Abstract
The parasitic nematode intestine is responsible for nutrient digestion and absorption, and many other processes essential for reproduction and survival, making it a valuable target for anthelmintic drug treatment. However, nematodes display extreme biological diversity (including occupying distinct trophic habitats), resulting in limited knowledge of intestinal cell/protein functions of fundamental or adaptive significance. We developed a perfusion model for isolating intestinal proteins in Ascaris suum (a parasite of humans and swine), allowing for the identification of over 1000 intestinal A. suum proteins (using mass spectrometry), which were assigned to several different intestinal cell compartments (intestinal tissue, the integral and peripheral intestinal membranes, and the intestinal lumen). A multi-omics analysis approach identified a large diversity of biological functions across intestinal compartments, based on both functional enrichment analysis (identifying terms related to detoxification, proteolysis, and host-parasite interactions) and regulatory binding sequence analysis to identify putatively active compartment-specific transcription factors (identifying many related to intestinal sex differentiation or lifespan regulation). Orthologs of A. suum proteins in 15 other nematodes species, five host species, and two outgroups were identified and analyzed. Different cellular compartments demonstrated markedly different levels of protein conservation; e.g. integral intestinal membrane proteins were the most conserved among nematodes (up to 96% conservation), whereas intestinal lumen proteins were the most diverse (only 6% conservation across all nematodes, and 71% with no host orthologs). Finally, this integrated multi-omics analysis identified conserved nematode-specific intestinal proteins likely performing essential functions (including V-type ATPases and ABC transporters), which may serve as promising anthelmintic drug or vaccine targets in future research. Collectively, the findings provide valuable new insights on conserved and adaptive features of nematode intestinal cells, membranes and the intestinal lumen, and potential targets for parasite treatment and control.
The phylum Nematoda is one of the most diverse phyla, containing animal species that have been extraordinarily successful in adapting to a wide variety of niches, including free-living (terrestrial and marine) and parasitic (plant and animal). The nonparasitic model nematode Caenorhabditis elegans is one of the most well studied animal model organisms, and has been of great value for many aspects of biomedical sciences as well as phylum-specific studies, including studies on parasitic nematodes. Infections by parasitic nematodes cause extensive suffering in humans and animals, as well as major losses in agricultural production, because of disease and the cost of implementing control programs (1). Calculations of the aggregate burden of nematode diseases in Disability Adjusted Life Years among humans indicate a tremendous global impact of these pathogens and a need for improved control or prevention (2). As a nonproliferating and nonregenerative tissue, the nematode intestine is an attractive target for developing strategies for control and prevention of parasitic nematode infections (3). In addition to nutrient digestion and absorption (3, 4), the nematode intestine is responsible for many other processes including innate immunity to microbial infection (5), defense against environmental toxins, reproduction, stress responses (6), and ion transport (7). The multiple roles and functions of the intestine are further complicated by the varying functions in the different cellular compartments, which include the intestinal cells, the apical surface of the cells, the intestinal lumen and the pseudocoelom surrounding the intestine (4). The surface of the nematode apical intestinal membrane and the intestinal lumen are key points of surface interaction between the worm and the environment. The apical membrane is composed of microvilli, and the membrane has metabolic and cellular trafficking activity as well as mechanisms for innate immunity and interaction with the host immune system (3). Because of its role as an interface with the host, the apical intestinal membrane and lumen are important targets for the treatment and control of parasitic nematodes, and although the intestine has been an organ of interest for decades, only a small fraction of the proteins located in these compartments have been identified (4, 8).
It is of interest to delineate characteristics of the nematode intestine that are conserved among all nematodes and parasitic species in particular, so that research can prospectively focus on parasite functions with the broadest potential benefit to human and animal health. At the same time, it is recognized that exploitation of the vast array of trophic niches inhabited by nematodes is likely to leverage large influence on features of the intestine to undergo adaptions relative to these niches. Therefore, it is expected that diversity of intestinal features among nematodes may reflect adaptions of importance to individual species or lineages of nematodes. Nevertheless, the level of conservation and diversity of intestinal cell proteins remains to be determined, as does the degree to which conservation of proteins may vary among intestinal cell compartments. One hypothesis is that diverse trophic niches would place comparatively high selective pressure on proteins at more immediate interfaces with the outside environment. If true, then the greatest diversity in intestinal cell proteins is expected to associate with proteins located in the lumen and, possibly, the apical intestinal membrane.
Although the nonparasitic C. elegans and the parasitic Haemonchus contortus are the model organisms for which much of the research about the nematode intestine is based (4, 9), their small size (1 mm and 2–5 cm in length, respectively (10, 11)) limits the ability to directly investigate (by proteomic methods) proteins that constitute cellular compartments of the intestine. However, the parasitic roundworm Ascaris suum (size 15–40 cm; which also has a complete draft genome available (12, 13)) is large enough that the intestine can be physically manipulated for this purpose. The large size of the A. suum intestine has also supported the broad molecular characterization of intestinal cell functions based on gene expression studies using expressed sequence tags (3), microarray (14) and RNA-seq approaches (15). These studies analyzed the transcript expression level in whole intestinal tissue relative to other tissues but not protein expression or compartmentalization of proteins within intestinal tissue, although a proteomic study of A. suum proteins in nonintestinal fluids including perienteric and uterine fluids was recently reported (16).
Here, we utilize a combination of a perfusion model (see Experimental Procedures) and high-throughput protein mass spectrometry to analyze the protein composition of the A. suum intestine, by collecting samples from the intestinal tissue, lumen contents, and lumen contents after perfusing with urea. The pseudocoelomic fluid was also collected to account for possible contamination of the intestinal samples. These various intestinal samples enabled determination of the proteins that comprise specific compartments of the intestine, and the functional enrichment of the detected proteins provides deep insight into major functions that characterize each of the compartments. An in-depth multi-omics analysis (genome/transcriptome/proteome) of the identified intestine proteins and the orthologous proteins deduced from other species clarified the degree to which individual proteins are conserved among nematode species that span the phylum Nematoda and among host species.
EXPERIMENTAL PROCEDURES
Collection of Intestinal Samples
To initiate infections, weanling pigs (mixed breed and obtained from the Washington State University Swine Center) were orally infected with A. suum eggs cultured to contain infective larvae and provided by Dr. Joseph Urban (US Department of Agriculture, Beltsville, MD). Eggs originated from uteri dissected from adult female worms. Pigs were infected three times a week with 50 eggs per dose for 2 weeks (300 eggs). Swine were euthanized after 60 to 70 days to obtain adult worms from the host small intestine. Adult worms were collected into phosphate buffered saline (PBS1, pH7.4) at 37 °C for perfusion, or ice cold for collection of intestinal tissues. Animal protocols utilized here were approved by the Institutional Animal Care and Use Committee, Washington State University.
Cannulation and Collection of Intestinal Perfusates and Tissue Samples
The body walls of adult worms maintained in ice cold PBS were opened longitudinally with iris scissors to expose and dissect intestines from male and female worms used to generate whole intestinal homogenates or intestinal fractionations. Only female worms were used for purposes of fractionation and perfusion. Pseudocoelomic fluid was obtained from the body cavity of a female worm that was exposed in this manner. Different individual worms were used to generate each category of sample.
Intestinal perfusion samples were obtained by first severing the anterior end of female worms just below the pharynx, which exposed the intestine and intestinal lumen. A blunt ended, 25 gauge needle (cannula) was inserted into the anterior end of intestinal lumen, and superglue was used to attach the cannula to the anterior end of the worm. The cannula was then attached to a syringe containing perfusion solutions. To allow flow through the intestine, the posterior 1/6th of the worm was removed and approximately one cm of the body wall resected, to expose the posterior intestine. The exposed posterior segment of the intestine was placed on parafilm to collect perfusates. Perfusates were generated by gentle delivery of solution by syringe to the anterior lumen and collection of perfusate after passage through the intestine onto the parafilm. Placement of the posterior intestine on parafilm allowed perfusate collection with minimal contamination by pseudocoelomic fluid. The PBS perfusate was generated by perfusion with 300–500 μl PBS. Secondarily, a solution of 4 m urea in PBS (4 MU) was perfused to generate the 4 MU perfusate. The PBS and 4 MU samples were generated from a single worm following full development of the method. Perfusate samples were stored at −20 °C, a bicinchoninic acid assay (Micro BCA Protein Assay Kit, Thermo Scientific, Rockford, IL) was used to determine protein concentrations for all samples.
A third intestinal protein sample was obtained from adult female intestinal homogenates, and was sequentially fractionated by centrifugation first at 5000 × g to produce a supernatant and pellet (S1, P1). S1 was centrifuged at 50,000 × g to produce a pellet (5k to 50k pellet; designated P2), which was intended to be enriched for intestinal membrane proteins. In addition, whole intestinal protein preparations were generated by dissection of the intestine from adult male and female A. suum. The whole intestinal samples were cryopulverized using a Covaris CP02 instrument and homogenized in 2% SDS containing buffer (800 μl) in a Covaris S220X Adaptive Focused Acoustics Instrument. The protein mixture (∼500 μg) was loaded into a gel elution entrapment cartridge, four samples each from male and female worms were applied to the eight channels in the gel cartridge. The samples were separated into seven subsequent molecular weight fractions (F1 to F7), and the samples were transferred to spin cartridges (∼10,000 MWCO) and the SDS was removed as previously described (17). Pseudocoelomic fluid (designated PF) was obtained by opening the body wall of adult female worms and collecting fluid located in the pseudocoelom. Soluble proteins in each of these samples were processed for mass spectrometry.
High Resolution MS1 and MS2 Mass Spectrometry
Proteins were digested sequentially with endoprotease Lys-C (cleaving lysine at the C terminus) and trypsin as previously described (18). A solid phase extraction of peptides with microtips of C4 and porous graphite carbon (GlyGen) was performed on a Bravo automated liquid-handling robot prior to LC-MS. Peptide mixtures were analyzed using high-resolution nano-LC-MS on a hybrid mass spectrometer consisting of a linear quadrupole ion-trap and an Orbitrap (LTQ-Orbitrap XL, Thermo Fisher Scientific). Chromatographic separations were performed using a nanoLC 1D Plus™ (Eksigent) for gradient delivery and a cHiPLC-nanoflex (Eksigent) equipped with a 15 cm x 75 μm C18 column (ChromXP C18-CL, 3m, 120 Å, Eksigent). The liquid chromatograph was interfaced to the mass spectrometer with a nanospray source (PicoView PV550; New Objective). Mobile phases were 1% FA in water (A) and 1% FA in 60% ACN (B). After equilibrating the column in 98% solvent A (aqueous 1% FA) and 2% of solvent B (ACN containing 1% FA), the samples (5 μl) were injected from autosampler vials using the LC-system's autosampler at a flow rate of 500 nL/min followed by gradient elution (250 nL/min) with solvent B: isocratic at 2% B, 0–5 min; 2% B to 25% B, 5–110 min; 25% to 80%, 110–170 min; 80% to 2%, 170–175; and isocratic at 2% B, 175–190 min. For high resolution data directed analysis, the survey scans (m/z 350–2000) (MS1) were acquired at high resolution (60,000 at m/z = 400) in the Orbitrap in profile mode and the MS/MS spectra (MS2) were acquired at 7500 resolution in the Orbitrap in profile mode after fragmentation in the linear ion trap. The maximum injection times for the MS1 scan in the Orbitrap and the LTQ were both 500 ms, and the maximum injection times for the MSn scan in the Orbitrap and the LTQ were 800 ms and 5000 ms, respectively. The automatic gain control targets for the Orbitrap and the LTQ were 5 × 105 and 3 × 104, respectively for the MS1 scans and 2 × 105 and 1 × 104, respectively, for the MSn scans. The MS1 scans were followed by three MS2 events in the linear ion trap with collision activation in the ion trap (parent threshold = 10,000; isolation width = 4.0 Da; normalized collision energy = 30%; activation Q = 0.250; activation time = 30 ms). Dynamic exclusion was used to remove selected precursor ions (−0.20/+1.0 Da) for 90 s after MS2 acquisition. A repeat count of three, a repeat duration of 45 s, and a maximum exclusion list size of 500 was used. The following ion source parameters were used: capillary temperature 200 °C, source voltage 2.5 kV, source current 100 μA, and the tube lens at 79 V. The data were acquired using Xcalibur, version 2.0.7 (Thermo Fisher).
Data Processing and Analysis of Gel Plug Samples
Data files from the LC-MS analysis of tryptic peptides generated by in situ tryptic digestion of SDS-PAGE gel plugs were processed using MASCOT Distiller (Matrix Science, version 2.3.0.0) with the settings previously described (19). The data were searched with Mascot (Matrix Science, London, UK; version 2.1.6) against the deduced A. suum proteome (12) (18,542 proteins) as well as the Sus scrofa proteome (Uniprot, downloaded Sept. 2012; 23,118 proteins) to identify potential host contamination. The search was conducted using trypsin as the endoprotease, allowing nine missed cleavages, specifying carbamidomethylation of Cys residues as a fixed modification and oxidation of Met residues as a variable modification and setting a fragment ion mass tolerance of 0.800 Da and a parent ion tolerance of 20 ppm. The Protein and Peptide Prophet algorithms in Scaffold (version 3.6.4, Proteome Software Inc., Portland, OR) were used to qualify the database results. A probability of 50% was used to assign peptide sequences to MS2 spectra using the Peptide Prophet algorithm (20). A probability of 95% was used to assign proteins with the Protein Prophet algorithm (21), with a 0.1% false discovery rate according to Scaffold.. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Database Searches of In-solution Digests
The LC-MS data files for both low resolution and high resolution MS2 were processed using MASCOT Distiller (Matrix Science, version 2.3.0.0) with previously described settings for both low resolution (19) and high resolution (22) MS2. Settings for high resolution spectra acquired in the Orbitrap were as follows: 1) MS processing: 200 data points per Da; sum aggregation method; maximum charge state, 3+; minimum number of peaks, 1; 2) MS/MS processing: 200 data points per Da; time domain aggregation method; minimum number of peaks, 10; use precursor charge as maximum; precursor charge and m/z, “try to re-determine charge from the parent scan (tolerance, 1.2 Da)”; charge defaults, 1; maximum charge state, 3) time domain parameters: minimum precursor mass, 300; maximum precursor mass, 16,000; precursor m/z tolerance for grouping, 0.01; maximum number of intermediate scans, 0; minimum number of scans in a group, 1; and 4) peak picking: maximum iterations, 500; correlation threshold, 0.60; minimum signal to noise, 2; minimum peak m/z, 50; maximum peak m/z, 100,000; minimum peak width, 0.002; maximum peak width, 0.2; expected peak width, 0.02. The resulting MS2 centroided files were used for database searching (see previous section) with MASCOT (Matrix Science, version 2.1.6). For low-resolution MS2 spectra, the search was conducted using trypsin as the specified protease, allowing nine missed cleavages, specifying carbamidomethylation of Cys residues as a fixed modification and oxidation of Met residues as a variable modification and setting a fragment ion mass tolerance of 0.800 Da and a parent ion tolerance of 20 ppm. For high resolution MS2 spectra, the search was conducted with no enzyme specificity and a fragment ion mass tolerance of 100 absolute milli-mass units. All other settings were the same as those used for searching the low resolution MS2 spectra. Scaffold (version Scaffold_ 3_00_03, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (20). The total number of spectra and unique proteins identified in each sample is shown in Table I, and the total number assigned per protein (as well as the breadth of coverage for each detected protein) are shown in supplemental Table S1. Information for individual peptides and spectra is provided in supplemental Table S2. Raw data, databases, scaffold output files, and search parameters are available on Peptide Atlas, accession # PASS00571.
Table I. The number of spectra and proteins identified in each of the 18 mass-spectrometry proteomics samples.
| Sample description | Sample label | Number of spectra mapped to proteins | Number of unique proteins identified |
|---|---|---|---|
| Pseudocoelomic fluid | Pseudo | 1,549 | 118 |
| Male intestinal tissue | M-F1 | 2,061 | 473 |
| M-F2 | 2,352 | 583 | |
| M-F3 | 1,799 | 569 | |
| M-F4 | 2,736 | 563 | |
| M-F5 | 2,625 | 522 | |
| M-F6 | 3,117 | 454 | |
| M-F7 | 3,019 | 374 | |
| Female intestinal issue | F-F1 | 3,003 | 562 |
| F-F2 | 2,898 | 638 | |
| F-F3 | 2,423 | 598 | |
| F-F4 | 3,045 | 563 | |
| F-F5 | 3,214 | 512 | |
| F-F6 | 3,145 | 435 | |
| F-F7 | 3,292 | 353 | |
| Female intestinal tissue, 5K-50K pellet | P2 pellet | 3,162 | 642 |
| Lumen wash, PBS | PBS perfusate | 2,910 | 134 |
| Lumen wash, PBS + 4 m urea | 4MU perfusate | 4,712 | 295 |
| All combined | All | 51,062 | 1,048 |
Functional Annotation
Interproscan (23–25) was used to determine associations of genes in the current A. suum genome (12) to Gene Ontology (GO) terms (26) and Interpro domains (27), as previously described (15). In addition, predicted proteins were searched against the KEGG database (28) using KAAS (29). Proteins with signal peptides and transmembrane domains were identified using the Phobius (30) and nonclassical secretion was predicted using SecretomeP 1.0 (31).
Intestinal Compartment Definition
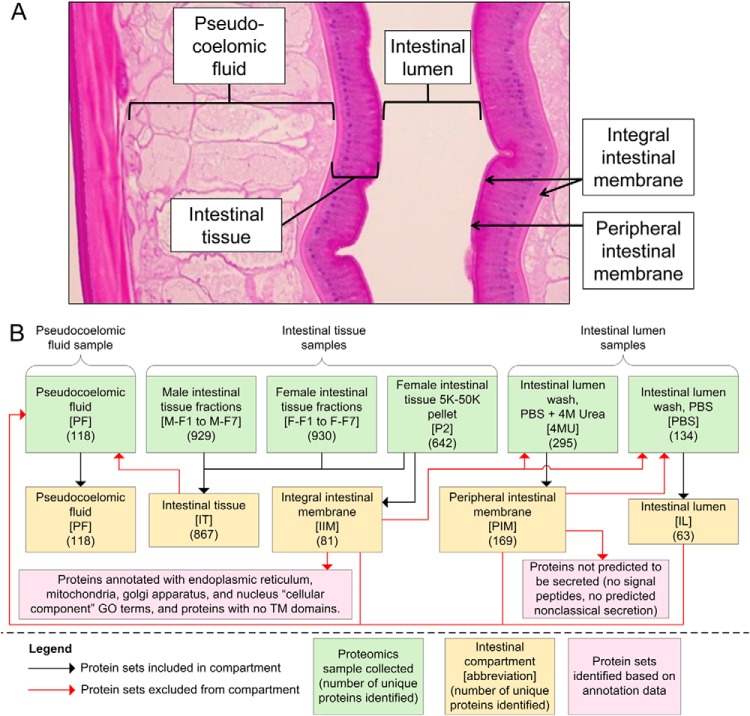
The proteins belonging to each of the intestinal compartments (Fig. 1A and 1B) were defined as follows: All of the proteins detected in the PF protein set were removed from all of the other compartment protein sets, because they represent a potential source of contamination. The intestinal tissue (IT) proteins included all of the unique proteins detected in the 14 total tissue molecular weight-fractions (M-F1 to M-F7, and F-F1 to F-7) as well as in the P2 pellet sample, because all of these are expected to be a part of the intestinal tissue. The integral intestinal membrane (IIM) protein set represents expected transmembrane proteins on either the basal or apical intestinal membranes. This compartment included the proteins in the P2 pellet fraction (which enriches for TM-domain containing proteins), while excluding (1) the proteins found in either of the perfusate samples, in order to reduce contamination from the peripheral membrane and the intestinal lumen (2) proteins without predicted transmembrane domains according to annotation by Phobius (30), and (3) proteins annotated with ‘cellular compartment’ Gene Ontology terms (23) related to the endoplasmic reticulum, mitochondria, Golgi apparatus, and nucleus, in order to reduce contamination from proteins embedded in these organelles rather than the external cellular membrane, which is the target of interest (GO:0005783, GO:0005789, GO:0005739, GO:0005741, GO:0000139, GO:0005840, GO:0005740, GO:0005635, GO:0016459). The peripheral intestinal membrane (PIM) proteins on the apical intestinal membrane represents proteins predicted to be peripheral to the lumen space, but apical relative to the intestinal cells. This included proteins in the 4MU perfusate, but not in the PBS perfusate because the 4MU urea treatment was expected to release these proteins from the apical surface. Only proteins with predicted signal peptides or nonclassical secretion sequences were considered to be in the PIM, because these are better candidates for proteins that are transported to the membrane. The intestinal lumen (IL) proteins were all of the PBS perfusate proteins with overlapping PF proteins removed. The stringent criteria used may have led to false negatives in some cases, that is, some proteins ascribed to PF might also occur in the IL. Nevertheless, the assignments were intended to minimize the number of false positives for each compartment.
Fig. 1.
Defining protein sets belonging to intestinal compartments. A, A longitudinal section of A. suum showing the physical location of each of the intestinal compartments included in the proteomics analysis. B, The proteomics sample combinations used to identify protein sets in each intestinal compartment. Black arrows indicate protein sets (green) which were included in the deduced compartment protein sets (yellow), and red arrows indicate proteins which were removed from the deduced protein sets. Numbers in parentheses indicate the number of proteins in each protein set.
Functional Enrichment
FUNC (32) (which considers the hierarchical structure of GO) was used to determine significant functional enrichment among the proteins present in each set, with a p ≤ 0.001 significance threshold (after FDR population correction; Table II). Only GO terms with the root “Molecular Function” term were considered for analysis, to avoid redundant functional information from other terms. Interpro domain enrichment was determined using a nonparametric binomial distribution test with a p ≤ 0.01 significance threshold (after FDR population correction; Table III). Only Interpro domains found in at least five proteins assigned to the compartment being tested were considered for enrichment testing.
Table II. “Molecular Function” Gene Ontology enrichment results for proteins detected in each intestinal compartment, relative to all detected proteins.
| Compartment | Gene Ontology Number | GO term annotation | Enrichment p value |
|---|---|---|---|
| Pseudo-coelomic fluid (PF) | GO:0004867 | Serine-type endopeptidase inhibitor activity | 6.00E-08 |
| GO:0005319 | Lipid transporter activity | 4.30E-04 | |
| GO:0003755 | Peptidyl-prolyl cis-trans isomerase activity | 9.70E-04 | |
| Intestinal tissue (IT) | GO:0003735 | Structural constituent of ribosome | 2.20E-06 |
| Integral intestinal membrane (IIM) | GO:0016758 | Transferase activity, transferring hexosyl groups | 1.50E-05 |
| Peripheral intestinal membrane (PIM) | GO:0046961 | Proton-transporting ATPase activity, rotational mechanism | 1.07E-05 |
| GO:0046933 | Hydrogen ion transporting ATP synthase activity, rotational | 1.66E-04 | |
| GO:0005544 | Calcium-dependent phospholipid binding | 2.99E-04 | |
| GO:0030170 | Pyridoxal phosphate binding | 5.91E-04 | |
| GO:0004075 | Biotin carboxylase activity | 7.71E-04 | |
| Intestinal lumen (IL) | GO:0004190 | Aspartic-type endopeptidase activity | 6.30E-05 |
| GO:0008237 | Metallopeptidase activity | 8.70E-05 | |
| GO:0004553 | Hydrolase activity, O-glycosyl compounds | 1.40E-04 |
Table III. Interpro Domain enrichment results for proteins detected in each intestinal compartment, relative to all detected proteins.
| Compartment | Interpro domain number | Interpro domain annotation | Enrichment p value |
|---|---|---|---|
| Pseudocoelomic fluid (PF) | IPR001534 | Transthyretin-like | 9.5E-07 |
| IPR000215 | Protease inhibitor I4, serpin | 2.3E-03 | |
| IPR023796 | Serpin domain | 2.3E-03 | |
| IPR007110 | Immunoglobulin-like | 6.2E-03 | |
| IPR023795 | Protease inhibitor I4, serpin, conserved site | 6.9E-03 | |
| IPR003599 | Immunoglobulin subtype | 6.9E-03 | |
| IPR003598 | Immunoglobulin subtype 2 | 6.9E-03 | |
| IPR013098 | Immunoglobulin I-set | 9.6E-03 | |
| Intestinal tissue (IT) | None | ||
| Integral intestinal membrane (IIM) | IPR002213 | UDP-glucuronosyl/UDP-glucosyltransferase | 3.3E-04 |
| IPR011527 | ABC transporter, transmembrane domain, type 1 | 1.1E-03 | |
| IPR003439 | ABC transporter-like | 1.1E-03 | |
| IPR003593 | ATPase, AAA+ type, core | 1.6E-03 | |
| IPR017871 | ABC transporter, conserved site | 2.6E-03 | |
| IPR001140 | ABC transporter, transmembrane domain | 2.6E-03 | |
| IPR017940 | ABC transporter, integral membrane type 1 | 2.6E-03 | |
| IPR001199 | Cytochrome b5 | 9.7E-03 | |
| Peripheral intestinal membrane (PIM) | None | ||
| Intestinal lumen (IL) | IPR002035 | von Willebrand factor, type A | 2.7E-06 |
| IPR001930 | Peptidase M1, alanine aminopeptidase/leukotriene A4 | 2.3E-05 | |
| IPR000322 | Glycoside hydrolase, family 31 | 4.6E-05 | |
| IPR016186 | C-type lectin-like | 5.3E-05 | |
| IPR016187 | C-type lectin fold | 7.0E-05 | |
| IPR001304 | C-type lectin | 2.0E-04 | |
| IPR014782 | Peptidase M1, membrane alanine aminopeptidase | 2.0E-04 | |
| IPR001969 | Peptidase aspartic, active site | 4.8E-04 | |
| IPR001461 | Peptidase A1 | 4.8E-04 | |
| IPR009007 | Peptidase aspartic, catalytic | 4.8E-04 | |
| IPR021109 | Peptidase aspartic | 4.8E-04 | |
| IPR011013 | Glycoside hydrolase-type carbohydrate-binding | 2.3E-03 | |
| IPR018497 | Peptidase M13, neprilysin, C-terminal | 2.5E-03 | |
| IPR008753 | Peptidase M13 | 2.5E-03 | |
| IPR000718 | Peptidase M13, neprilysin | 2.5E-03 | |
| IPR000519 | P-type trefoil | 8.5E-03 | |
| IPR017853 | Glycoside hydrolase, superfamily | 9.2E-03 |
Transcription Factor Binding Site Motif Enrichment
Transcription factor (TF) consensus binding site (motif) enrichment and annotation was performed similarly to our previous study on tissue-specific overexpression in A. suum (15), but with a much more expansive reference database containing rich annotation for C. elegans (33). A maximum length of 2000bp 5′ upstream regions (URs) were extracted for each gene based on the A. suum genome annotation, with shorter lengths where URs reached the end of a scaffold or overlapped with other coding sequences on either strand (12). In C. elegans, this UR length covers more than 91% of promoters (33), so this length was chosen to reduce background noise while maintaining a large proportion of promoters. Motif enrichment was performed using a discriminative motif analysis algorithm (DREME (34), using an 8-nucleotide maximum sequence search and 1000 generalized REs to increase accuracy), where the 5′ URs of the genes corresponding to the proteins within a compartment were compared with the 5′ URs of all other genes. Potential annotated transcription factors binding the discovered motifs were identified using TOMTOM using a custom consensus motif database (35). This database consisted of the JASPAR CORE nematoda (15 motifs) and vertebrate (205) motif databases (36), the UniProbe mouse (386) and worm (31) motif databases (37), the ELT-2 consensus motif (38), as well as all predicted C. elegans transcription factor binding motifs calculated from ChIP-seq data sets annotated as part of the modENCODE project (33, 39) (404, including multiple putative binding motifs for most TFs; Retrieved from http://www.broadinstitute.org/∼pouyak/motif-disc/worm/). BLASTP (40) was used to identify potential orthologs of these transcription factors in the A. suum genome.
Orthologous Clustering
Whole-proteome data from 23 eukaryotic species was collected. The data sets were comprised of 16 nematode species and seven host/outgroup species (Fig. 2). Data were obtained from the following sources: Homo sapiens, Mus musculus, Saccharomyces cerevisiae and Drosophila melanogaster were obtained from Ensembl (41) release 67. Bos taurus, Sus scrofa, and Ovis aries were from GenBank release 100, 103, and 100 respectively. For the nematodes, Caenorhabditis elegans, Pristionchus pacificus, Meloidogyne hapla, and Brugia malayi were from Wormbase (42) WS230; Trichinella spiralis, Ascaris suum, Haemonchus contortus, Dirofilaria immitis, and Necator americanus were from published data (43–47). Wuchereria bancrofti was downloaded from GenBank (Retrieved Feb 2012), and Strongyloides ratti and Trichuris muris were from the Sanger Institute; both retrieved Aug 2012); Meloidogyne incognita was from the French National Institute for Agricultural Research (INRA); Loa loa was from Broad Institute. Ancylostoma ceylanicum and Dictyocalus viviparus were from unpublished in-house sequencing data. Isoforms of these downloaded sequences were determined from coding genes, and only the longest isoforms were kept. For clustering, protein families (orthologous groups) across all of the deduced proteomes were defined using the Markov cluster algorithm available in the OrthoMCL package (48, 49), with an inflation factor of 1.5. In the discussion below, any reference to orthologous proteins is with respect to the identifications made by this algorithm. In the discussion below, any reference to orthologous proteins is with respect to the identifications made by this algorithm.
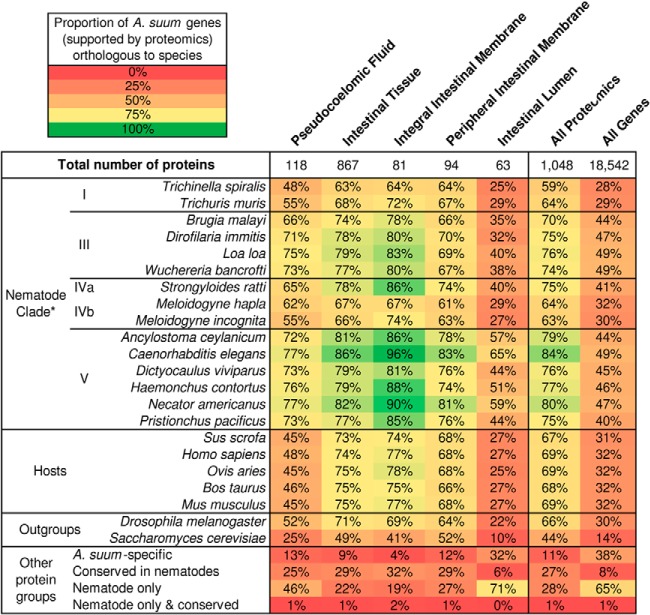
Fig. 2.
Conservation of A. suum intestinal proteins based on orthology to nematode, host and outgroup species. Numbers indicate the proportion of A. suum proteins within the protein sets which had predicted orthologs in each species and group listed. “Other protein groups” include A. suum-specific proteins (no orthologs in other species), nematode-conserved proteins (orthologs in all nematode species), nematode-only proteins (orthologs only in one or more nematode species, and not hosts or outgroups; proteins can be nematode-only if they only have a single ortholog in A. suum), and proteins which were both nematode-only and conserved. *Nematode clades are defined according to Blaxter et al. (100).
Drug Target Modeling
A. suum proteins were compared (BLAST with threshold 1e-05, 35% identity at over 50% fraction of length) to the protein sequences with known tertiary structures from the Protein Data Bank (PDB) (50) to identify homologs. The Chembl DrugEBIlity database (51) was then utilized to determine whether the PDB sequence was druggable (based upon the Lipinksi rule of five (52)). If a PDB chain is reported by the database to have a positive druggability score (including any of tractable, druggable or ensemble score), it is labeled as a druggable PDB structure. The druggability of the nematode proteins (i.e. their predicted ability to bind to a drug) was then determined based upon the BLAST match with the PDB sequence. Modeling of protein structures was also carried out for candidate genes using the I-TASSER Suite 2.1 (53). The alignments of protein sequence against the PDB template were applied as restraints of the target proteins related to the templates, with other parameters set to default.
Protein Physical Property Calculation
The isoelectric point, charge per amino acid and molecular weight calculations for each predicted protein in the proteome were calculated using the ProtParam package available in Perl Biotools (54) (Fig. 3).
Fig. 3.
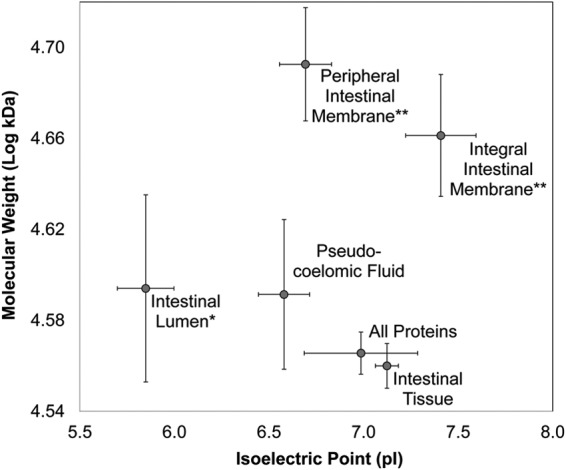
The average isoelectric point and molecular weight of the sets of proteins assigned to each intestinal compartment. * and ** = The isoelectric point and molecular weight differ significantly from entire set of detected proteins (respectively). Error bars indicate standard error.
Ethics Statement
The research involving use of swine was reviewed and approved by the Washington State University Institutional Animal Care and Use Committee, protocol #04097–004, approved on 12/19/2013. Guidelines are provided by the Federal Animal Welfare Act, USA.
RESULTS AND DISCUSSION
A total of 51,062 unique spectra obtained from the 18 samples were mapped to 1,048 unique predicted proteins (Table I; supplemental Table S1). The genes encoding all of the detected proteins were previously determined to be expressed in the A. suum intestine by RNA-Seq analysis (15) except for three (GS_14239 and GS_21581 in the PF, and GS_14331 in the PBS perfusate; The expression of these three genes was detected in in the head, pharynx, and/or testes). Proteins were assigned to intestinal compartments according to the method of collection and computational annotation of results (Fig. 1; see Experimental Procedures). For instance, intestinal lumen (IL) proteins were obtained by direct perfusion of the intestine with PBS (PBS perfusate). This simple approach lends high confidence that proteins obtained in this perfusate were located in the IL, with the exception of protein contaminants that likely originated from pseudocoelomic fluid (PF) during collection. For this reason, the 71 PBS perfusate proteins, which were also found in the PF fraction, were removed from the IL protein set, resulting in 63 high-confidence proteins being assigned to the IL compartment. The 4MU perfusate of the intestine was obtained after the PBS perfusate, and was intended to solubilize peripheral membrane proteins located on the apical intestinal membrane (AIM). Proteins unique to the 4MU perfusate (compared with the PBS perfusate) were assigned to the peripheral intestinal membrane (PIM) of the AIM. In order to remove potential contamination from cytoplasmic proteins in this set, proteins that were not predicted to be secreted (see Experimental Procedures) were excluded from in the PIM set, and PF proteins were removed as contamination as well, resulting in a set of 169 PIM proteins. It should be noted that there are potentially incomplete gene models in the draft A. suum genome (12), which may result in some signal peptide or nonclassical secretion sequences being missed, therefore resulting in false negatives for the PIM set given this filtering criteria. The integral intestinal membrane (IIM; 5k-50k pellet) protein set had the potential to contain proteins destined for the PIM, as well as the lumen, the PF, the basal intestinal membrane and other proteins associated with organellar membranes requiring exclusion of certain protein categories (see Experimental Procedures), so these potential sources of contamination were removed, resulting in a set of 81 proteins. The intestinal tissue (IT) protein set consisted of all of the collected whole-tissue fractions with PF proteins removed (leaving 867 total in the IT). The PF sample contained 118 detectable proteins. Although this integrated approach will not distinguish AIM from basal intestinal membrane proteins (other bioinformatics and experimental information can aid in this distinction), it is particularly important for identifying predicted integral membrane proteins, as integral membrane proteins associated with the AIM were not expected to be solubilized by PBS or 4MU. Overall, the filtering criteria were carefully chosen to make optimal use of the available data, controlling for sources of contamination while minimizing inclusion of false positives. Downstream analyses were based on the protein set definitions described above, and included evolutionary conservation analysis, functional comparisons (enrichment and depletion), transcriptional regulation analysis, and the identification of proteins of interest for developing novel therapeutics.
Protein Diversification Rates Vary Significantly Among Intestinal Compartments
Phylogenetic analysis of A. suum intestinal proteins was performed based on the ortholog conservation of the detected A. suum proteins in each intestinal compartment across fifteen other nematode species, five host species, and two outgroups (Fig. 2). In each compartment, the percentage of A. suum proteins with orthologs in each of the other species was used to measure phylogenetic conservation. For example, 57 of the 118 (48%) of the PF proteins in A. suum had orthologs in T. spiralis (Fig. 2). Large differences in conservation among intestinal compartments were observed, offering the ability to partition elements of the nematode intestine which appear to have more or less impact on species diversification within the Nematoda.
The IL proteins were the least conserved among the intestinal compartments, with an average of only 35.4% of the proteins having orthologs in other individual nematode species (compared with of 72.8% for all detected proteins; p = 1.5 × 10−8, binomial distribution test). Only 6% (4 IL proteins) had orthologs in all of the nematodes (significantly lower than 27% for all detected proteins; p = 3 × 10−5), as well as in all of the host species. These proteins were GS_03797 (a spliceosome protein), GS_02934 (a citrate cycle pathway protein), GS_10488 (a serine-type endopeptidase inhibitor) and GS_03147 (a purine metabolism pathway enzyme). Although each is widely conserved, the functions represented are expected to reflect cytoplasmic localization. It remains uncertain if detection of these proteins in the lumen has any functional significance to the nematode intestine.
In contrast, the 81 IIM proteins showed the highest conservation among nematodes (among the compartments studied), with an average of 81.0% being conserved in other nematode species (compared with 72.8% for all detected proteins; p = 0.0078), and 32% of the proteins being conserved across all nematodes. These results suggest that IL proteins are more significantly associated with phylogenetic diversity and species adaptation than other proteins, and that IIM proteins appear to serve more basic functions required by all nematodes. These findings are somewhat intuitive, because high levels of adaptability would seem essential for functions at the primary site for parasite interactions with host nutrients and other molecules obtained from the host, if organisms are to evolve and exploit highly diverse trophic niches (as the species included in our analysis do). Although the level of conservation was extended to outgroups as well, the low level of IL protein conservation among nematodes supports that proteins that comprise the IL compartment are more diverse among nematodes, and hence more adaptable among nematode species. The results provide the first clear quantitative values for exploring this relationship in the Nematoda, and suggest that different intestinal cell compartments have different levels of potential for elucidating broadly conserved characteristics with applications to parasite control.
We further examined functions that may reflect adaptive features located in the A. suum IL (the full list along with comprehensive annotation of the 63 proteins in this compartment is provided in supplemental Table S1). Orthologs for 35 of these proteins were identified in only five or fewer of the other nematode species, and no orthologs were found in other species for 20 of those (p = 1.1 × 10−6 for enrichment of A. suum-specific proteins, compared with all other detected proteins, binomial distribution test). Functional annotation (supplemental Table S1) was established for all but seven of the lumen proteins. Hence, the diversification of proteins observed in this compartment appears to involve protein orthologs of broadly established protein families, rather than de novo inventions of A. suum. Although a wide range of functions is represented by the lumen proteins, three functional categories dominated the subset of 20 lumen proteins that were specific (no apparent ortholog in other species) to A. suum: peptidases (three M1 peptidases and one aspartic peptidase); glycoside hydrolases (five); and type A Von Willebrand factor-containing proteins (three; associated with C-type lectin binding (55), and reported in the intestine of other nematode species (56–58)). Hence, these adult A. suum proteins may have relatively high importance in the adaptation of this species to the trophic niche presented by swine and human hosts. Alternatively, neutral (instead of adaptive) diversification of these proteins could explain the noted diversity, and cannot be excluded at this time. In either case, the results identified an intestinal compartment with proteins that show relatively high diversity among nematode species (in comparison to other compartments evaluated), and identify a distinct subset of protein functions that may have contributed disproportionately to this diversity.
Although the methods used do not distinguish more specific molecular details of adaptations, (as reported for intestinal hemoglobinase activity in hookworms (59)), they do provide a quantitative approach to deduce, at a higher level, protein subsets that appear to have collectively contributed more significantly to species diversification than other protein subsets.
Functional Enrichment Analyses Reveal Known and Novel Functions for Intestinal Compartments
Next, we evaluated the inferred functions of all intestinal proteins detected and in relation to intestinal compartments to which they were assigned. GO enrichment analysis was first used to identify notable protein functions from the various compartments.
The pseudocoelomic cavity is a fluid-filled cavity that surrounds the intestine and other internal organs within the external body wall (hypodermis and musculature) of the nematode, providing hydrostatic pressure and lubrication, as well as serving as a medium for intercellular signaling and nutrient transport (60). In our experimental design, the PF was obtained primarily as a control to assess contamination of intestinal perfusates by this fluid during sample collection, so information reported here is included primarily for the record. A more detailed proteomic analysis of the A. suum PF (with a focus on excretory/secretory products) can be found in a recent study (16). GO enrichment analysis of the PF proteins identified “lipid transporter activity” as being significantly enriched (GO:0005319; p = 4.3 × 10−4, Table II), which is consistent with previous studies showing that these proteins are abundant in the A. suum PF (16), and that lipid-transporting yolk proteins (vitellogenins) are secreted into the pseudocoelomic cavity of C. elegans after synthesis in the intestine (61). However, the most significantly enriched term was “serine-type endopeptidase inhibitor activity” (GO:0004867; p = 6.0 × 10−8), which has been associated with the PF of Ancylostoma species (62), and was not specifically noted in the previous study analyzing A. suum PF proteins (16). The most significantly enriched Interpro domain among the proteins isolated from the PF was the “transthyretin-like” (ttl) domain (IPR001534; p = 9.5 × 10−7; Table III), which is consistent with the previous discovery of these proteins specifically in the pseudocoelomic cavity of A. suum (16) and the parasitic nematode Ostertagia ostertagi (63). Proteins containing ttl domains have been associated with parasite–host interactions in several parasitic nematode species, though their exact function remains unknown (64).
The IIM proteins had a significantly higher average molecular weight than the complete protein set, likely due in part to their large number of transmembrane domains (p = 7 × 10−4 according to a t test; Fig. 3). In support of the methods used to derive the IIM set of proteins, the “cellular component: integral to membrane” and “biological process: transmembrane transport” GO terms were significantly enriched among detected proteins in this tissue (GO:0016021 and GO:0055085; p = 2.3 × 10−10 and 1.1 × 10−4, respectively). The only enriched “Molecular Function” (MF) GO term found for this compartment was “transferase activity, transferring hexosyl groups” (GO:0016758; p = 1.5 × 10−5; Table II), an activity common among genes highly or exclusively expressed in the nematode intestine, for a variety of purposes including maintaining bacterial flora, producing mucin and modulating fat storage (4). Interpro domain enrichment testing showed that “UDP-glucuronosyl/UDP-glucosyltransferase” domains were the most significantly enriched among IIM proteins (IPR002213; p = 3.3 × 10−4; Table III); UDP-glycosyltransferases are found in all living organisms, and in C. elegans they are primarily responsible for the detoxification of foreign compounds (65, 66). The IIM proteins were also enriched for many ABC transporter domain-containing proteins (Table III) that affect the absorption, distribution, and elimination of xenobiotics, including known anthelmintic drugs (67, 68). Multiple examples of P-glycoprotein ABC transporters are located on the AIM of C. elegans (4). Among the other significantly enriched Interpro domains was “Cytochrome b5” (IPR001199; p = 9.7 × 10−3), which is consistent with the previous observation of cytochrome b5 activity in the A. suum intestine (though not specifically in the intestinal membrane), where it functions to regenerate functional ferrous hemoglobin (69). Notably, cytochromes (including P450 and b5), UDP-transferases and ABC transporters represent components of each of a three-phase system, respectively, for detoxification of eukaryotic cells, including in the C. elegans intestine (70). These observations may indicate the existence of a related system in A. suum intestinal cells, in parallel to what is known for C. elegans.
The PIM proteins had a significantly higher average molecular weight than the complete protein set (p = 3.4 × 10−7), with an even higher average value than for the IIM proteins (Fig. 3). The top two most significantly enriched GO terms among the PIM proteins were “proton-transporting ATPase activity, rotational mechanism” and its sister term “hydrogen ion transporting ATP synthase activity, rotational” (GO:0046961 and GO:0046933; p = 1.1 × 10−5 and p = 1.7 × 10−4, respectively), both of which are associated with Vacuolar-type (V-type) ATPases, which regulate pH and ultimately nutrient absorption in the nematode intestine via proton pumping across the apical intestinal membrane surface (71). Here, seven of the eight V1 domain subunits (A-F, and H; based on KEGG classifications) were detected in the PIM. Although this V1 domain is found on the cytoplasm portion of the protein complex, it has been previously shown that urea sufficiently disrupts this protein complex to release V1 subunits from the cytoplasmic side of the membrane (72), hence potentially explaining the ability to detect them in our PIM sample (4MU perfusate). The membrane-bound V0 domain subunit C (forming the ring structure through which proteins pass) and subunit I, which are bound in but exposed on the apical surface, were found in the IIM protein set, suggesting that disruption of these membrane-bound subunits may have released the V1 subunit into the 4MU perfusate sample. Consideration should be made for the possibility that other PIM proteins described in this study may actually be embedded on the cytoplasmic side of the apical intestinal membrane, although GO annotations were used to minimize this contamination (Fig. 1B; see Experimental Procedures). Because the 4MU fraction was obtained by perfusion of the intestinal lumen, ATPase and other proteins contained in the set are expected to be associated with the AIM, and localization of V-type ATPases on the C. elegans AIM provides additional support for this likelihood (73).
The average isoelectric point of the 63 IL proteins was 5.84, significantly lower (more acidic) than the other detected proteins (p = 4.1 × 10−10; Fig. 3). This is consistent with the acidic environment of the intestinal lumen (estimated to be pH ∼ 5 in C. elegans (4), but not previously documented for A. suum). The proteins in the IL were significantly enriched for functions related to proteolysis and peptidase activity (Tables II and III) including “aspartic-type endopeptidase activity” (GO:0004190; p = 6.3 × 10−5), particularly from the A1 class (IPR001461; p = 4.8 × 10−4) and “metallopeptidase activity” (GO:0008237; p = 8.7 × 10−5), particularly from the M1 and M13 classes (IPR001930 and IPR008753; p = 2.3 × 10−5 and 2.5 × 10−3, respectively), reflecting the prominent role of digestion in this intestinal compartment. Although classified as IL proteins, many IL proteins were also detected in the 4MU sample, which can be explained if they originally exist on the AIM surface, possibly as peripheral membrane proteins, and then are released into the lumen. Consistent with previous research, transcripts encoding these classes of hydrolases were the most abundant in C. elegans relative to all intestinal transcripts and were highly abundant in the H. contortus intestine (58).
In the IT (compared with all detected proteins in all compartments), the only “Molecular Function” (MF) GO term that was significantly enriched was “structural constituent of ribosome” (GO:0003735; p = 2.2 × 10−6; Table II), likely because the membranes and intestinal lumen do not contain protein synthesis machinery.
These enrichment results demonstrate the value of the filtering criteria shown in Fig. 1B. Without the filtering, the PBS sample used to derive the IL protein set is enriched for seven different PF GO functions or IPR domains and fewer of the IL functions, with less significance (supplemental Table S3). None of the other unfiltered protein sets used to derive compartment protein sets showed any significance for any GO terms or IPR domains.
Functional Characterization of Specific Proteins of Interest
In addition to the broad functional characterization of all proteins in each compartment, specific proteins of interest in each intestinal compartment were identified and researched in further detail. These proteins were selected based on their annotations and classifications (e.g. proteins classified as “channel and transporter proteins” (74)) as well as their phylogenetic distribution (with a particular focus on proteins which were conserved across nematodes but absent in the host groups and outgroup species).
Of the 118 proteins detected in the PF, 54 (45.7%) had orthologs exclusively in nematodes, which is significantly higher than the 27.7% found across all detected proteins (p = 1 × 10−5; binomial distribution test). This result likely reflects functions of a fundamental and specialized nature for nematodes. The only “biological process” GO term that was significantly enriched among these 54 nematode-specific PF proteins was “oxygen transport” (GO:0015671, p = 7.2 × 10−4), confirming previous observations of nematode-specific oxygen transport proteins in this compartment (75).
A total of 150 proteins (14.3%) identified in this study were “channel and transporter” proteins according to hits to the Transporter Classification Database (TCDB), as outlined in the A. suum genome publication (12) (supplemental Table S1). The IIM proteins were significantly enriched (p = 7.4 × 10−6) for channels and transporters, with 28 out 81 (34.6%) having this annotation, including nine ATPase proteins (five of which were also annotated as ABC transmembrane transport proteins) and four NAD(P) transhydrogenase proteins. Four of the IIM channel/transport proteins only had orthologs among nematode species. Two of these were A. suum-specific: GS_21555, which had no additional annotation, and GS_15007, which was annotated with a “Sulfate transporter/antisigma-factor antagonist” (STAS) IPR domain (IPR002645). Proteins with STAS domains can transport a wide range of nutrients, and are specifically associated with salt absorption in the human intestine, but have not been analyzed in detail in nematodes (76). The third nematode-specific IIM channel/transport protein (GS_23107) had orthologs in eight other nematodes from clades III, IVa and V, including O-Acetyltransferase (OAC)-39 and OAC-55 in C. elegans (neither of which have been previously described in detail). This protein also contains “Nose resistant-to-fluoxetine protein” IPR domain (IPR006621), which is associated with drug and yolk transport across the basal intestinal membrane surface in C. elegans (77). Finally, the fourth nematode-specific IIM channel/transport protein (GS_08530) was conserved across all nematode species studied. GS_08530 was annotated with a “Major facilitator superfamily domain” Interpro domain (IPR020846) and the “transmembrane transporter activity” GO term (GO:0022857), and was orthologous to Organic Anion Transport-1 (OAT-1) in C. elegans (T01B11.7; also involved in xenobiotic elimination (78)).
Ten proteins containing the “von Willebrand factor, type A” (VWA) IPR domain (IPR002035) were found among all the intestinal compartments, including eight in the IL (for which there was significant enrichment; p = 2.7 × 10−6). VWA domains are predominantly associated with extracellular cell adhesion, and nematodes have separately evolved a novel set of VWA-containing proteins with C-type lectin domains (56). Accordingly, all eight of the IL VWA-containing proteins were specific to nematodes. Five of these were also annotated with “C-type lectin” IPR domains (IPR001304; also enriched in the IL; p = 2.0 × 10−4), three of which (GS_04559, GS_05739, and GS_12996) shared orthology to 14 C. elegans proteins including CLEC-60 and CLEC-61, which play a role in bacterial pathogen defense in the C. elegans intestine (79, 80). The three VWA-containing proteins without a “C-type lectin” IPR domain were all A. suum-specific, and may perform an expanded function in A. suum. The results here support the previous hypothesis that VWA-containing bacterial defense proteins are secreted into the IL (56), and present several A. suum-specific VWA-containing proteins (GS_16378, GS_08951, and GS_06178) and two unique to several parasitic nematodes (GS_17824 and GS_02845). Additionally, seven of the 21 IL proteins which were conserved in two or more nematodes but absent in all hosts and outgroup species were predicted to be involved in proteolysis (GO:0006508). Three of these 21 proteins (GS_19445, GS_15316, GS_14901) were predicted APR-1 orthologs (also known as necepsin-1, a well-characterized aspartic protease involved in blood feeding in nematodes (81)).
Collectively, the proteins present in the various A. suum intestinal compartments provided unique and deep insight on a wide range of proteins that are sited in the adult female A. suum intestinal lumen, on the AIM and likely inserted into the AIM as integral membrane proteins. Phylogenetic application of the results indicates relevance ranging from a single species (A. suum) up to all studied species spanning the phylum Nematoda. This extensive addition of inferred functions in these intestinal cell compartments was facilitated by methods that are enabled by the large size of the A. suum intestine. Although the perfusates provide the highest confidence on location to the apical side of the cell, membrane proteins in the IIM compartment may have a basal or apical membrane location. Nevertheless, some IIM proteins have orthologs known to reside on the AIM in other nematodes, whereas other IIM proteins have predicted functions consistent with a location sited at the AIM.
Transcriptional Regulation of Genes Encoding Intestinal Compartment-Specific Proteins
Enriched putative transcription factor (TF) consensus binding sequences (“motifs”) were identified among proteins assigned to each intestinal compartment, based on the maximum 2000bp upstream region (UR) sequences of the genes corresponding to each protein compartment, compared with the URs of all other genes (12) (using DREME (34); Fig. 4; See Experimental Procedures). Motifs were associated with putative transcription factors by comparing to predicted motifs from the C. elegans regulatory analysis portion of the modENCODE project, which included predicted consensus binding sequences for 71.4% of known C. elegans TFs (33). In addition, known C. elegans motifs from JASPAR (36) and UniProbe (37) were included in the search, as well as vertebrate and mouse (respectively) sequences from the same databases. Vertebrate sequences were included because (as the modENCODE project demonstrated (33)) orthologous transcription factors from vertebrates to nematodes maintain conserved sequence preferences, so these annotations were used when no significant C. elegans transcription factor matches were annotated (as previously performed for A. suum tissue-specific gene expression TF annotation, before the modENCODE database was available (15)). Fig. 4 shows the results of this TF search approach for the top five annotated motifs from each intestinal compartment (discussed below). The intestinal tissue, which contained the most total proteins, had the largest number of significantly enriched motifs at 47, although none were found for the smaller protein set in the intestinal lumen.
Fig. 4.
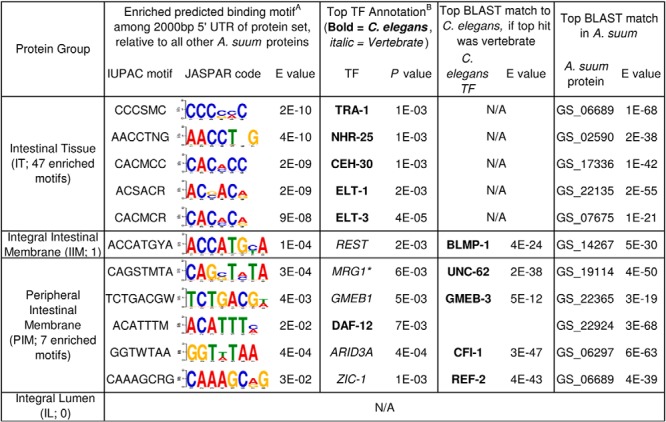
Identification of significantly enriched predicted binding motifs, their predicted transcription factor annotations in C. elegans (based either on direct annotation or homology in cases of vertebrate TF annotation), and their predicted orthologs in A. suum. A, motifs are annotated using DREME (34). B,TFs are annotated using TOMTOM (35).
The C. elegans transcription factors matched to the five most significantly enriched predicted motifs among the IT proteins were annotated as: (1) TRA-1, which is responsible for gender-specific differentiation of the C. elegans intestine, with mutants having hermaphrodite intestines in otherwise male bodies (82). A previous gene expression study in A. suum (14) found that the intestine expressed many more gender-specific genes (and had many more gender-specific functions) than other somatic tissues tested; (2) NHR-25, which is expressed in the C. elegans intestine and regulates an endocrine signaling cascade that ultimately modulates dietary fat uptake and de novo fat synthesis (83); (3) CEH-30, an anti-apoptotic gene regulating sexual identity and is transcriptionally repressed by TRA-1 in C. elegans (84) (the TF for the most significantly enriched motif in the intestinal tissue); (4) ELT-1, and (5) ELT-3, which are functionally redundant GATA transcription factors responsible for early intestine development in C. elegans (85), but only ELT-3 has been implicated in controlling the expression of age-related genes in the intestine (86).
The only significantly enriched motif in the IIM was annotated as C. elegans BLMP-1 (inferred from sequence similarity to vertebrate REST), an evolutionarily conserved TF regulating spatiotemporal cell migration patterns (87). BLMP-1 is expressed throughout the life cycle (including adult stage) in many C. elegans tissues including the intestine, and is essential for multiple cellular and developmental processes (87). In mice, BLMP-1 is responsible for the development of the intestinal epithelium from neonatal to adult stages, and most strongly affected gene expression in the brush border (the membrane exposed to the intestinal lumen) (88). Here, we find evidence for enrichment of BLMP-1 binding sites for proteins found in the integral intestinal membrane of A. suum, suggesting a conserved function in the intestine.
The C. elegans transcription factors matched to the five most significantly enriched predicted motifs among the PIM proteins were annotated as: (1) zk546.5, with unknown function. Instead, the second-best hit, UNC-62 was used (inferred from vertebrate MRG-1). UNC-62 is a developmental regulator which is expressed in diverse tissues, but its functions in the intestine play a particularly important role in modulating lifespan, via differential intestinal resource allocation between fertility and somatic functions (89); (2) GMEB-3 (inferred from vertebrate GMEB-1), has no previously described functional annotation in the literature; (3) DAF-12, like the most-significantly enriched UNC-62, is also an important modulator of lifespan, and has particularly high expression in the intestine (90); (4) CFI-1 (inferred from vertebrate ARID3A), which is not well understood, but is thought to plays a role in neuronal subtype differentiation, with detected expression and predicted function in the posterior intestine (91); (5) REF-2 (inferred from vertebrate ZIC-1), which is responsible for the regulation of TRA-2, which in turn regulates TRA-1, described previously as the most significantly enriched TF in the intestinal tissue (92). Interestingly, in A. suum, the top BLAST match for both TRA-1 and REF-2 is the same protein (GS_06689), suggesting the possibility that the complexity of this pathway may be reduced in A. suum, or that the true TRA-1 and REF-2 orthologs have similar enough sequences that they cannot be distinguished based only on sequence similarity.
Overall, this regulatory motif analysis presents a novel approach for functional annotation of genes encoding specific protein sets in a recently annotated nematode genome, using all relevant sequence and consensus binding motif information available. Based on upstream region sequence data, we not only identify homologs of TFs previously annotated as being involved in intestinal activity in nematodes, but we also identify several of the most significantly enriched TFs (TRA-1 and CEH-30 in the IT, and REF-2 in the PIM) as being involved in the same C, elegans pathway related to lifespan determination originating in the intestine.
Identifying Potential Control Targets
The confirmation of compartment specific intestinal proteins, combined with their annotation and phylogenetic distribution, allows for the identification of promising potential targets for anthelmintic drugs and vaccine development. In order to further identify proteins of interest as anthelmintic drug targets, all detected proteins were searched against the Protein Data Bank (PDB) (50) to identify homologs, and PDB structures were searched in the Chembl DrugEBIlity database (51), resulting in the identification of potentially druggable A. suum proteins, the identification of the putative drugs which may target them, and scores for both druggability and ENSEMBL (supplemental Table S1; See Experimental Procedures).
Druggable scores were assigned for 34.6% of all proteins, with druggable proteins being over-represented in both the PIM and IL (46% in each; p = 0.01 and 0.02, respectively). This database of druggability information can be used here to further prioritize target protein sets of interest. For example, all five A. suum-specific glycoside hydrolases in the IL were predicted to be druggable, making these a particularly interesting target. One protein of particular interest, GS_15316, was one of six druggable proteins detected in the IL which had no host orthologs, druggability scores of 1 and ENSEMBL scores of more than 0.5. This gene is an A. suum ortholog of necepsin-1 (also known as Na-apr-2), which is involved in host interaction and degradation of host molecules including hemoglobin, and was previously suggested to be a strong vaccine candidate (59, 93). Here, as previously described based on a reliable alignment structure (59), although the protein is relatively divergent, it has homology to the human cathepsin E protein (PDB structure 1TZS). Sequence alignments of all the 47 nematode orthologs of this gene (spanning all tested nematodes except for D. immitis and T. spiralis) showed that there is long stretch of insertion near the N terminus in comparison with the human Cathepsin sequence (for most of the nematode orthologs of GS_15316), close to the potential ligand binding site, that is likely to lead to differential drug binding between the host and nematode species, and which may be specifically exploited for targeting. The druggability information (Supplemental Table S1) will be useful in future research, providing a resource for downstream experimental testing.
In addition to targets known to be druggable because of homology with known drug targets in databases, nematode-specific proteins with no close human orthologs are also promising targets. For example, the V-type ATPase proteins found in the PIM (described above) have been considered to be promising anthelmintic drug targets, because they are conserved across nematodes (and distinct enough from host proteins for selective targeting), are present in all life cycle stages, and are critical for intestinal nutrient uptake, development, osmoregulation and detoxification (71). Three PIM proteins (GS_05894, GS_11702 and GS_13614) were also identified to have orthologs in every one of the 16 nematode species, but none in the host or outgroup species (i.e. nematode-specific and conserved; Fig. 2). GS_05894 is orthologous to C. elegans Profilin-2 (PFN-2; F35C8.6) which is the only C. elegans pfn ortholog found in the intestinal cell membrane, where it modulates cell-cell contact and cell structure through binding to actin, poly(l-proline) and phosphatidylinositol 4,5-bisphosphate micelles (94). This suggests that orthologs of PFN-2 could serve as a novel target for disrupting intestinal membrane integrity in parasitic nematodes while not affecting the mammalian host. Likewise, GS_11702 is orthologous to the C. elegans glycolytic and gluconeogenic protein “cofactor-independent phosphoglycerate mutase” (iPGM; F57B10.3a), which has been previously identified as being conserved across nematodes but distinct from mammalian PGM (95, 96). Although iPGM was previously considered an excellent potential drug target for the control of parasitic nematodes, studies have recently shown that it has limited druggability because of difficulty in accessing the active site (97). GS_13614 is orthologous to an unannotated C. elegans gene (F42C5.9). However, Interproscan on the A. suum sequence identified a “Pleckstrin homology (PH) domain” (IPR001849) which plays a crucial role in diverse functions, including intracellular signaling, nuclear transport and protein localization (98). Although pleckstrin domains target proteins to cellular membranes (99), they are most likely on the cytoplasmic side of the membrane. As inferred previously in our study, proteins closely associated with the cytoplasmic side of the AIM may have been released by the 4MU treatment used to derive the PIM protein set.
CONCLUSIONS
The ability to perfuse the A. suum intestine allowed us to directly investigate functions that localize to several compartments of the nematode intestine in a manner not previously achievable. Those compartments include the lumen (IL), and both integral (IIM) and peripheral membrane proteins (PIM) that are components of the AIM. We have generated for A. suum, and potentially many other nematodes, evidence for specific intestinal functions related to nutrient digestion and absorption, detoxification of environmental compounds, reproduction, antibacterial protection and physiological homeostasis, among many others. We provide inferred transcriptional factor activity and binding sites enriched among specific intestinal compartments, identifying enriched transcriptional regulation from sex-determinant and lifespan-determinant transcription factors. We also provide the first pan-phylum comparison among nematodes on the relative conservation of proteins that occupy distinct cellular compartments, which showed that A. suum IL proteins appear to have contributed disproportionately to species adaptations compared with other intestinal cell compartments. For each compartment, protein constituents, their detailed annotation, and their conservation among other species allow for the identification of proteins with functions of likely central importance for nematodes, proteins with novel functions not previously described in the nematode intestine, and proteins which may present promising anthelmintic/vaccine targets. The AIM proteins may hold particular significance for drug targeting because of their direct exposure to the surrounding environment including components of the host immune system. Among the most interesting proteins for further study are a number of IIM proteins that are apparent nematode-specific channel proteins and components of apparent detoxification systems. This rich and well-annotated proteomic data set presents many opportunities to clarify essential functions of nematode intestinal cells and develop methods by which those functions can be disrupted.
Supplementary Material
Acknowledgments
We thank Qi Wang for assistance with the protein modeling, physical property calculation, and OrthoMCL.
Footnotes
Author contributions: D.P.J. and M.M. designed research; B.A.R., R.T., and D.P.J. performed research; B.A.R. and M.M. analyzed data; B.A.R., D.P.J., and M.M. wrote the paper.
* This work was supported by NIH/NIGMS Grant No. GM097435 to MM.
 This article contains supplemental Tables S1 to S3.
This article contains supplemental Tables S1 to S3.
1 The abbreviations used are:
- IT
- intestinal tissue
- IL
- intestinal lumen
- PIM
- peripheral intestinal membrane
- IIT
- integral intestinal membrane
- PF
- pseudocoelomic fluid
- AIM
- apical intestinal membrane
- M-F1 to M-F7
- male intestinal tissue fractions
- F-F1 to F-F7
- female intestinal tissue fractions
- P2
- pellet from 5Kg to 50Kg centrifugation
- PBS
- PBS perfusate
- 4MU
- 4M urea + PBS perfusate
- UR
- Upstream region.
REFERENCES
- 1. Jasmer D. P., Goverse A., Smant G. (2003) Parasitic nematode interactions with mammals and plants. Annu. Rev. Phytopathol. 41, 245–270 [DOI] [PubMed] [Google Scholar]
- 2. Hotez P. J., Molyneux D. H., Fenwick A., Ottesen E., Ehrlich Sachs S., Sachs J. D. (2006) Incorporating a rapid-impact package for neglected tropical diseases with programs for HIV/AIDS, tuberculosis, and malaria. PLoS Med. 3, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yin Y., Martin J., Abubucker S., Scott A. L., McCarter J. P., Wilson R. K., Jasmer D. P., Mitreva M. (2008) Intestinal transcriptomes of nematodes: comparison of the parasites ascaris suum and haemonchus contortus with the free-living Caenorhabditis elegans. PLoS Negl. Trop. Dis. 2, e269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McGhee J. D. The C. elegans intestine. In: Community, T. C. e. R., ed. WormBook, WormBook [DOI] [PMC free article] [PubMed]
- 5. Schulenburg H., Kurz C. L., Ewbank J. J. (2004) Evolution of the innate immune system: the worm perspective. Immunol. Rev. 198, 36–58 [DOI] [PubMed] [Google Scholar]
- 6. Park B. J., Lee D. G., Yu J. R., Jung S. K., Choi K., Lee J., Kim Y. S., Lee J. I., Kwon J. Y., Singson A., Song W. K., Eom S. H., Park C. S., Kim D. H., Bandyopadhyay J., Ahnn J. (2001) Calreticulin, a calcium-binding molecular chaperone, is required for stress response and fertility in Caenorhabditis elegans. Mol. Biol. Cell 12, 2835–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nehrke K. (2003) A reduction in intestinal cell pHi due to loss of the Caenorhabditis elegans Na+/H+ exchanger NHX-2 increases life span. J. Biol. Chem. 278, 44657–44666 [DOI] [PubMed] [Google Scholar]
- 8. Jasmer D. P., Brown W. C., Parish S. (2011) Direct immunization of the abomasum or rectum of goats induces local lymph node responses against Haemonchus contortus mucosal antigens. Vaccine 29, 2938–2946 [DOI] [PubMed] [Google Scholar]
- 9. Laing R., Kikuchi T., Martinelli A., Tsai I. J., Beech R. N., Redman E., Holroyd N., Bartley D. J., Beasley H., Britton C., Curran D., Devaney E., Gilabert A., Hunt M., Jackson F., Johnston S., Kryukov I., Li K., Morrison A. A., Reid A. J., Sargison N., Saunders G., Wasmuth J. D., Wolstenholme A., Berriman M., Gilleard J. S., Cotton J. A. (2013) The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 14, R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Richmond J. (2009) Dissecting and recording from the C. Elegans neuromuscular junction. J. Vis. Exp. 24, e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gilleard J. (2013) Haemonchus contortus as a paradigm and model to study anthelmintic drug resistance. Parasitology 140, 1506–1522 [DOI] [PubMed] [Google Scholar]
- 12. Jex A. R., Liu S., Li B., Young N. D., Hall R. S., Li Y., Yang L., Zeng N., Xu X., Xiong Z., Chen F., Wu X., Zhang G., Fang X., Kang Y., Anderson G. A., Harris T. W., Campbell B. E., Vlaminck J., Wang T., Cantacessi C., Schwarz E. M., Ranganathan S., Geldhof P., Nejsum P., Sternberg P. W., Yang H., Wang J., Wang J., Gasser R. B. (2011) Ascaris suum draft genome. Nature 479, 529–533 [DOI] [PubMed] [Google Scholar]
- 13. Wang J., Mitreva M., Berriman M., Thorne A., Magrini V., Koutsovoulos G., Kumar S., Blaxter M. L., Davis R. E. (2012) Silencing of germline-expressed genes by DNA elimination in somatic cells. Dev. Cell 23, 1072–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Z., Gao X., Martin J., Yin Y., Abubucker S., Rash A. C., Li B. W., Nash B., Hallsworth-Pepin K., Jasmer D. P., Mitreva M. (2013) Gene expression analysis distinguishes tissue-specific and gender-related functions among adult Ascaris suum tissues. Mol. Genet. Genomics 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rosa B. A., Jasmer D. P., Mitreva M. (2014) Genome-wide tissue-specific gene expression, co-expression and regulation of co-expressed genes in adult nematode Ascaris suum. PLoS Negl. Trop. Dis. 8, e2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chehayeb J. F., Robertson A. P., Martin R. J., Geary T. G. (2014) Proteomic analysis of adult ascaris suum fluid compartments and secretory products. PLoS Negl. Trop. Dis. 8, e2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 18. Chen Z. W., Fuchs K., Sieghart W., Townsend R. R., Evers A. S. (2012) Deep amino acid sequencing of native brain GABAA receptors using high-resolution mass spectrometry. Mol. Cell. Proteomics 11, 011445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nittis T., Guittat L., LeDuc R. D., Dao B., Duxin J. P., Rohrs H., Townsend R. R., Stewart S. A. (2010) Revealing novel telomere proteins using in vivo cross-linking, tandem affinity purification, and label-free quantitative LC-FTICR-MS. Mol. Cell. Proteomics 9, 1144–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 21. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 22. Meyer M. R., Lichti C. F., Townsend R. R., Rao A. G. (2011) Identification of in vitro autophosphorylation sites and effects of phosphorylation on the Arabidopsis CRINKLY4 (ACR4) receptor-like kinase intracellular domain: insights into conformation, oligomerization, and activity. Biochemistry 50, 2170–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones P., Binns D., Chang H. Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., Pesseat S., Quinn A. F., Sangrador-Vegas A., Scheremetjew M., Yong S. Y., Lopez R., Hunter S. (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zdobnov E. M., Apweiler R. (2001) InterProScan – an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17, 847–848 [DOI] [PubMed] [Google Scholar]
- 25. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T. K., Bateman A., Bernard T., Binns D., Bork P., Burge S., de Castro E., Coggill P., Corbett M., Das U., Daugherty L., Duquenne L., Finn R. D., Fraser M., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., McMenamin C., Mi H., Mutowo-Muellenet P., Mulder N., Natale D., Orengo C., Pesseat S., Punta M., Quinn A. F., Rivoire C., Sangrador-Vegas A., Selengut J. D., Sigrist C. J., Scheremetjew M., Tate J., Thimmajanarthanan M., Thomas P. D., Wu C. H., Yeats C., Yong S. Y. (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, D306–D312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, D109–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., Kanehisa M. (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kall L., Krogh A., Sonnhammer E. L. (2007) Advantages of combined transmembrane topology and signal peptide prediction – the Phobius web server. Nucleic Acids Res. 35, W429–W432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bendtsen J. D., Jensen L. J., Blom N., Von Heijne G., Brunak S. (2004) Feature-based prediction of nonclassical and leaderless protein secretion. Protein Eng. Des. Sel. 17, 349–356 [DOI] [PubMed] [Google Scholar]
- 32. Prufer K., Muetzel B., Do H., Weiss G., Khaitovich P., Rahm E., Paabo S., Lachmann M., Enard W. (2007) FUNC: a package for detecting significant associations between gene sets and Ontological annotations. BMC Bioinformatics 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Araya C. L., Kawli T., Kundaje A., Jiang L., Wu B., Vafeados D., Terrell R., Weissdepp P., Gevirtzman L., Mace D., Niu W., Boyle A. P., Xie D., Ma L., Murray J. I., Reinke V., Waterston R. H., Snyder M. (2014) Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature 512, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bailey T. L. (2011) DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics 27, 1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gupta S., Stamatoyannopoulos J., Bailey T., Noble W. (2007) Quantifying similarity between motifs. Genome Biol. 8, R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bryne J. C., Valen E., Tang M. H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A. (2008) JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 36, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Newburger D. E., Bulyk M. L. (2009) UniPROBE: an online database of protein binding microarray data on protein–DNA interactions. Nucleic Acids Res. 37, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McGhee J. D., Sleumer M. C., Bilenky M., Wong K., McKay S. J., Goszczynski B., Tian H., Krich N. D., Khattra J., Holt R. A., Baillie D. L., Kohara Y., Marra M. A., Jones S. J., Moerman D. G., Robertson A. G. (2007) The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 302, 627–645 [DOI] [PubMed] [Google Scholar]
- 39. Muers M. (2011) Functional genomics: the modENCODE guide to the genome. Nat. Rev. Genet. 12, 80. [DOI] [PubMed] [Google Scholar]
- 40. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 41. Kinsella R. J., Kahari A., Haider S., Zamora J., Proctor G., Spudich G., Almeida-King J., Staines D., Derwent P., Kerhornou A., Kersey P., Flicek P. (2011) Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harris T. W., Baran J., Bieri T., Cabunoc A., Chan J., Chen W. J., Davis P., Done J., Grove C., Howe K., Kishore R., Lee R., Li Y., Muller H. M., Nakamura C., Ozersky P., Paulini M., Raciti D., Schindelman G., Tuli M. A., Auken K. V., Wang D., Wang X., Williams G., Wong J. D., Yook K., Schedl T., Hodgkin J., Berriman M., Kersey P., Spieth J., Stein L., Sternberg P. W. (2013) WormBase 2014: new views of curated biology. Nucleic Acids Res. 42, D789–D793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mitreva M., Jasmer D. P., Zarlenga D. S., Wang Z., Abubucker S., Martin J., Taylor C. M., Yin Y., Fulton L., Minx P., Yang S. P., Warren W. C., Fulton R. S., Bhonagiri V., Zhang X., Hallsworth-Pepin K., Clifton S. W., McCarter J. P., Appleton J., Mardis E. R., Wilson R. K. (2011) The draft genome of the parasitic nematode Trichinella spiralis. Nat. Genet. 43, 228–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jex A. R., Liu S., Li B., Young N. D., Hall R. S., Li Y., Yang L., Zeng N., Xu X., Xiong Z., Chen F., Wu X., Zhang G., Fang X., Kang Y., Anderson G. A., Harris T. W., Campbell B. E., Vlaminck J., Wang T., Cantacessi C., Schwarz E. M., Ranganathan S., Geldhof P., Nejsum P., Sternberg P. W., Yang H., Wang J., Wang J., Gasser R. B. (2011) Ascaris suum draft genome. Nature 479, 529–533 [DOI] [PubMed] [Google Scholar]
- 45. Laing R., Kikuchi T., Martinelli A., Tsai I. J., Beech R. N., Redman E., Holroyd N., Bartley D. J., Beasley H., Britton C., Curran D., Devaney E., Gilabert A., Hunt M., Jackson F., Johnston S. L., Kryukov I., Li K., Morrison A. A., Reid A. J., Sargison N., Saunders G. I., Wasmuth J. D., Wolstenholme A., Berriman M., Gilleard J. S., Cotton J. A. (2013) The genome and transcriptome of Haemonchus contortus, a key model parasite for drug and vaccine discovery. Genome Biol. 14, R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Godel C., Kumar S., Koutsovoulos G., Ludin P., Nilsson D., Comandatore F., Wrobel N., Thompson M., Schmid C. D., Goto S., Bringaud F., Wolstenholme A., Bandi C., Epe C., Kaminsky R., Blaxter M., Mäser P. (2012) The genome of the heartworm, Dirofilaria immitis, reveals drug and vaccine targets. FASEB J. 26, 4650–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tang Y. T., Gao X., Rosa B. A., Abubucker S., Hallsworth-Pepin K., Martin J., Tyagi R., Heizer E., Zhang X., Bhonagiri-Palsikar V., Minx P., Warren W. C., Wang Q., Zhan B., Hotez P. J., Sternberg P. W., Dougall A., Gaze S. T., Mulvenna J., Sotillo J., Ranganathan S., Rabelo E. M., Wilson R. K., Felgner P. L., Bethony J., Hawdon J. M., Gasser R. B., Loukas A., Mitreva M. (2014) Genome of the human hookworm Necator americanus. Nat. Genet. advance online publication 46, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fischer S., Brunk B. P., Chen F., Gao X., Harb O. S., Iodice J. B., Shanmugam D., Roos D. S., Stoeckert C. J., Jr. (2011) Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Current protocols in bioinformatics/editoral board, Andreas D. Baxevanis . [et al.] Chapter 6, Unit 6 12 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li L., Stoeckert C. J., Jr., Roos D. S. (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr., Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. (1977) The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 112, 535–542 [DOI] [PubMed] [Google Scholar]
- 51. Gaulton A., Bellis L. J., Bento A. P., Chambers J., Davies M., Hersey A., Light Y., McGlinchey S., Michalovich D., Al-Lazikani B., Overington J. P. (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lipinski C. A., Lombardo F., Dominy B. W., Feeney P. J. (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 [DOI] [PubMed] [Google Scholar]
- 53. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gasteiger E., Hoogland C., Gattiker A., Duvaud S. e., Wilkins M., Appel R., Bairoch A. (2005) Protein Identification and Analysis Tools on the ExPASy Server. In: Walker J., ed. The Proteomics Protocols Handbook, pp. 571–607, Humana Press [Google Scholar]
- 55. Rydz N., Swystun L. L., Notley C., Paterson A. D., Riches J. J., Sponagle K., Boonyawat B., Montgomery R. R., James P. D., Lillicrap D. (2013) The C-type lectin receptor CLEC4M binds, internalizes and clears von Willebrand factor and contributes to the variation in plasma von Willebrand factor levels. Blood 26, 5228–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Whittaker C. A., Hynes R. O. (2002) Distribution and evolution of von Willebrand/Integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell 13, 3369–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McGhee J. D., Fukushige T., Krause M. W., Minnema S. E., Goszczynski B., Gaudet J., Kohara Y., Bossinger O., Zhao Y., Khattra J., Hirst M., Jones S. J., Marra M. A., Ruzanov P., Warner A., Zapf R., Moerman D. G., Kalb J. M. (2009) ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev. Biol. 327, 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jasmer D. P., Mitreva M. D., McCarter J. P. (2004) mRNA sequences for Haemonchus contortus intestinal cathepsin B-like cysteine proteases display an extreme in abundance and diversity compared with other adult mammalian parasitic nematodes. Mol. Biochem. Parasitol. 137, 297–305 [DOI] [PubMed] [Google Scholar]
- 59. Williamson A. L., Brindley P. J., Abbenante G., Datu B. J. D., Prociv P., Berry C., Girdwood K., Pritchard D. I., Fairlie D. P., Hotez P. J., Zhan B., Loukas A. (2003) Hookworm aspartic protease, Na-APR-2, Cleaves human hemoglobin and serum proteins in a host-specific fashion. J. Infect. Dis. 187, 484–494 [DOI] [PubMed] [Google Scholar]
- 60. Altun Z. F., Hall D. H. (2007) Pericellular structures, WormAtlas, In WormAtlas [Google Scholar]
- 61. Fares H., Grant B. (2002) Deciphering endocytosis in Caenorhabditis elegans. Traffic 3, 11–19 [DOI] [PubMed] [Google Scholar]
- 62. Delaney A., Williamson A., Brand A., Ashcom J., Varghese G., Goud G. N., Hawdon J. M. (2005) Cloning and characterisation of an aspartyl protease inhibitor (API-1) from Ancylostoma hookworms. Int. J. Parasitol. 35, 303–313 [DOI] [PubMed] [Google Scholar]
- 63. Saverwyns H., Visser A., Van Durme J., Power D., Morgado I., Kennedy M. W., Knox D. P., Schymkowitz J., Rousseau F., Gevaert K., Vercruysse J., Claerebout E., Geldhof P. (2008) Analysis of the transthyretin-like (TTL) gene family in Ostertagia ostertagi – Comparison with other strongylid nematodes and Caenorhabditis elegans. Int. J. Parasitol. 38, 1545–1556 [DOI] [PubMed] [Google Scholar]
- 64. Jacob J., Vanholme B., Haegeman A., Gheysen G. (2007) Four transthyretin-like genes of the migratory plant-parasitic nematode Radopholus similis: members of an extensive nematode-specific family. Gene 402, 9–19 [DOI] [PubMed] [Google Scholar]
- 65. Hasegawa K., Miwa S., Tsutsumiuchi K., Miwa J. (2010) Allyl isothiocyanate that induces GST and UGT expression confers oxidative stress resistance on C. elegans, as demonstrated by nematode biosensor. PLoS One 5, e9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Huang F.-F., Chai C.-L., Zhang Z., Liu Z.-H., Dai F.-Y., Lu C., Xiang Z.-H. (2008) The UDP-glucosyltransferase multigene family in Bombyx mori. BMC Genomics 9, 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ménez C., Mselli-Lakhal L., Foucaud-Vignault M., Balaguer P., Alvinerie M., Lespine A. (2012) Ivermectin induces P-glycoprotein expression and function through mRNA stabilization in murine hepatocyte cell line. Biochem. Pharmacol. 83, 269–278 [DOI] [PubMed] [Google Scholar]
- 68. De Graef J., Demeler J., Skuce P., Mitreva M., Von Samson-Himmelstjerna G., Vercruysse J., Claerebout E., Geldhof P. (2013) Gene expression analysis of ABC transporters in a resistant Cooperia oncophora isolate following in vivo and in vitro exposure to macrocyclic lactones. Parasitology 140, 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hashimoto M., Takamiya S., Yokota T., Nakajima Y., Yamakura F., Sugio S., Aoki T. (2008) Ascaris suum cytochrome b5, an adult-specific secretory protein reducing oxygen-avid ferric hemoglobin. Arch. Biochem. Biophys. 471, 42–49 [DOI] [PubMed] [Google Scholar]
- 70. Oliveira R. P., Porter Abate J., Dilks K., Landis J., Ashraf J., Murphy C. T., Blackwell T. K. (2009) Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell 8, 524–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Knight A. J., Behm C. A. (2012) Minireview: the role of the vacuolar ATPase in nematodes. Exp. Parasitol. 132, 47–55 [DOI] [PubMed] [Google Scholar]
- 72. Graham L. A., Powell B., Stevens T. H. (2000) Composition and assembly of the yeast vacuolar H(+)-ATPase complex. J. Exp. Biol. 203, 61–70 [DOI] [PubMed] [Google Scholar]
- 73. Allman E., Johnson D., Nehrke K. (2009) Loss of the apical V-ATPase a-subunit VHA-6 prevents acidification of the intestinal lumen during a rhythmic behavior in C. elegans. Am. J. Physiol. Cell Physiol. 297, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Saier M. H., Jr., Reddy V. S., Tamang D. G., Vastermark A. (2014) The transporter classification database. Nucleic Acids Res. 42, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hoogewijs D., Geuens E., Dewilde S., Moens L., Vierstraete A., Vinogradov S., Vanfleteren J. (2004) Genome-wide analysis of the globin gene family of C. elegans. IUBMB Life 56, 697–702 [DOI] [PubMed] [Google Scholar]
- 76. Sharma A. K., Rigby A. C., Alper S. L. (2011) STAS domain structure and function. Cell Physiol. Biochem. 28, 407–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Choy R. K., Kemner J. M., Thomas J. H. (2006) Fluoxetine-resistance genes in Caenorhabditis elegans function in the intestine and may act in drug transport. Genetics 172, 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. George R. L., Wu X., Huang W., Fei Y.-J., Leibach F. H., Ganapathy V. (1999) Molecular Cloning and Functional Characterization of a Polyspecific Organic Anion Transporter from Caenorhabditis elegans. J. Pharmacol. Exp. Ther. 291, 596–603 [PubMed] [Google Scholar]
- 79. O'Rourke D., Baban D., Demidova M., Mott R., Hodgkin J. (2006) Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 16, 1005–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Irazoqui J. E., Troemel E. R., Feinbaum R. L., Luhachack L. G., Cezairliyan B. O., Ausubel F. M. (2010) Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6, e1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Williamson A. L., Lecchi P., Turk B. E., Choe Y., Hotez P. J., McKerrow J. H., Cantley L. C., Sajid M., Craik C. S., Loukas A. (2004) A multi-enzyme cascade of hemoglobin proteolysis in the intestine of blood-feeding hookworms. J. Biol. Chem. 279, 35950–35957 [DOI] [PubMed] [Google Scholar]
- 82. Szabo E., Hargitai B., Regos A., Tihanyi B., Barna J., Borsos E., Takacs-Vellai K., Vellai T. (2009) TRA-1/GLI controls the expression of the Hox gene lin-39 during C. elegans vulval development. Dev. Biol. 330, 339–348 [DOI] [PubMed] [Google Scholar]
- 83. Mullaney B. C., Blind R. D., Lemieux G. A., Perez C. L., Elle I. C., Faergeman N. J., Van Gilst M. R., Ingraham H. A., Ashrafi K. (2010) Regulation of C. elegans fat uptake and storage by Acyl-CoA Synthase-3 is dependent on NR5A family nuclear hormone receptor nhr-25. Cell Metab. 12, 398–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schwartz H. T., Horvitz H. R. (2007) The C. elegans protein CEH-30 protects male-specific neurons from apoptosis independently of the Bcl-2 homolog CED-9. Genes Dev. 21, 3181–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Fukushige T., Hawkins M. G., McGhee J. D. (1998) The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev. Biol. 198, 286–302 [PubMed] [Google Scholar]
- 86. Budovskaya Y. V., Wu K., Southworth L. K., Jiang M., Tedesco P., Johnson T. E., Kim S. K. (2008) An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134, 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Huang T. F., Cho C. Y., Cheng Y. T., Huang J. W., Wu Y. Z., Yeh A. Y., Nishiwaki K., Chang S. C., Wu Y. C. (2014) BLMP-1/Blimp-1 regulates the spatiotemporal cell migration pattern in C. elegans. PLoS Genet. 10, e1004428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Muncan V., Heijmans J., Krasinski S. D., Buller N. V., Wildenberg M. E., Meisner S., Radonjic M., Stapleton K. A., Lamers W. H., Biemond I., van den Bergh Weerman M. A., O'Carroll D., Hardwick J. C., Hommes D. W., van den Brink G. R. (2011) Blimp1 regulates the transition of neonatal to adult intestinal epithelium. Nat. Commun. 2, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Van Nostrand E. L., Sánchez-Blanco A., Wu B., Nguyen A., Kim S. K. (2013) Roles of the developmental regulator unc-62/Homothorax in limiting longevity in Caenorhabditis elegans. PLoS Genet. 9, e1003325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Antebi A., Yeh W. H., Tait D., Hedgecock E. M., Riddle D. L. (2000) daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 14, 1512–1527 [PMC free article] [PubMed] [Google Scholar]
- 91. Shaham S., Bargmann C. I. (2002) Control of neuronal subtype identity by the C. elegans ARID protein CFI-1. Genes Dev. 16, 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kuersten S., Segal S. P., Verheyden J., LaMartina S. M., Goodwin E. B. (2004) NXF-2, REF-1, and REF-2 Affect the choice of nuclear export pathway for tra-2 mRNA in C. elegans. Molecular Cell 14, 599–610 [DOI] [PubMed] [Google Scholar]
- 93. Hotez P. J., Ghosh K., Hawdon J. M., Narasimhan S., Jones B., Shuhua X., Sen L., Bin Z., Haechou X., Hainan R., Heng W., Koski R. A. (1999) Experimental approaches to the development of a recombinant hookworm vaccine. Immunol. Rev. 171, 163–171 [DOI] [PubMed] [Google Scholar]
- 94. Polet D., Lambrechts A., Ono K., Mah A., Peelman F., Vandekerckhove J., Baillie D. L., Ampe C., Ono S. (2006) Caenorhabditis elegans expresses three functional profilins in a tissue-specific manner. Cell Motil. Cytoskeleton 63, 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kumar S., Chaudhary K., Foster J. M., Novelli J. F., Zhang Y., Wang S., Spiro D., Ghedin E., Carlow C. K. S. (2007) Mining predicted essential genes of Brugia malayi for nematode drug targets. PLoS One 2, e1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhang Y., Foster J. M., Kumar S., Fougere M., Carlow C. K. S. (2004) Cofactor-independent phosphoglycerate mutase has an essential role in Caenorhabditis elegans and is conserved in parasitic nematodes. J. Biol. Chem. 279, 37185–37190 [DOI] [PubMed] [Google Scholar]
- 97. Crowther G. J., Booker M. L., He M., Li T., Raverdy S., Novelli J. F., He P., Dale N. R., Fife A. M., Barker R. H., Jr., Kramer M. L., Van Voorhis W. C., Carlow C. K., Wang M. W. (2014) Cofactor-independent phosphoglycerate mutase from nematodes has limited druggability, as revealed by two high-throughput screens. PLoS Negl. Trop. Dis. 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Blomberg N., Baraldi E., Sattler M., Saraste M., Nilges M. (2000) Structure of a PH domain from the C. elegans muscle protein UNC-89 suggests a novel function. Structure 8, 1079–1087 [DOI] [PubMed] [Google Scholar]
- 99. Cozier G. E., Carlton J., Bouyoucef D., Cullen P. J. (2004) Membrane targeting by pleckstrin homology domains. Curr. Top. Microbiol. Immunol. 282, 49–88 [DOI] [PubMed] [Google Scholar]
- 100. Blaxter M. L., De Ley P., Garey J. R., Liu L. X., Scheldeman P., Vierstraete A., Vanfleteren J. R., Mackey L. Y., Dorris M., Frisse L. M., Vida J. T., Thomas W. K. (1998) A molecular evolutionary framework for the phylum Nematoda. Nature 392, 71–75 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.