Abstract
Pleural effusion (PE), a tumor-proximal body fluid, may be a promising source for biomarker discovery in human cancers. Because a variety of pathological conditions can lead to PE, characterization of the relative PE proteomic profiles from different types of PEs would accelerate discovery of potential PE biomarkers specifically used to diagnose pulmonary disorders. Using quantitative proteomic approaches, we identified 772 nonredundant proteins from six types of exudative PEs, including three malignant PEs (MPE, from lung, breast, and gastric cancers), one lung cancer paramalignant PE, and two benign diseases (tuberculosis and pneumonia). Spectral counting was utilized to semiquantify PE protein levels. Principal component analysis, hierarchical clustering, and Gene Ontology of cellular process analyses revealed differential levels and functional profiling of proteins in each type of PE. We identified 30 candidate proteins with twofold higher levels (q<0.05) in lung cancer MPEs than in the two benign PEs. Three potential markers, MET, DPP4, and PTPRF, were further verified by ELISA using 345 PE samples. The protein levels of these potential biomarkers were significantly higher in lung cancer MPE than in benign diseases or lung cancer paramalignant PE. The area under the receiver-operator characteristic curve for three combined biomarkers in discriminating lung cancer MPE from benign diseases was 0.903. We also observed that the PE protein levels were more clearly discriminated in effusions in which the cytological examination was positive and that they would be useful in rescuing the false negative of cytological examination in diagnosis of nonsmall cell lung cancer-MPE. Western blotting analysis further demonstrated that MET overexpression in lung cancer cells would contribute to the elevation of soluble MET in MPE. Our results collectively demonstrate the utility of label-free quantitative proteomic approaches in establishing differential PE proteomes and provide a new database of proteins that can be used to facilitate identification of pulmonary disorder-related biomarkers.
The lungs are covered by parietal and visceral pleural membranes, including a small amount of fluid (10–20 ml) in the pleural cavity that helps the lungs expand and contract smoothly. Pleural effusions (PE)1, an accumulation of pleural fluid, contain proteins originating from the plasma filtrate and are released by inflammatory or epithelial cells. PE is triggered by a variety of etiologies, including malignancies and benign diseases such as pneumonia (PN), tuberculosis (TB), pulmonary embolism, heart failure, renal dysfunction, and autoimmune disease (1). Based on their biochemical characteristics, PEs are classified as transudative or exudative; determination of the PE type is a crucial step in the differential diagnosis and management of PEs. Transudative effusions, generally caused by systemic diseases, can be effectively distinguished from exudative PEs using the established modified Light's criteria (2, 3). However, further discrimination among different exudate types such as malignant and nonmalignant effusions (e.g. paramalignancies or acute and chronic inflammatory diseases) is sometimes diagnostically challenging because of similar biochemical and/or cellular profiles. For example, neutrophil-rich fluid is generally observed in patients with bacterial PN whereas lymphocytic effusions are generally observed in cancer or chronic inflammatory diseases such as TB (4).
PEs caused by cancer are generally divided into two categories, malignant (MPE) and paramalignant (PMPE). MPEs result when cancer cells metastasize to the pleural cavity (stage IV), wherein exfoliated malignant cells are observed in pleural fluid by cytological examination or detected in percutaneous pleural biopsy, thoracoscopy, thoracotomy, or at autopsy (5). PMPE occurs in cancer patients with no evidence of tumor invasion in the pleural space and may be caused by airway obstruction with lung collapse, lymphatic obstruction, or the systemic effects of cancer treatment (5). A high percentage of MPEs (>75%) arise from lung, breast, and ovarian cancer or lymphoma/leukemia. Lung cancer is a major etiology underlying MPE (6); however, only ∼40–87% patients with MPE can be accurately diagnosed upon initial examination (7). Inaccurate diagnosis of MPE and PMPE underestimates or overestimates the disease stage and leads to inappropriate therapy. Thus, it is important to identify a specific and powerful biomarker to distinguish MPE from benign diseases and PMPE.
Notably, tumor-proximal body fluids are promising sources for biomarker discovery because they represent a reservoir of in vivo tumor-secreted proteins without a large dynamic range or complexity of plasma or serum (8). Tumor-proximal fluids include PEs, nipple aspirate, stool, saliva, lavage, and ascites fluid. Previously, we utilized the powerful analytical capability of high-abundance protein depletion followed by one-dimensional SDS-PAGE combined with nano-LC-MS/MS (GeLC-MS/MS) for biomarker discovery to generate a comprehensive MPE proteome data set from 13 pooled nonsmall cell lung cancer (NSCLC) patients (9). Because a variety of pathological conditions can lead to exudative effusions, generating different PE proteomic profiles would accelerate discovery of potential PE biomarkers that can be used to discriminate between malignant and nonmalignant pulmonary disorders. The aim of this study is to establish differential PE proteomes from six types of exudative PEs, including three MPEs (from NSCLC, breast, and gastric cancers), one PMPE from NSCLC, and two benign diseases (TB and PN), using a label-free semiquantitative proteomics approach. Our results were verified by clinical validation of three potential biomarkers using an enzyme-linked immunosorbent assay (ELISA; Fig. 1).
Fig. 1.
Biomarker discovery strategy for identifying differentially expressed proteins from six pleural effusion (PE) types. The strategy comprised prefractionation by removal of high-abundance proteins, GeLC-MS/MS, comparative analysis of the six PE proteomes based on spectral counts, proteome clustering, functional classification of differentially expressed proteins, and selection and validation of biomarker candidates by ELISA.
EXPERIMENTAL PROCEDURES
Patient Population and Clinical Specimens
This study was approved by the Institutional Review Board for Research Ethics at the Chang Gung Memorial Hospital, Linkou, Tao-Yuan, Taiwan. Written informed consent was received from all patients prior to sample collection. Medical records of patients were reviewed, and all patient identities were protected. All PE samples were obtained from patients subjected to PE aspiration at Chang Gung Memorial Hospital, Linkou, Tao-Yuan, Taiwan. Patients with PMPE were radiologically monitored regularly over 6 months to exclude the possibility of occult malignancy within the effusion. For biomarker discovery, we used 60 PEs: 10 lung adenocarcinoma MPEs, 10 lung adenocarcinoma PMPEs, 10 TB PEs, 10 PN PEs, 10 gastric cancer (GC) PEs, and 10 breast cancer (BC) PEs. Demographics of these 60 patients are summarized in supplemental Table S1. To validate potential biomarkers by ELISA, 345 PE samples from six types of PE were used: 109 MPEs and 43 PMPEs from NSCLC, 61 TB, 68 PN, 45 breast cancer, and 19 gastric cancer. Demographics of these individuals, including age, gender, and smoking behavior are summarized in supplemental Table S2. PE samples were centrifuged at 2000 × g for 15 min at 4 °C. The cell-free supernatants were transferred to a new tube with a protease inhibiter mixture (Roche, Mannheim, Germany, cat. no. 11836145001) and stored at −80 °C until analysis. To detect the protein expressions in lung cancer tissues by Western blotting, four pairs of specimens of surgically resected primary lung adenocarcinoma lesions and adjacent noncancerous tissues were obtained from four patients (two stage IA and two stage IV). The fresh frozen tissues were stored at −80 °C until analysis.
PE Sample Preparation
PE samples (40 μl) from each patient (n = 60) were depleted of six high-abundance proteins (albumin, IgG, IgA, transferrin, antitrypsin, and haptoglobin) using a Multiple Affinity Removal System affinity column (Hu-6HC, 4.6 × 50 mm; Agilent Technologies, Wilmington, DE) via ÄKTA purifier-10 fast performance LC (GE Healthcare, Uppsala, Sweden). ÄKTA binding and elution procedures were performed according to the manufacturer's instructions (Agilent Technologies). Unbound fractions were collected, concentrated, and desalted by centrifugation in Amicon Ultra-4 tubes (molecular weight cut off, 3 kDa; Millipore, Billerica, MA). The resulting protein concentration was determined by Bradford protein assay (Bio-Rad Laboratories, Inc., Hercules, CA).
1D-SDS-PAGE and In-gel Protein Digestion
After depletion of high-abundance proteins, equal protein amounts obtained from each PE sample type were pooled (10 patients/group), and 40 μg protein samples were resolved on 10% SDS-PAGE and stained by Coomassie Brilliant Blue G-250 (AppliChem GmbH, Darmstadt, Germany). The entire gel lane was cut into 30 pieces and subjected to in-gel tryptic digestion as described previously (9). Briefly, gel pieces were destained in 50 mm NH4HCO3/ACN (3:2, v/v) three times for 25 min each and then dehydrated in ACN and dried in a SpeedVac. In-gel proteins were reduced with 10 mm dithiothreitol in 25 mm NH4HCO3 at 56 °C for 45 min, allowed to stand at room temperature (RT) for 10 min, and then alkylated with 55 mm iodoacetamide in the dark for 30 min at RT. After the proteins were digested by sequencing grade modified porcine trypsin (1:100; Promega, Madison, WI) overnight at 37 °C, peptides were extracted from the gel with ACN to a final concentration of 50%, dried in a SpeedVac, and then stored at −20 °C until further use.
Reverse-Phase LC-MS/MS
After trypsin digestion, each peptide mixture was reconstituted in high-performance LC buffer A (0.1% formic acid; Sigma, St. Louis, MO), loaded into the trap column (Zorbax 300SB-C18, 0.3 × 5 mm; Agilent Technologies) at a flow rate of 20 μl/min, and washed with buffer A at a flow rate of 20 μl/min for 10 min. Desalted peptides were then separated by 10 cm analytic C18 column (75 μm inner diameter; New Objective, Woburn, MA). The peptides were eluted by a linear gradient of 5–30% buffer B (99.9% ACN containing 0.1% formic acid) for 47 min, 30–45% buffer B for 5 min, 45–95% buffer B for 2 min, and 95% buffer B for 4 min at a flow rate of 0.25 μl/min. The LC setup was coupled with a 2D-linear ion trap LTQ-Orbitrap MS (Thermo Fisher, San Jose, CA) operated by Xcalibur 2.0.7 software (Thermo Fisher). The MS full-scan was performed over a range of 400–2000 Da and at a resolution of 60,000 at m/z 400. Internal calibration was performed using a (Si(CH3)2O)6H+ ion signal at m/z 445.120025 as a lock mass (10). The data-dependent procedure that alternated between one MS scan followed by 10 MS/MS scans for the 10 most abundant precursor ions in the MS survey scan was applied. The m/z values selected for MS/MS were dynamically excluded for 40 s; the electrospray voltage applied was 1.8 kV. Both MS and MS/MS spectra were acquired using a microscan with maximum fill times of 1000 and 150 ms for MS and MS/MS analyses, respectively. Automatic gain control was used to prevent overfilling the ion trap; 5 × 103 ions were accumulated in the ion trap for generation of MS/MS spectra. For triplicate GeLC-MS/MS analysis, PE samples were subjected to three independent 1D-SDS-PAGE in-gel protein digestion followed by reverse-phase LC-MS/MS analysis.
Database Searching
Resulting MS/MS spectra were searched using the Mascot algorithm (version 2.2.06, Matrix Science, London, U.K.) against the Swiss-Prot database (released Jun 15, 2010, selected for Homo sapiens, 20367 entries). The search parameters were set as follows: carbamidomethylation (C) as the fixed modification, oxidation (M) as variable modification, 10 ppm for MS tolerance, 0.5 Da for MS/MS tolerance, and one for missing cleavage. Validation of MS/MS-based peptides and protein identification were completed with Scaffold proteome software (version 3.3.2, Proteome Software Inc., Portland, OR), in which peptide and protein threshold cut offs were at a minimum of 95.0% with a minimum of two peptides.
Calculation of Spectral Counts and Bioinformatic Analysis
To generate comparative PE proteome databases, label-free semiquantitation of protein levels was determined by spectral counts. The number of spectra assigned for each protein were exported from the Scaffold software. The total spectral counts were calculated by totaling the spectral counts obtained in each experiment (30 LC-MS/MS runs). The normalized spectral count of each protein in the experiment was obtained by dividing the spectral count of a given protein by the total spectral counts of the experiment. After normalization, a q value was used to evaluate significant differences between different types of PEs. First, we determined the p value by β-binomial modeling (11), followed by the calculation of a q value according to Storey et al. (12); a q < 0.05 between different PE sample types was considered significantly different. The fold change was determined by dividing the average spectrum count from any two different PE sample types. We failed to identify all proteins in all experiments (triplicate for each PE sample type); unidentified proteins or missing values in a particular example were assigned a spectral count of one to avoid dividing by zero and to prevent overestimation of fold changes. After quantification analysis, differentially expressed proteins of interest were converted into Swiss-Prot accession numbers and uploaded into MetaCore Version 6.13 build 61585 (GeneGo, St. Joseph, MI) for Gene Ontology (GO) of cellular processes analysis.
Cluster Analysis of PE Proteomes
All spectral counts were imported into Microsoft Excel and transformed to Z-scores, a common normalization approach used in microarray data analysis (13). Z-scores were calculated as Z = (X − μx)/σx, where X represents individual spectral counts, μx represents the mean of spectral counts for an identified protein across different PE types, and σx is the standard deviation associated with μx. A spreadsheet containing the Z-scores was uploaded to Partek® Genome Suite (Partek Inc., St. Louis, MO) and analyzed using principal component analysis (PCA) as well as a two-way hierarchical clustering algorithm (HCL); the parameter used in HCL was according to Pearson distance and Ward's aggregation method. All quantified proteins were arranged into mock phylogenetic trees (dendrograms), wherein the y axis displays PE triplicates and the x axis shows proteins.
ELISA
Hepatocyte growth factor receptor (MET) and dipeptidyl peptidase IV (DPP4) protein levels in PEs were determined by sandwich ELISA (R&D Systems, Minneapolis, MN). Receptor-type tyrosine-protein phosphatase F (PTPRF) levels in PEs were determined by homemade ELISA as described by Whitmore (14). Briefly, monoclonal rat anti-human PTPRF antibodies (MAB3004; R&D Systems), polyclonal goat anti-human PTPRF antibodies (AF3004; R&D Systems), and purified NSO-derived recombinant human PTPRF (aa27–1251; R&D Systems) were used. Monoclonal rat anti-human PTPRF antibodies were coated on ELISA plates (250 ng per well) overnight at 4 °C, followed by six washes using wash buffer (0.05% Tween 20 in PBS) and blocking with reagent diluents (1% bovine serum albumin in PBS) for 2 h at room temperature (RT). PE samples with 1:50 dilution in reagent diluents and various amounts of recombinant PTPRF (standards; 0.3125–20 ng/ml) were added to the wells followed by incubation for 1 h at RT. After another six washes, polyclonal goat anti-human PTPRF antibodies (1:400) were added and incubated on a shaker for 1 h at RT. After washing, horseradish peroxidase (HRP)-labeled donkey anti-goat IgG antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) were added to wells and incubated for 1 h at RT. After six washes, tetramethylbenzidine substrate was added to wells for 10 min on a shaker in the dark, and the reaction was finally stopped with the addition of 2 N H2SO4. The resulting signals were measured by SpectraMax® M5 Multi-Mode Microplate Reader (Molecular Devices, Sunnyvale, CA) at 540 and 450 nm, respectively. The performance of the ELISA assay was examined by coefficient of variation (CV%) values determined in the intra-plate and inter-plate of ELISA assays using five individual PE samples. The results are shown in supplemental Fig. S1. All of the ELISA assays were performed twice for each sample. In addition, four PE samples were used as the internal standards in every ELISA plate, and all the protein levels determined in each batch were normalized based on these internal standards.
Cell Culture
CL1–0 and CL1–5 cells were kindly provided by Dr. P.C. Yang (Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan, Republic of China). The CL1 cell line was established from a 64-year-old man with a poorly differentiated adenocarcinoma, and the subpopulations CL1–0 and CL1–5 from CL1 cells were selected according to their differential invasiveness and metastatic ability in vitro and in vivo (15). The human breast cell line, MDA-MB231 was obtained from the American Type Culture Collection (ATCC, Rockville, MD). CL1–0 and CL1–5 cells were cultured in RPMI 1640 (Invitrogen, Grand Island, NY) supplemented with 10% FBS. MDA-MB231 cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS. All cells were cultured at 37 °C with a humidified atmosphere of 95% air/5% CO2. Conditioned media (CM) from the various cancer cell lines were collected and processed as previously described (16).
Lung Cancer Tissue Extraction
The fresh frozen tissues were dissolved in 200 μl of cold lysis buffer (20 mm Tris-HCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 50 mm NaF, 200 mm sodium pyrophosphate, 1 mm sodium orthovanadate, protease and inhibiter mixture [Roche]), and the protein extractions were prepared using a Precellys 24 homogenizer according to the manufacturer's instructions (Bertin Technology, France, MD).
Western blot
Protein samples prepared from CL1–0, CL1–5, and MD-AMB 231 cancer cell lines were separated on a 7.5% SDS-PAGE gel, then transferred onto a PVDF membrane, blocked with skim milk for 1 h, and incubated for 16 h at 4 °C with goat anti-MET polyclonal (R&D systems, cat. BAF358) or rabbit anti-MET monoclonal (Spring Bioscience, Pleasanton, CA, cat. M3440) antibodies. Bound primary antibodies were detected by HRP-labeled donkey anti-goat (Santa Cruz Biotechnology, Santa Cruz, CA) or HRP-labeled donkey anti-rabbit IgG secondary antibodies (GE Healthcare), respectively. Target proteins were visualized using an enhanced chemiluminescence system (Millipore).
Statistical Analysis
All data were processed using SPSS 12.0 (SPSS Inc., Chicago, IL). All continuous variables were expressed as the mean ± standard deviation (S.D.). To compare protein levels in different PE types, we used the nonparametric Mann-Whitney U test and linear regression to analyze variations in ELISA results for different clinical parameters; a p < 0.05 was considered statistically significant. Receiver-operator characteristic (ROC) curves were constructed by plotting sensitivity versus 1-specificity, and the areas under curves (AUC) were analyzed by the Hanley and McNeil method (17). The optimal cut off for establishing an accuracy score in each case was determined using Youden's index (J) (18). The ROC and AUC values of combined biomarker candidates were calculated using binary logistic regression. Briefly, we applied the binary logistic regression to calculate the probability of combined biomarkers according to their protein concentrations, and then the probability was used to obtain the ROC and AUC values for combined biomarkers. To analyze the AUC performance of individual marker and combined markers, we applied PanelComposer, a web-based panel construction tool developed by Professor Young-Ki Paik's research team for multivariate analysis of disease biomarker candidates (19). In this method, pairwise comparison between a panel of biomarkers and the individual proteins is performed using the Mann-Whitney U test and represented with a p value.
RESULTS
Generation of Proteomic Data sets from Six PE types
To accelerate discovery of potential PE biomarkers, we performed quantitative PE proteomics analysis (immunodepletion and GeLC-MS/MS) on samples from patients with six different PE types, including MPE and PMPE from NSCLC, TB, PN, breast cancer, and gastric cancer (Fig. 1). Validation of depletion efficacy and homogeneity of fractionated PE samples are shown in supplemental Fig. S2. Ten samples from each PE type were pooled and subjected to 1D-SDS-PAGE analysis and in-gel protein digestion followed by reverse-phase LC-MS/MS analysis; spectral searching identified 772 nonredundant proteins with high confidence (95.0% minimum peptide probability, 95.0% minimum protein probability and a minimum of two peptides). The reliability of the PE data sets was confirmed by the protein and peptide false discovery rate (FDR; supplemental Table S3). The highest FDRs for protein and peptide identification were 0.74% and 0.13%, respectively. The overlap of identified proteins between any two of three independent LC-MS/MS experiments for each type of PE was ∼84% (supplemental Fig. S3). Detailed information for proteins identified in triplicate experiments of six pleural effusion samples is shown in supplemental Table S4; this information includes name, gene symbol, protein probability, best peptide probability, unique peptide number, unique spectral number, spectral counts, sequence coverage, best Mascot ion score, and peptide sequence. A list of 772 nonredundant proteins identified in the different PE sample types is summarized in supplemental Table S5. The overlap and intersection of the PE proteomes were analyzed. A total of 721 proteins were identified in NSCLC-MPE or benign disease (TB and PN); 472 were common in these two types of PEs (supplemental Fig. S4A). There were 559 proteins identified in NSCLC-MPE or NSCLC-PMPE. Of these, 387 were detected in both types of PEs, yielding an overlap of 69.2% (supplemental Fig. S4B). We identified 633 proteins in NSCLC or nonlung cancer (BC and GC), and 477 were overlapping in these two PE types (supplemental Fig. S4C). Furthermore, the identified proteins of five PE types (NSCLC-MPE, TB, PN, NSCLC-PMPE, and nonlung cancer) were analyzed for the overlap. Among the 772 unique proteins identified in the present study, 363 (47.0%) were common to all types of PEs (supplemental Fig. S4D).
PCA and HCL Analysis of Identified Proteins
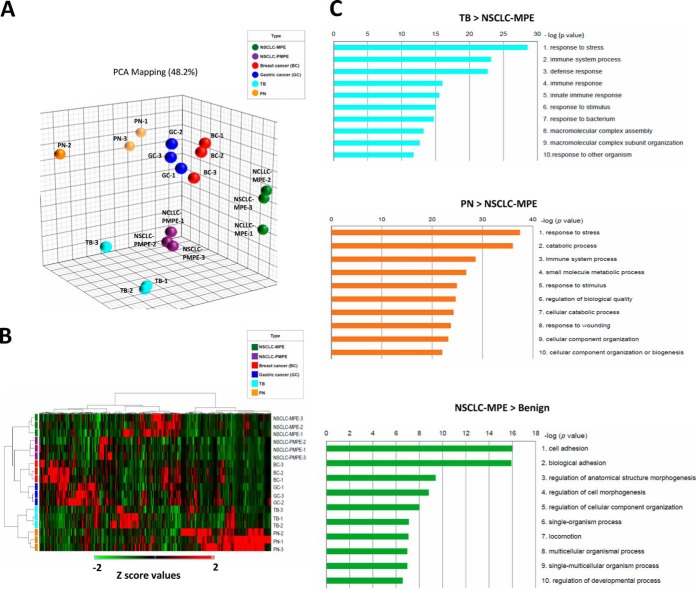
To generate differential proteome data sets, we used spectral counts to build label-free semiquantitative proteomic data sets for the six PE types. To examine the accuracy of quantification and reproducibility, we analyzed the correlation (r value) and CV% values among the three independent GeLC-MS/MS experiments. For each PE type, the r value between any two of three independent GeLC-MS/MS experiments was higher than 0.987 (Supplemental Fig. S5). The average CV% values of 772 quantified proteins obtained from the three independent GeLC-MS/MS experiments of six PE types was 20.87%, and the CV% value for each quantified protein was shown in supplemental Table S6. For the PCA and HCL analysis, 772 proteins were totally included; none was excluded. The different PE types clearly separated into six groups in addition to the grouping of triplicate experiments of each PE type in PCA (Fig. 2A). The resulting heat map and HCL dendrogram also showed a similar separation of the six PE types as well as a clustering of triplicate experiments (Fig. 2B). Notably, the PEs from different cancers were clustered together (lung, breast, and gastric cancers) but were separated from benign diseases (TB and PN), demonstrating distinct protein profiling in each PE type. Furthermore, these results support the high reproducibility of our triplicate experiments, suggesting that GeLC-MS/MS combined with spectral counting is a reliable method for generating both quantitative and qualitative PE proteomic data sets.
Fig. 2.
Hierarchical cluster analysis (HCL) of 772 proteins identified in six pleural effusion (PE) types. All spectral counts obtained from Scaffold 3 software were transformed to Z-scores, uploaded to the Partek® Genome Suite, and analyzed using principal component analysis (PCA) and a two-way HCL algorithm. A, PCA score-plot showing a distinct cluster separation of PE samples from nonsmall cell lung cancer malignant PE (NSCLC-MPE; green), nonsmall cell lung cancer paramalignant PE (NSCLC-PMPE; purple), breast cancer (BC; red), gastric cancer (GC; blue), tuberculosis (TB; cyan), and pneumonia (PN; orange). The plots show the first three principal components (PC), including PC#1: 23.6%, PC#2: 13.9%, and PC#3: 10.8% of total variables. B, HCL of PE proteomes was performed via unsupervised hierarchical classification, and distance trees were constructed from all identified proteins. PE types are shown in rows, and proteins are shown in columns. The heat map scale of Z-scores ranges from −2 (green) to 2 (red) with a midpoint of 0 (black). C, Functional classification of PE proteins using MetaCoreTM software based on universal Gene Ontology (GO) annotation terms. Proteins were linked to at least one annotation term within the GO cellular process categories.
Comparison of Differentially Expressed Proteins in Pulmonary Disorders
Multiple lines of evidence indicate a link between inflammation and cancer. Additionally, inflammatory microenvironments produced by chronic infections have been suggested to lead to cancer-related inflammation. Consequently, discovery of specific PE biomarkers that distinguish between lung cancer and benign diseases is necessary to assist in the diagnosis of pulmonary disorders. Therefore, we focused our analysis on differentially expressed proteins in MPE from NSCLC (NSCLC-MPE), TB, and PN. We identified 153 differentially expressed proteins with twofold changes (q<0.05) between NSCLC-MPE and TB (63 up- and 90 down-regulated in NSCLC-MPE; supplemental Fig. S6; supplemental Table S7) and 237 differentially expressed proteins with twofold changes (q<0.05) between NSCLC-MPE and PN (42 up- and 195 down-regulated in MPE; supplemental Fig. S6; supplemental Table S8). The pathophysiological status and potential pulmonary disorder diagnostic value for these proteins was assessed using Metacore bioinformatic software to analyze the GO of cellular processes involved in chronic (TB) and acute (PN) inflammatory diseases. The top three most significant cellular processes for the 90 up-regulated TB compared with NSCLC-MPE proteins were those related to stress, immune system process, and defense response (Fig. 2C) whereas for the 195 up-regulated PN compared with NSCLC-MPE proteins, the processes were response to stress and catabolic and immune processes. However, the three most significant cellular processes of the 75 up-regulated proteins in NSCLC-MPE compared with benign diseases (TB and PN) (Fig. 3) were related to cell adhesion, biological adhesion, and regulation of anatomical structure morphogenesis. These results further confirm that integration of multiple quantitative PE proteomes coupled with bioinformatic analysis is altogether feasible and invaluable to the discovery of potential pulmonary disorder biomarkers. Detailed information pertaining to GO cellular processes, p values and the protein list are summarized in supplemental Table S9.
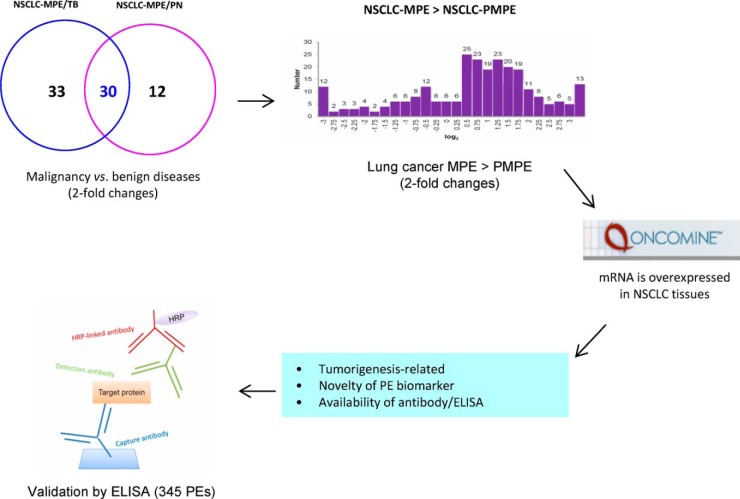
Fig. 3.
Strategy used to select malignancy-related PE biomarker candidates. Thirty proteins with higher levels in nonsmall cell lung cancer malignant pleural effusions (NSCLC-MPE) compared with benign diseases (tuberculosis [TB] and pneumonia [PN]) were identified and further selected by comparative analysis of nonsmall cell lung cancer malignant (NSCLC-MPE) and nonsmall cell lung cancer paramalignant pleural effusion (NSCLC-PMPE) proteome data sets, integration with NSCLC tissue transcriptome data sets, functional classification, consideration of novelty, and availability of commercial antibodies/ELISA kits.
Selection of Potential MPE Biomarkers for NSCLC
To identify malignancy-related proteins in NSCLC, we first combined 63 proteins that were increased in NSCLC-MPE compared with TB (Fig. 3; supplemental Table S7) with 42 proteins that were increased in NSCLC-MPE relative to PN (Fig. 3; supplemental Table S8). Accordingly, we identified 30 potential proteins that had higher protein levels (twofold change, q<0.05) in NSCLC-MPE compared with these two benign diseases (Table I). To narrow down the possible candidates that could be used for efficient NSCLC diagnostics, we established the following protein candidate selection criteria: First, we selected proteins with a twofold increase (q<0.05) in NSCLC-MPE relative to NSCLC-PMPE (supplemental Fig. S6C); discrimination between MPE and PMPE has profound implications in the therapy and prognosis of lung cancer. Second, we stipulated that mRNA levels of candidates should be up-regulated in NSCLC tissues compared with normal tissues to establish expression levels of secreted proteins in primary cancer and noncancerous tissues. Third, the biological functions of candidates were correlated to tumorigenesis. Finally, candidates had to represent novel PE biomarkers for NSCLC and have commercially available antibodies or ELISA kits (Fig. 3). Following application of the first selection criteria, 25 candidates were identified as NSCLC biomarkers (Table I; supplemental Table S10). Next, 10 mRNA data sets of lung adenocarcinoma deposited on the Oncomine 4.4.4.4 database, a cancer microarray database with web-based data-mining characteristics (20), were examined. These data sets included Beer (21), Bhattacharjee (22), Garber (23), Hou (24), Landi (25), Okayama (26), Selamat (27), Stearman (28), Su (29), and Yamagata (30) lung. We observed that 19 of the 25 candidate proteins were dysregulated in human cancers (supplemental Table S10), which suggests that our integrated approach provides a reliable method for identifying multiple potential NSCLC biomarkers. Among these 25 candidate proteins, the mRNA levels of 11 candidates were overexpressed in NSCLC tissues compared with noncancerous tissues in at least five data sets (p < 0.05; Table I; supplemental Table S10). Upon application of the remaining selection criteria, three of the 11 candidates were selected for further verification by ELISA: MET, PTPRF, and DPP4.
Table I. List of 30 candidate proteins with twofold higher levels (q<0.05) in NSCLC-MPE than two benign PEs.
| Protein name | Gene symbol | Swiss-Prot Acc. No. | M.W. (kDa) | NSCLC-MPE/TB log2 ratio | NSCLLC-MPE/PN log2 ratio | NSCLC-MPE/NSCLC-PMPE log2 ratio | mRNA Data set numbera | Up-regulated mRNA data set number (p value < 0.05)b |
|---|---|---|---|---|---|---|---|---|
| Mucin-1 | MUC1 | MUC1_HUMAN | 122 | 4.77 | 4.70 | 3.33 | 10 | 4 |
| Pulmonary surfactant-associated protein B | SFTPB | PSPB_HUMAN | 42 | 3.80 | 1.86 | 2.40 | 9 | 1 |
| Mimecan | OGN | MIME_HUMAN | 34 | 3.71 | 1.19 | 0.57 | 6 | 0 |
| Receptor-type tyrosine-protein phosphatase F | PTPRF | PTPRF_HUMAN | 213 | 3.62 | 3.99 | 2.44 | 10 | 8 |
| CD166 antigen | ALCAM | CD166_HUMAN | 65 | 3.41 | 3.34 | 3.21 | 10 | 0 |
| Carcinoembryonic antigen-related cell adhesion molecule 5 | CEACAM5 | CEAM5_HUMAN | 77 | 3.11 | 3.03 | 0.31 | 9 | 9 |
| Kunitz-type protease inhibitor 1 | SPINT1 | SPIT1_HUMAN | 58 | 2.92 | 2.85 | 2.72 | 7 | 6 |
| Polymeric immunoglobulin receptor | PIGR | PIGR_HUMAN | 83 | 2.38 | 1.92 | 2.74 | 8 | 2 |
| Intelectin-1 | ITLN1 | ITLN1_HUMAN | 35 | 2.26 | 2.18 | 1.08 | 5 | 0 |
| Sushi, von Willebrand factor type A, EGF and pentraxin domain-containing protein 1 | SVEP1 | SVEP1_HUMAN | 390 | 2.22 | 1.81 | 0.99 | 8 | 2 |
| Membrane primary amine oxidase | AOC3 | AOC3_HUMAN | 85 | 2.22 | 1.32 | 0.67 | 9 | 0 |
| 3,2-trans-enoyl-CoA isomerase, mitochondrial | DCI | D3D2_HUMAN | 33 | 2.18 | 2.11 | 1.97 | 9 | 5 |
| Antileukoproteinase | SLPI | SLPI_HUMAN | 14 | 2.06 | 1.23 | 1.85 | 9 | 0 |
| Interleukin-1 receptor accessory protein | IL1RAP | IL1AP_HUMAN | 65 | 2.05 | 1.97 | 1.84 | 8 | 4 |
| Thy-1 membrane glycoprotein | THY1 | THY1_HUMAN | 18 | 2.00 | 1.92 | 2.50 | 9 | 8 |
| CD9 antigen | CD9 | CD9_HUMAN | 25 | 1.93 | 1.85 | 1.72 | 10 | 3 |
| Mucin-5B | MUC5B | MUC5B_HUMAN | 591 | 1.89 | 1.82 | 1.69 | 9 | 7 |
| Carbonic anhydrase 3 | CA3 | CAH3_HUMAN | 30 | 1.89 | 1.40 | 2.24 | 10 | 0 |
| Cartilage intermediate layer protein 1 | CILP | CILP1_HUMAN | 133 | 1.74 | 3.20 | 3.07 | 7 | 7 |
| Neogenin | NEO1 | NEO1_HUMAN | 160 | 1.66 | 1.16 | 2.64 | 10 | 0 |
| Hepatocyte growth factor receptor | MET | MET_HUMAN | 156 | 1.66 | 1.58 | 1.45 | 9 | 5 |
| Claudin-4 | CLDN4 | CLD4_HUMAN | 22 | 1.65 | 1.57 | 1.44 | 8 | 8 |
| Tyrosine-protein kinase-like 7 | PTK7 | PTK7_HUMAN | 118 | 1.64 | 1.56 | 1.43 | 10 | 7 |
| Intercellular adhesion molecule 1 | ICAM1 | ICAM1_HUMAN | 58 | 1.48 | 1.67 | 3.47 | 8 | 0 |
| Deleted in malignant brain tumors 1 protein | DMBT1 | DMBT1_HUMAN | 261 | 1.48 | 1.41 | 1.27 | 7 | 0 |
| Latent-transforming growth factor beta-binding protein 2 | LTBP2 | LTBP2_HUMAN | 195 | 1.38 | 3.84 | 1.31 | 9 | 0 |
| Thrombospondin-4 | THBS4 | TSP4_HUMAN | 106 | 1.33 | 3.18 | 1.62 | 9 | 6 |
| Alpha-amylase 1 | AMY1A | AMY1_HUMAN | 58 | 1.27 | 1.19 | 1.06 | 5 | 2 |
| Mesothelin | MSLN | MSLN_HUMAN | 69 | 1.25 | 1.18 | -0.37 | 9 | 0 |
| Dipeptidyl peptidase 4 | DPP4 | DPP4_HUMAN | 88 | 1.12 | 1.01 | 1.65 | 9 | 7 |
a The data were taken from the Oncomine 4.4.4.4 Research Edition (https://www.oncomine.org/resource/login.html).
b The regulation of expression indicate the mRNA levels in NSCLC tissues Compared to noncancerous tissues.
Validation of MET, PTPRF, and DPP4 as Potential PE Biomarkers for NSCLC
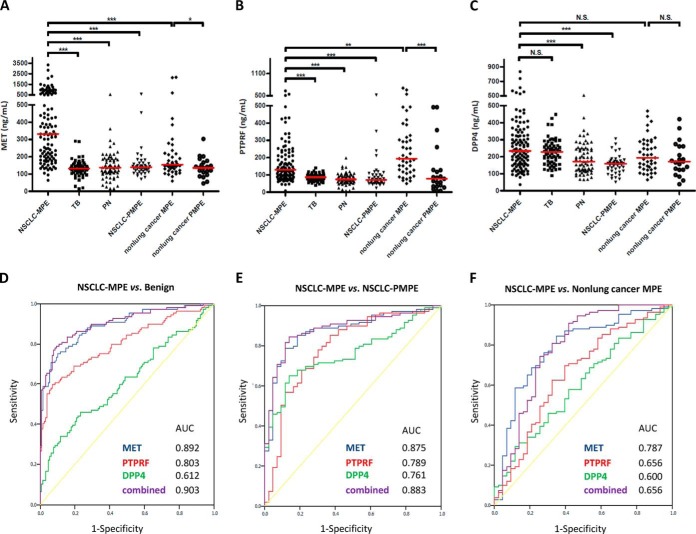
Clinical verification of MET, PTPRF, and DPP4 was conducted using 345 PE samples from six PE types (supplemental Table S2; Table II). The average CV% values of ELISA assays for duplicate 345 PE samples of MET, PTPFR, and DPP4 were 1.75%, 3.23%, and 2.15%, respectively. The mean protein concentrations of MET, PTPRF, and DPP4 in NSCLC-MPE were 527.71 ± 542.56, 165.95 ± 114.73, and 264.46 ± 141.62 ng/ml, respectively (Fig. 4A–4C; Table II; supplemental Table S11). PE levels of these three proteins in PN (MET, 146.29 ± 81.53 ng/ml; PTPRF, 77.27 ± 32.50 ng/ml; DPP4, 188.59 ± 88.83 ng/ml) were significantly lower (p < 0.0001) than in NSCLC-MPE. PE levels of MET and PTPRF in TB (MET, 132.76 ± 47.15 ng/ml; PTPRF, 86.30 ± 20.36 ng/ml) were also significantly lower than in NSCLC-MPE. However, DPP4 levels in NSCLC-MPE and TB (264.46 ± 141.62 ng/ml compared with 227.75 ± 75.88 ng/ml) were not significantly different. Nevertheless, results demonstrated that PE levels of MET, PTPRF, and DPP4 were elevated in NSCLC-MPE compared with benign lung diseases (TB and PN). Statistical analysis showed that malignancy was an independent factor of MET, PTPRF, and DPP4 levels (all p < 0.001) in 238 lung-disease PEs (supplemental Table S12). There was no significant correlation between the PE level of MET and sex, age, or smoking history; a similar result was also observed with PTPRF. However, both age and malignancy were independent factors of DPP4 level (p = 0.017 and p < 0.001, respectively) in patients with lung diseases. Thus, these findings collectively indicate that MET and PTPRF are effective PE biomarkers that distinguish NSCLC-MPE from benign inflammatory lung diseases. The ROC curves discriminating between NSCLC-MPE and benign diseases revealed that the AUC values of MET, PTPRF, and DPP4 were 0.892, 0.803, and 0.612, respectively (p < 0.05 for all three proteins; Fig. 4D; Table III). The combination of these three markers exhibited higher diagnostic capacity than any marker alone (AUC = 0.903; Fig. 4D; Table III). To analyze the AUC performance of MET and combined markers, we applied PanelComposer, a web-based panel construction tool developed by Professor Young-Ki Paik's research team for multivariate analysis of disease biomarker candidates (19). Supplemental Table S13 shows the comparison of the effectiveness of combined markers (MET, PTPRF, and DPP4) compared with the MET protein alone in distinguishing NSCLC-MPE from benign diseases, NSCLC-PMPE, or nonlung MPE. We observed that the AUC of A (combined markers) in distinguishing NSCLC-MPE from benign diseases (TB, PN) or NSCLC-PMPE was better than that of B (MET), and the significance of the difference is denoted by the p value. For example, with a given specificity of 90%, the sensitivities of using MET, PTPRF, and DPP4 alone to distinguish NSCLC-MPE from benign diseases were 73.39%, 60.55%, and 29.36%, respectively. Significantly, combining the three markers enhanced the sensitivity for NSCLC-MPE detection (78.90%, supplemental Table S13) compared with an individual marker. When applying any one of the three markers with a given cut-off value to discriminate NSCLC-MPE from benign diseases, the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were 88.07%, 65.89%, 68.57%, and 86.73%, respectively (supplemental Fig. S7).
Table II. Relations between PE levels of three potential biomarkers and clinical characteristics in 345 patients with PE.
| Variables | No. | MET (ng/ml)a | p valueb | PTPRF (ng/ml)a | p valueb | DPP4 (ng/ml)a | p valueb |
|---|---|---|---|---|---|---|---|
| Age | |||||||
| ≥ 60 years | 190 | 227.40 ± 394.23 | 0.880 | 124.71 ± 101.45 | 0.529 | 207.83 ± 107.07 | 0.049 |
| < 60 years | 155 | 286.28 ± 373.97 | 143.48 ± 126.76 | 231.01 ± 115.68 | |||
| Gender | |||||||
| Male | 185 | 281.95 ± 389.76 | 0.660 | 110.99 ± 79.11 | 0.008 | 214.76 ± 105.51 | 0.647 |
| Female | 160 | 280.75 ± 380.06 | 158.76 ± 139.69 | 222.27 ± 118.16 | |||
| Smoking status | |||||||
| Ex-/current smoker | 131 | 298.91 ± 432.20 | 0.989 | 117.12 ± 88.38 | 0.173 | 210.05 ± 103.18 | 0.374 |
| Never | 214 | 270.67 ± 353.19 | 142.95 ± 125.98 | 223.26 ± 116.18 | |||
| Lung disease | |||||||
| TB and PN | 129 | 139.89 ± 67.58 | <0.001c | 81.54 ± 27.71 | <0.001c | 207.11 ± 84.94 | 0.003c |
| NSCLC-PMPE | 43 | 157.57 ± 88.75 | <0.001d | 96.33 ± 88.17 | <0.001d | 154.31 ± 54.00 | <0.001d |
| NSCLC-MPE | 109 | 527.71 ± 542.56 | 165.95 ± 114.73 | 264.46 ± 141.62 | |||
| MPE | |||||||
| Lung cancer | 109 | 527.71 ± 542.56 | <0.001 | 165.95 ± 114.73 | 0.003 | 264.46 ± 141.62 | 0.054 |
| Nonlung cancer | 43 | 274.51 ± 432.80 | 244.66 ± 162.94 | 213.61 ± 100.38 | |||
| Nonlung cancer | |||||||
| MPE | 43 | 274.51 ± 432.80 | 0.048 | 244.66 ± 162.94 | <0.001 | 213.61 ± 100.38 | 0.345 |
| PMPE | 21 | 139.73 ± 57.31 | 126.90 ± 147.29 | 187.20 ± 98.03 | |||
| Lung cancer histology | |||||||
| Adenocarcinoma | 128 | 476.34 ± 517.56 | <0.001 | 151.48 ± 112.86 | 0.051 | 246.46 ± 138.42 | 0.005 |
| Nonadenocarcinoma | 24 | 138.55 ± 31.31 | 118.36 ± 105.90 | 163.10 ± 62.20 |
a The data are presented as the mean ± S.D.
b Mann-Whitney U test.
c The p value presents the difference between NSCLC-MPE and benign diseases (TB and PN).
d The p value presents the difference between NSCLC-MPE and NSCLC-PMPE.
Fig. 4.
Validation of three biomarker candidates by ELISA. The pleural effusion (PE) levels of A, MET, B, PTPRF, and C, DPP4 from 345 patients with lung cancer, benign diseases (TB and PN), and nonlung cancers (breast and gastric cancers) were determined by sandwich ELISA. Horizontal lines represent median values. ***p < 0.05, indicates statistical significance using the nonparametric Mann-Whitney U test. N.S., not significant. Receiver-operator characteristic (ROC) curve analysis was used to examine the diagnostic efficacy of the protein candidates. ROC curve analyses of MET (blue line), PTPRF (red line), DPP4 (green line), and a combination of all three proteins (purple line) for discrimination D, between nonsmall cell lung cancer malignant PE (NSCLC-MPE) and benign diseases (TB and PN), E, between NSCLC-MPE and nonsmall cell lung cancer paramalignant PE (NSCLC-PMPE), and F, between NSCLC-MPE and nonlung cancer MPE. AUC, area under the ROC curve.
Table III. The AUC, specificity and sensitivity of the diagnostic efficacy of three potential PE biomarkers.
| Category | MET |
PTPRF |
DPP4 |
MET & PTPRF & DPP4 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | Specificity/sensitivity | Cut off (ng/ml) | AUC | Specificity/sensitivity | Cut off (ng/ml) | AUC | Specificity/sensitivity | Cut off (ng/ml) | AUC | Specificity/sensitivity | Cut off (prob.d) | |
| NSCLC-MPE vs. Benigna | 0.892 | 0.899/0.752 | 200.755 | 0.803 | 0.930/0.596 | 116.716 | 0.612 | 0.767/0.459 | 257.793 | 0.903 | 0.907/0.789 | 0.421 |
| NSCLC-MPE vs. NSCLC-PMPE | 0.875 | 0.884/0.789 | 186.093 | 0.789 | 0.628/0.853 | 78.113 | 0.761 | 0.860/0.651 | 191.036 | 0.883 | 0.860/0.844 | 0.561 |
| NSCLC-MPE vs. nonlung cancer MPEb | 0.787 | 0.651/0.844 | 172.520 | 0.656 | 0.605/0.697 | 170.104 | 0.600 | 0.884/0.294 | 316.047 | 0.656 | 0.581/0.908 | 0.565 |
| NSCLC-MPE vs. all other tested pleural typesc | 0.870 | 0.852/0.761 | 199.020 | 0.709 | 0.674/0.688 | 100.745 | 0.642 | 0.784/0.459 | 257.793 | 0.840 | 0.881/0.706 | 0.299 |
a Tuberculosis and pneumonia.
b Malignant pleural effusions obtained from breast and gastric cancers.
c Pleural effusions obtained from tuberculosis, pneumonia, NSCLC-PMPE, breast and gastric cancers.
d Probability.
Next, we examined the potential application of these candidates in discriminating NSCLC-MPE from NSCLC-PMPE. The protein concentrations of MET, PTPRF, and DPP4 in NSCLC-PMPE were 157.57 ± 88.75, 96.33 ± 88.17, and 154.31 ± 54.00 ng/ml, respectively. The protein levels of these three candidates in NSCLC-MPE and NSCLC-PMPE were significantly different (p < 0.05; Table II; supplemental Table S11 and supplemental Table S15). The AUC values of MET, PTPRF, and DPP4 in distinguishing NSCLC-MPE from NSCLC-PMPE were 0.875, 0.789, and 0.761, respectively (p < 0.05 in all three proteins; Fig. 4E; Table III). The AUC that best discriminates between NSCLC-MPE and benign diseases or NSCLC-PMPE is MET, with cut offs of 200.755 and 186.093 ng/ml, respectively; corresponding specificity and sensitivity are shown in Table III. Unexpectedly, the AUC values of a combination of any two or all three candidates either did not improve or improved only slightly relative to MET alone in distinguishing between NSCLC-MPE and benign diseases or NSCLC-PMPE (supplemental Table S14). To confirm the reliable validation for these three potential PE biomarkers used in distinguishing malignant (NSCLC-MPE) from nonmalignant pulmonary disorders (TB, PN, and NSCLC-PMPE), we performed the ELISA assays using an independent sample set, including 59 NSCLC-MPE, 30 benign diseases, and 28 NSCLC-PMPE. We examined the clinical characteristics between the three sample groups before performing the ELISA verification. Supplemental Table S17 shows that there was no significant difference (p < 0.05) in age, sex and gender between NSCLC-MPE and NSCLC-PMPE or benign disease (TB and PN) patients. Consistent with the results shown in Table II, MET, PTPFR, and DPP4 were validated as the potential PE biomarkers for NSCLC-MPE using the second independent cohort (Table IV). Notably, we observed that the levels of these three potential PE biomarkers were significantly associated with malignancy (lung MPE compared with TB and PN; lung MPE compared with lung PMPE) and lung cancer histology (adenocarcinoma compared with nonadenocarcinoma). Although the sample population of adenocarcinoma (n = 181) and nonadenocarcinoma (n = 58) is asymmetrical in the current study (Table II and Table IV), we used multivariate analysis to analyze the independent factors of these three potential biomarkers. Table V shows that both MPE (p < 0.001) and histology (p = 0.032) were independent factors of MET levels; however, whether a patient had MPE was the only independent factor of PTPRF and DPP4 levels. Based on these results, we conclude that the three PE protein levels may be used as potential NSCLC biomarkers. However, we emphasize that MET was a potential PE biomarker used in distinguishing adenocarcinoma MPE, the most common histology type of NSCLC-MPE, from nonmalignant pulmonary diseases.
Table IV. Relations between PE levels of three marker candidates and clinical characteristics in new independent cohort (n = 117) with PE.
| Variables | No. | MET (ng/ml)a | p valueb | PTPRF (ng/ml) | p valueb | DPP4 (ng/ml) | p valueb |
|---|---|---|---|---|---|---|---|
| Lung disease | |||||||
| TB, PN | 30 | 144.78 ± 41.07 | <0.001c | 72.23 ± 26.32 | <0.001c | 195.71 ± 105.62 | 0.114c |
| PMPE | 28 | 156.05 ± 86.87 | <0.001d | 92.27 ± 74.37 | <0.001d | 167.73 ± 83.31 | 0.003d |
| MPE | 59 | 483.58 ± 527.98 | 174.23 ± 163.05 | 293.26 ± 421.60 | |||
| Lung cancer histology | |||||||
| Adenocarcinoma | 53 | 516.63 ± 546.64 | <0.001 | 174.66 ± 165.95 | 0.02 | 308.47 ± 442.20 | <0.001 |
| Nonadenocarcinoma | 34 | 168.57 ± 112.62 | 106.05 ± 93.62 | 166.17 ± 79.53 |
a The data are presented as the mean ± S.D.
b Mann-Whitney U test.
c The p value presents the difference between NSCLC-MPE and benign diseases (TB and PN).
d The p value presents the difference between NSCLC-MPE and NSCLC-PMPE.
Table V. Univariate and multivariate analyses of MET, PTPRF, and DPP4 levels in patients with NSCLC in first and second cohort (n = 239).
| Variables | No. | Univariate analysis |
Multivariate analysis |
Univariate analysis |
Multivariate analysis |
Univariate analysis |
Multivariate analysis |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MET (ng/ml)a | p valueb | p valuec | Beta (95% CI) | PTPRF (ng/ml) | p valueb | p valuec | Beta (95% CI) | DPP4 (ng/ml) | p valueb | p valuec | Beta (95% CI) | ||
| Age | |||||||||||||
| ≥ 60 years | 151 | 387.19 ± 467.18 | 0.688 | – | – | 150.51 ± 130.29 | 0.508 | – | – | 244.97 ± 282.64 | 0.756 | – | – |
| < 60 years | 88 | 440.13 ± 502.55 | 140.52 ± 115.70 | 232.61 ± 130.83 | |||||||||
| Gender | |||||||||||||
| Male | 145 | 409.49 ± 489.68 | 0.589 | – | – | 147.68 ± 126.02 | 0.954 | – | – | 237.56 ± 281.00 | 0.249 | – | – |
| Female | 94 | 402.34 ± 467.61 | 145.52 ± 123.97 | 244.81 ± 150.50 | |||||||||
| Smoking status | |||||||||||||
| Ex-/current smoker | 106 | 415.08 ± 514.17 | 0.928 | – | – | 146.87 ± 128.01 | 0.904 | – | – | 237.08 ± 320.16 | 0.085 | – | – |
| Never | 133 | 399.98 ± 453.07 | 146.81 ± 122.97 | 243.07 ± 143.42 | |||||||||
| Lung cancer | |||||||||||||
| MPE | 168 | 512.21 ± 536.31 | <0.001 | <0.001 | –264.58 (−414.62∼−114.53) | 168.85 ± 133.27 | <0.001 | <0.001 | –71.43 (−111.76∼−31.09) | 274.57 ± 273.67 | <0.001 | 0.022 | –91.07 (−168.72∼−13.42) |
| PMPE | 71 | 156.97 ± 87.39 | 94.73 ± 82.47 | 159.60 ± 66.86 | |||||||||
| Lung cancer histology | |||||||||||||
| Adenocarcinoma | 181 | 486.96 ± 524.97 | <0.001 | 0.032 | 175.60 (15.66∼335.53) | 158.27 ± 130.59 | 0.001 | 0.811 | 5.22 (−37.77∼48.21) | 264.61 ± 266.10 | <0.001 | 0.272 | 46.29 (−36.48∼129.05) |
| Nonadenocarcinoma | 58 | 156.14 ± 89.23 | 111.14 ± 98.17 | 164.90 ± 72.28 | |||||||||
a The data are presented as the mean ± S.D.
b Mann-Whitney U test.
c Linear regression.
The Specificity of Potential Biomarker Used in Lung Cancer MPE Diagnosis
To examine whether increased PE levels of MET, PTPRF, and/or DPP4 were specific to NSCLC, we performed ROC analysis to measure the PE levels of candidates used to discriminate NSCLC-MPE from nonlung cancer MPE (breast and gastric cancers, n = 43). We observed that the concentrations of MET, PTPRF, and DPP4 in nonlung cancer MPEs were 274.51 ± 432.80, 244.66 ± 162.94, and 213.61 ± 100.38 ng/ml, respectively (Fig. 4 and Table II). Notably, significant alternation of MET and PTPRF in lung malignancy (n = 109) compared with other malignancies (n = 43) was observed (Fig. 4A–4C; Table II; supplemental Table S16). The AUC values of MET, PTPRF, and DPP4 used to distinguish between NSCLC-MPE and nonlung cancer MPE were 0.787, 0.656, and 0.600, respectively (Fig. 4F; Table III). In addition, we showed that the AUC values of MET, PTPRF, and DPP4 used to distinguish NSCLC-MPE from all other tested pleural types (TB, PN, NSCLC-PMPE, BC, and GC) were 0.870, 0.709, and 0.642, respectively. These results imply that MET may be a specific lung cancer MPE biomarker; however, we require more samples from nonlung cancer MPE to warrant this conclusion.
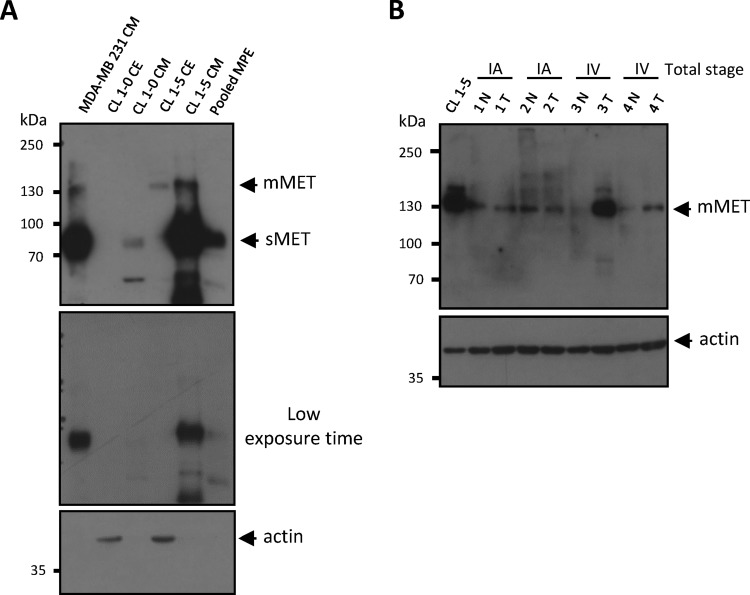
Considering its potential as a PE biomarker for lung cancer, we investigated the source of MET production in PEs. The MET protein contains a disulfide link between the α- (50 kDa) and β-subunits (145 kDa), forming a α/β-heterodimer (31–33). Notably, soluble MET (sMET) is generated via ectodomain shedding, in which the β-subunit is proteolytically cleaved and released (34). We confirmed that the molecular weight of the MET isoform detected in our PE sample was equivalent to sMET (90 kDa; Fig. 5A). Because met gene overexpression has been reported in lung cancer (35, 36), we posited that increased sMET results from overexpression and/or proteolysis of membrane-bound MET (mMET) in lung cancer tissues. To confirm this hypothesis, we examined the expression of mMET in lung cancer cell lines and tissues. Western blot analysis showed that mMET levels in crude extracts and sMET in conditioned media of CL1–5 lung adenocarcinoma cells (high malignancy) were higher than in CL1–0 lung adenocarcinoma cells (low malignancy; Fig. 5A). Advanced stage cancer tissues (stage IV) also showed high levels of mMET compared with adjacent normal tissue (Fig. 5B). These results suggest that sMET was derived from cancerous cells and tissues and supports the positive correlation we observed between MET levels and cancer malignancy.
Fig. 5.
Detection of MET expression in lung cancer tissues and cell lines by Western blot. A, Cell extracts (CE; 50 μg) and conditioned media (CM; 20 μg) prepared from two lung adenocarcinoma cell lines (CL1–0 and CL1–5) and one pooled malignant pleural effusion (pooled MPE) sample with high-abundance protein depletion (20 μg) were resolved on 7.5% SDS-PAGE. Both membrane-bound MET (mMET) and soluble MET (sMET) were detected by Western blot using a goat anti-MET polyclonal antibody (R&D system, cat. BAF358). CM prepared from breast cancer cell line MDA-MB 231 was used as a positive control of sMET. Actin was used as a loading control. B, CE obtained from the CL1–5 cell line (20 μg) or lung adenocarcinoma tissues (50 μg) were separated by SDS-PAGE. mMET was detected by Western blot using a rabbit anti-MET monoclonal antibody (Spring Bioscience, cat. M3440). Actin was used as a loading control. T, tumor parts. N, adjacent normal tissues.
The PE protein Levels as a Useful Adjunct if Combined with Cytological Evaluation in Diagnosis of NSCLC with Pleural Cavity Metastasis
Considering that cytological examination is one of the gold standards for diagnosis of pleural cavity metastasis in NSCLC, it is worthwhile to examine the potential clinical applications of PE biomarkers if combined with cytological examination. We observed that the sensitivity of cytological examination for NSCLC-MPE diagnosis in the present study was 73.80%, indicating that only 124 of 168 NSCLC-MPE were examined as positive cytological samples. When any one of three biomarkers with a given cut-off value (MET: 186.093 ng/ml; PTPRF: 78.113 ng/ml; DPP4: 191.036 ng/ml) was applied for NSCLS-MPE diagnosis, the sensitivity was 93.45% (157/168, supplemental Fig. S8A). This result indicates that 39 of 44 NSCLC-MPE samples missed by cytological examination were rescued by PE biomarkers. When cytological examination was combined with PE biomarkers, the sensitivity was 97.02% ([(124 + 39)/168]*100% = 97.02%) (supplemental Fig. S8A). We also showed that the sensitivity of repeated cytological examination, adjunctive methods (pleural biopsy, pleural seeding nodules) and any one of three markers in the diagnosis of these 44 NSCLC-MPE missed by cytological examination was 77.3% (34/44), 22.7% (10/44), and 88.6% (39/44), respectively (supplemental Fig. S8B). In addition, if we applied the same given cut-off value of three PE biomarkers described above to discriminate NSCLC-MPE from all other tested pleural types in the current study (TB, PN, NSCLC-PMPE, BC, and GC), the sensitivity, specificity, PPV and NPV were 93.45%, 27.55%, 42.43%, and 88.04%, respectively (supplemental Fig. S9). These results collectively suggest that cytological examination (100% specificity and appropriate sensitivity) combined with PE biomarkers (high sensitivity and NPV) would improve the overall clinical diagnostic efficacy of NSCLC-MPE.
Notably, the levels of MET and PTPRF in NSCLC-MPE patients with positive cytology results (n = 124) were significantly higher than in patients with negative cytology results (false negative, n = 44) (Table VI). Significantly, the protein levels of these three potential PE markers (MET, PTPRF, and DPP4) in NSCLC-MPE patients with negative cytology results (n = 44) were significantly higher than the levels in NSCLC-PMPE (true negative, n = 71) (Table VI). These results suggest that the PE protein markers were more discriminated in effusions in which the cytology sample was positive. Accordingly, we propose that PE protein levels would provide a useful adjunct if combined with cytological evaluation in diagnosis of NSCLC with pleural cavity metastasis.
Table VI. The protein levels of MET, PTPRF, and DPP4 in 239 NSCLC patients.
| Category | No. | MET (ng/ml)a | p valueb | PTPRF (ng/ml)a | p valueb | DPP4 (ng/ml)a | p valueb |
|---|---|---|---|---|---|---|---|
| NSCLC-PMPE | 71 | 156.97 ± 87.39 | <0.001c | 94.73 ± 82.47 | <0.001c | 159.60 ± 66.86 | <0.001c |
| NSCLC-MPE with positive cytology result | 124 | 555.49 ± 563.23 | 0.016d | 187.16 ± 147.30 | 0.005d | 292.50 ± 312.69 | 0.299d |
| NSCLC-MPE with negative cytology result | 44 | 390.26 ± 434.79 | 117.26 ± 56.84 | 224.04 ± 87.48 |
a The data are presented as the mean ± S.D.
b Mann-Whitney U test.
c The p value presents the difference between NSCLC-MPE with negative cytology result and NSCLC-PMPE.
d The p value presents the difference between NSCLC-MPE with negative cytology result and NSCLC-MPE with positive cytology result.
DISCUSSION
Lung cancer is the most common malignancy in humans and the leading cause of cancer-related death worldwide (37). Previous studies collectively demonstrate the utility of MPE proteomics in biomarker discovery for human cancers (8). The first proteomics study on MPE was published in 2004; in this study, Bard et al. focused on identification of protein components derived from MPE exosomes in three cancer types (mesothelioma, lung, and breast cancer) by MALDI-TOF. This study reported 18 proteins identified in exosomes isolated from the PE of one lung cancer patient (38). Previously, we created a comprehensive MPE proteome data set with 482 proteins and established the clinical relevance of potential biomarkers in NSCLC (9). Wang et al. recently also identified 16 differentially expressed proteins between lung adenocarcinoma and benign inflammatory PEs by two-dimensional difference gel electrophoresis combined with MALDI-TOF (39). We identified/quantified 15 of 16 (93.75%) differently expressed proteins reported by Wang et al., and only one protein, Jumonji domain containing five, has not been identified in our PE proteome (supplemental Table S18). The present study is the first comprehensive, label-free, quantitative proteomic study of six types of exudative PEs with 772 identified/quantified proteins. Our results established differentially expressed PE proteomes from patients with malignancy (lung, breast, and gastric cancers) and nonmalignancy (TB, PN, and paramalignancy). To the best of our knowledge, the most comprehensive PE data set with more than 1300 proteins was recently generated by Mundt et al. (40). The authors used immunodepletion (Top-14) and narrow-range immobilized pH gradient/high-resolution isoelectric focusing (pH 4–4.25), followed by LC-MS/MS, to perform the PE proteome from mesothelioma, lung adenocarcinoma, and benign pleurisy patients. Therefore, application of multidimensional protein fractionation technology should be necessary to improve the number of identified PE proteins in the near future.
The four proteins (AHSG, AGN, CST3, and IGFBP2) reported as the potential PE biomarkers in our previous NSCLC-MPE data set (9) were also identified/quantified in the current study (supplemental Tables S4 and S5). Consistent with our previous findings, the label-free quantification of these potential PE markers revealed that the protein levels in malignant PE were higher than the levels in nonmalignant PE (TB, PN, and PMPE), although the average ratio of NSCLC-MPE/PN for IGFBP2 was 0.97 (supplemental Table S19). Current bioinformatic analyses revealed distinct expression profiles and biological processes in lung cancer and inflammatory diseases. GO cellular process analysis supports the pathophysiological status of pulmonary disorders herein, including TB, PN, and malignancy. We also verified the PE levels of three potential biomarkers using two cohorts of clinical samples. Our results support the novelty of these three PE biomarkers and the utility of currently established PE proteomes for biomarker discovery in pulmonary disorders.
The three biomarker protein candidates (MET, PTPRF, and DPP4) identified herein were selected based on five criteria (Fig. 3) and validated by ELISA analysis of two independent individual sample sets. MET, encoded by the met proto-oncogene, is a prototypical member of the subfamily of receptor tyrosine kinases, predominantly expressed in epithelia (33). The main ligand that activates MET is hepatocyte growth factor (HGF) also known as a scatter factor (41, 42). Mature MET is expressed on the cell surface to facilitate ligand binding and activation of related transduction molecules through auto-phosphorylation of tyrosine residues in tyrosine kinase and juxtamembrane domains. The HGF/MET pathway regulates important processes involved in cellular development, including differentiation, proliferation, motility, the cell-cycle, and cell death (43, 44). Inappropriate HGF signaling has been observed to dysregulate proliferation, motility, and invasion in several human malignancies (45). Specifically, treatment of lung cancer cells with HGF to activate the MET pathway has been reported to stimulate cellular motility, migration, and invasion (35, 36, 46).
According to our results shown in Fig. 5, we proposed that MET overexpression in advanced stage NSCLC cells elevated soluble MET in PEs. Regardless of the underlying mechanisms, our results were consistent with recent findings by Fu et al., who reported that the shedding of the MET ectodomain was detected in human plasma, and sMET correlated with MET expression levels and tumor size in NSCLC. The authors concluded that a high MET expression led to an increased potential for tumor metastasis and consequently a poor prognosis (47). These phenomena were similar to those shown in a breast cancer study in which MET shedding correlated with malignancy in cultured cells and tumor burden in tumor xenograft mouse models (48). Conversely, Yang et al. demonstrated the beneficial effects of high sMET concentrations in human plasma, showing that the overall median plasma concentration of sMET in patients with gastric cancer was lower than in controls, and sMET levels decreased as the onset of cancer drew nearer (49). This study also investigated the interactions between CagA-related genes and sMET protein concentration in the development of gastric cancer, suggesting that the genetic background of different cancer types may influence the application of MET concentration in the diagnosis/prognosis of human cancers.
A second biomarker candidate in this study was PTPRF, a member of the receptor protein tyrosine phosphatase type IIa subfamily, which exclusively comprise extracellular Ig domains and fibronectin III repeats (50). This phosphatase family has been implicated in several signaling pathways by regulating receptors such as EGFR, RET, and MET (51). PTPRF is expressed in various cell types, including epithelial cells, neuronal cells, and fibroblasts (52), and plays an important role in cell adhesion and migration by directly interacting with integrins in focal adhesions (53). In addition, PTPRF expression was observed to be associated with the potential for metastasis in the well-characterized 13762NF rat mammary adenocarcinoma clones (54).
Another protein candidate herein was DPP4, also known as CD26, a widely distributed 110 kDa transmembrane glycoprotein with peptidase activity. DPP4 is expressed as a cell surface antigen in melanocytes, epithelial cells, and lymphocytes (55–57), is a functional receptor for collagen, and is essential for normal immune function (58). Moreover, there are significant levels of DPP4 activity in plasma, serum (sCD26), and urine (59). Similar to sMET, sCD26 originates from the shedding of surface-bound CD26. Overexpression of DPP4 is observed in various human cancers, including thyroid, breast, prostate, and ovarian cancers and is involved in tumor development, invasion, and metastasis (60, 61). However, previous studies have also observed that sCD26 was deficient in total homogenates of colon, kidney, lung, and liver tumors (62, 63) as well as in different transformed and cancer-derived cell lines (64).
To further explore the clinical application of PE markers, we examined the positive correlation between PE and serum protein levels obtained from the same individuals; the levels of MET and DPP4 in PE and serum positively correlated (p < 0.05; n = 36, supplemental Fig. S10). It is notable that MET levels in PEs (475.879 ± 650.286 ng/ml) were higher than levels in paired serum samples (228.711 ± 66.725 ng/ml), supporting the benefits of using PEs for pulmonary disease-related biomarker discovery. Although future work is warranted to identify and validate additional protein biomarker candidates using a large cohort of PE and serum/plasma samples, the 30 proteins identified as associated with malignancy in the present study (Table I) are viable candidates to establish a useful panel of biomarkers used in diagnosis and/or prognosis of NSCLC.
Supplementary Material
Acknowledgments
We thank Lichieh Julie Chu for analyzing the overlap of proteins identified in five PE types. We would like to thank all members of the Chang Gung Memorial Hospital Cancer Center (Linkou, Tao-Yuan, Taiwan) for clinical patient sample and tissue collection and management. We also thank the Taiwan Lung Cancer Tissue/Specimen and Information Resource Center (MOST 103-2325-B-400-011) at the National Health Research Institute, Taiwan, for samples/data support. This center was supported by grants from the National Research Program for Biopharmaceuticals of the Ministry of Science and Technology, Taiwan.
Footnotes
Author contributions: C.C.W. and C.J.Y. designed research; P.J.L. and C.D.C. performed research; C.L.W., Y.C.W., J.S.Y., and Y.S.C. contributed new reagents or analytic tools; P.J.L., C.D.C., C.L.W., C.W.H., C.W.L., and L.H.H. analyzed data; P.J.L., C.C.W., and C.J.Y. wrote the paper.
* This work was supported by grants from the Chang Gung Medical Research Fund (CMRPD1C0091-2, CMRPG3A0661, CMRPD2B0053, and CLRPD190013), the Ministry of Science and Technology, Taiwan (101-2320-B-182-035-MY3, 102-2320-B-182-029-MY3, and 103-2325-B-182-007), and the Ministry of Education, Taiwan (EMRPD1C0021).
 This article contains supplemental Figs S1 to S10 and Tables S1 to S19.
This article contains supplemental Figs S1 to S10 and Tables S1 to S19.
1 The abbreviations used are:
- PE
- pleural effusion
- NSCLC
- nonsmall cell lung cancer
- TB
- tuberculosis
- PN
- pneumonia
- BC
- breast cancer
- GC
- gastric cancer
- MPE
- malignant pleural effusion
- PMPE
- paramalignant pleural effusion
- GeLC-MS/MS
- one-dimensional SDS-PAGE combined nano-LC-MS/MS
- FDR
- false discovery rate
- MET
- hepatocyte growth factor receptor
- PTPRF
- receptor type tyrosine protein phosphatase F
- DPP4
- dipeptidyl peptidase IV
- RT
- room temperature
- PCA
- principle component analysis
- HCL
- hierarchical clustering algorithm
- HRP
- horseradish peroxidase
- SD
- standard deviation
- ROC
- receiver-operator characteristic
- AUC
- areas under curves
- PPV
- positive predictive value
- NPV
- negative predictive value
- sMET
- soluble MET
- mMET
- membrane-bound MET
- EGFR
- epidermal growth factor receptor
- HGF
- hepatocyte growth factor
- sCD26
- soluble CD26
- CV
- coefficient of variation.
REFERENCES
- 1. Andrews C. O., Gora M. L. (1994) Pleural effusions: pathophysiology and management. Ann. Pharmacother. 28, 894–903 [DOI] [PubMed] [Google Scholar]
- 2. Light R. W. (1997) Diagnostic principles in pleural disease. Eur. Respir. J. 10, 476–481 [DOI] [PubMed] [Google Scholar]
- 3. Lee Y. C., Light R. W. (2004) Management of malignant pleural effusions. Respirology 9, 148–156 [DOI] [PubMed] [Google Scholar]
- 4. Porcel J. M., Light R. W. (2006) Diagnostic approach to pleural effusion in adults. Am. Fam. Physician. 73, 1211–1220 [PubMed] [Google Scholar]
- 5. Sahn S. A. (1997) Pleural diseases related to metastatic malignancies. Eur. Respir. J. 10, 1907–1913 [DOI] [PubMed] [Google Scholar]
- 6. Johnston W. W. (1985) The malignant pleural effusion. A review of cytopathologic diagnoses of 584 specimens from 472 consecutive patients. Cancer 56, 905–909 [DOI] [PubMed] [Google Scholar]
- 7. Maskell N. A., Butland R. J., and Pleural Diseases Group, S. o. C. C. B. T. S. (2003) BTS guidelines for the investigation of a unilateral pleural effusion in adults. Thorax 58, ii8–ii17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schaaij-Visser T. B., de Wit M., Lam S. W., Jiménez C. R. (2013) The cancer secretome, current status and opportunities in the lung, breast and colorectal cancer context. Biochim. Biophys. Acta 1834, 2242–2258 [DOI] [PubMed] [Google Scholar]
- 9. Yu C. J., Wang C. L., Wang C. I., Chen C. D., Dan Y. M., Wu C. C., Wu Y. C., Lee I. N., Tsai Y. H., Chang Y. S., Yu J. S. (2011) Comprehensive proteome analysis of malignant pleural effusion for lung cancer biomarker discovery by using multidimensional protein identification technology. J. Proteome. Res. 10, 4671–4682 [DOI] [PubMed] [Google Scholar]
- 10. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 11. Ong K. C., Indumathi V., Poh W. T., Ong Y. Y. (2000) The diagnostic yield of pleural fluid cytology in malignant pleural effusions. Singapore Med. J. 41, 19–23 [PubMed] [Google Scholar]
- 12. Storey J. D., Tibshirani R. (2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheadle C., Vawter M. P., Freed W. J., Becker K. G. (2003) Analysis of microarray data using Z score transformation. J. Mol. Diagn. 5, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Whitmore T. E., Peterson A., Holzman T., Eastham A., Amon L., McIntosh M., Ozinsky A., Nelson P. S., Martin D. B. (2012) Integrative analysis of N-linked human glycoproteomic data sets reveals PTPRF ectodomain as a novel plasma biomarker candidate for prostate cancer. J. Proteome Res. 11, 2653–2665 [DOI] [PubMed] [Google Scholar]
- 15. Chu Y. W., Yang P. C., Yang S. C., Shyu Y. C., Hendrix M. J., Wu R., Wu C. W. (1997) Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am. J. Respir. Cell Mol. Biol. 17, 353–360 [DOI] [PubMed] [Google Scholar]
- 16. Wang C. L., Wang C. I., Liao P. C., Chen C. D., Liang Y., Chuang W. Y., Tsai Y. H., Chen H. C., Chang Y. S., Yu J. S., Wu C. C., Yu C. J. (2009) Discovery of retinoblastoma-associated binding protein 46 as a novel prognostic marker for distant metastasis in nonsmall cell lung cancer by combined analysis of cancer cell secretome and pleural effusion proteome. J. Proteome Res. 8, 4428–4440 [DOI] [PubMed] [Google Scholar]
- 17. Hanley J. A., McNeil B. J. (1983) A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology 148, 839–843 [DOI] [PubMed] [Google Scholar]
- 18. Youden W. J. (1950) Index for rating diagnostic tests. Cancer 3, 32–35 [DOI] [PubMed] [Google Scholar]
- 19. Jeong S. K., Na K., Kim K. Y., Kim H., Paik Y. K. (2012) PanelComposer: a web-based panel construction tool for multivariate analysis of disease biomarker candidates. J. Proteome Res. 11, 6277–6281 [DOI] [PubMed] [Google Scholar]
- 20. Rhodes D. R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A. M. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beer D. G., Kardia S. L., Huang C. C., Giordano T. J., Levin A. M., Misek D. E., Lin L., Chen G., Gharib T. G., Thomas D. G., Lizyness M. L., Kuick R., Hayasaka S., Taylor J. M., Iannettoni M. D., Orringer M. B., Hanash S. (2002) Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 8, 816–824 [DOI] [PubMed] [Google Scholar]
- 22. Bhattacharjee A., Richards W. G., Staunton J., Li C., Monti S., Vasa P., Ladd C., Beheshti J., Bueno R., Gillette M., Loda M., Weber G., Mark E. J., Lander E. S., Wong W., Johnson B. E., Golub T. R., Sugarbaker D. J., Meyerson M. (2001) Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. U.S.A. 98, 13790–13795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garber M. E., Troyanskaya O. G., Schluens K., Petersen S., Thaesler Z., Pacyna-Gengelbach M., van de Rijn M., Rosen G. D., Perou C. M., Whyte R. I., Altman R. B., Brown P. O., Botstein D., Petersen I. (2001) Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl. Acad. Sci. U.S.A. 98, 13784–13789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hou J., Aerts J., den Hamer B., van Ijcken W., den Bakker M., Riegman P., van der Leest C., van der Spek P., Foekens J. A., Hoogsteden H. C., Grosveld F., Philipsen S. (2010) Gene expression-based classification of nonsmall cell lung carcinomas and survival prediction. PloS One 5, e10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Landi M. T., Dracheva T., Rotunno M., Figueroa J. D., Liu H., Dasgupta A., Mann F. E., Fukuoka J., Hames M., Bergen A. W., Murphy S. E., Yang P., Pesatori A. C., Consonni D., Bertazzi P. A., Wacholder S., Shih J. H., Caporaso N. E., Jen J. (2008) Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PloS One 3, e1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okayama H., Kohno T., Ishii Y., Shimada Y., Shiraishi K., Iwakawa R., Furuta K., Tsuta K., Shibata T., Yamamoto S., Watanabe S., Sakamoto H., Kumamoto K., Takenoshita S., Gotoh N., Mizuno H., Sarai A., Kawano S., Yamaguchi R., Miyano S., Yokota J. (2012) Identification of genes up-regulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 72, 100–111 [DOI] [PubMed] [Google Scholar]
- 27. Selamat S. A., Chung B. S., Girard L., Zhang W., Zhang Y., Campan M., Siegmund K. D., Koss M. N., Hagen J. A., Lam W. L., Lam S., Gazdar A. F., Laird-Offringa I. A. (2012) Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 22, 1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stearman R. S., Dwyer-Nield L., Zerbe L., Blaine S. A., Chan Z., Bunn P. A., Jr., Johnson G. L., Hirsch F. R., Merrick D. T., Franklin W. A., Baron A. E., Keith R. L., Nemenoff R. A., Malkinson A. M., Geraci M. W. (2005) Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am. J. Pathol. 167, 1763–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Su L. J., Chang C. W., Wu Y. C., Chen K. C., Lin C. J., Liang S. C., Lin C. H., Whang-Peng J., Hsu S. L., Chen C. H., Huang C. Y. (2007) Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap resampling scheme. BMC Genomics 8, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yamagata N., Shyr Y., Yanagisawa K., Edgerton M., Dang T. P., Gonzalez A., Nadaf S., Larsen P., Roberts J. R., Nesbitt J. C., Jensen R., Levy S., Moore J. H., Minna J. D., Carbone D. P. (2003) A training-testing approach to the molecular classification of resected nonsmall cell lung cancer. Clin. Cancer Res. 9, 4695–4704 [PubMed] [Google Scholar]
- 31. Prat M., Crepaldi T., Gandino L., Giordano S., Longati P., Comoglio P. (1991) C-terminal truncated forms of Met, the hepatocyte growth factor receptor. Mol. Cell. Biol. 11, 5954–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Giordano S., Ponzetto C., Di Renzo M. F., Cooper C. S., Comoglio P. M. (1989) Tyrosine kinase receptor indistinguishable from the c-met protein. Nature 339, 155–156 [DOI] [PubMed] [Google Scholar]
- 33. Giordano S., Di Renzo M. F., Narsimhan R. P., Cooper C. S., Rosa C., Comoglio P. M. (1989) Biosynthesis of the protein encoded by the c-met proto-oncogene. Oncogene 4, 1383–1388 [PubMed] [Google Scholar]
- 34. Galvani A. P., Cristiani C., Carpinelli P., Landonio A., Bertolero F. (1995) Suramin modulates cellular levels of hepatocyte growth factor receptor by inducing shedding of a soluble form. Biochem. Pharmacol. 50, 959–966 [DOI] [PubMed] [Google Scholar]
- 35. Sadiq A. A., Salgia R. (2013) MET as a possible target for nonsmall-cell lung cancer. J. Clin. Oncol. 31, 1089–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salgia R. (2009) Role of c-Met in cancer: emphasis on lung cancer. Semin. Oncol. 36, S52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kawakami M., Morita S., Sunohara M., Amano Y., Ishikawa R., Watanabe K., Hamano E., Ohishi N., Nakajima J., Yatomi Y., Nagase T., Fukayama M., Takai D. (2013) FER overexpression is associated with poor postoperative prognosis and cancer-cell survival in nonsmall cell lung cancer. Int. J. Clin. Exp. Pathol. 6, 598–612 [PMC free article] [PubMed] [Google Scholar]
- 38. Bard M. P., Hegmans J. P., Hemmes A., Luider T. M., Willemsen R., Severijnen L. A., van Meerbeeck J. P., Burgers S. A., Hoogsteden H. C., Lambrecht B. N. (2004) Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am. J. Respir. Cell Mol. Biol. 31, 114–121 [DOI] [PubMed] [Google Scholar]
- 39. Wang Z., Wang C., Huang X., Shen Y., Shen J., Ying K. (2012) Differential proteome profiling of pleural effusions from lung cancer and benign inflammatory disease patients. Biochim. Biophys. Acta 1824, 692–700 [DOI] [PubMed] [Google Scholar]
- 40. Mundt F., Johansson H. J., Forshed J., Arslan S., Metintas M., Dobra K., Lehtiö J., Hjerpe A. (2014) Proteome screening of pleural effusions identifies galectin 1 as a diagnostic biomarker and highlights several prognostic biomarkers for malignant mesothelioma. Mol. Cell. Proteomics 13, 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bottaro D. P., Rubin J. S., Faletto D. L., Chan A. M., Kmiecik T. E., Vande Woude G. F., Aaronson S. A. (1991) Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 251, 802–804 [DOI] [PubMed] [Google Scholar]
- 42. Abella J. V., Peschard P., Naujokas M. A., Lin T., Saucier C., Urbé S., Park M. (2005) Met/Hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Mol. Cell. Biol. 25, 9632–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Feng Y., Thiagarajan P. S., Ma P. C. (2012) MET signaling: novel targeted inhibition and its clinical development in lung cancer. J. Thorac. Oncol. 7, 459–467 [DOI] [PubMed] [Google Scholar]
- 44. Gao C. F., Vande Woude G. F. (2005) HGF/SF-Met signaling in tumor progression. Cell Res. 15, 49–51 [DOI] [PubMed] [Google Scholar]
- 45. Birchmeier C., Birchmeier W., Gherardi E., Vande Woude G. F. (2003) Met, metastasis, motility, and more. Nat. Rev. Mol. Cell Biol. 4, 915–925 [DOI] [PubMed] [Google Scholar]
- 46. Landi L., Minuti G., D'Incecco A., Cappuzzo F. (2013) Targeting c-MET in the battle against advanced nonsmall-cell lung cancer. Curr. Opin. Oncol. 25, 130–136 [DOI] [PubMed] [Google Scholar]
- 47. Fu L., Guo W., Liu B., Sun L., Bi Z., Zhu L., Wang X., Liu B., Xie Q., Li K. (2013) Shedding of c-Met ectodomain correlates with c-Met expression in nonsmall cell lung cancer. Biomarkers 18, 126–135 [DOI] [PubMed] [Google Scholar]
- 48. Athauda G., Giubellino A., Coleman J. A., Horak C., Steeg P. S., Lee M. J., Trepel J., Wimberly J., Sun J., Coxon A., Burgess T. L., Bottaro D. P. (2006) c-Met ectodomain shedding rate correlates with malignant potential. Clin. Cancer Res. 12, 4154–4162 [DOI] [PubMed] [Google Scholar]
- 49. Yang J. J., Yang J. H., Kim J., Ma S. H., Cho L. Y., Ko K. P., Shin A., Choi B. Y., Kim H. J., Han D. S., Eun C. S., Song K. S., Kim Y. S., Chang S. H., Shin H. R., Kang D., Yoo K. Y., Park S. K. (2013) Soluble c-Met protein as a susceptible biomarker for gastric cancer risk: a nested case-control study within the Korean Multicenter Cancer Cohort. Int. J. Cancer 132, 2148–2156 [DOI] [PubMed] [Google Scholar]
- 50. Craig S. E., Brady-Kalnay S. M. (2011) Tumor-derived extracellular fragments of receptor protein tyrosine phosphatases (RPTPs) as cancer molecular diagnostic tools. Anticancer Agents Med. Chem. 11, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stewart K., Uetani N., Hendriks W., Tremblay M. L., Bouchard M. (2013) Inactivation of LAR family phosphatase genes Ptprs and Ptprf causes craniofacial malformations resembling Pierre-Robin sequence. Development 140, 3413–3422 [DOI] [PubMed] [Google Scholar]
- 52. Streuli M. (1996) Protein tyrosine phosphatases in signaling. Curr. Opin. Cell Biol. 8, 182–188 [DOI] [PubMed] [Google Scholar]
- 53. Zheng W., Lennartsson J., Hendriks W., Heldin C. H., Hellberg C. (2011) The LAR protein tyrosine phosphatase enables PDGF beta-receptor activation through attenuation of the c-Abl kinase activity. Cell. Signal. 23, 1050–1056 [DOI] [PubMed] [Google Scholar]
- 54. Levea C. M., McGary C. T., Symons J. R., Mooney R. A. (2000) PTP LAR expression compared to prognostic indices in metastatic and nonmetastatic breast cancer. Breast Cancer Res. Treat. 64, 221–228 [DOI] [PubMed] [Google Scholar]
- 55. Iwata S., Morimoto C. (1999) CD26/dipeptidyl peptidase IV in context. The different roles of a multifunctional ectoenzyme in malignant transformation. J. Exp. Med. 190, 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cordero O. J., Ayude D., Nogueira M., Rodriguez-Berrocal F. J., de la Cadena M. P. (2000) Preoperative serum CD26 levels: diagnostic efficiency and predictive value for colorectal cancer. Br. J. Cancer 83, 1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cordero O. J., Imbernon M., Chiara L. D., Martinez-Zorzano V. S., Ayude D., de la Cadena M. P., Rodriguez-Berrocal F. J. (2011) Potential of soluble CD26 as a serum marker for colorectal cancer detection. World J. Clin. Oncol. 2, 245–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. De Meester I., Korom S., Van Damme J., Scharpé S. (1999) CD26, let it cut or cut it down. Immunol. Today 20, 367–375 [DOI] [PubMed] [Google Scholar]
- 59. Scharpé S., De Meester I., Vanhoof G., Hendriks D., van Sande M., Van Camp K., Yaron A. (1988) Assay of dipeptidyl peptidase IV in serum by fluorometry of 4-methoxy-2-naphthylamine. Clin. Chem. 34, 2299–2301 [PubMed] [Google Scholar]
- 60. Amatya V. J., Takeshima Y., Kushitani K., Yamada T., Morimoto C., Inai K. (2011) Overexpression of CD26/DPPIV in mesothelioma tissue and mesothelioma cell lines. Oncol. Rep. 26, 1369–1375 [DOI] [PubMed] [Google Scholar]
- 61. Yamada K., Hayashi M., Madokoro H., Nishida H., Du W., Ohnuma K., Sakamoto M., Morimoto C., Yamada T. (2013) Nuclear localization of CD26 induced by a humanized monoclonal antibody inhibits tumor cell growth by modulating of POLR2A transcription. PloS One 8, e62304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ten Kate J., Wijnen J. T., Boldewijn J., Khan P. M., Bosman F. T. (1985) Immunohistochemical localization of adenosine deaminase complexing protein in intestinal mucosa and in colorectal adenocarcinoma as a marker for tumour cell heterogeneity. Histochem. J. 17, 23–31 [DOI] [PubMed] [Google Scholar]
- 63. Ten Kate J., Dinjens W. N., Meera Khan P., Bosman F. T. (1986) Adenosine deaminase complexing protein in cancer studies. Anticancer Res. 6, 983–988 [PubMed] [Google Scholar]
- 64. Ten Kate J., van den Ingh H. F., Khan P. M., Bosman F. T. (1986) Adenosine deaminase complexing protein (ADCP) immunoreactivity in colorectal adenocarcinoma. Int. J. Cancer 37, 479–485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.