Abstract
The mutational status of the immunoglobulin heavy chain variable region defines two clinically distinct forms of chronic lymphocytic leukemia (CLL) known as mutated (M-CLL) and unmutated (UM-CLL). To elucidate the molecular mechanisms underlying the adverse clinical outcome associated with UM-CLL, total proteomes from nine UM-CLL and nine M-CLL samples were analyzed by isobaric tags for relative and absolute quantification (iTRAQ)-based mass spectrometry. Based on the expression of 3521 identified proteins, principal component analysis separated CLL samples into two groups corresponding to immunoglobulin heavy chain variable region mutational status. Computational analysis showed that 43 cell migration/adhesion pathways were significantly enriched by 39 differentially expressed proteins, 35 of which were expressed at significantly lower levels in UM-CLL samples. Furthermore, UM-CLL cells underexpressed proteins associated with cytoskeletal remodeling and overexpressed proteins associated with transcriptional and translational activity. Taken together, our findings indicate that UM-CLL cells are less migratory and more adhesive than M-CLL cells, resulting in their retention in lymph nodes, where they are exposed to proliferative stimuli. In keeping with this hypothesis, analysis of an extended cohort of 120 CLL patients revealed a strong and specific association between UM-CLL and lymphadenopathy. Our study illustrates the potential of total proteome analysis to elucidate pathogenetic mechanisms in cancer.
Chronic lymphocytic leukemia (CLL)1 is the most common adult leukemia in Western countries. It is characterized by the clonal expansion of antigen-experienced B cells with a distinctive immunophenotype (1, 2). The disease runs a highly variable clinical course, with some patients surviving for decades without treatment and others dying from drug-resistant disease within a year of presentation (3).
Many biological variables have been identified in CLL that correlate with clinical outcome. Among these variables, the somatic mutational status of the immunoglobulin heavy chain variable region (IGHV) gene expressed by the malignant clone has a unique biological importance as it is the only prognostic biomarker that is fixed at the initiation of clonal expansion, inherited by the entire malignant clone, and stable over time (4–6). Furthermore, because somatic hypermutation is tightly regulated during B cell development, IGHV mutational status provides insight into the clonogenic cell of origin in CLL. Specifically, cases of CLL with mutated IGHV genes (M-CLL) are thought to arise from a memory B cell that has encountered a T cell-dependent antigen, whereas those with unmutated IGHV genes (UM-CLL) are thought to arise from a B cell that has reacted to a T-independent antigen (7). Importantly, IGHV status is a strong and independent predictor of outcome in CLL, with M-CLL being associated with a favorable outcome and UM-CLL being associated with early disease progression and shorter survival (4–6).
Gene expression profiling studies have shown that, irrespective of their IGHV mutational status, CLL cells have an mRNA signature similar to that of memory B cells (8, 9). Although supervised clustering has shown that a number of genes are differentially expressed in M-CLL and UM-CLL (8, 9), the molecular mechanisms responsible for the more aggressive clinical course of UM-CLL remain incompletely understood. This may reflect the inability of mRNA profiling to detect differences in protein expression due to post-transcriptional regulation (10).
The advent of two-dimensional gel electrophoresis and mass spectrometry (MS) in the mid-1990s represented a technological breakthrough in profiling gene expression at the protein level (11, 12). Application of this technique to the question of how M-CLL and UM-CLL differ from one another has revealed a limited number of differentially expressed proteins (the details of which are provided in supplemental Table S1) (13–18). However, these studies have failed to provide a convincing explanation for the adverse clinical outcome associated with UM-CLL.
The ability of two-dimensional gel electrophoresis and MS to detect differences in protein expression is limited by the fact that they provide only limited coverage of the proteome and suffer from poor reproducibility (19). Recently, more powerful gel-free techniques for proteomic analysis have been developed, such as isobaric tags for relative and absolute quantification (iTRAQ)-based MS. Here, we describe the application of iTRAQ-based MS to analyze the total proteome of nine M-CLL and nine UM-CLL samples. This approach has enabled us to generate the largest quantity of proteomic information for CLL to date and, in particular, to directly compare the functions of differentially expressed proteins between UM-CLL and M-CLL cells through a systems biology approach. Our findings strongly support the idea that M-CLL and UM-CLL are biologically distinct and suggest that the adverse outcome associated with UM-CLL reflects the propensity of malignant cells to be retained in lymph nodes, where they are induced to proliferate. In keeping with this observation, we found a strong and specific association between UM-CLL and lymphadenopathy.
EXPERIMENTAL PROCEDURES
Sample Selection
All samples used for this study were obtained with informed consent and with the approval of the North West 2 Research Ethics Committee–Liverpool Central. Samples were characterized for IGHV mutational status as described below. Because the extent of IGHV mutation in CLL varies continuously from 0 to >10%, a cutoff value of 2% was applied to distinguish M-CLL from UM-CLL. This cutoff was used in the original studies describing the prognostic importance of IGHV mutation (4, 5) and continues to be applied (20). Nine cases were selected to represent extremely low levels of IGHV mutation, and nine were selected to represent extremely high levels. Care was taken to ensure that other biological variables of prognostic significance were evenly balanced between the two groups. The clinical features of the 18 CLL samples used for this study are shown in Table 1.
Table I. Clinical features of the 18 CLL samples subjected to iTRAQ-MS.
| Clinical feature | M-CLL (n = 9) | UM-CLL (n = 9) | p |
|---|---|---|---|
| Age at diagnosis (median), years | 67 | 70 | 0.694 |
| Gender (males/females) | 3/6 | 6/3 | 0.347 |
| Prior therapy (treated/untreated)a | 3/6 | 2/7 | 0.620 |
| Leukocyte count at time of sampling (median), ×109/liter | 78.4 | 135.8 | 0.423 |
| High-risk chromosomal abnormalities (17p− and/or 11q−; yes/no)b | 2/7 | 2/7 | 1.000 |
| IGHV (median), %c | 7.48 | 0.34 | 0.00004 |
The statistical significance of the difference (p values) between the two groups was determined using a two-tailed Mann-Whitney U test for parametric data and Fisher's exact test for nonparametric data, respectively.
a Prior therapy consisted of various combinations of glucocorticoid, chlorambucil, fludarabine, or fludarabine plus cyclophosphamide.
b CLL samples were tested by interphase fluorescence in situ hybridization for del17p13 (17p−), del11q23 (11q−), trisomy 12 (12+), and del13q14 (13q−). 17p and 11q− are regarded as high-risk chromosomal abnormalities.
c IGHV refers to somatic mutation in the IGHV gene of CLL cells compared with the gene sequence of the nearest germ line, where <2% was classed as UM-CLL and ≥2% was classed as M-CLL.
Preparation of CLL Samples
Venous blood was drawn from CLL patients into tubes containing sodium heparin at a final concentration of 10 units/1 ml of blood. Mononuclear cells were isolated by centrifugation of blood over Lymphoprep (Axis-Shield PoC AS, Oslo, Norway) within 4 h of sampling and stored at −150 °C within 2 h of separation. Analysis for recurrent chromosomal abnormalities was performed as described previously (21). All mononuclear cell samples used in the study contained >90% CD19+ cells.
IGHV Mutational Analysis
Total RNA was extracted from CLL cells using an RNeasy mini kit (Qiagen, Crawley, UK). 1 μg of RNA was reverse-transcribed with Moloney murine leukemia virus reverse transcriptase (Promega, Southampton, UK) and an oligo(dT)15 primer. Aliquots of the resulting cDNAs were used to identify clonally expressed IGHV in six PCRs, each with one of the six 5′-leader primers (22) and three mixed 3′-primers specific for the immunoglobulin heavy chain isotypes α, γ, and μ (23), respectively. The 50 microliter PCR containing 20 pmol of each primer, 1.5 mM MgCl2, 100 μm each dNTP, and 2.5 units of Taq polymerase (Promega) was performed for 30 cycles by using a touch-down protocol, with the annealing temperature reducing from 63 to 57 °C over the first 12 cycles. The PCR products were visualized following electrophoresis. The amplified clonal IGHV gene was purified using the Wizard SV Gel and PCR Clean-up system (Promega), followed by Sanger sequencing from both directions with the same primers used in PCR. The IGHV gene usage and extent of somatic hypermutation were determined by comparison to the nearest germ line counterpart sequence in the international ImMunoGeneTics information system with IMGT/V-QUEST.
Protein Extraction and iTRAQ Labeling
Protein was extracted from CLL cells by sonication on ice in 500 mM triethylammonium bicarbonate with 0.1% SDS. Protein concentrations were determined using Bradford assay reagent (Sigma-Aldrich, Gillingham, UK). Labeling with iTRAQ reagents was then carried out according to the AB SCIEX (Framingham, MA) protocol for an 8plex procedure as described previously (24), but with the following modifications. Briefly, 100 μg of protein from each sample was reduced with tris(2-carboxyethyl)phosphine hydrochloride and capped with methyl methanethiosulfate before overnight digestion with trypsin (Promega). Peptides were then labeled with isobaric tags, pooled, diluted to 5 ml with 10 mM potassium dihydrogen phosphate and 25% acetonitrile (ACN), and acidified to pH <3 with phosphoric acid.
Sample Prefractionation and Desalting
Samples were fractionated on a PolySULFOETHYL A strong cation-exchange column (200 × 4.6 mm, 5 μm, 300 Å; Poly LC, Columbia, MD) at 1 ml/min using a gradient from 10 mM potassium dihydrogen phosphate and 25% (w/v) ACN to 0.5 m potassium chloride, 10 mM potassium dihydrogen phosphate, and 25% (w/w/v) ACN over 75 min. Fractions of 2 ml were collected and dried by centrifugation under vacuum (SpeedVac, Eppendorf UK, Stevenage, UK). Fractions were reconstituted in 1 ml of 0.1% trifluoroacetic acid and desalted using an mRP high-recovery protein column (4.6 × 50 mm; Agilent, Berkshire UK) on a VISION workstation (Applied Biosystems/Life Technologies, Paisley, UK) prior to MS analysis.
MS Analysis of iTRAQ Samples
Desalted fractions were reconstituted in 40 microliters of 0.1% formic acid, and 5 microliter aliquots were delivered into a TripleTOF 5600 system (AB SCIEX) via an Eksigent NanoUltra cHiPLC system (AB SCIEX) mounted with a microfluidic trap and an analytical column (15 cm × 75 micrometer) packed with ChromXP C18-CL (3 μm). A NanoSpray III source was fitted with a 10 μm inner diameter PicoTip emitter (New Objective, Woburn, MA). The trap column was washed with 2% ACN and 0.1% formic acid for 10 min at 2 microliter/min before switching in-line with the analytical column. A gradient of 2–50% ACN and 0.1% (v/v) formic acid over 90 min was applied to the column at a flow rate of 300 nl/min. Spectra were acquired automatically in positive ion mode using information-dependent acquisition powered by Analyst TF 1.5.1 software (AB SCIEX). Up to 25 MS/MS spectra were acquired per cycle (10 Hz) using a threshold of 100 counts/s and with dynamic exclusion for 12 s. The rolling collision energy was increased automatically by selecting the iTRAQ check box in Analyst and manually by increasing the collision energy intercepts by 5.
iTRAQ Data Analysis
Data were searched using ProteinPilot 4.2 and the Paragon Algorithm (AB SCIEX) against the Swiss-Prot database (2013-2, 40,646 human entries), with methyl methanethiosulfate as a fixed modification of cysteine residues and biological modifications allowed. Mass tolerance for precursor and fragment ions was 10 ppm. The data were also searched against a reversed decoy database, and only proteins lying within a 1% global false discovery rate were taken forward for analysis (25, 26). Each 8plex iTRAQ-MS experiment consisted of three M-CLL samples, three UM-CLL samples, a replicate sample, and a common reference sample consisting of a pool of all 18 samples. Quantitation of proteins was relative to the common pooled sample present in each iTRAQ-MS experiment. iTRAQ data for proteins identified by two or more peptides with at least 90% confidence of correct sequence assignment or by a single peptide with at least 99% confidence were log2-transformed, batch-corrected using the Batch Remover tool in the Partek Genomics Suite (version 6.3, Partek, St. Louis, MO), and included in subsequent analyses. The MS proteomic data have been deposited in the ProteomeXchange Consortium (27) via the PRIDE partner repository with the data set identifiers PXD001512, PXD001515, and PXD001516.
Principal Component Analysis (PCA)
PCA was performed using the Partek Genomics Suite (version 6.3) to assess variance across the sample set. A correlation method, which adjusts the data to be standardized to a mean of zero and an S.D. of 1, was used for dispersion matrix. Eigenvector scaling was performed orthogonal to the original variables and scaled to unity.
Statistical Analysis
Statistical significance of the difference in the levels of expression of proteins between UM-CLL and M-CLL samples was determined using Student's t test (two-tailed and unpaired), and proteins whose levels of expression were significantly different (p < 0.05) were included in subsequent analyses. Statistical analysis was conducted using the R computational environment (28).
Heat Maps
To detect outliers, hierarchical agglomerative clustering with complete linkage and Euclidean distance measure was employed on the selected data, and heat maps were drawn using the heatmap.2 package in R (28).
Computational Functional Analysis
Proteins found to have significantly higher or lower levels of expression in UM-CLL samples by iTRAQ-MS (p < 0.05) were subjected to computational functional analysis. Proteins were functionally classified using the Protein Analysis Through Evolutionary Relationships (PANTHER) classification system. Pathway analysis was conducted using the GeneGo pathway maps in the MetaCore database (version 6.14, build 61508; Thomson Reuters, New York, NY). The Pathway Maps tool was used to enrich for pathways, and p values were calculated based on a hypergeometric distribution, with the default database used as the background. Significant pathway enrichment was defined as a false discovery rate-corrected p < 0.05.
Western Blotting
Proteins were separated on an SDS-polyacrylamide gel and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore, Bedford, MA), which were probed with rat monoclonal MNDA (myeloid cell nuclear differentiation antigen), rabbit monoclonal LEF-1 (lymphoid enhancer-binding factor 1), and rabbit polyclonal TCL-1 (T cell leukemia/lymphoma 1) antibodies (all from Cell Signaling Technologies, New England Biolabs, Herts, UK), and mouse monoclonal β-actin antibody (Sigma-Aldrich). Immunoreactivity was detected by incubation with horseradish peroxidase-conjugated goat anti-rat IgG, anti-rabbit IgG, or anti-mouse IgG antibodies (Santa Cruz Biotechnology, Insight Biotechnology, Middlesex, UK) and an enhanced chemiluminescence kit (Millipore, Watford, UK), with subsequent digitization using an Image Reader LAS-1000 (Fujifilm, Tokyo, Japan). For quantification of the data, the images were further analyzed on the same instrument using 2D Densitometry Aida Image Analyzer software (Fujifilm). Mann-Whitney tests were used to determine statistical significance of differences in the median values of optical density of the signals corresponding to proteins of interest in CLL samples between UM-CLL and M-CLL cases.
Transmigration Assay and CCR7 Expression
Transmigration assays were carried out as described previously (29), but with the following modifications. Briefly, 5 × 105 CLL cells were added to the inserts of Transwell plates (5 μm pore size;Corning B.V., Amsterdam, The Netherlands). The CCL21 (CC chemokine ligand 21; 1 μg/ml; R&D Systems Europe, Oxford, UK) was then added to the bottom wells. The cells were incubated for 4 h at 37 °C with 5% CO2. After incubation, the transmigrated cells from three technical replicates were counted, and the migration index was calculated (migration index = number of cells migrating in the presence of chemokine divided by number of cells migrating in the absence of chemokine). The expression of the CCL21 receptor (CCR7) in CLL cells was measured by FACS (Becton Dickinson, Oxford, UK).
RESULTS
Quantitative Proteomic Analysis Separates CLL Samples Based on IGHV Mutational Status
To generate quantitative information on the whole proteome of CLL cells, mononuclear cells from nine M-CLL and nine UM-CLL samples containing >90% CLL cells were subjected to iTRAQ-based MS (Table 1). The two groups were well balanced in other respects, including age at diagnosis (median of 67 versus 70 years), prior therapy (37% versus 22%), and adverse chromosomal abnormalities (22% versus 22%), although fewer M-CLL than UM-CLL patients were male (33% versus 67%).
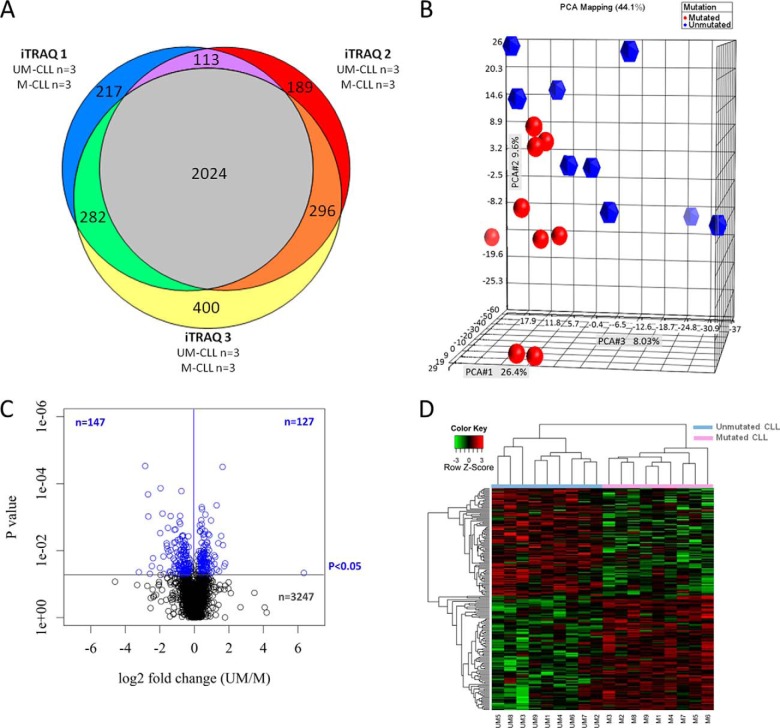
A total of 3521 proteins were identified within a 1% global false discovery rate and with a high confidence of correct peptide sequence assignment (Fig. 1A and the full list of proteins provided in the supplemental data). Of these, 2024 proteins were identified in all three separate iTRAQ-MS experiments (Fig. 1A). To determine whether protein expression was similar between UM-CLL and M-CLL samples, we subjected the data to PCA. As shown in Fig. 1B, PCA showed a distinction between UM-CLL and M-CLL samples based on protein expression.
Fig. 1.
Bioinformatic analysis of proteins identified by iTRAQ-MS. A, Venn diagram showing the number of proteins reproducibly identified between the three separate iTRAQ-MS experiments. In total, 3521 proteins were identified, 2024 of which were identified in all three experiments, and 2715 proteins were identified in two or more experiments. B, PCA showing the separation of UM-CLL and M-CLL samples based on relative protein levels. C, volcano plot of the entire protein data set obtained by iTRAQ-based MS showing differences in protein expression between M-CLL and UM-CLL according to magnitude and p value (t test). Protein expression was remarkably similar in the two CLL subsets, with 92% (n = 3247) of identified proteins sharing similar levels of expression across the sample cohort (p > 0.05). However, >270 proteins were differentially expressed between the two CLL subsets, giving a p value of <0.05, with 147 proteins being expressed at lower levels and 127 proteins expressed at higher levels in UM-CLL compared with M-CLL. D, heat map showing levels of differentially expressed proteins for which relative quantitative values were obtained for all 18 CLL cases (n = 186). Hierarchical clustering of CLL cases based on the relative expression of these proteins generated two clusters comprising the nine cases of UM-CLL and the nine cases of M-CLL, respectively.
A two-tailed t test was used to determine the statistical significance of differences in protein expression between UM-CLL and M-CLL samples. As shown in Fig. 1C, protein expression was largely comparable between the two CLL subsets, with 92% of proteins sharing similar levels of expression across the sample cohort. However, we found 274 proteins that were differentially expressed in UM-CLL and M-CLL samples (p < 0.05) (supplemental Table S2). Among them, 127 proteins were expressed at higher levels and 147 proteins at lower levels in UM-CLL samples compared with M-CLL counterparts (Fig. 1C). Relative quantification of 186 of 274 differentially expressed proteins was obtained across all 18 samples. As shown in Fig. 1D, hierarchical clustering of CLL samples based on the expression of these 186 differentially expressed proteins clearly separated cases into two distinct groups corresponding to IGHV mutational status, with no samples identified as outliers.
Verification of iTRAQ-MS Data by Western Blotting
To validate the iTRAQ-MS data, we sought to verify the differential expression of a small number of proteins by Western blotting. We selected three differentially expressed proteins as follows: TCL-1 (30, 31), MNDA (32), and LEF-1 (33). The expression of TCL-1, MNDA, and LEF-1 as measured by Western blotting varied among the 18 samples (supplemental Fig. S1A) and correlated with IGHV mutational status with a pattern of differential expression entirely consistent with the iTRAQ-MS data (supplemental Fig. S1B).
UM-CLL Cells Are Characterized by Defective Cell Migration Pathways
We next used GeneGo pathway maps from MetaCore to identify pathways enriched by the 274 proteins that were differentially expressed in the two subsets of CLL samples (p < 0.05). The enrichment analysis identified 169 signaling pathways (p < 0.05) (supplemental Table S3). Remarkably, 26 of the top 50 most significantly altered pathways were associated with cell migration/adhesion. In total, 43 cell migration/adhesion pathways (Table 2) were significantly enriched by 39 differentially expressed proteins (Table 3). As all of these 39 proteins were involved in lymphocyte entry into, transit within, or exit from the lymphoid tissues (Fig. 2) and 35 of them were expressed at significantly lower levels in UM-CLL samples, these results suggest that UM-CLL cells have reduced migratory properties compared with their M-CLL counterparts.
Table II. Chemotaxis, cell adhesion, and cytoskeletal remodeling pathway maps enriched by proteins found to be differentially expressed in M-CLL versus UM-CLL.
| Enriched pathways | p | FDR | No. of differentially expressed proteins in pathway | Total proteins in pathway |
|---|---|---|---|---|
| 1. Cytoskeleton remodeling_Regulation of actin cytoskeleton by Rho GTPases | <0.001 | 0.000 | 7 | 23 |
| 2. Chemotaxis_Inhibitory action of lipoxins on IL-8- and leukotriene B4-induced neutrophil migration | <0.001 | 0.000 | 8 | 51 |
| 3. Inhibitory action of lipoxins on neutrophil migration | <0.001 | 0.000 | 8 | 57 |
| 4. Cell adhesion_Chemokines and adhesion | <0.001 | 0.000 | 10 | 100 |
| 5. Cell adhesion_IL-8-dependent cell migration and adhesion | <0.001 | 0.000 | 6 | 33 |
| 6. Chemotaxis_Leukocyte chemotaxis | <0.001 | 0.000 | 8 | 75 |
| 7. Immune response_Immunological synapse formation | <0.001 | 0.001 | 7 | 59 |
| 8. Cell adhesion_Histamine H1 receptor signaling in interruption of cell barrier integrity | <0.001 | 0.001 | 6 | 45 |
| 9. Cytoskeleton remodeling_Integrin outside-in signaling | <0.001 | 0.002 | 6 | 49 |
| 10. Cytoskeleton remodeling_Cytoskeleton remodeling | <0.001 | 0.002 | 8 | 102 |
| 11. CCR4-dependent immune cell chemotaxis in asthma and atopic dermatitis | <0.001 | 0.002 | 5 | 34 |
| 12. Development_S1PR1 signaling via β-arrestin | <0.001 | 0.002 | 5 | 34 |
| 13. Chemotaxis_CCR4-induced chemotaxis of immune cells | <0.001 | 0.002 | 5 | 34 |
| 14. Mechanism of action of CCR4 antagonists in asthma and atopic dermatitis (Variant 1) | <0.001 | 0.002 | 5 | 34 |
| 15. Immune response_CXCR4 signaling via second messenger | <0.001 | 0.002 | 5 | 34 |
| 16. Chemotaxis_C5a-induced chemotaxis | <0.001 | 0.005 | 5 | 43 |
| 17. Immune response_MIF-induced cell adhesion, migration and angiogenesis | <0.001 | 0.006 | 5 | 46 |
| 18. Chemotaxis_Lipoxin inhibitory action on fMLP-induced neutrophil chemotaxis | <0.001 | 0.006 | 5 | 46 |
| 19. Development_S1PR2 and S1PR3 in cell proliferation and differentiation | <0.001 | 0.006 | 4 | 26 |
| 20. Development_Thromboxane A2 pathway signaling | 0.001 | 0.007 | 5 | 49 |
| 21. Cytoskeleton remodeling_TGF, Wnt, and cytoskeletal remodeling | 0.001 | 0.010 | 7 | 111 |
| 22. Immune response_CCL2 signaling | 0.001 | 0.010 | 5 | 54 |
| 23. Cell adhesion_Integrin inside-out signaling | 0.001 | 0.011 | 5 | 56 |
| 24. Chemotaxis_CXCR4 signaling pathway | 0.001 | 0.012 | 4 | 34 |
| 25. Development_c-Kit ligand signaling pathway during hemopoiesis | 0.001 | 0.014 | 5 | 61 |
| 26. Cell adhesion_Role of tetraspanins in integrin-mediated cell adhesion | 0.002 | 0.015 | 4 | 37 |
| 27. Cytoskeleton remodeling_α1A-Adrenergic receptor-dependent inhibition of PI3K | 0.002 | 0.019 | 3 | 19 |
| 28. Cytoskeleton remodeling_Role of PKA in cytoskeleton reorganization | 0.002 | 0.019 | 4 | 40 |
| 29. Development_S1PR3 signaling pathway | 0.003 | 0.023 | 4 | 43 |
| 30. Chemotaxis_CCL2-induced chemotaxis | 0.007 | 0.042 | 4 | 56 |
| 31. Muscle contraction_S1PR2-mediated smooth muscle contraction | 0.008 | 0.044 | 3 | 30 |
| 32. Cytoskeleton remodeling_RalA regulation pathway | 0.008 | 0.044 | 3 | 30 |
| 33. Cytoskeleton remodeling_Reverse signaling by ephrin B | 0.008 | 0.046 | 3 | 31 |
| 34. Cell adhesion_α4 integrins in cell migration and adhesion | 0.011 | 0.057 | 3 | 34 |
| 35. G-protein signaling_S1PR2 signaling | 0.012 | 0.059 | 3 | 35 |
| 36. Cell adhesion_Tight junctions | 0.013 | 0.062 | 3 | 36 |
| 37. Development_S1PR1 signaling pathway | 0.022 | 0.088 | 3 | 44 |
| 38. Development_S1PR4 signaling pathway | 0.036 | 0.122 | 2 | 22 |
| 39. Immune response_IL-33 signaling pathway | 0.043 | 0.134 | 3 | 57 |
| 40. Immune response_IL-18 signaling | 0.048 | 0.144 | 3 | 60 |
| 41. Immune response_IL-17 signaling pathways | 0.048 | 0.144 | 3 | 60 |
| 42. Cell adhesion_Cadherin-mediated cell adhesion | 0.049 | 0.144 | 2 | 26 |
| 43. Development_Cross-talk between VEGF and angiopoietin 1 signaling pathways | 0.049 | 0.144 | 2 | 26 |
FDR, false discovery rate; MIF, migration inhibitory factor; fMLP, formylmethionylleucylphenylalanine.
Table III. 39 individual proteins differentially expressed in M-CLL versus UM-CLL (p < 0.05) that are involved in chemotaxis, cell adhesion, and cytoskeletal remodeling pathways.
| Swiss-Prot accession No. | Gene | Protein name | Fold change (UM/M) | p |
|---|---|---|---|---|
| P68032 | Actin cytoskeletal | Actin, α cardiac muscle 1a | −1.5 | 0.011 |
| P63261 | Actin cytoskeletal | Actin, cytoplasmic 2 | −2.7 | 0.014 |
| O15143 | Arp2/3 | Actin-related protein 2/3 complex subunit 1B | −1.4 | 0.038 |
| O15144 | Arp2/3 | Actin-related protein 2/3 complex subunit 2 | −1.8 | 0.001 |
| O15145 | Arp2/3 | Actin-related protein 2/3 complex subunit 3 | −1.6 | 0.014 |
| P61158 | Arp2/3 | Actin-related protein 3 | −1.6 | 0.031 |
| P62330 | ARF6 | ADP-ribosylation factor 6 | −1.8 | 0.013 |
| P35611 | Adducin 1 (α) | -αAdducin | −1.6 | 0.004 |
| P63010 | Actin cytoskeletal | AP-2 complex subunit β | −1.5 | 0.010 |
| Q96CW1 | AP complex 2 medium (μ) chain | AP-2 complex subunit μ | 2.2 | 0.014 |
| P16070 | CD44 | CD44 antigen | 1.4 | 0.012 |
| O43639 | GRB4/NCK2 | Cytoplasmic protein NCK2a | −1.3 | 0.045 |
| Q92608 | DOCK2 | Dedicator of cytokinesis protein 2 | −1.8 | 0.037 |
| Q02750 | MEK1/2 | Dual specificity mitogen-activated protein kinase kinase 1 | −1.4 | 0.007 |
| P50570 | Dynamin-2 | Dynamin-2 | −1.3 | 0.048 |
| P02675 | Fibrinogen | Fibrinogen β chain | −6.0 | 0.049 |
| P02679 | Fibrinogen γ | Fibrinogen γ chain | −9.3 | 0.044 |
| P21333 | Filamin | Filamin-A | −3.0 | 0.006 |
| P04899 | G protein αi family | Guanine nucleotide-binding protein Gi subunit α-2 | −1.6 | 0.006 |
| P20036 | MHC class II | HLA class II histocompatibility antigen, DP α1 chain | 2.0 | 0.020 |
| O14920 | IKK-β | Inhibitor of nuclear factor κB kinase subunit β | −1.3 | 0.017 |
| P20701 | αLα integrin | Integrin αL | −2.1 | 0.023 |
| P11215 | αMβ integrin | Integrin αM | −2.6 | 0.010 |
| P05107 | ITGB2/βL integrin | Integrin β2 | −2.3 | 0.046 |
| Q9UJU2 | Tcf/LEF-1 | Lymphoid enhancer-binding factor 1 | 1.2 | 0.019 |
| P49137 | MAPKAP | MAPK-activated protein kinase 2a | −1.4 | 0.043 |
| P60660 | MELC/myosin | Myosin light polypeptide 6 | −1.8 | 0.023 |
| P24844 | MRLC/myosin | Myosin regulatory light polypeptide 9 | −5.5 | 0.029 |
| P35579 | Myosin | Myosin-9 | −2.9 | 0.002 |
| Q00653 | NF-κB | Nuclear factor NF-κB p100 subunit | −1.3 | 0.035 |
| Q14289 | FAK2/Pyk2 | Protein-tyrosine kinase 2β | −1.4 | 0.038 |
| Q7LDG7 | CalDAG-GEFI | RAS guanyl-releasing protein 2 | −2.2 | 0.006 |
| P08575 | CD45 | Receptor-type tyrosine-protein phosphatase C | −1.7 | 0.009 |
| Q92888 | ARHGEF1 | Rho guanine nucleotide exchange factor 1 | −1.5 | 0.049 |
| P51812 | p90RSK | Ribosomal protein S6 kinase α3 | −1.5 | 0.006 |
| P42229 | STAT5 | Signal transducer and activator of transcription 5A | −1.9 | 0.028 |
| Q9Y490 | Talin | Talin-1 | −3.5 | 0.015 |
| P07996 | Thrombospondin-1 | Thrombospondin-1 | −5.2 | 0.017 |
| P50552 | VASP | Vasodilator-stimulated phosphoprotein | −1.8 | 0.050 |
All proteins were identified by two or more peptides at ≥90% confidence and were present in two or more iTRAQ experiments unless indicated otherwise.
a Proteins were identified by a single peptide at ≥99% confidence and were present in two or more iTRAQ experiments.
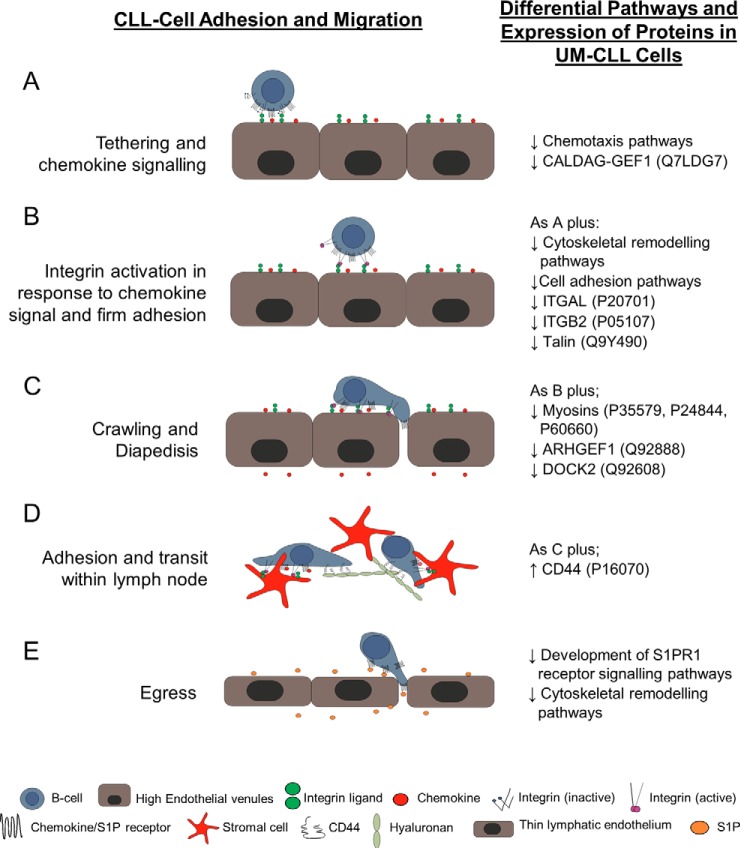
Fig. 2.
Schematic diagram illustrating factors involved in lymphocyte migration into, retention within, and egress from lymph nodes. Migration into, within, and through lymphoid tissues is a complex multistep process. The major steps are illustrated here, along with the pathways/proteins that are altered in UM-CLL versus M-CLL. On initial contact with the endothelium, the lymphocytes become loosely tethered (A). If the cell encounters chemokine presented on the endothelial cell surface, it then becomes firmly adherent in a process involving integrin activation and chemokine signaling (B). The cell then crawls along the endothelium until it reaches an intercellular junction, where it undergoes diapedesis in response to chemokine (C); this process also requires integrin activation. Once within the lymphoid tissue (D), two different mechanisms mediate the adherence of CLL cells within the tissues; these involve α4β1 and CD44 binding to their respective ligands, fibronectin and hyaluronan. Adhesion mediated by both substrata is influenced by chemokine signaling. The final step of transit though the lymph node is egress (E), a process that is entirely dependent on S1PR1 and is independent of integrins.
The lymphocyte chemotaxis pathway, which is essential for migration into and transit within lymphoid tissues (34), was significantly altered (p < 0.001) in UM-CLL cells. Of particular note, eight of the nine differentially expressed proteins in this pathway were expressed at significantly lower levels in UM-CLL samples (Fig. 3). These underexpressed proteins included the Rap (Ras-related protein) activator CalDAG-GEFI (RAS guanyl-releasing protein 2; Q7LDG7), which is involved in integrin activation (35, 36); both chains of the αLβ2 integrin (P20701 and P05107), which is required for the migration of lymphocytes into lymph nodes; and talin (Q9Y490), which is important in maintaining the high-affinity binding state of αLβ2 (37). Collectively, these observations strongly suggest that UM-CLL is associated with impaired Rap1-dependent αLβ2-mediated migration.
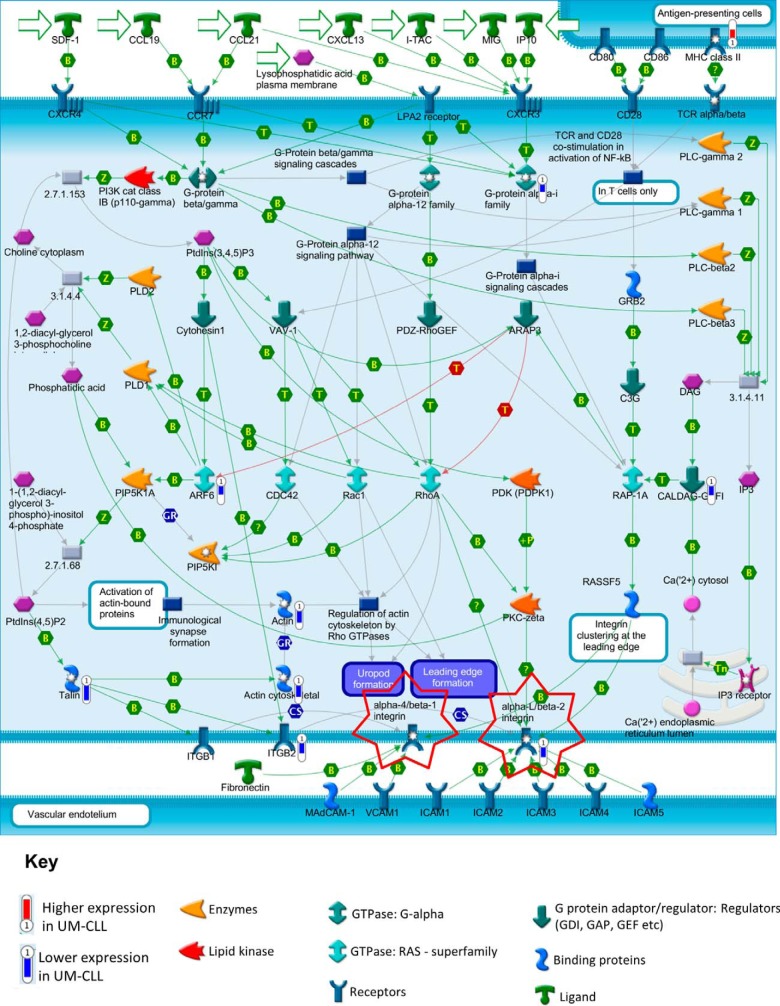
Fig. 3.
Enrichment of leukocyte chemotaxis pathway by proteins found to be differentially expressed in M-CLL versus UM-CLL. One of the migration pathways most enriched by the GeneGo MetaCore pathway maps using proteins differentially expressed between UM-CLL and M-CLL was the leukocyte chemotaxis pathway (p < 0.001). This pathway directs leukocyte movement to lymphatic organs and also allows them to migrate to sites of infection and/or inflammation via either αLβ2 or α4β1 integrins (highlighted). Within this pathway, nine proteins were found to be differentially expressed: MHC class II (P20036, p = 0.02), G-protein αi family (P04899, p = 0.006), CalDAG-GEFI (Q7LDG7, p = 0.006), ARF6 (P62330, p = 0.013), actin (P68032, p = 0.011), actin cytoskeletal (P63261, p = 0.014), talin (Q9Y490, p = 0.015), ITGB2 (P05107, p = 0.046), and αLβ2 (P20701, p = 0.023; P05107, p = 0.046). MHC class II had a higher expression in UM-CLL (fold changes represented by a red thermometer). However, the other eight differentially expressed proteins had a lower expression in UM-CLL (fold changes represented by blue thermometers) compared with M-CLL. This suggests that migration into the tissue microenvironments via the αLβ2 integrin pathway may be dysfunctional in UM-CLL.
We also found that CD44 (P16070), which facilitates the adhesion of CLL cells within the lymph node microenvironment (38), was expressed at significantly higher levels in UM-CLL cells, whereas two proteins that regulate S1PR1 (sphingosine 1-phosphate receptor 1) signaling, dynamin-2 (P50570) and G-protein αi (P04899) (39, 40), were expressed at significantly lower levels. Because egress of lymphocytes from lymph nodes is absolutely dependent on S1PR1 (41), these findings suggest that the latter process is also dysfunctional in UM-CLL. In keeping with the GeneGo MetaCore pathway analysis, PANTHER analysis indicated that many of the proteins that were expressed at reduced levels in UM-CLL cells are involved in cytoskeletal remodeling (supplemental Fig. S2, A and B).
UM-CLL Cells Retain Their Ability to Migrate toward CCL21
The proteomic data suggest that UM-CLL cells have migration defects that affect both lymph node entry and exit. To formulate a hypothesis that could be validated at the clinical level, we examined one of the key steps involved in the entry of CLL cells into lymph nodes, namely, migration toward CCL21. To ascertain the functional integrity of this aspect of lymph node entry in light of our published observations that integrin activation on CLL cells is defective (29, 42, 43), CLL cells were examined for their ability to migrate in the absence of adhesion using Transwell assays. Although the UM-CLL cells tended to migrate less than the M-CLL cells, no significant differences were observed (p = 0.713) (supplemental Fig. S3). These data therefore support the notion that UM-CLL cells at least partly retain their ability to migrate toward CCL21 and are therefore able to enter the lymph node environment.
Patients with UM-CLL Have More Lymphadenopathy than Those with M-CLL
On the basis of our proteomic and functional data, we hypothesized that UM-CLL cells are retained in lymph nodes due to a global defect in migration coupled with impaired S1PR1-mediated egress and increased adhesion to hyaluronan within the tissue via CD44. To test this hypothesis, we examined the clinical records of patients with M-CLL (n = 78) and UM-CLL (n = 42) for documentation of lymph node enlargement. To be able to analyze lymphadenopathy independently of overall tumor burden, cases of CLL used for this comparison were selected to have similar levels of blood involvement; this was achieved by setting an upper limit for the blood leukocyte count of 130 × 109/liter. In keeping with our hypothesis, twice as many patients with UM-CLL compared with M-CLL had documented lymphadenopathy (50% versus 24%; p < 0.01) (Table 4). As expected, there was no significant difference in the leukocyte count between the two groups selected for this comparison (medians of 37 × 109/liter and 28 × 109/liter, respectively; p > 0.05). This indicates that the increased lymphadenopathy observed in UM-CLL is not simply a nonspecific manifestation of increased tumor burden but instead reflects the selective retention of UM-CLL cells in lymph nodes.
Table IV. Relationship between IGHV mutational status and lymphadenopathy in 120 CLL patients.
| Clinical feature | M-CLL (n = 78) | UM-CLL (n = 42) | p |
|---|---|---|---|
| Age (median), years | 67 | 69 | 0.589 |
| Gender (males/females) | 44/34 | 20/22 | 0.443 |
| Prior therapy (treated/untreated) | 3/61 | 1/32 | 1.000 |
| Leukocyte count (median), ×109/liter | 28.25 | 37.35 | 0.072 |
| Lymphadenopathy ≥1.5 cm (yes/no) | 19/59 | 21/21 | 0.0078 |
The extended cohort comprised all locally stored CLL cases for which IGHV mutational status was known, information on lymphadenopathy available, and leukocyte count ≤130 × 109/liter. The statistical significance of the difference (p values) between the two groups was determined using a two-tailed Mann-Whitney U test for parametric data and Fisher's exact test for nonparametric data, respectively. All information presented relates to the time of sample collection.
Other Pathways Enriched by Differentially Expressed Proteins
Although the most striking difference between M-CLL and UM-CLL cells identified by proteomic analysis was the defect in cell migration pathways in the IGHV-unmutated group, other differences were also observed. For example, PANTHER analysis showed that the majority of proteins that were overexpressed in UM-CLL cells are involved in nucleic acid binding and RNA splicing factor activity (supplemental Fig. S2, C and D), suggesting higher levels of transcriptional and translational activities in these cells. Furthermore, GeneGo MetaCore analysis showed that pathways that promote cell survival and proliferation in UM-CLL cells were significantly enriched (supplemental Table S3). These pathways included the immune response pathway involving B cell receptor signaling (p = 0.006) (supplemental Fig. S4), the endoplasmic reticulum stress response pathway (p = 0.035) (supplemental Fig. S5), and the Wnt signaling pathway (p = 0.006), where LEF-1, a critical transcription factor in this pathway (44), was significantly overexpressed in UM-CLL cells (p = 0.019 and 0.004 for iTRAQ-based MS and Western blot confirmation, respectively).
DISCUSSION
The aim of this study was to elucidate pathogenetic mechanisms responsible for the adverse clinical course of UM-CLL by comparing the proteome of M-CLL and UM-CLL cells. Using iTRAQ-MS, we analyzed the total proteome of 18 primary CLL samples and identified 3521 proteins, making this the largest proteomic study hitherto conducted in CLL. In agreement with previous mRNA profiling studies (8, 9), overall protein expression in the nine UM-CLL and nine M-CLL samples was largely comparable, with 92% of identified proteins sharing similar levels of expression. However, almost 8% of proteins were differentially expressed. This contrasts with the much lower proportion of genes (<1%) that were found to be differentially expressed in mRNA profiling studies (8, 9, 45). It can therefore be deduced that post-transcriptional regulation is an important determinant of gene expression in CLL and that differences in gene expression between M-CLL and UM-CLL are substantially greater at the protein than mRNA level. In keeping with this observation, it is noteworthy that PCA clearly separated CLL samples into two groups corresponding to IGHV mutational status.
After verifying the differential expression of three proteins (i.e., TCL-1, MNDA, and LEF-1), the iTRAQ-MS data were subjected to two different forms of computational analyses to explore the functional implications of the 274 differentially expressed proteins. Strikingly, both GeneGo MetaCore pathway analysis and PANTHER analysis of individual proteins revealed that a high proportion of differentially expressed proteins were involved in cell migration processes and that almost all of these were underexpressed in UM-CLL. It can therefore be concluded that UM-CLL cells have a global defect in migration.
With regard to specific migration pathways that are defective in UM-CLL, the results of the computational analysis may at first appear contradictory. Thus, although UM-CLL cells underexpressed proteins involved in egress from lymph nodes (dynamin-2 and G-protein αi, which activate S1PR1), they also underexpressed proteins involved in lymph node entry (αLβ2 integrin and CalDAG-GEFI, which activate αLβ2 via Rap1, and talin, which stabilizes the integrin in its active conformation). UM-CLL cells also overexpressed CD44, which contributes to the retention of CLL cells in lymph nodes by mediating adhesion to hyaluronan following engagement of CD40 by CD154-expressing T cells in the lymph node microenvironment (38). The overall impact of these different pathway defects clearly depends on which are functionally most important. Pertinent to this consideration is our previous demonstration that Rap1-dependent αLβ2 activation is defective in CLL but that this defect can be overcome if α4β1 is coexpressed (43). Other groups have shown that surface α4β1 expression is higher in UM-CLL (46). We therefore speculated that the αLβ2 defect in UM-CLL cells is counteracted by their increased surface expression of α4β1. In keeping with this idea, we found that UM-CLL cells at least partially retain their ability to migrate toward CCL21, which is required for lymph node entry (42, 47). This led us to formulate the hypothesis that in UM-CLL, defective lymph node entry is more than compensated for by enhanced retention and impaired egress, resulting in the selective accumulation of the malignant cells in lymph nodes. To test this hypothesis, we related lymph node enlargement to IGHV status in a cohort of 120 patients and observed that lymphadenopathy was twice as common in patients with UM-CLL compared with those with M-CLL. The association between UM-CLL and lymph node enlargement was not simply a reflection of increased tumor burden, as the M-CLL and UM-CLL patients used for this comparison had similar levels of blood involvement.
Retention of CLL cells in lymph nodes is likely to have profound implications for disease pathogenesis, given that the lymph node microenvironment provides crucial proliferative stimuli (48–50) and that lymph node enlargement is associated with adverse clinical outcome (51–53). In keeping with this concept, PANTHER analysis revealed that the majority of proteins that were overexpressed in UM-CLL cells had nucleic acid binding and RNA splicing factor activity. It is also of interest that UM-CLL cells displayed a pattern of protein expression indicating increased activity in B cell receptor signaling, endoplasmic reticulum stress response, and Wnt signaling pathways. These properties of UM-CLL cells could be intrinsic to their differentiation/maturation status. Alternatively, they could represent changes induced by extrinsic stimuli during their delayed passage through the lymph node microenvironment that are retained after the cells have re-entered the blood stream.
The concept that UM-CLL cells are preferentially retained in lymph nodes, where they are exposed to proliferative stimuli, suggests that therapeutic strategies that displace CLL cells from lymph nodes may be particularly effective in UM-CLL. In agreement with this prediction, clinical studies have shown that the overall response to ibrutinib, an inhibitor of Bruton tyrosine kinase that redistributes CLL cells from tissues into the blood by preventing tissue adhesion and homing (54–56), is significantly higher in patients with UM-CLL (77%) compared with those with M-CLL (33%) (p = 0.005) (57). Very similar findings have been observed with idelalisib (58), a selective inhibitor of the phosphoinositide 3′-kinase p110 delta isoform that mobilizes CLL cells into the blood by interfering with microenvironmental interactions (59, 60).
In conclusion, we have shown that quantitative analysis of the total proteome by iTRAQ-MS was able to separate individual CLL cases according to IGHV status and explain the more aggressive clinical behavior of UM-CLL and its particular sensitivity to treatments that induce anatomical displacement from the lymph node microenvironment. More generally, and in accordance with the ability of proteomic analysis to detect alterations in gene expression resulting from both transcriptional and post-transcriptional mechanisms (61, 62), the study illustrates the considerable potential of iTRAQ-MS coupled with a systems biology approach to elucidate pathogenetic mechanisms and indicate therapeutic strategies in cancer.
Supplementary Material
Acknowledgments
We are grateful to Prof. Gerry Cohen for critical review of the manuscript.
Footnotes
Author contributions: J.Z., N.R.K., and A.R.P. designed research; G.L.E. performed research; G.L.E., J.Z., P.V.J., N.R.K., and A.R.P. analyzed data; G.L.E., J.Z., R.E.J., K.J.T., P.V.J., N.R.K., and A.R.P. wrote the paper; R.E.J. generated MS data; K.J.T. generated migration assay data; K.L. and G.G.J. characterized clinical features of patient samples; M.O. provided patient samples and clinical data; K.P. contributed to the generation of MS data.
* This work was supported by the North West Cancer Research (UK), Leukaemia & Lymphoma Research (UK) and the Liverpool Cancer Research UK Centre Research Development Fund.
 This article contains supplemental Tables S1–S3, supplemental Figs. S1–S5, and supplemental data.
This article contains supplemental Tables S1–S3, supplemental Figs. S1–S5, and supplemental data.
1 The abbreviations used are:
- CLL
- chronic lymphocytic leukemia
- M-CLL
- IGHV-mutated CLL
- UM-CLL
- IGHV-unmutated CLL
- IGHV
- immunoglobulin heavy chain variable region
- ACN
- acetonitrile
- MS
- mass spectrometry
- iTRAQ
- isobaric tags for relative and absolute quantification
- PANTHER
- Protein Analysis Through Evolutionary Relationships
- MNDA
- myeloid cell nuclear differentiation antigen
- PCA
- principal component analysis.
REFERENCES
- 1. Chiorazzi N., Rai K. R., Ferrarini M. (2005) Chronic lymphocytic leukemia. N. Engl. J. Med. 352, 804–815 [DOI] [PubMed] [Google Scholar]
- 2. Zenz T., Mertens D., Küppers R., Döhner H., Stilgenbauer S. (2010) From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat. Rev. Cancer 10, 37–50 [DOI] [PubMed] [Google Scholar]
- 3. Zenz T., Häbe S., Denzel T., Mohr J., Winkler D., Bühler A., Sarno A., Groner S., Mertens D., Busch R., Hallek M., Döhner H., Stilgenbauer S. (2009) Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood 114, 2589–2597 [DOI] [PubMed] [Google Scholar]
- 4. Damle R. N., Wasil T., Fais F., Ghiotto F., Valetto A., Allen S. L., Buchbinder A., Budman D., Dittmar K., Kolitz J., Lichtman S. M., Schulman P., Vinciguerra V. P., Rai K. R., Ferrarini M., Chiorazzi N. (1999) Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 94, 1840–1847 [PubMed] [Google Scholar]
- 5. Hamblin T. J., Davis Z., Gardiner A., Oscier D. G., Stevenson F. K. (1999) Unmutated Ig VH genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 94, 1848–1854 [PubMed] [Google Scholar]
- 6. Cramer P., Hallek M. (2011) Prognostic factors in chronic lymphocytic leukemia–what do we need to know? Nat. Rev. Clin. Oncol. 8, 38–47 [DOI] [PubMed] [Google Scholar]
- 7. Seifert M., Sellmann L., Bloehdorn J., Wein F., Stilgenbauer S., Dürig J., Küppers R. (2012) Cellular origin and pathophysiology of chronic lymphocytic leukemia. J. Exp. Med. 209, 2183–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klein U., Tu Y., Stolovitzky G. A., Mattioli M., Cattoretti G., Husson H., Freedman A., Inghirami G., Cro L., Baldini L., Neri A., Califano A., Dalla-Favera R. (2001) Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J. Exp. Med. 194, 1625–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosenwald A., Alizadeh A. A., Widhopf G., Simon R., Davis R. E., Yu X., Yang L., Pickeral O. K., Rassenti L. Z., Powell J., Botstein D., Byrd J. C., Grever M. R., Cheson B. D., Chiorazzi N., Wilson W. H., Kipps T. J., Brown P. O., Staudt L. M. (2001) Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J. Exp. Med. 194, 1639–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lian Z., Wang L., Yamaga S., Bonds W., Beazer-Barclay Y., Kluger Y., Gerstein M., Newburger P. E., Berliner N., Weissman S. M. (2001) Genomic and proteomic analysis of the myeloid differentiation program. Blood 98, 513–524 [DOI] [PubMed] [Google Scholar]
- 11. Duncan M. W., Hunsucker S. W. (2005) Proteomics as a tool for clinically relevant biomarker discovery and validation. Exp. Biol. Med. 230, 808–817 [DOI] [PubMed] [Google Scholar]
- 12. Rabilloud T., Chevallet M., Luche S., Lelong C. (2010) Two-dimensional gel electrophoresis in proteomics: past, present and future. J. Proteomics 73, 2064–2077 [DOI] [PubMed] [Google Scholar]
- 13. Cochran D. A., Evans C. A., Blinco D., Burthem J., Stevenson F. K., Gaskell S. J., Whetton A. D. (2003) Proteomic analysis of chronic lymphocytic leukemia subtypes with mutated or unmutated Ig VH genes. Mol. Cell. Proteomics 2, 1331–1341 [DOI] [PubMed] [Google Scholar]
- 14. Scielzo C., Ghia P., Conti A., Bachi A., Guida G., Geuna M., Alessio M., Caligaris-Cappio F. (2005) HS1 protein is differentially expressed in chronic lymphocytic leukemia patient subsets with good or poor prognoses. J. Clin. Invest. 115, 1644–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rees-Unwin K. S., Faragher R., Unwin R. D., Adams J., Brown P. J., Buckle A. M., Pettitt A., Hutchinson C. V., Johnson S. M., Pulford K., Banham A. H., Whetton A. D., Lucas G., Mason D. Y., Burthem J. (2010) Ribosome-associated nucleophosmin 1: increased expression and shuttling activity distinguishes prognostic subtypes in chronic lymphocytic leukaemia. Br. J. Haematol. 148, 534–543 [DOI] [PubMed] [Google Scholar]
- 16. Barnidge D. R., Jelinek D. F., Muddiman D. C., Kay N. E. (2005) Quantitative protein expression analysis of CLL B cells from mutated and unmutated IgVH subgroups using acid-cleavable isotope-coded affinity tag reagents. J. Proteome Res. 4, 1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alsagaby S. A., Khanna S., Hart K. W., Pratt G., Fegan C., Pepper C., Brewis I. A., Brennan P. (2014) Proteomics-based strategies to identify proteins relevant to chronic lymphocytic leukemia. J. Proteome Res. 13, 5051–5062 [DOI] [PubMed] [Google Scholar]
- 18. Perrot A., Pionneau C., Nadaud S., Davi F., Leblond V., Jacob F., Merle-Béral H., Herbrecht R., Béné M. C., Gribben J. G., Bahram S., Vallat L. (2011) A unique proteomic profile on surface IgM ligation in unmutated chronic lymphocytic leukemia. Blood 118, e1–e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Issaq H., Veenstra T. (2008) Two-dimensional polyacrylamide gel electrophoresis (2D-PAGE): advances and perspectives. BioTechniques 44, 697–698, 700 [DOI] [PubMed] [Google Scholar]
- 20. Hallek M., Fischer K., Fingerle-Rowson G., Fink A. M., Busch R., Mayer J., Hensel M., Hopfinger G., Hess G., von Grünhagen U., Bergmann M., Catalano J., Zinzani P. L., Caligaris-Cappio F., Seymour J. F., Berrebi A., Jäger U., Cazin B., Trneny M., Westermann A., Wendtner C. M., Eichhorst B. F., Staib P., Bühler A., Winkler D., Zenz T., Böttcher S., Ritgen M., Mendila M., Kneba M., Döhner H., Stilgenbauer S., International Group of Investigators, German Chronic Lymphocytic Leukaemia Study Group (2010) Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet 376, 1164–1174 [DOI] [PubMed] [Google Scholar]
- 21. Carter A., Lin K., Sherrington P. D., Atherton M., Pearson K., Douglas A., Burford A., Brito-Babapulle V., Matutes E., Catovsky D., Pettitt A. R. (2006) Imperfect correlation between p53 dysfunction and deletion of TP53 and ATM in chronic lymphocytic leukaemia. Leukemia 20, 737–740 [DOI] [PubMed] [Google Scholar]
- 22. Campbell M. J., Zelenetz A. D., Levy S., Levy R. (1992) Use of family specific leader region primers for PCR amplification of the human heavy chain variable region gene repertoire. Mol. Immunol. 29, 193–203 [DOI] [PubMed] [Google Scholar]
- 23. Fais F., Ghiotto F., Hashimoto S., Sellars B., Valetto A., Allen S. L., Schulman P., Vinciguerra V. P., Rai K., Rassenti L. Z., Kipps T. J., Dighiero G., Schroeder H. W., Jr., Ferrarini M., Chiorazzi N. (1998) Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Invest. 102, 1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kitteringham N. R., Abdullah A., Walsh J., Randle L., Jenkins R. E., Sison R., Goldring C. E., Powell H., Sanderson C., Williams S., Higgins L., Yamamoto M., Hayes J., Park B. K. (2010) Proteomic analysis of Nrf2 deficient transgenic mice reveals cellular defence and lipid metabolism as primary Nrf2-dependent pathways in the liver. J. Proteomics 73, 1612–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shilov I. V., Seymour S. L., Patel A. A., Loboda A., Tang W. H., Keating S. P., Hunter C. L., Nuwaysir L. M., Schaeffer D. A. (2007) The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6, 1638–1655 [DOI] [PubMed] [Google Scholar]
- 26. Tang W. H., Shilov I. V., Seymour S. L. (2008) Nonlinear fitting method for determining local false discovery rates from decoy database searches. J. Proteome Res. 7, 3661–3667 [DOI] [PubMed] [Google Scholar]
- 27. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. RDevelopment Core Team (2005) R: A Language and Environment for Statistical Computing, The R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 29. Till K. J., Spiller D. G., Harris R. J., Chen H., Zuzel M., Cawley J. C. (2005) CLL, but not normal, B cells are dependent on autocrine VEGF and α4β1 integrin for chemokine-induced motility on and through endothelium. Blood 105, 4813–4819 [DOI] [PubMed] [Google Scholar]
- 30. Herling M., Patel K. A., Khalili J., Schlette E., Kobayashi R., Medeiros L. J., Jones D. (2006) TCL1 shows a regulated expression pattern in chronic lymphocytic leukemia that correlates with molecular subtypes and proliferative state. Leukemia 20, 280–285 [DOI] [PubMed] [Google Scholar]
- 31. Mansouri M. R., Sevov M., Aleskog A., Jondal M., Merup M., Sundström C., Osorio L., Rosenquist R. (2010) IGHV3–21 gene usage is associated with high TCL1 expression in chronic lymphocytic leukemia. Eur J Haematol 84, 109–116 [DOI] [PubMed] [Google Scholar]
- 32. Joshi A. D., Hegde G. V., Dickinson J. D., Mittal A. K., Lynch J. C., Eudy J. D., Armitage J. O., Bierman P. J., Bociek R. G., Devetten M. P., Vose J. M., Joshi S. S. (2007) ATM, CTLA4, MNDA, and HEM1 in high versus low CD38 expressing B-cell chronic lymphocytic leukemia. Clin. Cancer Res. 13, 5295–5304 [DOI] [PubMed] [Google Scholar]
- 33. Erdfelder F., Hertweck M., Filipovich A., Uhrmacher S., Kreuzer K. A. (2010) High lymphoid enhancer-binding factor-1 expression is associated with disease progression and poor prognosis in chronic lymphocytic leukemia. Hematol. Rep. 2, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cathcart M. K. (2009) Signal-activated phospholipase regulation of leukocyte chemotaxis. J. Lipid Res. 50, (suppl.) S231–S236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawasaki H., Springett G. M., Toki S., Canales J. J., Harlan P., Blumenstiel J. P., Chen E. J., Bany I. A., Mochizuki N., Ashbacher A., Matsuda M., Housman D. E., Graybiel A. M. (1998) A Rap guanine nucleotide exchange factor enriched highly in the basal ganglia. Proc. Natl. Acad. Sci. U.S.A. 95, 13278–13283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kinashi T., Katagiri K. (2004) Regulation of lymphocyte adhesion and migration by the small GTPase Rap1 and its effector molecule, RAPL. Immunol. Lett. 93, 1–5 [DOI] [PubMed] [Google Scholar]
- 37. Calderwood D. A. (2004) Integrin activation. J. Cell Sci. 117, 657–666 [DOI] [PubMed] [Google Scholar]
- 38. Girbl T., Hinterseer E., Grössinger E. M., Asslaber D., Oberascher K., Weiss L., Hauser-Kronberger C., Neureiter D., Kerschbaum H., Naor D., Alon R., Greil R., Hartmann T. N. (2013) CD40-mediated activation of chronic lymphocytic leukemia cells promotes their CD44-dependent adhesion to hyaluronan and restricts CCL21-induced motility. Cancer Res. 73, 561–570 [DOI] [PubMed] [Google Scholar]
- 39. Windh R. T., Lee M. J., Hla T., An S., Barr A. J., Manning D. R. (1999) Differential coupling of the sphingosine 1-phosphate receptors Edg-1, Edg-3, and H218/Edg-5 to the Gi, Gq, and G12 families of heterotrimeric G proteins. J. Biol. Chem. 274, 27351–27358 [DOI] [PubMed] [Google Scholar]
- 40. Rakhit S., Pyne S., Pyne N. J. (2000) The platelet-derived growth factor receptor stimulation of p42/p44 mitogen-activated protein kinase in airway smooth muscle involves a G-protein-mediated tyrosine phosphorylation of Gab1. Mol. Pharmacol. 58, 413–420 [DOI] [PubMed] [Google Scholar]
- 41. Matloubian M., Lo C. G., Cinamon G., Lesneski M. J., Xu Y., Brinkmann V., Allende M. L., Proia R. L., Cyster J. G. (2004) Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 427, 355–360 [DOI] [PubMed] [Google Scholar]
- 42. Till K. J., Lin K., Zuzel M., Cawley J. C. (2002) The chemokine receptor CCR7 and α4 integrin are important for migration of chronic lymphocytic leukemia cells into lymph nodes. Blood 99, 2977–2984 [DOI] [PubMed] [Google Scholar]
- 43. Till K. J., Harris R. J., Linford A., Spiller D. G., Zuzel M., Cawley J. C. (2008) Cell motility in chronic lymphocytic leukemia: defective Rap1 and αLβ2 activation by chemokine. Cancer Res. 68, 8429–8436 [DOI] [PubMed] [Google Scholar]
- 44. Petropoulos K., Arseni N., Schessl C., Stadler C. R., Rawat V. P., Deshpande A. J., Heilmeier B., Hiddemann W., Quintanilla-Martinez L., Bohlander S. K., Feuring-Buske M., Buske C. (2008) A novel role for Lef-1, a central transcription mediator of Wnt signaling, in leukemogenesis. J. Exp. Med. 205, 515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haslinger C., Schweifer N., Stilgenbauer S., Döhner H., Lichter P., Kraut N., Stratowa C., Abseher R. (2004) Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J. Clin. Oncol. 22, 3937–3949 [DOI] [PubMed] [Google Scholar]
- 46. Gattei V., Bulian P., Del Principe M. I., Zucchetto A., Maurillo L., Buccisano F., Bomben R., Dal-Bo M., Luciano F., Rossi F. M., Degan M., Amadori S., Del Poeta G. (2008) Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood 111, 865–873 [DOI] [PubMed] [Google Scholar]
- 47. Park E. J., Peixoto A., Imai Y., Goodarzi A., Cheng G., Carman C. V., von Andrian U. H., Shimaoka M. (2010) Distinct roles for LFA-1 affinity regulation during T-cell adhesion, diapedesis, and interstitial migration in lymph nodes. Blood 115, 1572–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Smit L. A., Hallaert D. Y., Spijker R., de Goeij B., Jaspers A., Kater A. P., van Oers M. H., van Noesel C. J., Eldering E. (2007) Differential Noxa/Mcl-1 balance in peripheral versus lymph node chronic lymphocytic leukemia cells correlates with survival capacity. Blood 109, 1660–1668 [DOI] [PubMed] [Google Scholar]
- 49. van Gent R., Kater A. P., Otto S. A., Jaspers A., Borghans J. A., Vrisekoop N., Ackermans M. A., Ruiter A. F., Wittebol S., Eldering E., van Oers M. H., Tesselaar K., Kersten M. J., Miedema F. (2008) In vivo dynamics of stable chronic lymphocytic leukemia inversely correlate with somatic hypermutation levels and suggest no major leukemic turnover in bone marrow. Cancer Res. 68, 10137–10144 [DOI] [PubMed] [Google Scholar]
- 50. Herishanu Y., Pérez-Galán P., Liu D., Biancotto A., Pittaluga S., Vire B., Gibellini F., Njuguna N., Lee E., Stennett L., Raghavachari N., Liu P., McCoy J. P., Raffeld M., Stetler-Stevenson M., Yuan C., Sherry R., Arthur D. C., Maric I., White T., Marti G. E., Munson P., Wilson W. H., Wiestner A. (2011) The lymph node microenvironment promotes B-cell receptor signaling, NF-κB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood 117, 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rai K. R., Sawitsky A., Cronkite E. P., Chanana A. D., Levy R. N., Pasternack B. S. (1975) Clinical staging of chronic lymphocytic leukemia. Blood 46, 219–234 [PubMed] [Google Scholar]
- 52. Binet J. L., Lepoprier M., Dighiero G., Charron D., D'Athis P., Vaugier G., Beral H. M., Natali J. C., Raphael M., Nizet B., Follezou J. Y. (1977) A clinical staging system for chronic lymphocytic leukemia: prognostic significance. Cancer 40, 855–864 [DOI] [PubMed] [Google Scholar]
- 53. Swerdlow S. H., Campo E., Harris N. L., Jaffe E. S., Pileri S. A., Stein H., Thiele J., W V. J. (2008) WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, IARC Press, Lyon, France [Google Scholar]
- 54. de Rooij M. F., Kuil A., Geest C. R., Eldering E., Chang B. Y., Buggy J. J., Pals S. T., Spaargaren M. (2012) The clinically active BTK inhibitor PCI-32765 targets B-cell receptor- and chemokine-controlled adhesion and migration in chronic lymphocytic leukemia. Blood 119, 2590–2594 [DOI] [PubMed] [Google Scholar]
- 55. Ponader S., Chen S. S., Buggy J. J., Balakrishnan K., Gandhi V., Wierda W. G., Keating M. J., O'Brien S., Chiorazzi N., Burger J. A. (2012) The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood 119, 1182–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Advani R. H., Buggy J. J., Sharman J. P., Smith S. M., Boyd T. E., Grant B., Kolibaba K. S., Furman R. R., Rodriguez S., Chang B. Y., Sukbuntherng J., Izumi R., Hamdy A., Hedrick E., Fowler N. H. (2013) Bruton tyrosine kinase inhibitor ibrutinib (PCI-32765) has significant activity in patients with relapsed/refractory B-cell malignancies. J. Clin. Oncol. 31, 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Byrd J. C., Furman R. R., Coutre S. E., Flinn I. W., Burger J. A., Blum K. A., Grant B., Sharman J. P., Coleman M., Wierda W. G., Jones J. A., Zhao W., Heerema N. A., Johnson A. J., Sukbuntherng J., Chang B. Y., Clow F., Hedrick E., Buggy J. J., James D. F., O'Brien S. (2013) Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 369, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Furman R. R., Sharman J. P., Coutre S. E., Cheson B. D., Pagel J. M., Hillmen P., Barrientos J. C., Zelenetz A. D., Kipps T. J., Flinn I., Ghia P., Eradat H., Ervin T., Lamanna N., Coiffier B., Pettitt A. R., Ma S., Stilgenbauer S., Cramer P., Aiello M., Johnson D. M., Miller L. L., Li D., Jahn T. M., Dansey R. D., Hallek M., O'Brien S. M. (2014) Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 370, 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hoellenriegel J., Meadows S. A., Sivina M., Wierda W. G., Kantarjian H., Keating M. J., Giese N., O'Brien S., Yu A., Miller L. L., Lannutti B. J., Burger J. A. (2011) The phosphoinositide 3′-kinase delta inhibitor, CAL-101, inhibits B-cell receptor signaling and chemokine networks in chronic lymphocytic leukemia. Blood 118, 3603–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fiorcari S., Brown W. S., McIntyre B. W., Estrov Z., Maffei R., O'Brien S., Sivina M., Hoellenriegel J., Wierda W. G., Keating M. J., Ding W., Kay N. E., Lannutti B. J., Marasca R., Burger J. A. (2013) The PI3-kinase delta inhibitor idelalisib (GS-1101) targets integrin-mediated adhesion of chronic lymphocytic leukemia (CLL) cell to cndothelial and marrow stromal sells. PLoS ONE 8, e83830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim M. S., Pinto S. M., Getnet D., Nirujogi R. S., Manda S. S., Chaerkady R., Madugundu A. K., Kelkar D. S., Isserlin R., Jain S., Thomas J. K., Muthusamy B., Leal-Rojas P., Kumar P., Sahasrabuddhe N. A., Balakrishnan L., Advani J., George B., Renuse S., Selvan L. D., Patil A. H., Nanjappa V., Radhakrishnan A., Prasad S., Subbannayya T., Raju R., Kumar M., Sreenivasamurthy S. K., Marimuthu A., Sathe G. J., Chavan S., Datta K. K., Subbannayya Y., Sahu A., Yelamanchi S. D., Jayaram S., Rajagopalan P., Sharma J., Murthy K. R., Syed N., Goel R., Khan A. A., Ahmad S., Dey G., Mudgal K., Chatterjee A., Huang T. C., Zhong J., Wu X., Shaw P. G., Freed D., Zahari M. S., Mukherjee K. K., Shankar S., Mahadevan A., Lam H., Mitchell C. J., Shankar S. K., Satishchandra P., Schroeder J. T., Sirdeshmukh R., Maitra A., Leach S. D., Drake C. G., Halushka M. K., Prasad T. S., Hruban R. H., Kerr C. L., Bader G. D., Iacobuzio-Donahue C. A., Gowda H., Pandey A. (2014) A draft map of the human proteome. Nature 509, 575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wilhelm M., Schlegl J., Hahne H., Moghaddas Gholami A., Lieberenz M., Savitski M. M., Ziegler E., Butzmann L., Gessulat S., Marx H., Mathieson T., Lemeer S., Schnatbaum K., Reimer U., Wenschuh H., Mollenhauer M., Slotta-Huspenina J., Boese J. H., Bantscheff M., Gerstmair A., Faerber F., Kuster B. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.