Abstract
Upon entry into mammalian host cells, the pathogenic bacterium Francisella must import host cell arginine to multiply actively in the host cytoplasm. We identified and functionally characterized an arginine transporter (hereafter designated ArgP) whose inactivation considerably delayed bacterial phagosomal escape and intracellular multiplication. Intramacrophagic growth of the ΔargP mutant was fully restored upon supplementation of the growth medium with excess arginine, in both F. tularensis subsp. novicida and F. tularensis subsp. holarctica LVS, demonstrating the importance of arginine acquisition in these two subspecies. High-resolution mass spectrometry revealed that arginine limitation reduced the amount of most of the ribosomal proteins in the ΔargP mutant. In response to stresses such as nutritional limitation, repression of ribosomal protein synthesis has been observed in all kingdoms of life. Arginine availability may thus contribute to the sensing of the intracellular stage of the pathogen and to trigger phagosomal egress. All MS data have been deposited in the ProteomeXchange database with identifier PXD001584 (http://proteomecentral.proteomexchange.org/dataset/PXD001584).
Francisella tularensis is a Gram-negative coccobacillus responsible for the zoonotic disease tularemia. This pathogen is considered as one of the most virulent microorganisms and has been categorized as a Category A select agent by the US Centers for Disease Control and Prevention. It is known to infect a broad range of animal species such as mammals including humans. F. tularensis is a facultative intracellular pathogen able to multiply in a variety of cell types but is thought to multiply in vivo preferentially in macrophages (1). Upon entry into macrophages, Francisella escapes within one hour from Francisella-containing phagosome to reach the cytosolic compartment where it multiplies actively (2). To multiply efficiently in its cytosolic niche, Francisella must evade the immune responses of the host (3) and possess dedicated virulence attributes (4). Francisella also needs to adapt its metabolism to the nutrients available in the infected host (5). It has, therefore, developed sophisticated tools recently designated as “nutritional virulence” attributes (6, 7).
Amino acids have been described as the essential nutrient source that Francisella has evolved to scavenge, by different means, during its intracellular life cycle (8). For examples, activation of the host macro-autophagy degradation machinery (9), and exploitation of host polypeptides such as glutathione (10), allow Francisella to obtain its essential amino acids. We recently demonstrated that Francisella possessed amino acid transporters that were required to fulfill some of its metabolic requirements during the different stages of its intracellular progression (11–13). The virulence of the intracellular pathogen Salmonella also depends on the simultaneous exploitation of numerous different host nutrients, including vitamins, carbohydrates, and amino acids (14). More generally, comparisons of the predicted nutrient utilization and biosynthetic pathways of other mammalian pathogens suggest that most of them share the capability to utilize multiple nitrogen and carbon sources.
Francisella genomes are predicted to encode 11 amino acid/polyamine/organocation (APC)1 family members (15), possibly involved in different amino acid uptake functions. Remarkably, only two of them were identified in at least four different genetic screens as potentially involved in bacterial virulence that is, the glutamate transporter GadC and FTN_0848 (in F. novicida). We have shown that GadC promoted bacterial resistance to the oxidative stress generated in the phagosomal compartment by fueling the tricarboxylic acid (TCA) cycle (13). FTN_0848 is the closest paralog of GadC in F. novicida and shares 33.4% amino acid identity with GadC. In addition, the gene FTN_0848 has been identified in multiple in vitro and in vivo screens, in both F. novicida and F. tularensis LVS (16–21), suggesting that it displays functions that could not be replaced by other Francisella transporter.
We demonstrate here that FTN-0848, which we have designated argP, encodes a high affinity arginine transporter (hence designated ArgP for simplification). Arginine is an essential amino acid that can only be obtained by intracellular Francisella via import from the host. We show that ArgP-mediated arginine uptake is crucial for efficient phagosomal escape, highlighting for the first time the importance of essential amino acids during early stage infection. The data presented further suggest that arginine limitation influences ribosomal protein synthesis.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Media, and Chemicals
F. tularensis subsp. novicida (F. novicida) strain U112, its ΔFPI derivative, F. tularensis subsp. holartica strain LVS, its ΔiglC derivative, and all the mutant strains constructed in this work, were grown as described in supplementary Materials and Methods. All bacterial strains, plasmids, and primers used in this study are listed in supplemental Table S2.
Details of the construction and characterization of mutant and complemented strains; macrophage preparation and infections, are described in supplementary Materials and Methods. Quantitative (q)RT-PCR (real-time PCR) was performed with gene-specific primers (supplemental Table S2), using an ABI PRISM 7700 and SYBR green PCR master mix (Applied Biosystems, Foster city, CA), are described in supplementary Materials and Methods.
Confocal and electron microscopy complete descriptions, real time cell death, and phagosome permeabilization assays, are described in supplementary Materials and Methods.
Stress Survival Assays
Oxidative Stress
Exponential-phase bacterial cultures in TSB broth were diluted to a final concentration of 108 bacteria ml−1 and subjected to 500 μm H2O2. The number of viable bacteria was determined by plating appropriate dilutions of bacterial cultures on Chocolate Polyvitex plates at the start of the experiment and after the indicated durations. Cultures (10 ml) were incubated at 37 °C with rotation (180 rpm) and aliquots were removed at indicated times, serially diluted and plated immediately.
pH Stress
Bacteria were incubated for 2 h in CDM at pH 4, the presence of various arginine concentrations (2.3 mm, 0.46 mm, and 0.23 mm). At 30 min intervals, bacteria were collected and numerated on TSB solid medium.
Bacteria were enumerated after 48 h incubation at 37 °C. Each point represents the average of three different plates. Experiments were repeated independently twice.
Evaluation of Nitrite Levels
NO levels in supernatants were evaluated with Griess test. Briefly, J774.1 cells were seeded at 2.105cells/well in 96 microplates. Cells were infected at an MOI of 1000 for 1 h with WT or ΔargP strains versus noninfected cells, with DMEM without red phenol. 100 μl of each cell culture supernatant were reacted with equal volumes of Griess reagent (1% sulfanilamide, 0.1% naphthylenediamine dihydrochloride, 2.5% H3PO4) at room temperature (RT) for 10 min away from light. Sodium nitrite was used to generate a standard curve for NO2− production, and peak absorbance was measured at 540 nm (Bio-Rad, iMark microplate absorbance reader). Levels present in cell medium of noninfected cells were subtracted from the values of experimental cultures.
l-NAME Effect on ΔargP Mutant
One day before infection, J774.1 cells were seeded at 2.105 cells/well in 12-well cell tissue plates, supplemented with l-NAME (NG-nitro-l-arginine methyl ester hydrochloride) at the concentration of 1 mm in DMEM. Cells were infected at an MOI 100 for 1 h. Bacteria were then counted after 1 h, 4 h, 10 h, and 24 h of infection as described previously. l-NAME was present all along the infection.
Arginine Transport Assays
Cells were grown in Chamberlain medium to mid-exponential phase and then harvested by centrifugation and washed twice with Chamberlain without amino acid. The cells were suspended at a final OD600 nm of 0.5 in the same medium containing 50 μg ml−1 of chloramphenicol. After 15 min of pre-incubation at 25 °C, uptake was started by the addition of l-[U-14C] arginine (Perkin Elmer), at various concentrations (14C-Arg ranging from 1 to 50 μm). The radiolabeled 14C-Arg was at a specific activity of 9.25 GBq.mmol−1. Samples (100 μl of bacterial suspension) were removed after 5 min and collected by vacuum filtration on membrane filters (Millipore type HA, 25 mm, 0.22 mm) and rapidly washed with Chamberlain without amino acid (2 × 5 ml). The filters were transferred to scintillation vials and counted in a Hidex 300 scintillation counter. The counts per minute (c.p.m.) were converted to picomoles of amino acid taken up per sample, using a standard derived by counting a known quantity of the same isotope under similar conditions.
Virulence Determination
All experimental procedures involving animals were conducted in accordance with guidelines established by the French and European regulations for the care and use of laboratory animals (Decree 87–848, 2001–2464, 2001–486, and 2001–131 and European Directive 2010/63/UE) and approved by the INSERM Ethics Committee (Authorization Number: 75–906).
Wild-type F. novicida and ΔargP mutant strains were grown in TSB to exponential growth phase and diluted to the appropriate concentrations. Six to eight week-old female BALB/c mice (Janvier, Le Genest St Isle, France) were intraperitoneally (intraperitoneal) inoculated with 200 μl of bacterial suspension. The actual number of viable bacteria in the inoculum was determined by plating dilutions of the bacterial suspension on chocolate plates. For competitive infections, wild-type F. novicida and mutant bacteria were mixed in 1:1 ratio and a total of 100 bacteria were used for infection of each of five mice. After 2 days, mice were sacrificed. Homogenized spleen and liver tissue from the five mice in one experiment were mixed, diluted, and spread on to chocolate agar plates. Kanamycin selection to distinguish wild-type and mutant bacteria was performed. Competitive index (CI) [(mutant output/WT output)/(mutant input/WT input)]. Statistical analysis for CI experiments was as described in (22). Macrophage experiments were analyzed by using the Student's unpaired t test.
MS/MS Protein Identification and Quantification
WT and ΔargP mutant strains were grown overnight in CDMmin. The next day, bacteria were resuspended at an OD600 of 0.1 i in CDM containing arginine at the concentration of 0.43 mm and cultivated for 2 h. The cells (50 ml) were harvested by centrifugation at 3000 × g for 20 min and resuspended in 2 ml of 5 mm Tris-HCl, pH 8. The bacterial suspensions were sonicated three times for 30 s at 4.0 output, 70% pulsed (Branson Sonifier 250). Lysates were centrifuged once again at 8000 × g for 6 min and supernatant were collected.
For label-free, relative quantitative analysis, 50 μg protein from each of the eight samples (WT and ΔargP, at 1 h and 2 h of growth, at 2.3 mm or 0.46 mm arginine) were precipitated in 80%, acetone and digested with 1 μg sequencing grade trypsin (Promega). Following an overnight digestion at 37 °C samples were vacuum dried, and resuspended in 100 μl of 10% acetonitrile, 0.1% formic acid, for LC-MS/MS.
For each run, 5 μl were injected in a nanoRSLC-Q Exactive PLUS (Dionex RSLC Ultimate 3000, Thermo Scientific, Waltham, MA). Peptides were separated on a reversed-phase liquid chromatographic column (Pepmap C18, Dionex). Chromatography solvents were (A) 0.1% formic acid in water, and (B) 80% acetonitrile and 0.08% formic acid. Peptides were eluted from the column with the following gradient 5% to 40% B (120 min) and 40% to 80% (10 min). At 131 min, the gradient returned to 5% to re-equilibrate the column for 30 min before the next injection. Two blanks, one short 40 min step gradient and one 60 min linear gradient, were run between triplicates to prevent sample carryover. Peptides eluting from the column were analyzed by data dependent MS/MS, using top-10 acquisition method. Briefly, the instrument settings were as follows: resolution was set to 70,000 for MS scans and 17,500 for the data dependent MS/MS scans in order to increase speed. The MS AGC target was set to 3.106 counts, whereas MS/MS AGC target was set to 5.104. The MS scan range was from 400 to 2000 m/z. MS and MS/MS scans were recorded in profile mode. Dynamic exclusion was set to 20 s duration. Each sample was analyzed in biological triplicate by nanoLC/MS/MS. Each biological replicate was acquired in three technical replicates.
Data Processing Following LC-MS/MS Acquisition
The MS files were processed with the MaxQuant software version 1.4.1.2 and searched with Andromeda search engine against the NCBI F. tularensis subsp. novicida database (release 28–04-2014; 1719 entries). To search parent mass and fragment ions, we set an initial mass deviation of 4.5 ppm and 0.5 Da, respectively. The minimum peptide length was set to seven amino acids and strict specificity for trypsin cleavage was required, allowing up to two missed cleavage sites. Carbamidomethylation (Cys) was set as fixed modification, whereas oxidation (Met) and N-term acetylation were set as variable modifications. The false discovery rates (FDRs) at the protein and peptide level were set to 1%. Scores were calculated in MaxQuant as described previously (Cox and Mann). The reverse and common contaminants hits were removed from MaxQuant output. Proteins were quantified according to the MaxQuant label-free algorithm using LFQ intensities (23, 24); protein quantification was obtained using at least two peptides per protein.
Statistical and bioinformatic analysis, including heatmaps, profile plots, and clustering, were performed with Perseus software (version 1.4.1.3) freely available at www.perseus-framework.org. For statistical comparison, we set two groups, each containing biological and technical triplicates, and we then filtered the data to keep only proteins with at least nine valid values out of 18. Next, the data were imputed to fill missing data points by creating a Gaussian distribution of random numbers with a standard deviation of 33% relative to the standard deviation of the measured values and 1.8 standard deviation downshift of the mean to simulate the distribution of low signal values. We performed a t test, with a permutation based testing correction that was controlled by using a false discovery rate threshold of 0.05, 250 randomizations. Hierarchical clustering of proteins that survived the test was performed in Perseus on logarithmized LFQ intensities after z-score normalization of the data, using Euclidean distances using default parameters and setting 12 clusters for the row tree.
All MS data have been deposited in the ProteomeXchange database with identifier PXD001584 (http://proteomecentral.proteomexchange.org/dataset/PXD001584). Annotated spectra can be consulted using MS-viewer (25) with the key a9omruthfd.
RESULTS
A New Member of the APC Superfamily of Transporter
The protein ArgP (FTN_0848) is highly conserved in all the F. tularensis subspecies and shares more than 99% identity with its orthologs in F. tularensis subsp. tularensis (FTT_0968c), F. tularensis subsp. holarctica (FTL_1233), and F. tularensis subsp. mediasiatica (FTM_0980). Secondary structure prediction (using the program HMMTM available at the internet site www.cbs.dtu.dk) indicates that the protein ArgP comprises 12 transmembrane helixes (not shown). The closest homolog in other bacterial species is the protein Lpg1917 of Legionella pneumophila (47.9% identity), an APC member of unknown function, also predicted to comprise 12 transmembrane helixes (http://www.membranetransport.org/protein.php?pOID=lpne1&pSynonym=lpg1917). High-resolution three-dimensional x-ray structures of several members of the APC superfamily have been published (26–32). ArgP shares only limited homology with these APC members, including the arginine/agmatine antiporter AdiC of E. coli (26% amino acid sequence identity (26).
To investigate the function of gene argP, we constructed a chromosomal deletion in F. novicida (FTN_0848) by allelic replacement (33). We checked that the deletion of argP did not have any polar effect on the expression of the flanking genes by qRT_PCR (supplemental Fig. S1). This was further confirmed by functional complementation (see below). The mutant showed wild-type growth kinetics when grown in broth, in chemically defined (34) or rich medium (supplemental Fig. S2A, S2B).
ArgP is Involved in Bacterial Multiplication and Virulence
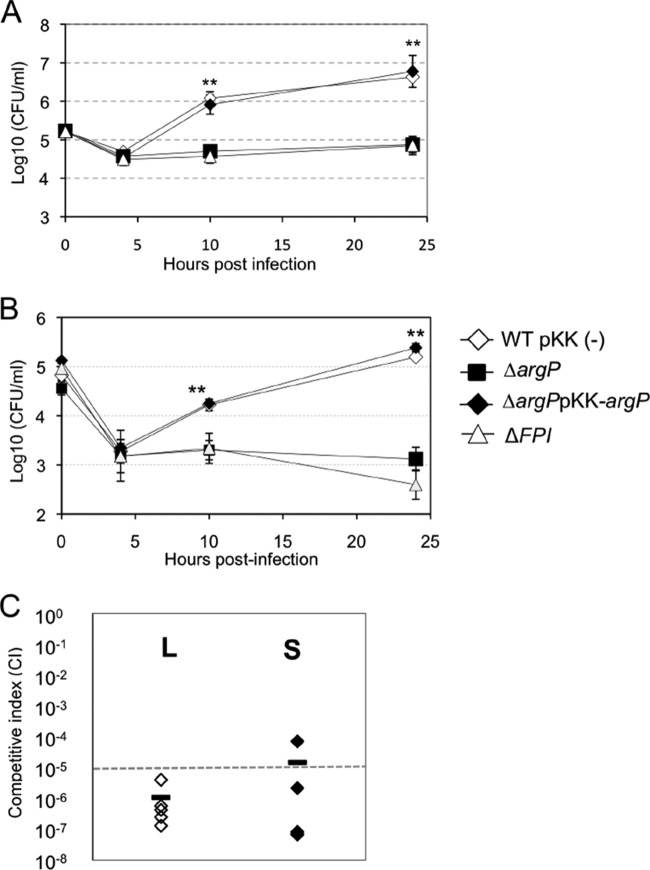
We monitored bacterial multiplication in J774.1 macrophage-like cells and bone marrow-derived macrophages (BMM) from BALB/c mice (Fig. 1A, B). Cells were incubated for 1 h with a multiplicity of infection (MOI) of 100, either with wild-type F. novicida, ΔargP mutant, ΔargP-complemented strain, or ΔFPI strain, a F. novicida mutant with a deletion of the entire Francisella pathogenicity island (16). In both cells types, the ΔargP and ΔFPI mutant strains showed a significant growth defect. At 10 h of infection, a 17-fold and 11-fold reduction of intracellular bacteria were recorded in J774.1 and BMM, respectively, as compared with wild-type F. novicida; and at 24 h, a 50-fold, and 112-fold reduction, respectively. Introduction of the complementing plasmid (pKK-214-argP) restored bacteria multiplication to wild-type levels, confirming the unique role of argP gene inactivation in the intracellular multiplication defect observed.
Fig. 1.
In vitro and in vivo properties of a Francisella ΔargP mutant. Intracellular replication of wild-type F. novicida (WT) carrying the empty plasmid pKK214 (WT pKK (-)), of the ΔargP mutant (ΔargP) and complemented strain (ΔargP pKK-argP), and of the ΔFPI mutant (ΔFPI), was monitored in J774.1 murine macrophage-like cells in A, and in murine bone marrow-derived macrophages (BMDM, B) over a 24 h-period. Results are shown as the averages of log10 (CFU/ml) ± standard deviations. Experiences were realized twice. ** p < 0.01 as determined by Student's t test. C, Virulence. Group of five BALB/c mice were infected intraperitoneally with 100 CFU of wild-type F. novicida and 100 CFU of ΔargP mutant strain. Bacterial burden was realized in liver (L: white diamonds, left column) and spleen (S: black diamonds, right column) of mice. The data represent the competitive index (CI) value (in ordinate) for CFU of mutant/wild-type of each mouse, 48 h after infection divided by CFU of mutant/wild type in the inoculum. Bars represent the geometric mean CI value.
We then evaluated the impact of argP deletion on bacterial virulence in BALB/c mice. For this, we performed a competition assay by injecting simultaneously WT and ΔargP mutant strain (100 bacteria of each) in 7-week-old females. Two days after infection, mice were sacrificed, liver and spleen were collected and serial dilutions were plated on chocolate agar plates with or without kanamycin. The competition index (CI) recorded (Fig. 1C) was very low in both target organs (10−6 in liver and 10−5 in spleen), demonstrating that argP plays an essential role in F. novicida virulence.
ArgP is the Major Arginine Transporter of F. novicida
Francisella is auxotroph for several amino acids because of inactive or incomplete biosynthetic pathways (35–37) and must therefore obtain these essential amino acids (E-AA) from the external medium via dedicated transporters. These include arginine, cysteine, histidine, lysine, and methionine. Francisella has also the capacity to import amino acids that it is able to synthesize that is, the nonessential amino acids (NE-AA, alanine, asparagine, glutamic acid, glutamine, glycine, phenylalanine, and tryptophan) and can thus rely either on biosynthesis or uptake to constitute its cytoplasmic pools.
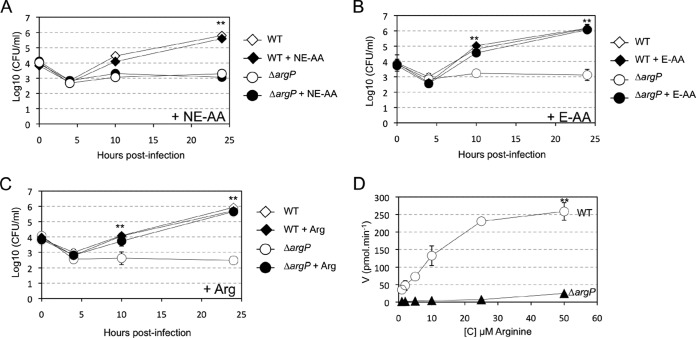
To identify which amino acid was imported by FTN_0848 (ArgP), we hypothesized that an excess of amino acid could be sufficient to bypass the specific defect of this transporter. Therefore, we first evaluated whether supplementation with an excess of each of these two different pools would restore multiplication of the ΔargP mutant in J774.1 macrophages (Fig. 2). Supplementation with a pool of NE-AA did not alleviate the growth defect of the ΔargP mutant (Fig. 2A). In contrast, wild-type growth was restored by supplementation with a pool of E-AA (Fig. 2B). The same experiment was then performed by individually supplementing the cell culture medium with each of the E-AA. Only supplementation with arginine could reverse the multiplication defect of the ΔargP mutant in J774.1 (Fig. 2C). The fact that the ΔargP mutant did not display any growth defect in CDM (supplemental Fig. S2) is likely because of the elevated concentration of arginine in the synthetic medium (2.3 mm as compared with 0.4 mm in standard DMEM).
Fig. 2.
Arginine is required for mutant multiplication in macrophages. Amino acid supplementation. J774.1 cells were infected with wild-type F. novicida (WT) or ΔargP mutant for 24 h, in medium supplemented (+AA) or not, with a pool of essential E-AA amino acids, A, or NE-AA, B, each at a final concentration of 1.5 mm. C, Arginine supplementation. J774.1 cells were infected with wild-type F. novicida (WT), or ΔargP mutant for 24 h with medium, supplemented (+AA) or not, with arginine 1.5 mm (+ Arg). D, Arginine transport assay. Kinetics of uptake of 14C radiolabeled arginine by wild-type F. novicida (WT, open circles) and ΔargP mutant (ΔargP, closed triangles), at substrate concentrations ranging from 1 to 50 μm. Ordinate, picomoles (pmol) of arginine taken up per minute (per sample of app. 2.5 × 109 bacteria). Abcissa, final concentration of arginine tested.
We also measured bacterial growth in CDMmin supplemented with reduced arginine concentrations (0.46 mm and 0.23 mm). At 0.23 mm arginine, growth of wild-type F. novicida and ΔargP mutant was almost abolished (Supplemental Fig. S2C, S2D). At 0.46 mm arginine, growth of the wild-type strain was not affected (as compared with that in 2.3 mm arginine), during the first 5 h, whereas that of the ΔargP mutant was slightly delayed. However, both strains reached approximately the same final OD600 after 8 h of growth.
To directly demonstrate the role of ArgP in arginine uptake, we measured incorporation of 14C-labeled arginine (14C-Arg) in wild-type F. novicida and compared it to that in ΔargP mutant (Fig. 2D). Bacteria grown to mid-exponential phase in CDM were tested. Uptake was measured after 5 min incubation with 14C-Arg. Remarkably, incorporation of 14C-Arg was completely abrogated in the ΔargP mutant, at all the arginine concentrations tested (1 to 50 μm), confirming that ArgP is a genuine high affinity arginine transporter (with an apparent Km in the10 μm range).
Impact of argP Inactivation on the Bacterial Proteome
In order to get further insight into the importance of arginine uptake in Francisella metabolism, we used high-resolution LC-MS to quantitatively compare the proteomes of wild-type F. novicida and ΔargP mutant bacteria, under arginine-limiting conditions. In standard chemically defined medium (CDM containing 2.3 mm arginine), growth of the ΔargP mutant is identical that of the wild-type (supplemental Fig. S2A) whereas a 10-fold reduction of the amount of arginine (0.23 mm) almost completely abrogates growth of both wild-type and mutant strains (not shown). We therefore chose to compare the proteomes of wild-type and ΔargP mutant, after 2 h of growth in CDM containing only a fivefold reduced amount of arginine (0.46 mm arginine). Under these conditions, growth of the two strains was not (or only mildly, for the ΔargP mutant) affected (Fig. S2C, 2D).
In total, 1225 different proteins (out of 1731 proteins predicted to be encoded by the F. tularensis subsp. novicida U112 genome (38) were identified across the 18 samples analyzed that is, three independent biological replicates, each in technical triplicate, for each strain (see “Experimental Procedures”). For statistical analysis, we kept only the proteins detected in at least nine samples that is, 842 proteins with quantification values across samples. A t test identified 37% of the proteins whose amount significantly differed between the two strains that is, 309 proteins.
These 309 proteins were submitted to bi-clustering analysis and represented in the heat-map (Fig. 3). The column tree revealed that the protein profiles of the wild-type strain and of the ΔargP mutant were different. The raw tree shows the proteins whose amount varied between the two strains were neatly divided in two categories: (1) proteins that were in lower amounts in the ΔargP mutant; and (2) those that were in higher amounts, as compared with the wild-type strain (supplemental Table S1). One hundred and forty-one proteins, present in lower amounts in the ΔargP mutant, were comprised within five clusters (32, 61, 175, 199, 259, and 268; Fig. 3). Remarkably, half of the proteins within cluster 61 (68 proteins) were ribosomal proteins (including 50% of the 30S ribosomal proteins). This cluster also comprised several proteins involved in protein synthesis (Ef-Ts, Ef-Tu, peptide chain release factor 1, translation initiation factor IF-3) or export (preprotein translocase SecA and Trigger factor). Cluster 199 (25 proteins) contained eight additional ribosomal protein (one from the 30S and seven from the 50S subunit) and two proteins of the general secretion machinery (SecB and SecD). Cluster 346 (39 proteins) was composed of a wide variety of enzymes and contained three additional 30S ribosomal proteins as well as several proteins involved in protein synthesis or export. Finally, cluster 32 and 175 (34 and 26 proteins, respectively), each contained seven additional ribosomal proteins. Altogether, the 38 ribosomal proteins, present in cluster 61 to 175 (Fig. 3), correspond to 79% of the ribosomal proteins analyzed in this study: 16 proteins belonged to the 30S subunit, and 22 to the 50S subunit (Table I). One hundred and sixty-eight proteins were present in higher amounts in the ΔargP mutant (supplemental Table S1). These belonged to six clusters (241, 244, 296, 150, 90, 122), also regrouping a wide variety of predicted functions and more than 40 hypothetical proteins of unknown function.
Fig. 3.
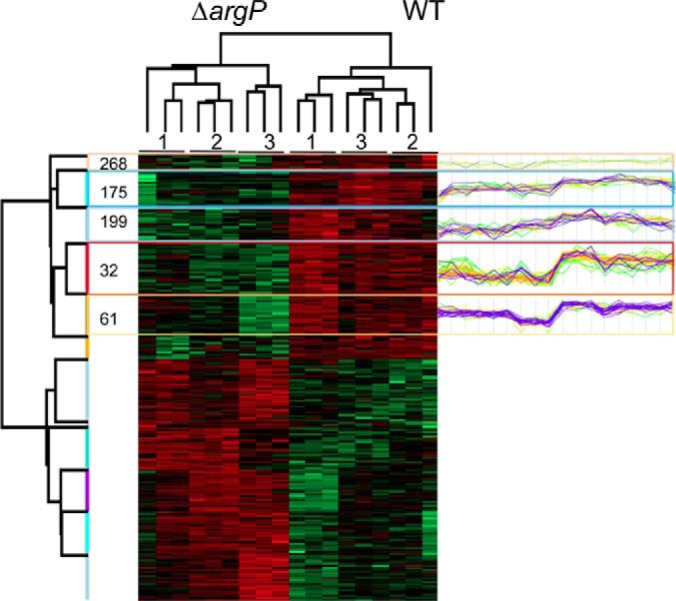
Hierarchical clustering and heatmap. Data were obtained by proteomic analyses of wild-type F. novicida and ΔargP mutant strains grown in chemically defined medium (CDM) containing a limiting concentration of arginine (0.46 mm). Samples were collected at 2 h of growth. For each strain, ΔargP mutant or wild-type (WT), three biological replicates were performed (indicated as 1, 2, 3, on top of the figure) and each replicate was tested in triplicate. 842 proteins with quantification values across samples were analyzed. 309 proteins whose amount significantly differed between the two strains were submitted to bi-clustering analysis and represented in the heat-map. The column tree shows that the protein profiles of the wild-type strain and of the ΔargP mutant were different. The proteins belong to two categories: (1) proteins present in lower amounts in the ΔargP mutant (141 proteins, in green); and (2) proteins present in higher amounts (168 proteins, in red). Five protein clusters (268, 175, 199, 32, 61), containing ribosomal proteins, are represented as profile plots to the right of the figure (highlighted in blue). For a detailed list of proteins, see supplemental Table S1.
Table I. The 38 ribosomal proteins present in reduced amounts in the ΔargP mutant, under limiting arginine conditions. The column Cluster indicates the cluster numbers defined in the Heatmap of Fig. 4. The last column indicates the percent of arginine (number of arginine residues/ polypeptide) contained in each ribosomal protein.
| Gene | Gene name | Gene product | Cluster | % arginine residues |
|---|---|---|---|---|
| FTN_1562 | rpsP | 30S ribosomal protein S16 | 268 | 6 |
| FTN_0278 | rpmE | 50S ribosomal protein L31 | 175 | 3.9 |
| FTN_0262 | rpsK | 30S ribosomal protein S11 | 175 | 8.5 |
| FTN_0261 | rpsM | 30S ribosomal protein S13 | 175 | 13.5 |
| FTN_0249 | rplN | 50S ribosomal protein L14 | 175 | 9 |
| FTN_0246 | rplP | 50S ribosomal protein L16 | 175 | 12.4 |
| FTN_0245 | rpsC | 30S ribosomal protein S3 | 175 | 7.2 |
| FTN_0236 | rpsG | 30S ribosomal protein S7 | 175 | 10.2 |
| FTN_1571 | rplA | 50S ribosomal protein L1 | 199 | 3.9 |
| FTN_1559 | rplS | 50S ribosomal protein L19 | 199 | 10.4 |
| FTN_1289 | rpsI | 30S ribosomal protein S9 | 199 | 12.4 |
| FTN_0254 | rplF | 50S ribosomal protein L6 | 199 | 3.9 |
| FTN_0251 | rplE | 50S ribosomal protein L5 | 199 | 6.7 |
| FTN_0250 | rplX | 50S ribosomal protein L24 | 199 | 5.7 |
| FTN_0241 | rplW | 50S ribosomal protein L23 | 199 | 6.1 |
| FTN_0239 | rplC | 50S ribosomal protein L3 | 199 | 4.8 |
| FTN_1572 | rplK | 50S ribosomal protein L11 | 32 | 2.8 |
| FTN_1007 | rplY | 50S ribosomal protein L25 | 32 | 3.3 |
| FTN_0951 | rpsF | 30S ribosomal protein S6 | 32 | 6.3 |
| FTN_0263 | rpsD | 30S ribosomal protein S4 | 32 | 9.2 |
| FTN_0258 | rplO | 50S ribosomal protein L15 | 32 | 5.6 |
| FTN_0252 | rpsL | 30S ribosomal protein S14 | 32 | 7 |
| FTN_0227 | rpsB | 30S ribosomal protein S2 | 32 | 3.3 |
| FTN_1570 | rplJ | 50S ribosomal protein L10 | 61 | 5.8 |
| FTN_1335 | rpmF | 50S ribosomal protein L32 | 61 | 6.3 |
| FTN_1288 | rplM | 50S ribosomal protein L13 | 61 | 5.3 |
| FTN_0916 | rpsU | 30S ribosomal protein S21 | 61 | 18.5 |
| FTN_0676 | rpmA | 50S ribosomal protein L27 | 61 | 11.4 |
| FTN_0675 | rplU | 50S ribosomal protein L21 | 61 | 6.7 |
| FTN_0608 | rpsO | 30S ribosomal protein S15 | 61 | 11.4 |
| FTN_0265 | rplQ | 50S ribosomal protein L17 | 61 | 11 |
| FTN_0257 | rpmD | 50S ribosomal protein L30 | 61 | 6.6 |
| FTN_0256 | rpsE | 30S ribosomal protein S5 | 61 | 4.8 |
| FTN_0247 | rpmC | 50S ribosomal protein L29 | 61 | 7.6 |
| FTN_0243 | rpsS | 30S ribosomal protein S19 | 61 | 5.4 |
| FTN_0242 | rplB | 50S ribosomal protein L2 | 61 | 10.2 |
| FTN_0235 | rpsL | 30S ribosomal protein S12 | 61 | 5.8 |
| FTN_0159 | rpsA | 30S ribosomal protein S1 | 61 | 3.8 |
Altogether, the 842 proteins belonged to multiple pathways, according to the KEGG pathway database for F. novicida (available at http://www.genome.jp/kegg-bin/show_organism?menu_type=pathway_maps&org=ftn). Of note, all the ribosomal proteins, as well as all the proteins involved in branched-chain amino acid (BCAA) biosynthesis, were present only in lower or unchanged amounts in the ΔargP mutant (supplemental Fig. S3).
ArgP is Involved in Phagosomal Escape
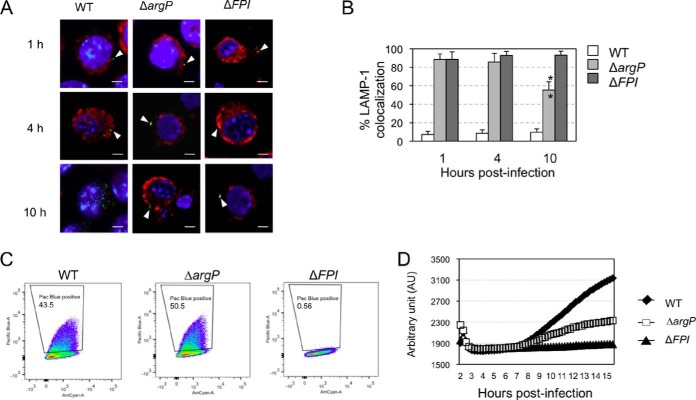
We followed the subcellular localization of the ΔargP mutant in infected cells by confocal microscopy. Intracellular localization of bacteria or LAMP-1 (used as a specific marker of phagosomes) was analyzed using specific antibodies and their colocalization was monitored at three time-points (1 h, 4 h, and 10 h postinfection; Fig. 4A). Quantification of each colocalization was performed with the Image J software (Fig. 4B). In cells infected with wild-type F. novicida, colocalization of bacteria with LAMP-1 was only 8% at 1 h and remained in the same range throughout the infection, showing that most bacteria escape the phagosome rapidly. In contrast, the ΔFPI mutant strain showed high colocalization with LAMP-1 throughout the infection (87% at 1 h, 90% at 4 h, and 91% at 10 h). In this assay, the ΔargP strain showed a severe delayed phagosomal escape. Indeed, at 4 h, 85% LAMP-1 colocalization was still observed. Partial phagosomal escape was detected only at 10 h of infection (with less than 55% bacteria still associated with LAMP-1). Taken together, these results demonstrate that argP gene is necessary for the bacteria to reach cytosol.
Fig. 4.
Subcellular localization of the ΔargP mutant. A, Confocal microscopy. J774 cells were incubated for 1 h with wild-type F. novicida (WT), ΔargP or ΔFPI strains and their colocalization with the phagosomal marker LAMP1 was observed by confocal microscopy. The phagosomes of J774 cells were labeled with anti-LAMP1 antibody (1/100 final dilution). Cell nuclei were labeled with DAPI. Bacteria were labeled with primary mouse monoclonal antibody anti-F. novicida (1/500 final dilution). A, The color images represent wild-type F. novicida (WT), ΔargP, ΔFPI, strains (green); phagosomes (red); and nuclei (blue); cytosolic bacteria appear in green (white arrowheads). Scale bars at the bottom right of each panel correspond to 10 μm. B, Quantification of bacteria/phagosome colocalization at 1 h, 4 h, and 10 h for wild-type F. novicida (WT), ΔargP and ΔFPI strains. **p < 0.01 (as determined by Student's t test). C, CCF4. The cytosol of bone marrow-derived infected macrophages was loaded with CCF4, as described previously (39). Wild-type F. novicida naturally expresses a β-lactamase able to cleave the CCF4. Bacterial escape into the host cytosol is thus associated with a shift of the CCF4 probe, upon excitation at 405 nm to a shift of fluorescence from green (535 nm) to blue (450 nm). After infection (at a multiplicity of infection of 100) with wild-type F. novicida (WT), ΔargP mutant and ΔFPI mutant, 43.5%, 50.5% and 0.56% of Pacific blue-positive cells were recorded, respectively. D, Cell death. The cell death kinetics of infected BMM (from BALB/c mice) was followed by monitoring propidium iodide (PI) incorporation in real time. PI fluorescence was measured every 15 min on a microplate fluorimeter (Tecan Infinite 1000). Wild-type F. novicida (WT), ΔargP and ΔFPI mutant strains.
We next tested the ability of the ΔargP mutant to induce vacuolar membrane rupture in macrophages by using a CCF4 assay, as previously described (39). Briefly, after 2 h of infection with wild-type F. novicida, ΔargP or ΔFPI strains, cells were loaded for 1 h with CCF4, a fluorescence resonance energy transfer (FRET) probe that is retained in the cytosol following the action of host esterases. F. novicida naturally secretes β-lactamase, a molecule able to cleave the CCF4 substrate. Once CCF4 is lysed, emission spectrum turns from 535 nm (green) to 450 nm (blue) (see “Experimental Procedures”). Wild-type F. novicida and ΔargP strains showed the same amount of cleaved CCF4 whereas there was no FRET signal for ΔFPI strain (Fig. 4C).
These data suggest that, although still confined in the phagosomal compartment after 3 h infection (as revealed by confocal microscopy), the ΔargP mutant is able to partially breach the phagosomal membrane, thus leading to the release of β-lactamase in the host cytosol.
Finally, to rule out a possible cytotoxicity of the ΔargP mutant, cell death kinetics were performed in infected bone marrow-derived macrophages (BMM) (13). As expected, delayed cell death was observed (measured by propidium iodide incorporation) in BMM infected with wild-type F. novicida (Fig. 4D) as a result of normal intracellular bacterial multiplication whereas the ΔFPI has no effect on BMM death. The ΔargP mutant reached an intermediate level of cell death (i.e. ∼1/3 that of the WT) after 16 h of infection. These results demonstrate that the intracellular multiplication defect of the ΔargP mutant could not be attributed to increased cytotoxicity.
Role of ArgP in Stress Defense
Upon entry into cells, Francisella resides transiently in a phagosomal compartment that transiently acidifies and acquires reactive oxygen species. We therefore first examined the ability of wild-type and ΔargP mutant strains to survive under acid or oxidative stress conditions. Bacteria were exposed either to pH 4 (supplemental Fig. S4A) or to 500 μm H2O2 (supplemental Fig. S4B). Under the pH condition tested, the viability of two strains was unaffected in standard CDM (containing 2.3 mm arginine), as well as in CDM containing limiting concentrations of arginine (0.46 mm or 0.23 mm). The ΔargP mutant was also as sensitive as wild-type F. novicida after 1 h of H2O2 challenge. Thus, the arginine transporter ArgP is not involved in acid or oxidative stress resistance, at least in the conditions tested, confirming the peculiar properties of the APC family members in Francisella.
We next evaluated a possible contribution of ArgP in the control of reactive nitrogen species production by infected macrophages (supplemental Fig. S5). The amount of NO, generated in infected J774.1 macrophages, was quantified using the Griess test (supplemental Fig. S5A). After 1 h, 24 h, and 48 h of infection, we found no difference of NO production between cells infected either by wild-type F. novicida or ΔargP mutant strain. We also tested whether reducing the amount of NO by infected macrophages would impact the intracellular behavior of the ΔargP mutant either by chemically inactivating iNOS synthase (using the irreversible inhibitor l-NG-Nitroarginine methyl ester) or by using iNOS-KO BMM. We did not observe any restoration of multiplication of ΔargP mutant in any of two conditions (Supplemental Fig. S5B, C). Altogether, these assays suggest that ArgP is not involved in the control of NO production in infected macrophages.
The Autophagic Pathway Captures Cytosolic ΔargP Mutant Bacteria
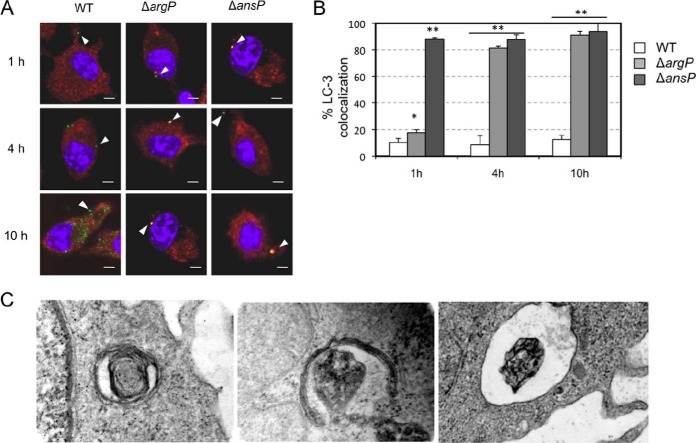
Celli and coworkers have recently demonstrated (40) that Francisella mutants unable to multiply in the host cytosol (independently of the type of genetic alteration responsible for this defect) were captured and processed by the classical LC3-dependent autophagic pathway. This prompted us to evaluate the physiological status of cytosolic ΔargP mutant bacteria by determining their capture by the classical autophagy pathway. J774.1 cells were infected with wild-type F. novicida or ΔargP mutant over a 1 h, 4 h, and 10 h period. LC3 recruitment to F. tularensis was measured by examining colocalization of bacteria with LC3.
Interestingly, at 1 h, there was no or very low colocalization (Fig. 5A, B). In contrast, 80% colocalization was recorded with ΔargP mutant at 4 h, and 91% at 10 h. For wild-type F. novicida, colocalization between bacteria and LC3 remained very low throughout the assay (13% at 10 h) whereas with the ΔansP mutant (11), used as a cytosolic nonreplicating control, 90% colocalization was already recorded at 1 h and remained very high at 10 h. Transmission electron microscopy analyses of J774-infected macrophages at 10 h post infection revealed individual replication-deficient bacteria within double-membrane structures resembling autophagosomes (Fig. 5C), consistent with the observations made by fluorescence microscopy. Hence, ΔargP replication-deficient bacteria are captured within vacuoles displaying autophagic features, suggesting that they were metabolically inactive. Thus, it is likely that a rapid phagosomal escape is critical for maintaining sufficient metabolic activity.
Fig. 5.
Cytosolic ΔargP mutant bacteria are LC3-positive. A, Confocal microscopy. J774.1 cells were incubated for 1 h with wild-type F. novicida (WT), ΔargP or ΔFPI strains and their colocalization with the autophagy marker LC3 was observed by confocal microscopy. B, Quantification at 1 h, 4 h, and 10 h for WT, ΔargP and ΔFPI strains. **p < 0.01 (as determined by Student's t test). C, Electron microscopy. Thin section electron microscopy of J774 cells infected with F. novicida ΔansP mutant strain at 10 h. The two left panels show individual bacteria surrounded by multilamellar structures; the right panel, an intact phagosome.
F. tularensis LVS also Relies on ArgP for its Intracellular Survival
In order to evaluate whether the properties of ArgP in the subspecies novicida could be extended to other subspecies, we also constructed a chromosomal deletion mutant in F. tularensis LVS (ΔFTL_1233) and evaluated the impact of the mutation in synthetic medium as well as in J774.1 macrophages. As for F. novicida ΔargP mutant, F. tularensis LVS ΔargP showed a severe multiplication defect in J774.1 macrophages after 24 h of infection (more than a 500-fold) as compared with wild-type LVS strain (in standard DMEM; supplemental Fig. S6A). The growth defect was abolished in the complemented strain.
Notably, in standard CDM, the LVS ΔargP mutant showed a significant multiplication defect (supplemental Fig. S6B). However, this defect was partially compensated upon supplementation with a 10-fold excess arginine (23 mm) and wild-type growth was restored upon supplementation with a twenty-fold excess arginine (46 mm). As in CDM, supplementation of the cell culture medium with 23 mm arginine partially restored intracellular multiplication of the ΔargP mutant in J774.1; and 46 mm arginine restored wild-type multiplication. Altogether, these results fully confirmed the importance of the transporter ArgP in F. tularensis intracellular multiplication.
DISCUSSION
Arginine is an essential modulator of the cellular immune response during infection, notably via the generation of nitrogen reactive species in macrophages, for the elimination of invading bacterial and parasitic pathogens (41, 42) and references therein). Arginine has also been shown to be important for the pathogen itself (43). In this study, we show that inactivation of the transporter ArgP severely impairs Francisella phagosomal escape, revealing that intracellular bacteria must import arginine to access to their cytoplasmic replicative niche. By using high-resolution mass spectrometry, we found that arginine limitation affected in particular ribosomal protein biogenesis in the ΔargP mutant. These data suggest possible links between ribosomal proteins amounts and phagosomal escape.
Importance of Arginine Uptake in Bacterial Phagosomal Escape
Macrophages are able to synthesize their own arginine (44) and to import it via constitutive and inducible cationic amino acid transporters (CAT-1 or SLC7A1 and CAT-2, or SLC7A2, respectively) (45). Macrophages may either use arginine to produce potentially harmful nitrogen reactive species (NO, via NO synthases) or convert it to ornithine and urea via type 1 arginase (supplemental Fig. S7A). Despite its apparent simplicity, the balance between the two processes is often quite complex. For example, it has been shown recently that two intracellular bacterial pathogens, Mycobacterium bovis and Listeria monocytogenes, induced early expression of iNOS, delayed expression of Arg1 upon infection of primary macrophages and boosted arginine import for NO synthesis (43). However, at a later stage of infection, macrophages re-import citrulline (initially expelled during the production of NO) to regenerate their intracellular pool of arginine via the action of the arginosuccinate synthase Ass1, thus maintaining sufficient NO production to control bacterial multiplication (supplemental Fig. S7A). Salmonella typhimurium has also been shown to reduce the production of the host iNOS through induction of the type 2 arginase in the spleen of infected mice (46) and to induce arginine uptake through enhanced expression of the genes encoding the cationic amino acids transporters CAT1 and CAT2 (41). However, these transporters are recruited preferentially to the Salmonella-containing vacuole (SCV), thus, fueling arginine into the SCV and supporting bacterial growth of in this compartment. Of note, a recent study demonstrated that, in C57BL/6 mice, the major arginine transporter CAT-2 was nonfunctional (42) because of a deletion in the promoter region of the corresponding gene. As a consequence, the intracellular arginine concentration in macrophages from C57BL/6 mice is significantly lower than that in macrophages from BALB/c mice. Of note, the kinetics of intracellular multiplication of wild-type F. novicida in bone marrow-derived macrophages (BMM) from C57BL/6 mice was comparable to that in BMM from BALB/c mice (data not shown), suggesting that the intracellular concentration of arginine in these cells is sufficient to promote bacterial growth.
As recalled earlier, Francisella is auxotroph for arginine and therefore can only obtain this amino acid from its environment. The fact that ArgP inactivation severely impaired phagosomal escape implies that arginine utilization is required at this early stage of the intracellular life cycle of the pathogen. Intra-macrophagic growth of the ΔargP mutant was fully restored upon supplementation of the growth medium with excess arginine, in both F. tularensis subsp. novicida and F. tularensis subsp. holarctica LVS, demonstrating the importance of arginine acquisition in these two subspecies.
Arginine Uptake is Critical for Protein Synthesis
We could cover 70% of Francisella predicted proteins, representing ∼20% more than the coverage of previously published proteomic analyses ((47) and references therein). Statistical analysis of our proteomic data identified one group of 309 proteins whose amount significantly differed between wild-type and ΔargP mutant, grown under arginine limiting conditions. This group could be further subdivided into: (1) proteins present in lower amount in the ΔargP mutant, which contained the majority of the ribosomal proteins identified (79%); and (2) proteins present in higher amount in the ΔargP mutant.
In Francisella, the metabolic pathways leading to or coming from arginine are predicted to be inactive (supplemental Fig. S7B). This implies that the bacterium relies on host arginine sources (oligopeptides and/or amino acids) for its intracellular multiplication. Because arginine is likely used primarily for protein synthesis, one could have expected that all protein syntheses would be reduced in the ΔargP mutant under arginine limitation (including the ribosomal proteins). However, a majority of proteins (533/842 i.e. 63% of the analyzed proteome) did not vary significantly in the ΔargP mutant, thus indicating that the reduction of the ribosomal proteins did not have a general impact on all protein syntheses. At any rate, indirect effects cannot be excluded. For example, proteins involved in purine and pyrimidine metabolism (supplemental Fig. S3) were either in lower (19 proteins), higher (13 proteins), or unchanged amounts (25 proteins) in the ΔargP mutant, suggesting complex effects of arginine limitation on these pathways.
Repression of ribosomal protein synthesis in response to stresses, such as nutritional limitation, has been observed in all kingdoms of life ((48) and references therein). Yet, the regulatory pathways controlling ribosomal protein syntheses are complex and not fully elucidated (49). The concept of a “ribosome code” that influences translation, initially proposed (50), has been recently supported by a number of studies in different eukaryotic models (51). The phagosome into which Francisella transiently resides is a dynamic entity whose size shrinks progressively until bacterial escape. The concentration of available nutrients is thus likely to vary considerably during the short lifespan of this compartment. It is reasonable to assume that the arginine concentration available to Francisella also decreases progressively in the phagosome during its maturation. This, in turn, should lead to a reduction of the bacterial arginine cytoplasmic pool itself. It is tempting to propose that intracellular Francisella may respond to variations of arginine availability in the phagosome (and consequently in its own cytoplasm) by regulating its ribosomal protein biogenesis (Fig. 6). The links between this process and phagosomal membrane disruption remain to be elucidated.
Fig. 6.
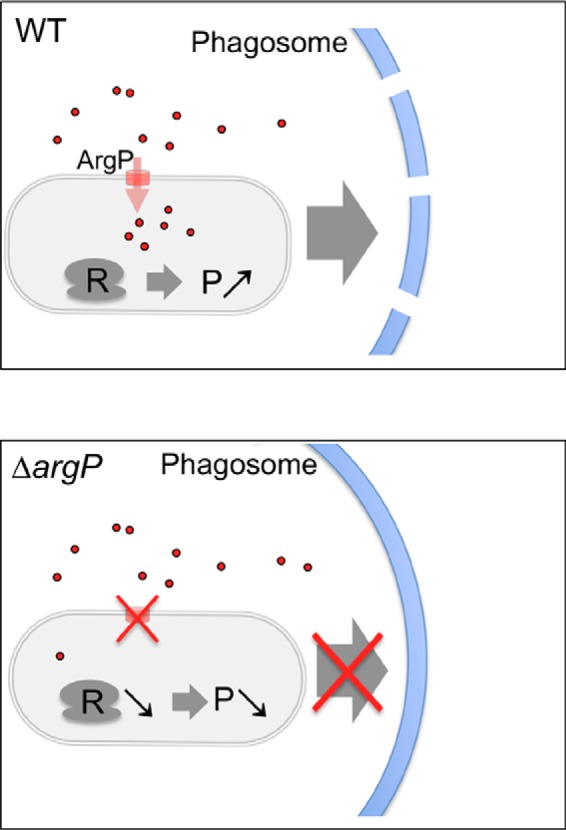
Hypothetical model for arginine sensing in the phagosome.
Francisella responds to variations of arginine availability in the phagosomal environment by regulating ribosomal protein biogenesis. In wild-type Francisella (WT, upper panel), import of arginine mediated by the transporter ArgP, promotes ribosomal protein synthesis (R), a pre-requisite for efficient protein (P) syntheses, a pre-requisite for membrane disruption and cytosolic escape. In the absence of ArgP (lower panel), ribososomal protein synthesis is down-regulated (R ), impairing thus protein syntheses (P
), impairing thus protein syntheses (P ), leading to impaired bacterial escape.
), leading to impaired bacterial escape.
Unlike other cytosolic pathogens, as L. monocytogenes that is equipped with the pore-forming listeriolysin O to disrupt the phagosomal membrane (52), Francisella genomes do not encode any cytolysin-like molecule. Instead, several enzymatic activites (e.g. phosphatases (53)) as well as metabolic or stress resistance functions (e.g. biotin biogenesis, oxidative stress (13, 54)) have been shown to contribute, directly or indirectly, to escape. This study reveals that ArgP-dependent arginine acquisition constitutes a novel player in phagosomal egress.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD001584. We thank the PRIDE team (http://www.ebi.ac.uk/pride/) for deposition of the mass spectrometry proteomics data.
Supplementary Material
Acknowledgments
We thank Dr A. Sjöstedt for providing the Francisella strains U112 and LVS.
Footnotes
Author contributions: E.R. and A.C. designed research; E.R., G.G., I.C.G., C.C., M.D., and M.R. performed research; E.R., G.G., I.C.G., C.C., M.D., M.R., T.H., M.B., and A.C. analyzed data; E.R., G.G., I.C.G., T.H., M.B., and A.C. wrote the paper.
* This study was funded by INSERM, CNRS and Université Paris Descartes Paris V. Elodie Ramond was funded by a fellowship from the “Région Ile de France” and Gael Gesbert by a fellowship from the Délégation Générale à l'Armement (DGA).
 This article contains supplemental Figs. S1 to S6, Tables S1 and S2, Experimental Procedures, and References.
This article contains supplemental Figs. S1 to S6, Tables S1 and S2, Experimental Procedures, and References.
1 The abbreviations used are:
- APC
- Amino acid/polyamine/organocation
- BMM
- Bone marrow derived macrophages
- CCF
- Coumarin-cephalosporin-fluorescein
- CDM
- Chemically defined medium
- CFU
- Colony forming unit
- CI
- Competition index
- DMEM
- Dubelcco's modified eagle medium
- E-AA
- Essential amino acid
- FCP
- Francisella-containing phagosome
- FDR
- False discovery rate
- FPI
- Francisella pathogenicity Island
- FRET
- Fluorescence resonance energy transfer
- iNOS
- Inducible nitric oxyde synthase
- KEGG
- Kyoto encyclopedia of genes and genomes
- L-NAME
- L-NG-Nitroarginine methyl ester
- LAMP
- Lysosomal associated membrane protein
- LC-MS/MS
- Liquid chromatography coupled to tandem mass spectrometry
- LFQ
- Label free quantification
- LVS
- Live vaccine strain
- MOI
- Multiplicity of infection
- NE-AA
- Non essential amino acid
- NO
- Nitric oxide
- OD
- Optical density
- qRT-PCR
- Quantitative real-time polymerase chain reaction
- TCA
- Tricarboxylic acid
- TSB
- Tryptic soy broth.
REFERENCES
- 1. Sjostedt A., ed. (2011) Francisella tularensis and tularemia, Fontiers Media SA, (eBook) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Celli J., Zahrt T. C. (2013) Mechanisms of Francisella tularensis intracellular pathogenesis. Cold Spring Harb. Perspect. Med. 3, a010314–a010314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones C. L., Napier B. A., Sampson T. R., Llewellyn A. C., Schroeder M. R., Weiss D. S. (2012) Subversion of host recognition and defense systems by Francisella spp. Microbiol. Mol. Biol. Rev. 76, 383–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ramond E., Gesbert G., Barel M., Charbit A. (2012) Proteins involved in Francisella tularensis survival and replication inside macrophages. Future Microbiol. 7, 1255–1268 [DOI] [PubMed] [Google Scholar]
- 5. Meibom K. L., Charbit A. (2010) The unraveling panoply of Francisella tularensis virulence attributes. Curr. Opin. Microbiol. 13, 11–17 [DOI] [PubMed] [Google Scholar]
- 6. Abu Kwaik Y., Bumann D. (2013) Microbial quest for food in vivo: “nutritional virulence” as an emerging paradigm. Cell Microbiol. 15, 882–890 [DOI] [PubMed] [Google Scholar]
- 7. Santic M., Abu Kwaik Y. (2013) Nutritional virulence of Francisella tularensis. Front. Cell Infect. Microbiol. 3, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raghunathan A., Shin S., Daefler S. (2010) Systems approach to investigating host-pathogen interactions in infections with the biothreat agent Francisella. Constraints-based model of Francisella tularensis. BMC Syst. Biol. 4, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Steele S., Brunton J., Ziehr B., Taft-Benz S., Moorman N., Kawula T. (2013) Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog. 9, e1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alkhuder K., Meibom K. L., Dubail I., Dupuis M., Charbit A. (2009) Glutathione provides a source of cysteine essential for intracellular multiplication of Francisella tularensis. PLoS Pathog. 5, e1000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gesbert G., Ramond E., Rigard M., Frapy E., Dupuis M., Dubail I., Barel M., Henry T., Meibom K., Charbit A. (2014) Asparagine assimilation is critical for intracellular replication and dissemination of Francisella. Cell Microbiol. 16, 434–449 [DOI] [PubMed] [Google Scholar]
- 12. Gesbert G., Ramond E., Tros F., Dairou J., Frapy E., Barel M., Charbit A. (2015) Importance of branched-chain amino acid utilization in Francisella intracellular adaptation. Infect. Immun. 83, 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ramond E., Gesbert G., Rigard M., Dairou J., Dupuis M., Dubail I., Meibom K., Henry T., Barel M., Charbit A. (2014) Glutamate utilization couples oxidative stress defense and the tricarboxylic acid cycle in Francisella phagosomal escape. PLoS Pathog. 10, e1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Steeb B., Claudi B., Burton N. A., Tienz P., Schmidt A., Farhan H., Mazé A., Bumann D. (2013) Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog. 9, e1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wong F. H., Chen J. S., Reddy V., Day J. L., Shlykov M. A., Wakabayashi S. T., Saier M. H., Jr. (2012) The amino acid-polyamine-organocation superfamily. J. Mol. Microbiol. Biotechnol. 22, 105–113 [DOI] [PubMed] [Google Scholar]
- 16. Weiss D. S., Brotcke A., Henry T., Margolis J. J., Chan K., Monack D. M. (2007) In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U.S.A. 104, 6037–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peng K., Monack D. M. (2010) Indoleamine 2,3-dioxygenase 1 is a lung-specific innate immune defense mechanism that inhibits growth of Francisella tularensis tryptophan auxotrophs. Infect. Immun. 78, 2723–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Su J., Yang J., Zhao D., Kawula T. H., Banas J. A., Zhang J. R. (2007) Genome-wide identification of Francisella tularensis virulence determinants. Infect. Immun. 75, 3089–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahlund M. K., Ryden P., Sjöstedt A., Stöven S. (2010) Directed screen of Francisella novicida virulence determinants using Drosophila melanogaster. Infect. Immun. 78, 3118–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moule M. G., Monack D. M., Schneider D. S. (2010) Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog. 6, e1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Llewellyn A. C., Jones C. L., Napier B. A., Bina J. E., Weiss D. S. (2011) Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS One 6, e24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brotcke A., Weiss D. S., Kim C. C., Chain P., Malfatti S., Garcia E., Monack D. M. (2006) Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 74, 6642–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luber C. A., Cox J., Lauterbach H., Fancke B., Selbach M., Tschopp J., Akira S., Wiegand M., Hochrein H., O'Keeffe M., Mann M. (2010) Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity 32, 279–289 [DOI] [PubMed] [Google Scholar]
- 24. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies, and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 25. Baker P. R., Chalkley R. J. (2014) MS-viewer: a web-based spectral viewer for proteomics results. Mol. Cell. Proteomics 13, 1392–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao X., Lu F., Zhou L., Dang S., Sun L., Li X., Wang J., Shi Y. (2009) Structure and mechanism of an amino acid antiporter. Science 324, 1565–1568 [DOI] [PubMed] [Google Scholar]
- 27. Shaffer P. L., Goehring A., Shankaranarayanan A., Gouaux E. (2009) Structure and mechanism of a Na+-independent amino acid transporter. Science 325, 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Singh S. K., Yamashita A., Gouaux E. (2007) Antidepressant binding site in a bacterial homolog of neurotransmitter transporters. Nature 448, 952–956 [DOI] [PubMed] [Google Scholar]
- 29. Weyand S., Shimamura T., Yajima S., Suzuki S., Mirza O., Krusong K., Carpenter E. P., Rutherford N. G., Hadden J. M., O'Reilly J., Ma P., Saidijam M., Patching S. G., Hope R. J., Norbertczak H. T., Roach P. C., Iwata S., Henderson P. J., Cameron A. D. (2008) Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322, 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perez C., Koshy C., Yildiz O., Ziegler C. (2012) Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP. Nature 490, 126–130 [DOI] [PubMed] [Google Scholar]
- 31. Schulze S., Köster S., Geldmacher U., Terwisscha van Scheltinga A. C., Kühlbrandt W. (2010) Structural basis of Na(+)-independent and cooperative substrate/product antiport in CaiT. Nature 467, 233–236 [DOI] [PubMed] [Google Scholar]
- 32. Watanabe A., Choe S., Chaptal V., Rosenberg J. M., Wright E. M., Grabe M., Abramson J. (2010) The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature 468, 988–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lauriano C. M., Barker J. R., Nano F. E., Arulanandam B. P., Klose K. E. (2003) Allelic exchange in Francisella tularensis using PCR products. FEMS Microbiol. Lett. 229, 195–202 [DOI] [PubMed] [Google Scholar]
- 34. Chamberlain R. E. (1965) Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13, 232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Larsson P., Oyston P. C., Chain P., Chu M. C., Duffield M., Fuxelius H. H., Garcia E., Halltorp G., Johansson D., Isherwood K. E., Karp P. D., Larsson E., Liu Y., Michell S., Prior J., Prior R., Malfatti S., Sjöstedt A., Svensson K., Thompson N., Vergez L., Wagg J. K., Wren B. W., Lindler L. E., Andersson S. G., Forsman M., Titball R. W. (2005) The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37, 153–159 [DOI] [PubMed] [Google Scholar]
- 36. Larsson P., Elfsmark D., Svensson K., Wikström P., Forsman M., Brettin T., Keim P., Johansson A. (2009) Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 5, e1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meibom K. L., Charbit A. (2010) Francisella tularensis metabolism and its relation to virulence. Front. Microbiol. 1, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rohmer L., Fong C., Abmayr S., Wasnick M., Larson Freeman T. J., Radey M., Guina T., Svensson K., Hayden H. S., Jacobs M., Gallagher L. A., Manoil C., Ernst R. K., Drees B., Buckley D., Haugen E., Bovee D., Zhou Y., Chang J., Levy R., Lim R., Gillett W., Guenthener D., Kang A., Shaffer S. A., Taylor G., Chen J., Gallis B., D'Argenio D. A., Forsman M., Olson M. V., Goodlett D. R., Kaul R., Miller S. I., Brittnacher M. J. (2007) Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8, R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nothelfer K., Dias Rodrigues C., Bobard A., Phalipon A., Enninga J. (2011) Monitoring Shigella flexneri vacuolar escape by flow cytometry. Virulence 2, 54–57 [DOI] [PubMed] [Google Scholar]
- 40. Chong A., Wehrly T. D., Child R., Hansen B., Hwang S., Virgin H. W., Celli J. (2012) Cytosolic clearance of replication-deficient mutants reveals Francisella tularensis interactions with the autophagic pathway. Autophagy 8, 1342–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Das P., Lahiri A., Lahiri A., Chakravortty D. (2010) Modulation of the arginase pathway in the context of microbial pathogenesis: a metabolic enzyme moonlighting as an immune modulator. PLoS Pathog. 6, e1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sans-Fons M. G., Yeramian A., Pereira-Lopes S., Santamaria-Babi L. F., Modolell M., Lloberas J., Celada A. (2013) Arginine transport is impaired in C57Bl/6 mouse macrophages as a result of a deletion in the promoter of Slc7a2 (CAT2), and susceptibility to Leishmania infection is reduced. J. Infect. Dis. 207, 1684–1693 [DOI] [PubMed] [Google Scholar]
- 43. Qualls J. E., Subramanian C., Rafi W., Smith A. M., Balouzian L., DeFreitas A. A., Shirey K. A., Reutterer B., Kernbauer E., Stockinger S., Decker T., Miyairi I., Vogel S. N., Salgame P., Rock C. O., Murray P. J. (2012) Sustained generation of nitric oxide and control of mycobacterial infection requires argininosuccinate synthase 1. Cell Host Microbe 12, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Morris S. M., Jr. (2007) Arginine metabolism: boundaries of our knowledge. J. Nutr. 137, 1602S–1609S [DOI] [PubMed] [Google Scholar]
- 45. Closs E. I., Boissel J. P., Habermeier A., Rotmann A. (2006) Structure and function of cationic amino acid transporters (CATs). J. Membr. Biol. 213, 67–77 [DOI] [PubMed] [Google Scholar]
- 46. Lahiri A., Das P., Chakravortty D. (2008) Arginase modulates Salmonella induced nitric oxide production in RAW264.7 macrophages and is required for Salmonella pathogenesis in mice model of infection. Microbes Infect. 10, 1166–1174 [DOI] [PubMed] [Google Scholar]
- 47. Lenco J., Tambor V., Link M., Klimentova J., Dresler J., Peterek M., Charbit A., Stulik J. (2014) Changes in proteome of the Deltahfq strain derived from Francisella tularensis LVS correspond with its attenuated phenotype. Proteomics 14, 2400–2409 [DOI] [PubMed] [Google Scholar]
- 48. Conrad M., Schothorst J., Kankipati H. N., Van Zeebroeck G., Rubio-Texeira M., Thevelein J. M. (2014) Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 38, 254–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Henras A. K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65, 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Komili S., Farny N. G., Roth F. P., Silver P. A. (2007) Functional specificity among ribosomal proteins regulates gene expression. Cell 131, 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xue S., Barna M. (2012) Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13, 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kayal S., Charbit A. (2006) Listeriolysin O: a key protein of Listeria monocytogenes with multiple functions. FEMS Microbiol. Rev. 30, 514–529 [DOI] [PubMed] [Google Scholar]
- 53. Mohapatra N. P., Soni S., Rajaram M. V., Strandberg K. L., Gunn J. S. (2013) Type A Francisella tularensis acid phosphatases contribute to pathogenesis. PLoS One 8, e56834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Napier B. A., Meyer L., Bina J. E., Miller M. A., Sjöstedt A., Weiss D. S. (2012) Link between intraphagosomal biotin and rapid phagosomal escape in Francisella. Proc. Natl. Acad. Sci. U.S.A. 109, 18084–18089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.