved that endogenous FBXO3 and p53 bound to myc-tagged mDia2 (Fig. 3D). Third, we could also show that endogenous mDia2 and p53, but not mDia1 or mDia3, associated with mildly overexpressed HA-tagged FBXO3 (Fig. 3E). Fourth and to give insight into the topology of these interactions, we co-expressed Flag-tagged p53 and HA-tagged FBXO3 at levels comparable to and far below the endogenous counterparts, respectively, either alone or with heavily overexpressed myc-tagged mDia2 (supplemental Fig. S2D). Because the association between p53 and FBXO3 was not outcompeted by mDia2, which instead could be specifically detected in both anti-Flag and anti-HA immunocomplexes (Fig. 3F left and right, respectively), it is likely that FBXO3 holds mDia2 and p53 in a complex.
Fig. 3.
mDia2 forms a complex with FBXO3 and p53. A, mDia2 and p53 coprecipitate. 293T cells were transfected with Flag-tagged p53 (1.5 μg) and myc-tagged mDia2 (8.5 μg). Total cell lysates (1 mg) were immunoprecipitated with anti-myc, anti-Flag and control (ctr) antibodies as described in the Experimental Procedures. Lysate (2%) and immunocomplexes (IP) were separated by SDS-PAGE and immunoblotted as indicated. B, FBXO3 and p53 coprecipitate. 293T cells were transfected with Flag-tagged p53 (1.5 μg) and HA-tagged FBXO3 (8.5 μg). Immunoprecipitations with anti-HA, anti-Flag and control (ctr) antibodies was performed as described in the Experimental Procedures. FBXO3 and p53 were detected as indicated. The lower bands in the IP lanes of the anti-p53 blot represent cross-reaction with the IgGs. C, Endogenous p53, FBXO3 and mDia2 form a complex. Total cell lysates (2 mg) were immunoprecipitated with anti-p53 and control (ctr) antibodies. Lysate (2%) and immunocomplexes (IP) were separated by SDS-PAGE and immunoblotted as indicated. D, Endogenous FBXO3 and p53 coprecipitate with myc-mDia2. 293T cells were transfected with either myc-tagged mDia2 (+) or the corresponding empty vector (−). Immunoprecipitation with anti-myc antibodies was performed as in A. Lysate (2%) and immunocomplexes (IP) were separated by SDS-PAGE and immunoblotted as indicated. Anti-myc and anti-mDia2 antibodies detected overexpressed and total mDia2, respectively. Selected short and long exposures of the anti-myc blot are illustrated (short exp. and long exp., respectively). In the long exposure, anti-myc cross-reacting bands are visible in the control IP lane. E, Endogenous mDia2 and p53 coprecipitate with HA-FBXO3. 293T cells were transfected with either HA-tagged FBXO3 (+) or the corresponding empty vector (−). Immunoprecipitation with anti-HA antibodies was performed as in B. Lysate (2%) and immunocomplexes (IP) were separated by SDS-PAGE and immunoblotted as indicated. Anti-HA and anti-FBXO3 antibodies detected overexpressed and total FBXO3, respectively. In the anti-mDia3 blot, some residual mDia2 signal is visible as a faint (and shifted) band in the IP lane. F, mDia2, FBXO3 and p53 form a complex. 293T cells were transfected with Flag-tagged p53 (1,5 μg) and HA-tagged FBXO3 (8,5 μg), with either myc-tagged mDia2 (10 μg) (+) or empty vector (−). Immunoprecipitations were performed as in A and B. mDia2, FBXO3 and p53 were detected as indicated. A–F: one of two experiments that were performed with similar results is shown.
mDia2 Regulates p53 Transcriptional Activity Independently of Its Actin Nucleation Abilities and Conformation Through FBXO3
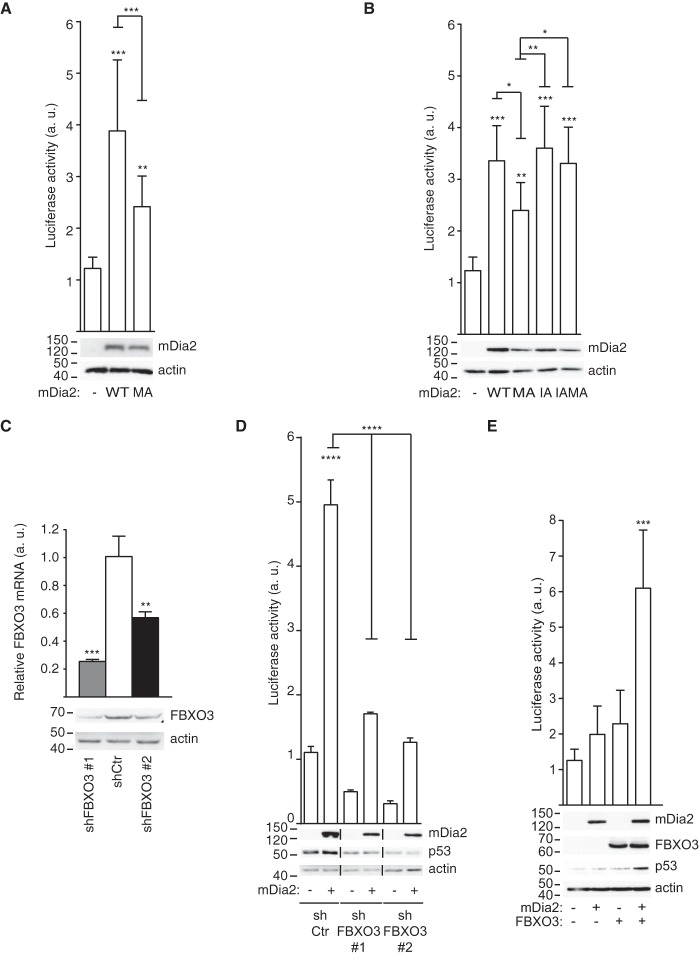
The interaction of mDia2 with FBXO3 and p53 prompted us to assess the effects of mDia2 on p53-dependent gene transcription. Wild-type and constitutively active mDia2, or an empty vector as a control, were transfected in 293T cells along with a plasmid having a minimal p53-responsive promoter located upstream of the luciferase gene (39). Strikingly, wild-type mDia2 did not only enhance the transcriptional activity of p53 but it also displayed a more prominent stimulatory action than the MA mutant (Fig. 4A). A point mutation abrogating actin nucleation (I704-to-A, hereafter referred to as IA) (3) made constitutively active mDia2 indistinguishable from the wild type (IAMA versus WT, Fig. 4B). In keeping with wild-type mDia2 attaining the closed conformation, the IA mutant activated p53 as efficiently as the wild type (IA versus WT, Fig. 4B). The notion that mDia2 plays a scaffolding role is further corroborated by the C-terminal region being required and sufficient for mDia2 to increase the transcriptional activity of p53 (supplemental Fig. S3A). The C-terminal region of mDia2 activated p53 also upon introduction of the MA mutation (supplemental Fig. S3A), thus ruling out that binding to endogenous mDia2 could account for these observations. Additionally, the inability of wild-type mDia1 to affect p53 activity (supplemental Fig. S3B) ruled out that the effects of mDia2 are caused by the sequestration of Rho GTPases or general Drf-binding proteins.
Fig. 4.
FBXO3 is required for mDia2 to stimulate p53-mediated gene transcription in a conformation- and actin-nucleation-independent manner. A, Wild-type mDia2 increases the transcriptional activity of p53 more efficiently than its MA mutant. 293T cells were cotransfected with the reporter plasmid and the empty vector (−), Flag-tagged mDia2 WT (WT) or mDia2 MA (MA). Luciferase activity (arbitrary units, a.u.) was measured and plotted as described in the Experimental Procedures. Data represent mean ± S.D. (n = 12; One-way ANOVA). mDia2 expression was confirmed using anti-Flag antibodies and actin served as a loading control. B, Actin-nucleation-deficient mDia2 activates p53 as efficiently as wild-type mDia2. Cells were transfected and luciferase activity measured and plotted as in A (n = 9; One-way ANOVA). I704 was replaced by A in both wild-type and MA mDia2 to generate IA and IAMA, respectively. C, Generation of control and FBXO3 knockdown cells. 293T cells were infected with either control (shCtr) or FBXO3-targeting (shFBXO3) viruses. Total RNA and lysates were obtained as indicated in the Experimental Procedures. Bar graph: RT-qPCR shows the relative FBXO3 levels (Relative FBXO3 mRNA, arbitrary units (a. u.)) (n = 3; Repeated t Test). Blots: Total cell lysates (30 μg) were immunoblotted as indicated with actin providing a loading control. FBXO3 is reduced by 51.2 ± 3% (shFBXO3 #1) after normalization against actin. One of three experiments that were performed with similar results is shown. D, mDia2-mediated activation of p53 requires FBXO3. Control (shCtr) and FBXO3 (shFBXO3) knockdown cells were cotransfected with the reporter plasmid along with the empty vector (−) or Flag-tagged wild-type mDia2 (+) and luciferase activity measured and plotted as in A (n = 9; One-way ANOVA). E, mDia2 and FBXO3 jointly enhance p53 expression and activity. U2OS cells were cotransfected with the reporter plasmid, Flag-tagged wild-type mDia2 (+) or the empty vector (−), and either HA-tagged FBXO3 (+) or its corresponding empty vector (−). Luciferase activity was measured and plotted as in A (n = 15; One-way ANOVA). p53, mDia2 and FBXO3 expression was confirmed using anti-p53, anti-Flag and anti-HA antibodies, respectively.
These results show that auto-inhibited mDia2, so far regarded as biologically inert, has an unexpected actin-independent role in the control of p53′s transcriptional activity. This new p53 regulatory pathway does not sense the conformation of mDia2, but it is rather negatively affected by the actin-nucleation activity of mDia2. Of note, FBXO3 plays a crucial role in this pathway because mDia2 failed to affect p53 transcriptional activity in FBXO3 knockdown cells (Fig. 4C, 4D). As p53 activation by mDia2 was abrogated in two FBXO3 knockdown cells with different FBXO3 residual levels, a threshold FBXO3 expression may be required for this p53 regulatory pathway to be functional.
We verified the generality of the above observations in U2OS cells, which express both mDia2 and FBXO3 at low levels (supplemental Fig. S4A) and have an intact p53 regulatory circuitry (40). In good agreement with the above findings, (1) the overexpression of either mDia2 or FBXO3 alone failed to activate p53 in U2OS cells (Fig. 4E), (2) mDia2 readily stimulated p53 transcriptional activity when co-expressed with FBXO3, as measured with a reporter based on either the HDM2 promoter (Fig. 4E) or the p21/WAF1 promoter (data not shown).
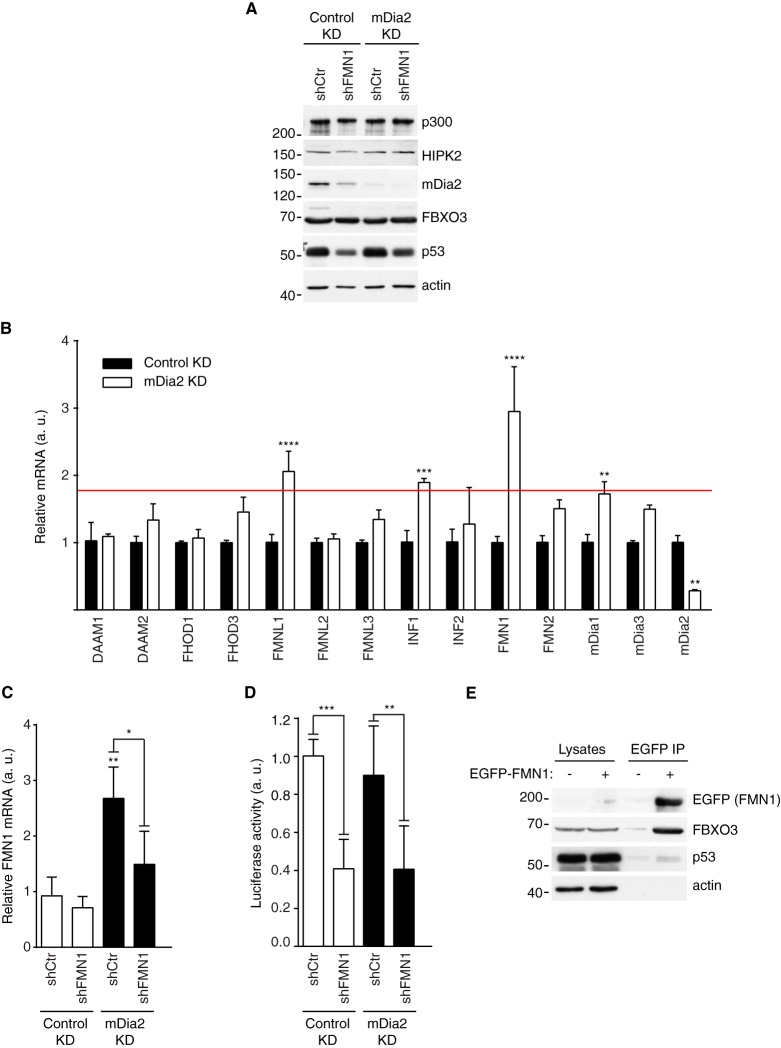
mDia2 and Formin 1 Cooperate in Regulating p53
In order to measure the activity of p53 in cells devoid of mDia2, we took advantage of shRNA and lentiviral infection because all tested siRNAs resulted in partial (less than 50%) and very transient down-regulation of mDia2 (supplemental Fig. S3C and unpublished observations). We obtained mDia2 knockdown cells having mDia2 protein levels reduced by about 75% compared with the control knockdown population (Fig. 5A). Silencing of mDia2 did not alter either the expression of FBXO3, HIPK2, p300, and p53 (Fig. 5A). The activity of p53 in 293T cells did not change upon mDia2 knockdown (not shown and Fig. 5D), thus suggesting that other Formins might cooperate with mDia2 in regulating p53.
Fig. 5.
mDia2 and FMN1 cooperate in the regulation of p53. A, Generation of control, mDia2, FMN1 and mDia2-FMN1 knockdown cells. 293T cells were infected with either control (Control KD) or mDia2-targeting (mDia2 KD) viruses. These cells were further infected with either control (shCtr) or FMN1-targeting (shFMN1) viruses. Total cell lysates were immunoblotted as indicated. One of two similar experiments is shown. B, Formin-expression landscape in control (Control KD) and mDia2 knockdown (mDia2 KD) cells. RT-qPCR analyses were performed using the indicated Formin-specific primers to determine relative expression levels (Relative mRNA, arbitrary units (a. u.)) (n = 6; Two-way ANOVA). Cut-off red line is based on the fact that mDia1 silencing in mDia2 KD cells did not affect p53 activity (not shown). C, FMN1 silencing in mDia2 KD cells restores normal FMN1 expression. Cells were generated and total RNA obtained as in A and Fig. 4C, respectively. RT-qPCR shows the relative FMN1 levels (Relative FMN1 mRNA, arbitrary units (a. u.)) (n = 6; One-way ANOVA). FMN1 down-regulation in the Control KD cells is not evident because of its very low basal expression. D, mDia2 and FMN1 cooperate in regulating p53 activity. Cells were plated, transfected and the luciferase activity measured and plotted as in B (n = 6; One-way ANOVA). Similar results were obtained with a second FMN1-specific hairpin (not shown). E, Endogenous FBXO3 and p53 coprecipitate with EGFP-FMN1. 293T cells were transfected with either EGFP-tagged FMN1 (+) or the corresponding empty vector (−). Immunoprecipitation with GFP-trap beads was performed as described in the Experimental Procedures. Lysate (2%) and immunocomplexes (IP) were separated by SDS-PAGE and immunoblotted as indicated.
We then measured and compared the expression of human Formins in control and mDia2 knockdown 293T cells. These experiments indicated that the messengers of Inverted Formin 1 (INF1), Formin-like 1 (FMNL1), and Formin 1 (FMN1) showed the highest increase in the absence of mDia2 (Fig. 5B), suggesting that these Formins might compensate mDia2 down-regulation. Hence, we knocked down INF1, FMNL1, and FMN1, both individually and in combination with mDia2, verified their silencing by RT-qPCR (Fig. 5C and data not shown) and assessed the transcriptional activity of p53. Although INF1 and FMNL1 turned out not to be involved in p53 regulation (data not shown), silencing of FMN1 significantly reduced p53-transcriptional activity in both control and mDia2 knockdown 293T cells (Fig. 5D). The fact that FMN1 down-regulation decreased the expression of mDia2 at the post-transcriptional level (Fig. 5A and data not shown, respectively) explains the effects on p53. Consistent with FMN1 being able to replace mDia2, EGFP-tagged FMN1 bound endogenous FBXO3 and p53, the former more efficiently than the latter (Fig. 5E). Interestingly, FMN1-paralog FMN2 was the most up-regulated Formin upon knockdown of mDia2 in U2OS cells (supplemental Fig. S4B). Thus, it is tempting to speculate that mDia2 and FMN proteins share the p53-regulatory function.
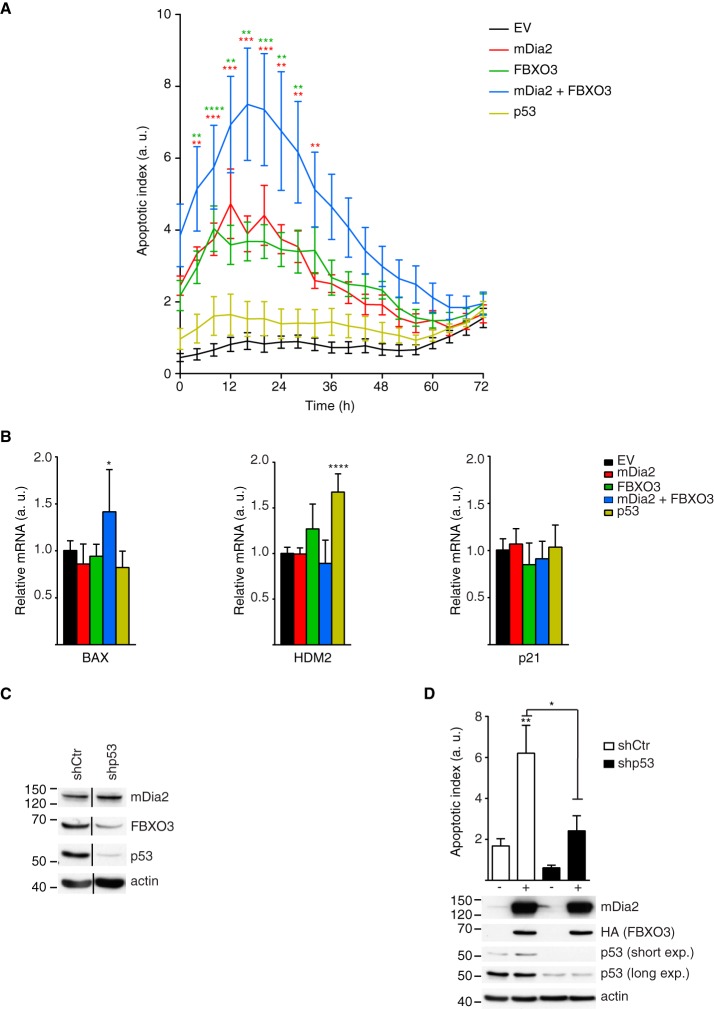
mDia2 and FBXO3 Regulate p53-dependent Apoptosis
p53 is a tumor suppressor with a crucial function in the cellular apoptotic programs (41) and the interactome of wild-type mDia2 associates with the cell death functional group (Fig. 1B). Therefore, we monitored apoptosis in U2OS cells transfected with mDia2 and FBXO3, either alone or in combination, and compared it to the control ones. These experiments revealed that the activity of Caspase-3/7 was significantly increased in the cells expressing either mDia2 or FBXO3 compared with the control ones (Fig. 6A). Most importantly, the highest apoptotic index was measured in cells co-expressing mDia2 and FBXO3 (Fig. 6A). As elevated p53 levels were insufficient to trigger apoptosis in U2OS cells (Fig. 6A), there appears that mDia2 and FBXO3 jointly promote p53 activation. Consistent with this notion, (1) mDia2 and FBXO3 were present in the nuclear compartment both in control and etoposide-treated cells (supplemental Fig. S5), (2) knockdown of FBXO3, FMN1, and FMN1-mDia2 resulted in decreased p53 levels (Fig. 4D and Fig. 5A), whereas (3) co-expression of mDia2 and FBXO3 increased them (Fig. 4E).
Fig. 6.
Increased mDia2 and FBXO3 levels sensitize cells to p53-dependent apoptosis. A, Increased mDia2 and FBXO3 levels sensitize cells to apoptosis. U2OS cells were transfected with Flag-tagged wild-type mDia2 (mDia2) and HA-tagged FBXO3 (FBXO3), either alone or in combination, a control empty vector (EV), or wild-type Flag-tagged p53 (p53). Apoptotic index (a. u. = arbitrary units) versus time (h = hours) was obtained by measuring Caspase-3/7 activity as described in the supplemental Experimental Procedures. Data represent mean ± S.D. (n = 7; Two-way ANOVA), color-coded asterisks indicate statistical significance between the cells co-expressing mDia2 and FBXO3 and those expressing either mDia2 (red) or FBXO3 alone (green). All but p53-overexpressing cells exhibit a significantly increased apoptotic index compared with EV. B, Increased mDia2 and FBXO3 levels upregulate the expression of Bax. U2OS were transfected as in A and, 48 h later, total RNA was isolated. RT-qPCR analyses were performed using the indicated gene-specific primers. In each set, the relative mRNA levels of the EV served as a reference (n = 6; One-way ANOVA). C, Generation of control and p53 knockdown cells. U2OS cells were infected with either control (shCtr) or p53-targeting (shp53) lentiviruses to obtain stable populations. Total cell lysates were immunoblotted as indicated. Black lines mark removal of intervening lanes. One of two similar experiments is shown. D, mDia2 and FBXO3-induced apoptosis involves p53. Control and p53-knockdown U2OS cells were transfected with empty vector (−) or Flag-tagged wild-type mDia2 (mDia2) and HA-tagged FBXO3 (FBXO3) (+). The Apoptotic index was measured at the same time points corresponding to the peak in A. Data represent mean ± S.E.M. (n = 6; One-way ANOVA). The expression of mDia2 and FBXO3 was confirmed using anti-mDia2 and anti-HA antibodies, respectively. p53 levels were assessed with both short and long exposures (short exp. and long exp., respectively) and actin served as loading control.
We then profiled the expression of key endogenous p53-target genes under the same conditions that sensitized U2OS cells to apoptosis. In agreement with the activity of Caspase 3/7, we measured significantly higher messenger levels of the pro-apoptotic p53-target Bax only in the cells co-expressing mDia2 and FBXO3 (Fig. 6B). Conversely and at variance with the luciferase assays, neither the HDM2 nor the p21 messengers were significantly altered in these cells compared with the control ones (Fig. 6B). These discrepancies might be because of the percentage of transfected cells (20 ± 3%) being insufficient to measure a statistically significant effect in the total population. Anyway, these observations suggest that mDia2 and FBXO3 exert a joint regulatory effect on p53 that impact primarily on the p53-dependent pro-apoptotic pathway.
Indeed, the knockdown of p53 strongly attenuated the activation of Caspase 3/7 induced by the co-expression of mDia2 and FBXO3 in U2OS cells (Fig. 6C, 6D). The sum of these data, suggests that mDia2 and FBXO3 regulate the p53-dependent apoptotic program.
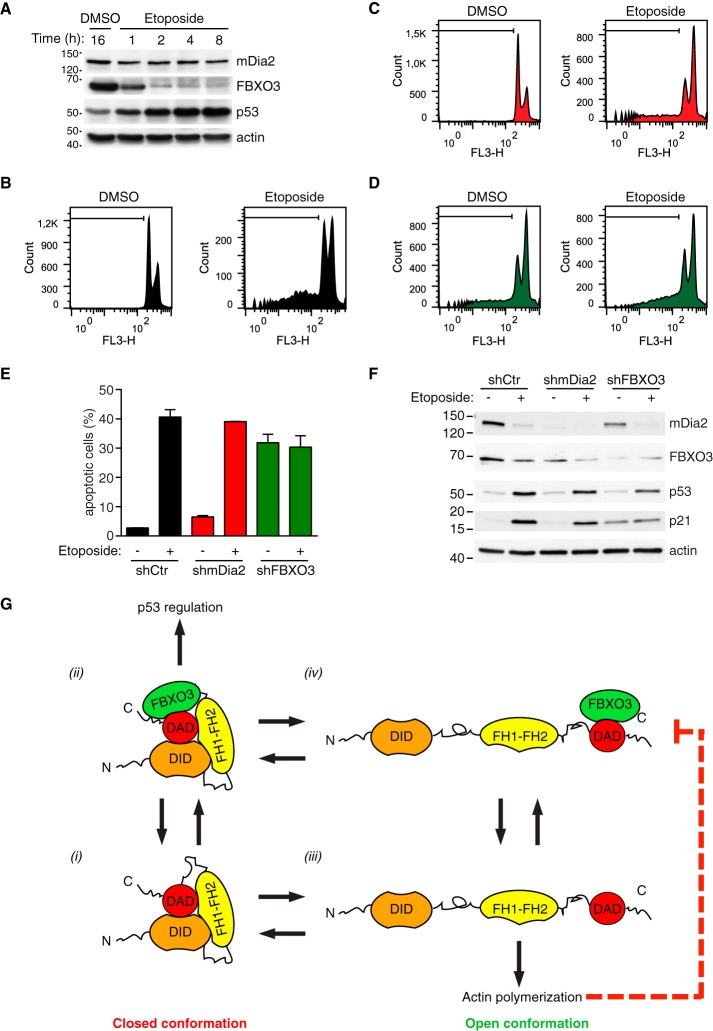
To further evaluate the role of mDia2 and FBXO3 in apoptotic cell death, U2OS were treated with etoposide, a topoisomerare II inhibitor that arrests cells in G2/M and induces apoptosis (42, 43). Time-course experiments indicated that addition of etoposide rapidly increased p53 expression before reducing FBXO3 expression (Fig. 7A). The correlation between down-regulation of mDia2 and FBXO3 and DNA fragmentation at both 24 and 48 h (not shown and Fig. 7B–7D, respectively) suggests that these proteins are no longer required when late-stage, irreversible apoptotic events have taken place. To assess the centrality of FBXO3 in this new pro-apoptotic pathway, we generated control, mDia2 knockdown and FBXO3 knockdown U2OS cells. As expected, etoposide caused control knockdown cells to accumulate in G2/M and increased the sub-G1 population (Fig. 7B and 7E, respectively). mDia2 knockdown cells responded as the control ones (Fig. 7C, 7E, and 7F), further suggesting that compensation by FMN proteins is at play (supplemental Fig. S4C). FBXO3 silencing resulted in a G2/M arrest, apoptosis and increased p21 expression without altering p53 levels (Fig. 7D–7F). However, etoposide did not further increase either apoptosis or p21 levels in FBXO3 knockdown cells (Fig. 7D–7F). These observations confirm that FBXO3 has a crucial role in the Formin-dependent pathway controlling the p53 apoptotic program and suggest that FBXO3 might affect apoptosis also independently of p53.
Fig. 7.
FBXO3 knockdown attenuates p53-mediated apoptosis upon DNA damage. A, Up-regulation of p53 is an early effect of etoposide. U2OS cells were treated with etoposide (20 μm) or DMSO and total cell lysates prepared at the indicated time (h = hour) and immunoblotted as indicated. One of two similar experiments is shown. B–D, DNA content in control, mDia2 knockdown and FBXO3-knockdown cells is affected by etoposide treatment. U2OS cells were infected with control (shCtr), mDia2-targeting (shmDia2) and FBXO3-targeting (shFBXO3) viruses. Cells were treated with etoposide (20 μm) or DMSO for 48 h. After fixation, cells were stained with propidium iodide and their DNA content (FL3-H, log scale) was analyzed by flow cytometry as indicated in the supplemental Experimental Procedures. In each histogram, a black line demarks the sub-G1 cell population. E, Knockdown of FBXO3 attenuates etoposide-induced apoptosis. Percentage of apoptotic cells from B–D are presented as mean ± S.D. F, p53 levels increase after DNA damage. Cells generated as in B–D were treated with either etoposide (+) or DMSO (−) for 24 h. Total lysates were separated by SDS-PAGE and immunoblotted as indicated. G, Proposed FBXO3-based mechanism for functional specification of mDia2's activity. Auto-inhibited mDia2 (closed conformation) can exist in an FBXO3-free (1) or an FBXO3-bound (2) state. In its FBXO3-bound state (2), mDia2 efficiently contributes to p53 regulation. Transition from the closed to the open conformation ((1) to (3) and (2) to (4)) unleashes mDia2's actin polymerization activity, which inhibits the activity of the mDia2-FBXO3 complex (4) on p53 (dashed red line).
DISCUSSION
We have capitalized on the identification of the interactomes of wild-type and constitutively active mDia2 to describe the widest landscape of mDia2's functions and disclose new principles regulating mDia2's activity.
Quantitative proteomics and bioinformatics have been instrumental for inferring four emergent properties of the mDia2 interactomes in an unbiased fashion.
First, the open conformation of mDia2 engages more protein-protein interactions than the closed one (Fig. 1C). Two nonmutually exclusive explanations account for these results: The open conformation might expose surfaces hidden in the closed, auto-inhibited state or simply employ the same binding surfaces, but with a decreased equilibrium dissociation constant for its partners.
Second, the overlap between the interactomes of wild-type and constitutively active mDia2 is 50.7%. Although we cannot formally rule out that a minor fraction of the wild type attains the open conformation, two observations argue against this possibility: (1) the overexpression of wild-type mDia2 fails to massively induce filopodia in a number of cell types (supplemental Fig. S1B and data not shown), and (2) wild-type mDia2 and its actin-polymerization-deficient mutant activate p53 in a similar manner (Fig. 4B). Thus, a conspicuous fraction of the identified interactions likely occurs independently of mDia2's conformation.
Third, mDia2 is embedded in protein networks accounting for its attributed functions. The mDia2-binding proteins could be clustered into six functional groups, four common to both the wild type and the MA and most likely regulating processes whose execution is not merely dictated by mDia2's conformation. However, it is worth noting that the cell assembly and organization functional group associated with actin- and microtubule-based processes and the cell death functional group characterize the interactome of constitutively active and wild-type mDia2, respectively. Finally, the remarkable absence of Cdc42 and Rho proteins from the mDia2 interactomes is likely because of Rho-GTPases having a relatively low affinity for mDia2, a low molecular weight and expression level, three features that reduce the number of tryptic peptides available for identification by MS. The minimal overlap between the mDia2 interactomes identified herein and the proteins binding to the FH2 domain of mDia2 ((16), supplemental Table S1 and supplemental Table S2) probably results from inherently different conformation, dimerization abilities and localization attained by full-length mDia2 and its isolated FH2 domain.
Fourth, a link between mDia2 and the UPS comes to light, as reported by the striking enrichment for proteins belonging to the protein ubiquitination pathway in the mDia2 interactomes. Importantly, the last two emergent properties highlight the descriptive and predictive power of our analyses.
A robust validation process and multidisciplinary functional analyses outlined new important concepts in the Drf field. The validations showed that the C-terminal region of mDia2 is a potential protein interaction hub and not only a surface implicated in auto-inhibition. Interestingly, microtubules bind to C-terminal region of mDia2 thereby inhibiting the actin nucleation activity (44). Hence, the proteins interacting with the C terminus of mDia2 may regulate both the conformation and the switch between actin- and microtubule-dependent functions of mDia2.
Molecular biology and biochemical analyses disclosed a new mode of action for mDia2. In the current view of the Formin activity cycle, the conversion from the auto-inhibited to the open conformation and the actin nucleation activity are prerequisites for Drfs' action (2). Yet, our findings depict a different mode of mDia2 regulation: mDia2 forms a complex with both FBXO3 and p53 and activates p53-dependent gene transcription in an actin-independent and conformation-insensitive manner. Consistently, we observed mDia2 and FBXO3 to be present in the nuclear compartment together with p53, both at the steady state and upon etoposide addition (supplemental Fig. S5). Remarkably, the actin-nucleation abilities of mDia2 oppose to p53 stimulation. As FBXO3 binds to the C terminus of mDia2, the lack of competition between FBXO3 and the DID for the C terminus of mDia2 would suffice to enable p53 regulation by mDia2 while preserving closed conformation (Fig. 7F). In keeping with this, (1) cells co-expressing FBXO3 and wild-type mDia2 did not show filopodia (data not shown) and (2) FBXO3 bound equally well to both wild-type and constitutively active mDia2 (Fig. 2 and supplemental Fig. S2B).
Assigning a role to auto-inhibited mDia2 challenges the dogma of its biological inactivity, calls for an update of the Formin activity cycle to incorporate actin- and conformation-independent functions, and raises the possibility that other auto-inhibited Formins possess neglected roles.
Our findings also suggest for the first time how mDia2's action is functionally specified (Fig. 7G). We observed that the mDia2-dependent regulation of p53 activity and expression requires FBXO3, whereas filopodium formation does not because an mDia2-deletion mutant lacking the C terminus induced filopodia as efficiently as the MA mutant (Fig. 4 and data not shown). Therefore, it is very likely that mDia2 achieves functional specificity upon interaction with key binding proteins.
It is tempting to speculate that the ability of mDia2 to regulate both the transcriptional activity and the expression of p53 contributes to its tumor-suppressor and antimetastatic roles (10, 45). In line with this idea, increasing the expression of mDia2 and FBXO3 was sufficient to sensitize U2OS cells to p53-dependent apoptosis. Whereas our results establish that FBXO3 plays an essential role in this process, mDia2 and FMN1 seem to cooperate in regulating p53. Our data also suggest that this function may extend to the other FMN subfamily member FMN2, which regulates p21 protein levels and cell-cycle arrest (46). Whatever the case, the mechanistic details of this new p53 regulatory pathway warrant future investigation.
In summary, the mDia2-interactomes and protein interaction maps identified herein promise to contain the master regulators of mDia2 and pave the way for a mechanistic dissection of the mDia2-dependent processes.
Supplementary Material
Acknowledgments
We thank all the people who generously provided us with reagents, K. de Lint and K. Jastrzebski for technical assistance, E. Argenzio and A. Sonnenberg for critically reading the manuscript.
Footnotes
Author contributions: T.I. and M.I. designed research; T.I., R.vd.K., S.S.G., A.A., and M.I. performed research; A.J.H. contributed new reagents or analytic tools; T.I., R.vd.K., S.S.G., A.A., and M.I. analyzed data; M.I. wrote the paper.
* This work was supported by a grant from the Cancer Genomics Centre (CGC 2009-2013) to M.I. A.F.M.A. was supported by the Netherlands Organization for Scientific Research (NWO) with a VIDI grant (723.012.102). This work is part of the project Proteins At Work, financed by the Netherlands Organisation for Scientific Research (NWO) as part of the National Roadmap Large-scale Research Facilities of the Netherlands (project number 184.032.201).
 This article contains supplemental Experimental Procedures, Figs. S1 to S5, and Tables S1 and S2.
This article contains supplemental Experimental Procedures, Figs. S1 to S5, and Tables S1 and S2.
1 The abbreviations used are:
- Drf
- Diaphanous-related formin
- BD
- Basic Domain
- FH
- Formin Homology Domain
- DAD
- Diaphanous Auto-regulatory Domain
- GBD
- GTPase-binding Domain
- DID
- Diaphanous Inhibitory Domain
- DD
- Dimerization Domain
- UPS
- Ubiquitin Proteasome System
- GST
- Glutathione S-Transferase
- AP
- Affinity Purification
- SAINT
- Significance Analysis of INTeractome
- UniProt
- The Universal Protein Resource.
REFERENCES
- 1. Schönichen A., Geyer M. (2010) Fifteen formins for an actin filament: a molecular view on the regulation of human formins. Biochim. Biophys. Acta 1803, 152–163 [DOI] [PubMed] [Google Scholar]
- 2. Chesarone M. A., DuPage A. G., Goode B. L. (2010) Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 11, 62–74 [DOI] [PubMed] [Google Scholar]
- 3. Bartolini F., Moseley J. B., Schmoranzer J., Cassimeris L., Goode B. L., Gundersen G. G. (2008) The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J. Cell Biol. 181, 523–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aspenström P. (2010) Formin-binding proteins: modulators of formin-dependent actin polymerization. Biochim. Biophys. Acta 1803, 174–182 [DOI] [PubMed] [Google Scholar]
- 5. Faix J., Grosse R. (2006) Staying in shape with formins. Dev. Cell 10, 693–706 [DOI] [PubMed] [Google Scholar]
- 6. Wallar B. J., Stropich B. N., Schoenherr J. A., Holman H. A., Kitchen S. M., Alberts A. S. (2006) The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J. Biol. Chem. 281, 4300–4307 [DOI] [PubMed] [Google Scholar]
- 7. Beli P., Mascheroni D., Xu D., Innocenti M. (2008) WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat. Cell Biol. 10, 849–857 [DOI] [PubMed] [Google Scholar]
- 8. Yang C., Czech L., Gerboth S., Kojima S., Scita G., Svitkina T. (2007) Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 5, e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eisenmann K. M., Harris E. S., Kitchen S. M., Holman H. A., Higgs H. N., Alberts A. S. (2007) Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr. Biol. 17, 579–591 [DOI] [PubMed] [Google Scholar]
- 10. Di Vizio D., Kim J., Hager M. H., Morello M., Yang W., Lafargue C. J., True L. D., Rubin M. A., Adam R. M., Beroukhim R., Demichelis F., Freeman M. R. (2009) Oncosome formation in prostate cancer: Association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 69, 5601–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lizárraga F., Poincloux R., Romao M., Montagnac G., Le Dez G., Bonne I., Rigaill G., Raposo G., Chavrier P. (2009) Diaphanous-related formins are required for invadopodia formation and invasion of breast tumor cells. Cancer Res. 69, 2792–2800 [DOI] [PubMed] [Google Scholar]
- 12. Narumiya S., Tanji M., Ishizaki T. (2009) Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 28, 65–76 [DOI] [PubMed] [Google Scholar]
- 13. Ji P., Jayapal S. R., Lodish H. F. (2008) Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat. Cell Biol. 10, 314–321 [DOI] [PubMed] [Google Scholar]
- 14. Wallar B. J., Deward A. D., Resau J. H., Alberts A. S. (2007) RhoB and the mammalian Diaphanous-related formin mDia2 in endosome trafficking. Exp. Cell Res. 313, 560–571 [DOI] [PubMed] [Google Scholar]
- 15. Watanabe S., Ando Y., Yasuda S., Hosoya H., Watanabe N., Ishizaki T., Narumiya S. (2008) mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol. Biol. Cell 19, 2328–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Daou P., Hasan S., Breitsprecher D., Baudelet E., Camoin L., Audebert S., Goode B. L., Badache A. (2014) Essential and nonredundant roles for Diaphanous formins in cortical microtubule capture and directed cell migration. Mol. Biol. Cell 25, 658–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Altelaar A. F., Munoz J., Heck A. J. (2013) Next-generation proteomics: Towards an integrative view of proteome dynamics. Nat. Rev. Genetics 14, 35–48 [DOI] [PubMed] [Google Scholar]
- 18. Choi H., Larsen B., Lin Z. Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z. S., Tyers M., Gingras A. C., Nesvizhskii A. I. (2011) SAINT: Probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Innocenti M., Gerboth S., Rottner K., Lai F. P., Hertzog M., Stradal T. E., Frittoli E., Didry D., Polo S., Disanza A., Benesch S., Di Fiore P. P., Carlier M. F., Scita G. (2005) Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 7, 969–976 [DOI] [PubMed] [Google Scholar]
- 20. Galovic M., Xu D., Areces L. B., van der Kammen R., Innocenti M. (2011) Interplay between N-WASP and CK2 optimizes clathrin-mediated endocytosis of EGFR. J. Cell Sci. 124, 2001–2012 [DOI] [PubMed] [Google Scholar]
- 21. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miki T., Okawa K., Sekimoto T., Yoneda Y., Watanabe S., Ishizaki T., Narumiya S. (2009) mDia2 shuttles between the nucleus and the cytoplasm through the importin-{alpha}/{beta}- and CRM1-mediated nuclear transport mechanism. J. Biol. Chem. 284, 5753–5762 [DOI] [PubMed] [Google Scholar]
- 23. Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., Jensen L. J., von Mering C. (2011) The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561–D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P. L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shima Y., Shima T., Chiba T., Irimura T., Pandolfi P. P., Kitabayashi I. (2008) PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation. Mol. Cell. Biol. 28, 7126–7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vasudevan S., Starostina N. G., Kipreos E. T. (2007) The Caenorhabditis elegans cell-cycle regulator ZYG-11 defines a conserved family of CUL-2 complex components. EMBO Rep. 8, 279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsuzawa S. I., Reed J. C. (2001) Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol. Cell 7, 915–926 [DOI] [PubMed] [Google Scholar]
- 28. Duda D. M., Scott D. C., Calabrese M. F., Zimmerman E. S., Zheng N., Schulman B. A. (2011) Structural regulation of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 21, 257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tai H. C., Besche H., Goldberg A. L., Schuman E. M. (2010) Characterization of the Brain 26S Proteasome and its Interacting Proteins. Front Mol Neurosci 3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rusten T. E., Vaccari T., Stenmark H. (2012) Shaping development with ESCRTs. Nat. Cell Biol. 14, 38–45 [DOI] [PubMed] [Google Scholar]
- 31. Abaza A., Soleilhac J. M., Westendorf J., Piel M., Crevel I., Roux A., Pirollet F. (2003) M phase phosphoprotein 1 is a human plus-end-directed kinesin-related protein required for cytokinesis. J. Biol. Chem. 278, 27844–27852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tojkander S., Gateva G., Schevzov G., Hotulainen P., Naumanen P., Martin C., Gunning P. W., Lappalainen P. (2011) A molecular pathway for myosin II recruitment to stress fibers. Curr. Biol. 21, 539–550 [DOI] [PubMed] [Google Scholar]
- 33. Jacobs D. T., Weigert R., Grode K. D., Donaldson J. G., Cheney R. E. (2009) Myosin Vc is a molecular motor that functions in secretory granule trafficking. Mol. Biol. Cell 20, 4471–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fukasawa K. (2008) P53, cyclin-dependent kinase and abnormal amplification of centrosomes. Biochim. Biophys. Acta 1786, 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Winston J. T., Koepp D. M., Zhu C., Elledge S. J., Harper J. W. (1999) A family of mammalian F-box proteins. Curr. Biol. 9, 1180–1182 [DOI] [PubMed] [Google Scholar]
- 36. Ward D. M., Vaughn M. B., Shiflett S. L., White P. L., Pollock A. L., Hill J., Schnegelberger R., Sundquist W. I., Kaplan J. (2005) The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 280, 10548–10555 [DOI] [PubMed] [Google Scholar]
- 37. DeWard A. D., Alberts A. S. (2009) Ubiquitin-mediated degradation of the formin mDia2 upon completion of cell division. J. Biol. Chem. 284, 20061–20069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emanuele M. J., Elia A. E., Xu Q., Thoma C. R., Izhar L., Leng Y., Guo A., Chen Y. N., Rush J., Hsu P. W., Yen H. C., Elledge S. J. (2011) Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Drost J., Mantovani F., Tocco F., Elkon R., Comel A., Holstege H., Kerkhoven R., Jonkers J., Voorhoeve P. M., Agami R., Del Sal G. (2010) BRD7 is a candidate tumour suppressor gene required for p53 function. Nat. Cell Biol. 12, 380–389 [DOI] [PubMed] [Google Scholar]
- 40. Bartkova J., Horejsi Z., Koed K., Krämer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J. M., Lukas C., Ørntoft T., Lukas J., Bartek J. (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 [DOI] [PubMed] [Google Scholar]
- 41. Bieging K. T., Attardi L. D. (2012) Deconstructing p53 transcriptional networks in tumor suppression. Trends Cell Biol. 22, 97–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hande K. R. (1998) Etoposide: four decades of development of a topoisomerase II inhibitor. Eur. J. Cancer 34, 1514–1521 [DOI] [PubMed] [Google Scholar]
- 43. Roos W. P., Kaina B. (2006) DNA damage-induced cell death by apoptosis. Trends Mol. Med. 12, 440–450 [DOI] [PubMed] [Google Scholar]
- 44. Gaillard J., Ramabhadran V., Neumanne E., Gurel P., Blanchoin L., Vantard M., Higgs H. N. (2011) Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol. Biol. Cell 22, 4575–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hager M. H., Morley S., Bielenberg D. R., Gao S., Morello M., Holcomb I. N., Liu W., Mouneimne G., Demichelis F., Kim J., Solomon K. R., Adam R. M., Isaacs W. B., Higgs H. N., Vessella R. L., Di Vizio D., Freeman M. R. (2012) DIAPH3 governs the cellular transition to the amoeboid tumour phenotype. EMBO Mol Med 4, 743–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yamada K., Ono M., Perkins N. D., Rocha S., Lamond A. I. (2013) Identification and functional characterization of FMN2, a regulator of the cyclin-dependent kinase inhibitor p21. Mol. Cell 49, 922–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.