Abstract
Dengue is the most common arboviral infection of humans and a public health burden in over 100 countries. Aedes aegypti mosquitoes stably infected with strains of the intracellular bacterium Wolbachia are resistant to dengue virus (DENV) infection and are being tested in field trials. To mimic field conditions, we experimentally assessed the vector competence of A. aegypti carrying the Wolbachia strains wMel and wMelPop after challenge with viremic blood from dengue patients. We found that wMelPop conferred strong resistance to DENV infection of mosquito abdomen tissue and largely prevented disseminated infection. wMel conferred less resistance to infection of mosquito abdomen tissue, but importantly did reduce the prevalence of mosquitoes with infectious saliva. A mathematical model of DENV transmission incorporating the dynamics of viral infection within humans and mosquitoes was fitted to the data collected. Model predictions suggested that wMel would reduce the basic reproduction number, R0, of DENV transmission by 66–75%. Our results suggest that establishment of wMelPop-infected A. aegypti at high frequency in a dengue endemic setting would result in complete abatement of DENV transmission. Establishment of wMel-infected A. aegypti is also predicted to have a substantial effect on transmission that would be sufficient to eliminate dengue in low or moderate transmission settings, but may be insufficient to achieve complete control in settings where R0 is high. These findings develop a framework for selecting Wolbachia strains for field releases and for calculating their likely impact.
Introduction
Dengue is an acute systemic viral infection (1). In 2010 there were an estimated 100 million apparent infections globally (2). The etiological agents of dengue are four dengue viruses (DENV-1-4), with transmission from human-to-human primarily by Aedes aegypti mosquitoes. Existing disease prevention strategies are based on reducing the mosquito vector population, yet this has been largely unsuccessful in halting dengue transmission in endemic countries.
A new entomological-based control method utilizes the phenotype of A. aegypti experimentally infected with strains (wMel and wMelPop) of the bacterial symbiont Wolbachia (3, 4). The heritable wMelPop infection of A. aegypti is characterized by widely disseminated and dense infection of mosquito tissues (3). wMelPop infection confers numerous phenotypic traits on A. aegypti including refractoriness to DENV infection (5), reduced lifespan (3), reduced viability of desiccated eggs (6) and reduced blood-feeding success (7). The heritable wMel infection of A. aegypti is associated with a relatively lower intensity of tissue infection yet is also able to confer complete resistance to disseminated DENV infection after laboratory challenge (4). The mechanism of virus interference is unknown, but could potentially be mediated by Wolbachia-triggered changes in immunoregulatory microRNA expression, elevation of reactive oxygen species or competition between DENV and Wolbachia for critical metabolic resources (8–10). Successful field-releases of wMel-A. aegypti have occurred in the northern Australian city of Cairns (11), providing proof of concept that stable, long-term establishment of Wolbachia in mosquito populations can be achieved.
The cost of developing a new operationalized vector control measure and testing its effectiveness in the field makes it a priority to try to predict the likely impact of the introduction of Wolbachia into A. aegypti populations on dengue transmission. However, previous vector competence studies of Wolbachia-infected A. aegypti had significant limitations in that they employed a single serotype of laboratory-passaged DENV that was spiked into animal or human blood to create infectious blood meals (4, 5). This model system probably does not accurately mimic a human DENV infection in that dengue viruses have evolved to efficiently transmit to mosquitoes via fresh blood meals from infected human hosts. We describe here vector competence studies that use viremic blood from dengue patients to blood feed field-derived Wolbachia-infected A. aegypti and thus provide “real-world” measures of vector competence.
More generally, translating laboratory studies of vector competence into an assessment of the potential effectiveness of Wolbachia in reducing dengue transmission to human populations requires an understanding of multiple interacting aspects of mosquito ecology and the biology of DENV infection. In addition to characterizing the invasion dynamics of Wolbachia into A. aegypti populations (the goal of field trials currently underway), we require better understanding of: (a) the development of DENV infection in mosquitoes (and how this is modified by Wolbachia); (b) the within-host dynamics of DENV infection in humans; and (c) DENV transmission from mosquitoes to humans and from humans to mosquitoes (and how this is modified by Wolbachia). Here, we begin to address these data needs by combining experimental characterization of the impact of Wolbachia infection on vector competence with mathematical modeling of the natural history of DENV infection in humans and vectors. By using more biologically realistic experimental and mathematical models than hitherto possible, we have generated estimates of the impact of Wolbachia strains on dengue transmission that can be used with greater confidence to inform future field trials in dengue endemic areas and to guide the development of additional Wolbachia strains in A. aegypti.
Results
Vector competence assessments of wMelPop-A. aegypti
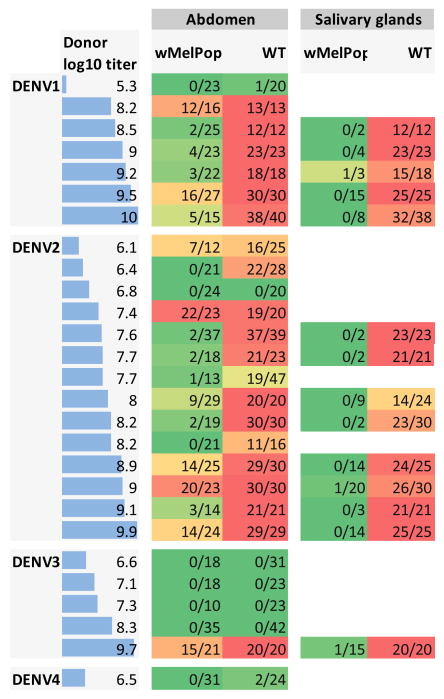
We measured the susceptibility of wMelPop-A. aegypti to DENV infection after human viremic blood feeding (n=27 independent feeds). wMelPop-A. aegypti were highly resistant to acquiring DENV as assessed by assaying their abdomen tissues compared with their wild-type (WT) counterparts (Figure 1). In a subset of mosquitoes with detectable virus in their abdomen, salivary glands were assayed for the presence of DENV infection. For WT mosquitoes, 90% (95% CI: 87–94%) of salivary glands contained DENV, while for wMelPop-infected mosquitoes, virus was detected in only 2.6% (95% CI: 0.5–7.6%) of the salivary glands tested (Figure 1). We did not explicitly fit mathematical models to the wMelPop data, as under the highly plausible assumption that infection of salivary glands is required for DENV transmission, the salivary gland data suggested at least 90% blocking of transmission.
Figure 1.
Susceptibility of WT and wMelPop-infected mosquitoes to DENV infection. Each row represents the results of feeding cohorts of WT and wMelPop infected mosquitoes on viremic blood collected from human dengue cases. The log10 viral titer (RNA copies/ml) in plasma in the donor blood is given in the first column (also indicated by the horizontal bars). Other columns indicate the numbers of mosquitoes with detectable abdomen or salivary gland infection over the total numbers fed on blood from that donor. Only mosquitoes with detectable abdominal infection, a pre-requisite for disseminated infection, were tested for salivary gland infection. Background color of table cells indicates the proportion of mosquitoes with detectable infection (0%=dark green to 100%=red).
Vector competence assessments of wMel-A. aegypti
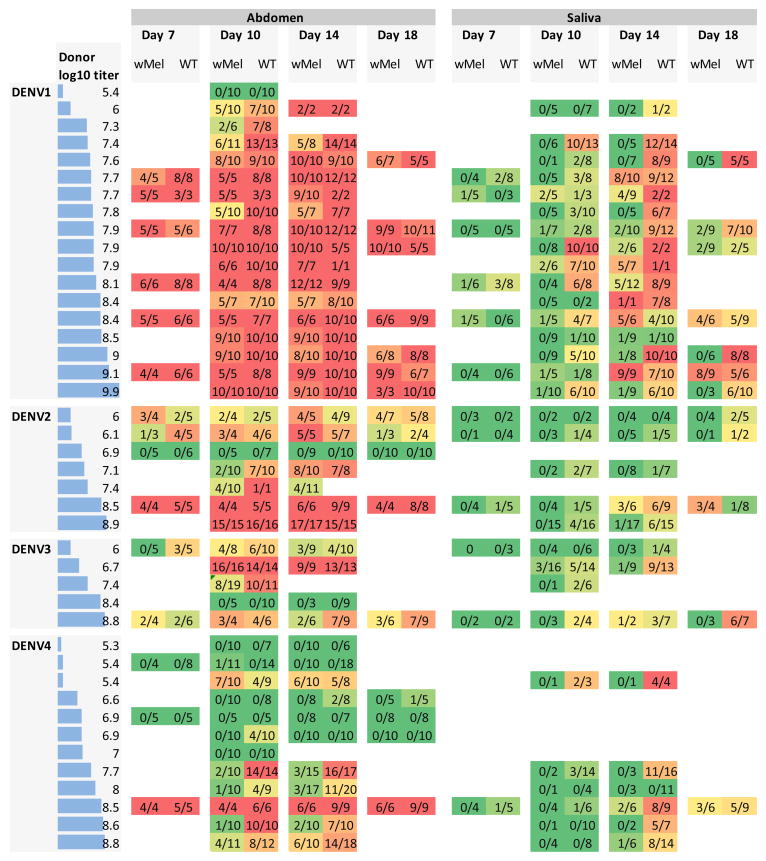
We postulated that wMel infection would confer lower levels of resistance to DENV infection in A. aegypti than wMelPop on the basis that wMel is present at lower tissue densities (11). To test this hypothesis, the prevalence of DENV-infected mosquito abdomens and saliva in WT and wMel-A. aegypti were measured after 42 independent human viremic blood feeds. Groups of mosquitoes were assessed at multiple time-points after viremic blood feeding to assess whether the phenotype of wMel-A. aegypti had a temporal component. The results, stratified by serotype, plasma viremia, time since blood meal and mosquito tissue type, are shown in Figure 2.
Figure 2.
Susceptibility of WT and wMel-infected mosquitoes to DENV infection. Each row represents the results of feeding cohorts of WT and wMel-infected mosquitoes on viremic blood collected from human dengue cases. The log10 viral titer (RNA copies/ml) in plasma in the donor blood is given in the first column (also indicated by the horizontal bars). Results indicate the numbers of mosquitoes with detectable abdomen or saliva infection over the total numbers fed on blood from that donor at four time points post-feeding (day 7, 10, 14 and 18). Background color of table cells indicates the proportion of mosquitoes with detectable infection (0%=dark green to 100%=red).
We used a non-parametric sign test (see Methods) to assess differences in infection rates between wMel and WT mosquitoes (Table 1). Note that for all data subsets examined, the number of paired observations for which infection rates in WT mosquitoes exceeded those in wMel infected mosquitos was always greater or equal to the number of pairs where the converse was true. Overall, the proportion of mosquitoes with DENV-infectious saliva was significantly lower in wMel-A. aegypti than in WT mosquitoes 10 and 14 days post-blood meal (p<0.005, Table 1), these being the two most data-rich time points.. Abdomen infections were significantly lower in wMel compared with WT day 14 post-blood meal (p=0.0044) and close to significant for day 10 (p=0.053). Two versions of saliva results are presented in Table 1. The ‘saliva conditional’ rows show results for the actual saliva samples tested, i.e. conditional upon detected abdominal infection. However, saliva was only tested in mosquitoes with dengue infection detected in abdominal tissue, since abdominal infection is a pre-requisite of more disseminated infection. The ‘saliva unconditional’ rows in Table 1 show results for saliva infection assuming that all mosquitoes with no detectable abdominal infection also had no detectable infection in saliva. This best summarizes all the available data on the impact of wMel infection on the probability of detecting infectious virus in saliva. The ‘saliva unconditional’ results in Table 1 show the most marked difference between DENV infection rates in wMel and WT groups, with significant differences (p<0.02) between the groups for each serotype-specific data subset, even including DENV3 – the least represented serotype in our dataset. This reflects the combined impact of wMel on both establishment of abdominal infection and dissemination of that infection to saliva.
Table 1.
Assessment of the differences in tissue infection rates in wMel vs. wild type mosquitoes using the sign test.
| Tissue | Serotype | Day | n pairs where pwMel<pwt | n pairs where pwMel=pwt | n pairs where pwMel>pwt | p-value (for accepting pwMel=pwt) |
|---|---|---|---|---|---|---|
| Abdomen | All | All | 41 | 55 | 14 | 0.0004 |
| All | 7 | 3 | 9 | 3 | 1 | |
| All | 10 | 16 | 20 | 6 | 0.053 | |
| All | 14 | 16 | 18 | 3 | 0.0044 | |
| All | 18 | 6 | 8 | 2 | 0.29 | |
| DENV1 | All | 17 | 25 | 5 | 0.017 | |
| DENV2 | All | 6 | 10 | 5 | 1 | |
| DENV3 | All | 6 | 4 | 2 | 0.29 | |
| DENV4 | All | 12 | 16 | 2 | 0.013 | |
|
| ||||||
| Saliva (conditional) | All | All | 57 | 14 | 13 | <0.0001 |
| All | 7 | 4 | 5 | 2 | 0.69 | |
| All | 10 | 22 | 7 | 2 | <0.0001 | |
| All | 14 | 22 | 2 | 6 | 0.0037 | |
| All | 18 | 9 | 0 | 3 | 0.15 | |
| DENV1 | All | 30 | 4 | 11 | 0.0043 | |
| DENV2 | All | 11 | 4 | 1 | 0.0063 | |
| DENV3 | All | 6 | 2 | 1 | 0.13 | |
| DENV4 | All | 10 | 4 | 0 | 0.002 | |
|
| ||||||
| Saliva (unconditional) | All | All | 59 | 16 | 10 | <0.0001 |
| All | 7 | 4 | 6 | 2 | 0.69 | |
| All | 10 | 22 | 7 | 2 | <0.0001 | |
| All | 14 | 24 | 3 | 3 | <0.0001 | |
| All | 18 | 9 | 0 | 3 | 0.15 | |
| DENV1 | All | 31 | 5 | 9 | 0.0007 | |
| DENV2 | All | 11 | 4 | 1 | 0.0063 | |
| DENV3 | All | 7 | 3 | 0 | 0.0156 | |
| DENV4 | All | 10 | 4 | 0 | 0.002 | |
The experimental data were treated as pairs of binomial observations corresponding to the proportions infected of the wMel and WT mosquito groups fed on a particular blood sample, which were sampled on a specific day. The table rows present the number of observation pairs for which the proportion of wMel mosquitoes infected was less than, equal to or greater than the proportion of WT mosquitoes infected for different data subsets. Subsets are shown which stratify the observations by the tissue type, DENV serotype and the day post infection that mosquitoes were assayed. The two-sided p-value is given, with p-values < 0.05 shown in italics.
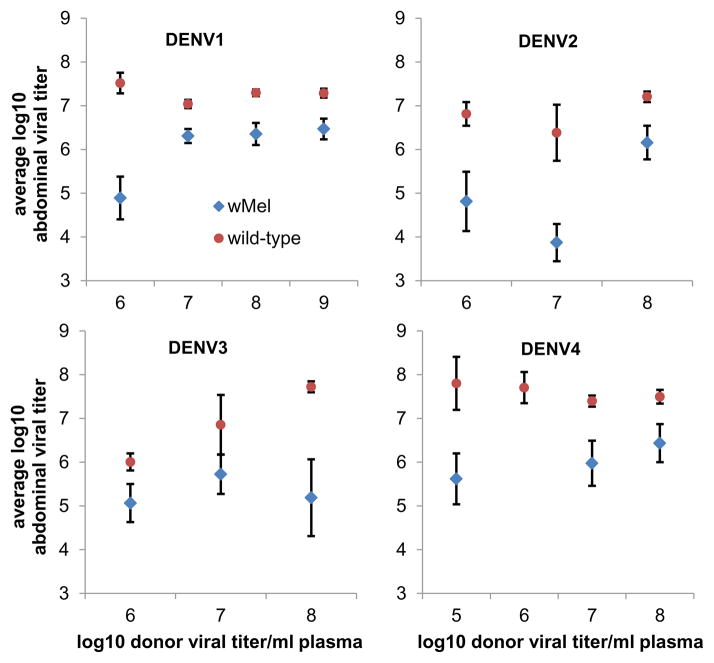
In addition, the concentration of DENV RNA in wMel-A. aegypti abdominal tissues for all serotypes was generally at least 10-fold lower than in WT mosquitoes (Figure 3), indicating wMel conferred partial protection against the fulminant DENV infection that was typical in WT mosquitoes. Collectively, these data, generated using physiologically relevant viremic blood meals, demonstrated significant but imperfect blocking of DENV infection by wMel.
Figure 3.
wMel attenuates DENV infection of abdomen tissues. Shown is the mean log10 titer (RNA copies/abdomen) of virus measured in mosquito abdomens (average over mosquitoes with detectable virus at any time point) of WT (circles) and wMel-infected (triangles) mosquitoes with DENV-infected abdomen tissues, binned by integer interval of log10 viral titer in the donor human blood. A–D show results for DENV1-4, respectively. Error bars show standard error of the mean.
We also tested for an effect of time since blood meal in the data presented in Figure 2. For the abdominal data, sign tests revealed no significant difference in the proportions of mosquitoes testing positive between day 7 and day 10 (p=0.09), day 10 and day 14 (p=0.11), or day 14 and day 18 (p=0.93). For the saliva data, there were significant differences between day 7 and day 10 (p=0.011), day 10 and day 14 (p<0.0001), but not between day 14 and day 18 (p=0.68).
Model fitting to empirical data of DENV infection in WT and wMel-A. aegypti
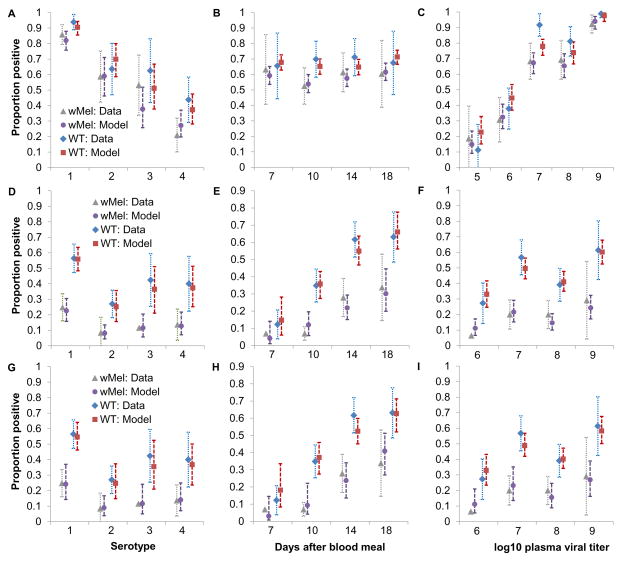
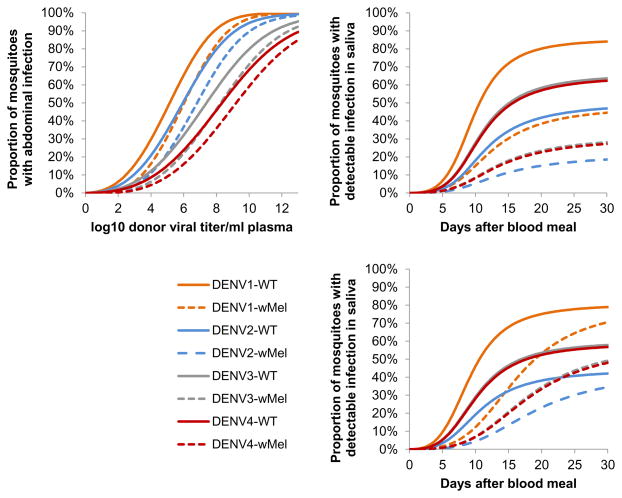
We developed mathematical models to replicate the phenotype of WT and wMel-A. aegypti. Figure 4 summarizes the fit of the abdomen and saliva infection models to the experimental data, illustrating that the models capture trends by serotype (Figure 4A, D, G), end time-point (Figure 4B, E, H) and donor plasma viral titer (Fig 4C, F, I). Both the abdomen and saliva models reproduce phenotypic differences between WT and wMel-A. aegypti. Model parameter estimates are listed in Table 2.
Figure 4.
Mosquito infection model fit to the empirical evidence of wMel-mediated blocking of DENV infection. (A–C) Observed (‘Data’) and median posterior fitted (‘Model’) proportions (with exact binomial confidence intervals) of WT and wMel-infected mosquitoes with detectable virus in abdomen, stratified by (A) serotype; (B) end time-point; (C) log10 donor plasma virus titer band. Panels D–F, as for panels AC, but showing the proportion of dengue-infected mosquitoes (i.e. with detectable virus in abdomen) that also had detectable infectious virus in saliva for the baseline model. Panels G–I, as for panels D–F but for the alternative saliva model.
Table 2.
Mathematical model parameter estimates
| Parameter | Description | Median estimate (95% crI)# | |
|---|---|---|---|
| Abdomen model | |||
| δwMel | Dose-response intercept for wMel-infected mosquitoes | −1.12 (−3.22, 0.33) | |
| θDENV1 | Infectious dose parameter for DENV-1 | 5.90 (4.53, 6.58) | |
| θDENV2 | Infectious dose parameter for DENV-2 | 6.78 (5.88, 7.66) | |
| θDENV3 | Infectious dose parameter for DENV-3 | 8.41 (7.17, 10.29) | |
| θDENV4 | Infectious dose parameter for DENV-4 | 9.50 (8.34, 12.27) | |
| γ | Dose response shape parameter | 2.88 (1.66, 3.97) | |
| ρabdomen | Over-dispersion parameter for abdomen model | 0.46 (0.38, 0.53) | |
| Saliva model§ | Baseline | Alternative | |
| εwMel | Scaling of infectious dose parameters for wMel-infected vs WT mosquitoes | 3.41 (0.66, 11.2) | Fixed at 1 |
| κ | Power on infectivity growth with time | 3.80 (1.99, 6.59) | 3.40 (2.02, 5.04) |
| βWT | Timescale of infectivity saturation in saliva of WT mosquitoes | 12.3 (9.5, 30.8) | 11.6 (8.7, 19.6) |
| βwMel | Timescale of infectivity saturation in saliva of wMel-infected mosquitoes | 12.8 (7.3, 32.5) | 20.7 (15.4, 40.9) |
| ϕDENV1 | Infectious dose parameter for DENV-1 | 0.52 (0.13, 0.81) | 0.60 (0.30, 0.97) |
| ϕDENV2 | Infectious dose parameter for DENV-2 | 1.57 (0.37, 2.99) | 1.79 (0.80, 3.44) |
| ϕDENV3 | Infectious dose parameter for DENV-3 | 0.94 (0.23, 2.15) | 1.11 (0.46, 2.33) |
| ϕDENV4 | Infectious dose parameter for DENV-4 | 0.99 (0.24, 1.95) | 1.13 (0.50, 2.32) |
| ρsaliva | Over-dispersion parameter for abdomen model | 0.19 (0.13, 0.27) | 0.19 (0.13, 0.27) |
Median estimates and 95% credible intervals of parameters of the mathematical models (equations 3 and 4) used to fit the abdomen and saliva infection data on wMel-infected and WT mosquitoes are shown. Time unit is days.
For the saliva model, estimates are shown for the best-fitting baseline model and an alternative model where the phenotypic effect of wMel infection is forced to act on the parameter β, determining EIP.
The mathematical model of abdomen infection adopted (see Materials and Methods) is a relatively simple dose response model depending solely on log10 viremia of the infecting blood meal, Wolbachia infection status and serotype. The impact of wMel infection was found to be best represented by a simple negative offset of the log10 viremia of the infecting blood meal, effectively meaning that the risk of DENV infection in wMel-infected mosquitoes fed on a blood meal with a certain viremia level was the same as in WT mosquitoes fed on blood with a viremia approximately 1 log10 less. Figure 5A illustrates the behavior of the best-fit abdominal infection model, highlighting the major differences in infectious dose seen between serotypes and the effect of wMel in partially blocking infection.
Figure 5.
Performance of the mosquito infection model. (A) Shown is the behavior of the abdominal infection model illustrating dependence of the probability of infection on viral titer in donor blood, serotype and Wolbachia infection status. (B) Shown is the behavior of the saliva infection model showing dependence of the probability of detectable infection in saliva (conditional upon abdominal infection) as a function of the days elapsed since the infecting blood meal, serotype and Wolbachia infection status. (C) Same as (B) but for the alternative saliva infection model where wMel infection affects only the EIP. All graphs show mean posterior predictions.
The model of saliva infection describes the development of detectable virus in saliva conditional upon abdominal infection having been established (see Materials and Methods) and, like the abdominal model, is also relatively simple depending only on time elapsed since the infecting blood meal, wMel infection status and serotype. No statistically significant dependence on viremia in the infecting blood meal could be resolved (assessed by comparison of the deviance information criterion), consistent with the patterns seen in Figure 2. wMel could have two phenotypic effects in the model: an overall reduction in the probability of detecting infectious virus (acting via a scaling of the infectious dose parameters), or a lengthening of the extrinsic incubation period [EIP] (acting via an increase in the time taken for infection to saturate in saliva). The former effect gives a level of inhibition that does not depend strongly on how much time has elapsed since the infecting blood meal, while the latter gives inhibition that decays. We estimated both effect sizes simultaneously in the baseline model, and the best fit estimates predicted that the sole effect of wMel infection was on scaling infectious dose, not on lengthening of the EIP. However, since the mode of effect has a potentially substantial effect on our overall estimates of the impact of wMel on DENV transmission, we also fitted an alternative model in which we forced all wMel infection to affect EIP only. This model fitted statistically significantly worse than the baseline model, but the qualitative quality of fit (Figure 4G–I) was very similar to that seen for the baseline model (Figure 4D–F). Figure 5B and 5C illustrate the differences between these two models in how inhibition acts, together with the marked differences between serotypes in the probability of infectious virus being detected in saliva.
For the abdomen model, the infectious dose parameters differ significantly between most pairs of serotypes; while the credible intervals for these parameters overlap, those for their ratios all have 95% credible intervals that do not include 1 (lower 95% bounds for θDENV2/θDENV1, θDENV3/θDENV1, θDENV4/θDENV1, θDENV3/θDENV2, θDENV4/θDENV2 of 1.004, 1.20, 1.32, 1.04, 1.17 respectively), with the exception of θDENV4/θDENV3 (95% range 0.94–1.48). For the saliva model, DENV-1 has a significantly lower infectious dose parameter than the other serotypes (lower 95% bounds for ϕDENV2/ϕDENV1, ϕDENV3/ϕDENV1, ϕDENV4/ϕDENV1 of 1.84, 1.10, 1.24, respectively), but differences between DENV-2, DENV-3 and DENV-4 are not statistically significant (95% ranges: ϕDENV3/ϕDENV2=0.30–1.28, ϕDENV4/ϕDENV2=0.33–1.19, ϕDENV4/ϕDENV3=0.53–2.19).
Given that the impact of wMel on DENV infection in A. aegypti depends on viral titer in the blood meal, the expected population impact of wMel will depend on the distribution of viral titers across DENV-infected human hosts, denoted ρh(v|τ) (see Materials and Methods). Supplementary Figure 1 shows our estimates of the distribution of human plasma viremia levels, fit using the model of ρh(v|τ) given in equation 2 in Materials and Methods. Substantial variation was seen between different patients infected with the same serotype and between serotypes. Of particular note are the higher peak viremias seen for DENV-1, earlier peaks seen for DENV-2 and the lower peak titers seen for DENV-3 and DENV-4. It should be noted that few data are available to characterize viremia around the time of peak titer, since few measurements were available before day 2 of illness. This leads to considerable uncertainty in early viral kinetics. We discuss the sensitivity of our results to this uncertainty below.
Predictions of wMel impact on DENV transmission
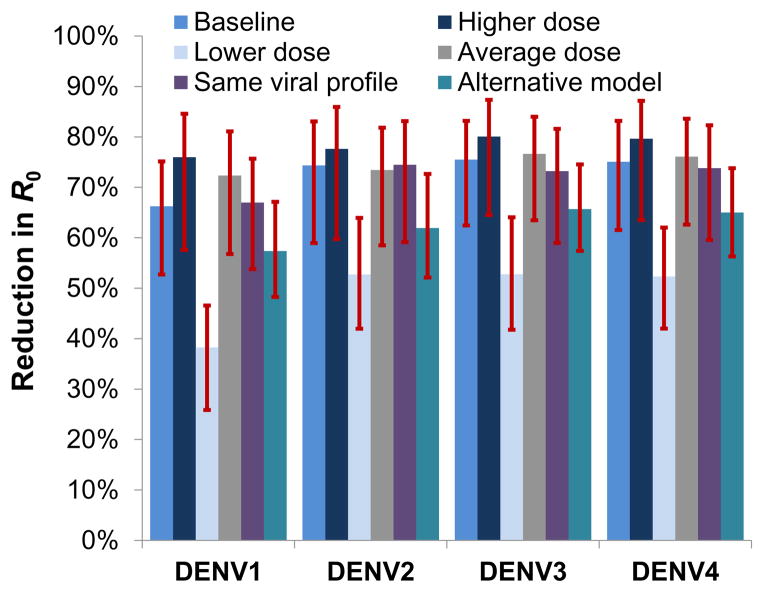
We use equation 1 (see Materials and Methods) to assess the overall impact of wMel infection on DENV transmission by combining the estimated posterior distributions for the dynamics of viral titer over time in infected humans, the probability that a mosquito will become infected on consuming a blood meal with a certain titer of virus and the development of infectivity in the mosquito. We represent impact on dengue transmission by the fractional reduction of the reproduction number, R0, of each serotype that would be caused by wMel infection of the entire mosquito population. Figure 6 shows the resulting posterior estimates of the reduction in R0 for each serotype. For the baseline scenario (which assumes mosquito infectivity to humans is directly proportional to the probability of detecting infectious virus in saliva), a 66–75% reduction is predicted, varying by serotype. While the credible intervals on the absolute estimates of transmission reduction overlapped across the serotypes, posterior estimates of the differences in reduction between DENV-1 and DENV-2/3/4 indicated that DENV-1 exhibited a significantly lower level of reduction than other serotypes (p<0.01)..
Figure 6.
Estimated reduction in transmissibility of DENV (quantified by serotype specific R0) caused by wMel infection. Median posterior estimates and 95% credible intervals are shown. ‘Baseline’ scenario: assumes data on infectious saliva translates directly to human infectiousness. ‘Higher/Lower dose’ scenarios: assume 10-fold higher/lower infectious dose for mosquito-to-human transmission than estimated using saliva infection model. ‘Average dose’: assumes same infectious dose for all serotypes (average across serotypes) for mosquito-to-human transmission. ‘Same viral profile’: uses a model of human viral kinetics that is the same for all serotypes. ‘Alternative model’: uses the alternative saliva infection model where wMel infection affects only the EIP.
Three other scenarios shown in Figure 6 illustrate the sensitivity of the predictions to assumptions about how the model of saliva infectivity is translated to estimates of mosquito to human infectivity. The ‘higher dose’ scenario assumed the infectious dose parameters in the saliva infectivity model (the parameters ϕS in equation 4 in Materials and Methods) need to be 10-fold larger than the estimated values to describe mosquito-human transmission probabilities. This scenario gave the greatest predicted reduction in transmission due to wMel infection due to the predicted slower growth of viral titers in saliva of wMel-infected mosquitoes. Conversely, assuming those infectious dose parameters (ϕS) are 10-fold lower than for mosquito-mosquito transmission (as quantified by our assay of saliva infectivity) resulted in substantially lower estimates of the impact of wMel infection on dengue transmission compared with the baseline scenario. However, it should be noted that this scenario gives unrealistically high per-bite probabilities of mosquito-human transmission, and thus very high (>10 for DENV-1) estimates of R0 for reasonable assumptions about mosquito numbers per person and the biting rate.
The ‘average dose’ scenario assumed there are no serotype differences in the dose parameter for mosquito-human transmission, implemented by specifying that the saliva model dose parameter for each serotype (ϕS) takes the mean of the serotype-specific estimates for each posterior distribution sample. The ‘same viral profile’ scenario ignored the differences in human viral kinetics between serotypes shown in Supplementary Figure 1 and uses a single model (see equation 2 in Materials and Methods) of ρh(v|τ) for all serotypes fitted to all the patient data shown in that figure. The estimated reductions in R0 due to wMel were very similar in both these scenarios to those obtained for the baseline scenario, highlighting that serotype differences in viremia kinetics do not explain the overall differences by serotype seen in Figure 6. Rather, the lower impact of wMel in DENV-1 is largely caused by the differences in infectious dose parameters for saliva and abdominal infection between serotypes (Figure 1).
The last ‘Alternative model’ scenario of Figure 6 shows results when the alternative saliva infection model is used, solely representing the impact of wMel as a lengthening of the EIP (Table 1 and Fig 5C). Under this model, the predicted impact of wMel on transmission was approximately 10% lower; i.e. a 57–66% reduction depending on serotype.
Discussion
We have experimentally characterized the phenotype of Wolbachia-infected A. aegypti mosquitoes challenged with viremic blood from symptomatic dengue patients. wMelPop conferred very strong resistance to DENV infection of the mosquito body and most importantly the salivary glands. wMel conferred an intermediate phenotype in which abdomen tissues were susceptible to DENV infection but dissemination was diminished as evidenced by a lower prevalence of mosquitoes with infectious saliva.
The profound level of virus blocking conferred by wMelPop infection is predicted to cause dramatic reductions in DENV transmission in settings where wMelPop is successfully and stably introduced. The impact of wMel on DENV transmission is more nuanced and serotype dependent; DENV-1 transmission is the least affected, with a predicted 66% reduction in R0 for the baseline scenario. For the other serotypes, higher estimated infectious dose parameters (compared with DENV-1) for both the abdominal and saliva infection models lead to larger predicted reductions in transmission of approximately 75%. To put these reductions in context, estimates of the basic reproduction number (R0) for dengue lie in the range 1.3–6.3 (12), with 2 to 5 being typical of endemic settings. A reduction of 66% is sufficient to eliminate dengue in a setting where R0=3, while a 75% reduction will achieve elimination for R0=4.
Our study highlights three effects of wMel infection on DENV infection in A. aegypti mosquitoes: an increase (compared with wild-type) in blood meal viremia required to achieve a certain probability of abdominal infection, a substantial reduction in the probability of detecting infectious virus in saliva, and a lengthening of the EIP. In our best-fit models, only the first two of these effects were found to be significant. However, an alternative saliva model which solely represented the impact of wMel in terms of an increased EIP gave an adequate (though statistically poorer) fit to the data, and predicted lower reductions in R0 than the baseline model. Additional data, particularly if it included time-points beyond 18 days, might more conclusively resolve the extent to which the impact of wMel is to reduce or just delay the onset of infectiousness in saliva. This issue is important for understanding the extent to which the estimated impact of wMel can be generalized to different settings: if wMel reduces the probability of mosquitoes being infectious independent of the time since infection, the reduction in R0 achieved is independent of adult mosquito survival. Conversely, if the main impact of wMel is to increase the EIP, this will have a larger effect on dengue transmission than that estimated here in situations where daily mosquito survival is lower than the relatively high 90% value we assumed.
Previous vector competence studies of Wolbachia-infected A. aegypti mosquitoes have employed in vitro passaged DENV strains that were spiked into animal or human blood before this mixture was presented to colony mosquitoes through membrane feeders (4, 5). The current study is distinguished from previous work in utilising fresh viremic blood samples from hospitalized dengue cases to mimic the virological challenge that A. aegypti mosquitoes experience when they feed on an infectious human case. In using viremic blood from hospitalized dengue cases, in whom peak viremia levels are significantly higher than in acute ambulatory (never hospitalized) cases in the same setting (13), we are likely being conservative in our experimental evaluation of wMel infected A. aegypti. Future experimental studies could examine susceptibility to DENV infection after blood-feeding on ambulatory dengue cases.
Our finding that wMelPop-A. aegypti do not develop disseminated infections with DENV is entirely consistent with the initial description of the vector competence phenotype of this strain (5). However, we found wMel-A. aegypti can develop infectious saliva after viremic blood feeding and this contrasts with the initial description by Walker et al who detected no infectious DENV-2 in the saliva of any of the 336 wMel-A. aegypti females used in artificial feeding experiments (4). There are methodological reasons why our results might differ: Walker et al used one lab strain of DENV-2 at a single concentration, employed cell culture to detect infectious virus in pooled saliva and used colony-sourced mosquitoes. Of these, we speculate that the virological differences are most important and that viremic blood from a human dengue case provides the most stringent and relevant challenge of the vector competence of Wolbachia-infected mosquitoes. This would underscore the importance of using clinical material for robust assessments of arboviral vector competence in general. Our data also highlights the importance of assessing vector competence at multiple time-points in order to characterize the impact on the dynamics of dengue infection in the mosquito. Whereas the wMelPop data presented here was all collected at a single time-point (12 days) post-blood meal, preliminary results from on-going work indicate comparable levels of inhibition of DENV infection at 14 and 18 days post-infection.
Our analysis suggested wMel could reduce the DENV force of infection by a degree which would have a highly significant public health impact – potentially achieving elimination in low to moderate transmission settings, albeit perhaps insufficient for complete control in high-transmission settings (especially for DENV-1). Yet a number of factors might lead to the field efficacy of wMel on DENV transmission differing from estimates presented here. First, while we did not collect data on the concentration of infectious DENV particles in mosquito saliva, it is a reasonable hypothesis that wMel reduces viral concentrations, which would lead to a larger reduction in transmission than that estimated here. Second, the effect of wMel on other aspects of mosquito behavior that impact on transmission, such as host-seeking, probing and blood feeding success rates, have yet to be investigated in a field setting and it is plausible that these could counteract the effect of wMel-mediated interference of virus transmission to mosquitoes. Finally, here we solely examined the impact of Wolbachia on the susceptibility of A. aegypti to DENV infection. In reality, wMel may modify A. aegypti fitness via decreased (or, less likely, increased) fecundity or longevity. Even small reductions in the lifespan of wMel-A. aegypti, as described previously (4), might cause reductions in dengue transmission.
A priori, that we found no statistically significant dependence on the level of viremia in the infecting blood meal in the mathematical model describing saliva infection might be viewed as surprising. However, the saliva model represents the probability of detecting infectious virus in saliva conditional upon abdominal infection being detectable. The limited association between mosquito abdomen viral titers and the blood meal viremia (Figure 3) suggests the primary influence of the level of viremia in blood is on the probability of establishing abdominal infection, but not on later dissemination once abdominal infection has been established.
Our study has several limitations. Quantification of the level of infectiousness of mosquito saliva along a continuous gradient, rather than just a binary measure of infectious status as described here, would allow impacts of reduction in DENV saliva titer due to wMel to be explored. However, we note that in vitro titration methods that work well for highly passaged reference DENV strains do not work well with clinical isolates. Further studies are also needed to understand the vector competence phenotype of Wolbachia-infected A. aegypti after challenge with DENV genotypes different from those currently circulating in Vietnam. We note that each serotype of DENV in circulation in southern Vietnam during the study period was comprised essentially of a single virus genotype (13) and thus our results are unlikely to be confounded by large fitness differences between viruses of the same serotype. Our mosquito studies were conducted with a single consistent set of environmental conditions: 27°C and 70% relative humidity. Previous experimental studies have noted shortening of the extrinsic incubation period (suggesting more rapid viral replication) as temperature is increased in the range 26–30°C. Thus, the impact of wMel on DENV transmission efficiency might also show some temperature dependence, although the direction and magnitude of such effects are not possible to predict a priori. Although it would be challenging (in cost and time) to repeat the clinical studies presented here for a wide range of environmental conditions, some exploration of the effect of temperature on wMel phenotype would be a worthwhile topic for future work.
Finally, there is an element of arbitrariness in the model structure. The relatively parsimonious biologically motivated model structures adopted allowed biologically reasonable extrapolation to low and high viremia and gave quality of fits to the data comparable with logistic regression with the same degrees of freedom. Future modeling efforts could move towards using a truly dynamic model of DENV infection within the mosquito.
We have determined that wMelPop confers on A. aegypti profound resistance to DENV infection. Establishment of wMelPop-infected A. aegypti at high frequency in a dengue endemic setting would result in complete abatement of DENV transmission, however, this might prove challenging given the fitness costs conferred by wMelPop infection. Establishment of wMel-infected A. aegypti, as has occurred in some communities in northern Australia (11), is also predicted to have a substantial effect on transmission, but may be insufficient to entirely control dengue in settings where the basic reproduction number is high. Other complementary interventions may therefore be needed to offset the lower efficacy of wMel in high transmission intensity settings, e.g. traditional vector control methods and new approaches such as using adult male Wolbachia-A. aegypti releases for population suppression. Additionally, dengue vaccines (14) might work in concert with a Wolbachia intervention to achieve long-term disease control. Finally, it will be desirable to evaluate other Wolbachia-A. aegypti strains, e.g. the well-established wAlbB- A. aegypti strain deserves evaluation in this viremic blood challenge system and in the field (15). The prospect of a “menu” of Wolbachia options, alongside other dengue interventions, could enable a bespoke approach to dengue control in a range of epidemiological and socioeconomic contexts.
Materials and Methods
Study design
This was a prospective observational study that used viremic blood from acute dengue cases to blood feed wild-type or Wolbachia-infected A. aegypti mosquitoes in the laboratory. The sample size was not pre-specified and instead was based on pragmatic considerations around the duration of the study, which spanned two dengue “seasons” (from June 2012 to December 2013). We pre-specified that data collection would stop in December 2013. We used biological replicates throughout the study; i.e. multiple blood samples from independent patients but infected with the same DENV serotype. We also used biological replicates of the mosquitoes with a. minimum 5 blood-fed mosquitoes per cohort.
Dengue patients were enrolled at the Hospital for Tropical Diseases (HTD), in Ho Chi Minh City, Vietnam. Patients were eligible for enrolment if (a) they were ≥1 year of age; (b) with less than 72hrs of fever; (c) they were clinically suspected of having dengue and had a positive NS1 rapid test. Exclusion criteria were (a) patients in intensive care unit; (b) patients with intellectual disabilities. The baseline features of the dengue cases from whom venous blood was used for vector competence studies are shown in Table S1. On the day of enrolment, venous blood (EDTA anticoagulant) was collected and split for mosquito feeding and for qRT-PCR measurement of DENV RNA concentrations in plasma using a validated, quantitative RT-PCR assay that has been described previously (16). All patients provided written informed consent to provide blood samples. The study protocol was reviewed and approved by the Scientific and Ethical committee of the HTD (reference number: CS/ND/09/24) and the Oxford Tropical Research Ethical Committee (reference number: OxTREC 20-09).
The pre-specified hypothesis was that Wolbachia-infected A. aegypti mosquitoes were more resistant to DENV infection. Hence the primary entomological endpoints of interest were the proportion of mosquitoes with infected abdomens or saliva. This was addressed by scoring mosquito tissues for the presence or absence of DENV infection using a molecular test and thence modeling the results as a basis to predict the wider epidemiological impact on DENV transmission. All laboratory assays to test for DENV infection were performed by technicians blinded to the clinical and virological details of the patient blood sample and the Wolbachia status of the mosquitoes. All data were submitted to a Good Clinical Laboratory Practice database and cleaned prior to data lock.
Viremic blood challenges of wild-type and Wolbachia-infected A. aegypti
Vector competence studies were performed with WT A. aegypti from Cairns, Australia and A. aegypti of the same origin but stably infected with wMel or wMelPop. The WT versus wMelPop A. aegypti studies were performed using eggs from outcrossed colonies maintained at Monash University, Australia. Colonies were maintained at population sizes of 400 with a 50:50 sex ratio. The WT versus wMel A. aegypti studies were performed with F2 generation adults and obtained by hatching eggs collected from field sites in Cairns, Australia (11). For all studies, up to 100 three to seven day-old female A. aegypti mosquitoes were starved for 24 hours before being membrane fed on fresh acute blood from laboratory-confirmed dengue patients. All blood samples were placed into glass membrane feeders within 1hr of the blood being collected and mosquitoes were allowed access to the blood for 1hr. Membrane feeders were water-jacketed and maintained at constant temperature during mosquito feeding (37°C). After cold-knockdown, fully engorged mosquitoes were selected and then maintained in an environmental chamber with 12:12 light:dark hours, 27°C and 70% relative humidity and access to 10% sucrose solution.
Detection of DENV in saliva and abdomen tissues
Infectious virus in mosquito saliva was detected by placing the proboscis of the de-winged and de-legged mosquito into the end of a filtered micropipette tip containing 6μl of sterile saliva medium (a 1:1 solution of 15% sucrose and inactivated fetal calf serum) for 30mins at room temperature. After 30mins, the 6μl of saliva medium was ejected and then drawn into a pointed glass capillary tube (tip diameter: < 0.3μm). The volume of saliva medium derived from one mosquito was then injected into the thorax of between 4–6 A. aegypti mosquitoes (4–7 days old, ~1μl injected per mosquito) and the injected mosquitoes maintained for 7 days in an environmental chamber with 12:12 light:dark hours, 28°C and 80% relative humidity. After 7 days, the injected mosquitoes for each saliva sample were killed, the bodies pooled, homogenized and tested by quantitative RT-PCR for DENV infection, with saliva samples scored as positive or negative depending on this result. Saliva samples were collected from all mosquitoes, but only saliva samples from mosquitoes with infected abdomens were evaluated for their infection status because pilot studies confirmed that abdomen infection was a pre-requisite for the saliva to contain infectious virus. After collection of saliva samples, the abdomen was dissected from the mosquito body. Dissected abdomens were suspended in 0.5ml of mosquito diluent (RPMI 1640 supplemented with 2% heat inactivated fetal calf serum, antibiotics and antimycotics). Individual mosquito abdomens were homogenized with 1mm Zirconia/Silica beads for 15 minutes at 30 Hz using a TissueLyser II (Qiagen). Mosquito tissues were scored as being DENV- infected using a quantitative, internally-controlled RT-PCR assay (16) on homogenized tissue and the results expressed as copies per tissue.
Detection of Wolbachia status in mosquito tissues by real time PCR
For quality control purposes, Wolbachia infection status was scored using a multiplex PCR assay on nucleic acid extracts from mosquito abdomens. A. aegypti ribosomal protein S17 (Ae-RpS17) was used as an internal control. Wolbachia strain wMel was detected with primers/probes specific to the WD0513 gene and wMelPop was detected with primers/probes specific to the polymorphic insertion sites of the IS5 at loci IS5-WD1310. Sequences of primers/probes for Wolbachia and DENV detection are shown in Table S2. The PCR was performed on a LightCycler480II machine using LightCycler480 Probes Master according to the manufacturer’s instructions.
Data release
See Supplementary Materials for the wMel and wMelPop data analyzed in this paper.
Statistical Analysis
Non-parametric assessment of the differences in tissue infection rates in wMel vs. wild type mosquitoes
We applied a standard sign test for paired data, treating the experimental data as pairs of binomial observations corresponding to the proportions infected of the wMel and WT mosquito groups fed on a particular blood sample which were sampled on a particular day. Rows in Table 1 present the number of observation pairs for which the proportion of wMel mosquitoes infected was less than, equal to or greater than the proportion of WT mosquitoes infected for different data subsets, designated npairs(pwMel<pwt), npairs(pwMel=pwt) and npairs(pwMel>pwt) respectively. If there was no difference between infection rates of wMel and WT mosquitoes, npairs(pwMel<pwt) would be expected to be drawn from a binomial distribution with p=0.5 and N=npairs(pwMel<pwt)+npairs(pwMel>pwt). The two-sided p-value in the final column of Table 1 is the probability of a sample from that distribution being equal to or more extreme than the observed value of npairs(pwMel<pwt).
Transmission model
Since the probability of a mosquito becoming infected with DENV from a blood meal depends strongly on the viral titer in that blood meal, quantitative assessment of the impact of Wolbachia on transmission requires a mathematical model that couples the dynamics of infection within human host with those in the vector. We found no previously published mathematical models of DENV transmission which included such coupling, so the framework presented below needed to be developed specifically for this study.
We define ρh(v|τ) to be the probability density that the plasma viral titre of a human host is v at time τ after infection; we model viral dynamics in humans probabilistically to represent the variation seen between individuals. We assume the probability that a mosquito taking a blood meal on that individual becomes infected depends on the viral titer in the blood at the time of feeding: let pi(v) be the probability that a vector becomes infected when feeding on a human with a plasma viral titer of v. If a mosquito becomes infected, then we assume its infectiousness to humans depends on the time elapsed from the infecting blood meal and the plasma viral titer of the blood meal. We define pm(v|t) to be the probability that a mosquito infected by taking a blood meal with viral titer v will infect another human host it bites time t later; this distribution captures the extrinsic incubation period (EIP).
Together these three distributions represent the complete transmission cycle; all that is additionally required to calculate the basic reproduction number (the average number of humans infections generated by a typical infected human in the absence of immunity), R0, for a serotype is the average number of female mosquitoes per human host, m, the mortality hazard for adult female mosquitoes, μ, and the biting rate of female mosquitoes, κ. Then
| (1) |
Here D is the maximum time to clearance of virus in humans. This equation is the standard definition of the reproduction number for a vector-borne disease, generalized to account for viral dose dependence in the mosquito.
We wish to estimate the distributions ρh(v|τ), pi(v), and pm(v|t) for each of the 4 DENV serotypes and for Wolbachia-infected and WT mosquitoes. However, the available data did not allow every parameter to be estimated independently for each combination of serotype and Wolbachia infection status, so it was necessary to assume that only a subset of parameters varied between serotypes or were affected by Wolbachia.
Our primary interest is the extent to which Wolbachia reduces transmission, as characterized by the ratio of R0 of a DENV serotype in a WT A. aegypti population to that in a Wolbachia infected A. aegypti population; values of m and κ in equation (1) are not needed for calculating this ratio. However, the assumed value of μ, the mortality hazard of adult mosquitoes, can affect estimates. A. aeqpti mortality is known to vary seasonally and by setting, with release-recapture studies typically giving daily survival probabilities below 85% (17–19). Since one possible phenotype of wMel on dengue replication in mosquitoes we explore below is a lengthening of the EIP, we conservatively assume daily survival is constant at its seasonal maximum of 90% (μ=of 0.1/day) (19). This results in a larger proportion of transmission being from older mosquitoes than assuming a lower value for daily survival, and hence reduces the potential impact of EIP lengthening on dengue transmission.
We estimate ρh(v|τ) from serial plasma viremia levels measured in 262 consecutively enrolled dengue cases in the IDAMS study (clinicaltrials.gov identifier NCT01550016) in Ho Chi Minh City, Vietnam. Of these 262 cases, 73 cases were hospitalized and 189 were managed entirely as ambulatory patients for the duration of their illness. The serial viremia measurements in these 189 ambulatory cases has been described previously (13), and the data are shown in Supplementary Figure 1. Here we use the following data fields for each measurement: study participant identifier, DENV serotype, the day of illness when the sample was collected, log base-10 viral titer/ml of plasma (measured with quantitative RT-PCR) in sample. We model viral kinetics in a human host with a simple biphasic exponential growth/decay function, where average (across all patients) viral titer at time t after infection is given by:
| (2) |
We assume individual patient log base-10 viral titers are drawn from a normal distribution with mean log[v(t)] and standard deviation σ; thus defining the distribution ρh(v|τ). Since the dates of infection are unknown, we estimate time of infection from the day of illness onset by assuming a fixed 7 day incubation period for dengue. Parameters a, b and c were fitted independently for each serotype, while σ was fitted assuming it to be the same for all serotypes.
Mosquito infection model
The probability that a mosquito feeding on blood with viral titer v will become infected, pi(v), is estimated from data on abdominal infection status in mosquitoes infected as part of this study. We used a simple dose-response model:
| (3) |
The single parameter δW was found to be sufficient to capture the phenotypic impact of Wolbachia. This parameter was assumed to be 0 for WT and was estimated for wMel infected mosquitoes. Its effect is to modify the infecting dose of virus by a fixed factor. The parameter θS determines the infectious dose and is estimated independently for each serotype S, while γ determines the slope of the dose response curve and was assumed not to vary by serotype. We do not model an effect of day of measurement (post mosquito feeding) for abdominal infection data as no significant differences were seen between the 7, 10, 14 and 18 day time points examined here.
In the absence of human challenge studies we lack direct measurements of mosquito infectiousness, pm(v|t); here we examine the closest proxy available, namely detection of infectious DENV in mosquito saliva. We define qm(v|t) to be the proportion of mosquitoes infected by taking a blood meal with viral titer v which will have detectable infection in saliva time t later. We assume the following functional form for qm(v|t):
| (4) |
This semi-mechanistic form gives power-law (~(t/βW)κ) temporal growth of saliva infection for small t. This growth saturates at a time governed by parameter βW; thus this parameter governs the extrinsic incubation period. Since we needed to use this model outside the observed range of 7 ≤ t ≤ 18 days, it was important to choose a functional form for the time dependence of saliva infection status that was well-behaved and biologically plausible for both small and large t. The model above gives close to zero probability of detectable infection for small t (< 7 days), and a probability that plateaus at large t (> 18 days). Similar to the abdominal infection model, the serotype-specific parameters ϕS govern the infectious dose. A dose-response shape parameter (akin to γ in equation 3) was also examined but found to result in over-fitting, with estimates having 95% credible intervals overlapping 1.
Two parameters, βW and εW, specify the phenotypic impact of Wolbachia for the saliva infection model. Hence Wolbachia can affect either or both the proportion of mosquitoes ever developing infectiousness in saliva, or the rate at which saliva infectiousness increases (and thus the extrinsic incubation period). The former is estimated separately for WT and wMel infected mosquitoes, while the latter scales the infectious dose parameters for wMel versus WT, and hence has value 1 for WT and is estimate for wMel infected mosquitoes.
When both βW and εW, were fitted (our baseline model), estimates for βWT and βwMel were nearly identical, with substantial overlap of the 95% credible intervals. Thus nearly the entire phenotypic effect was attributed to εW – representing a net reduction in the probability of infection in saliva in wMel versus WT mosquitoes, irrespective of the time elapsed since the infecting blood-meal (Figure 5B). However, since the lower credible of εW was just below 1 for the baseline model, we fitted a three simpler models, assuming: A. βWT =βwMel and εW =1 (i.e. no phenotypic effect of wMel); B. βWT =βwMel (i.e. phenotypic effect of wMel acting solely via εW); C. εW =1 (i.e. phenotypic effect of wMel acting solely via a difference between βWT and βwMel – effectively a lengthening of the EIP due to wMel infection). Model B had the highest DIC, with the baseline model (with both βW and εW fitted) next (DIC difference from B of 1.1), followed by model C (DIC difference from B of 2.4) and model A much worse (DIC difference from B of 47). Since the phenotypic effect of wMel infection substantively affects overall estimates of the impact of Wolbachia on transmission, we choose to present the estimates for model C (where εwMel=1) as an alternative to the baseline model. This alternative model (Figure 5C) fitted the data qualitatively well (Figure 4), albeit worse than the baseline model (difference in DIC=1.4). While model B had the highest DIC, the small numerical difference compared with the model fitting both βW and εW meant we opt to retain the latter as our baseline, as it best represents the uncertainty in the phenotypic effect of wMel infection and is slightly more pessimistic than model B in the estimates of the impact of wMel on the R0 of dengue.
The saliva infection model shows no dependence on the plasma viral titer of the infecting blood meal; including such dependence did not significantly improve model fit, reflecting the lack of obvious viral titer dependence seen in the raw saliva infection data shown in Figure 1. For example, substituting a term (ωW + log v)/ϕS for 1/εWϕS in equation 4, fitting ωwMel and assuming ωwMel=0 (akin to the abdominal model) increased the DIC by 2.3 relative to the baseline model. Furthermore, the central estimate for ωwMel was unreasonably large in magnitude (−6.1) given log10 donor viral titers only varied in the range 5.3–9.9, meaning this model variant was approximating the behavior of the functionally simpler baseline model with no improvement in fit.
Our default (and the simplest) approach to relating pm(v|t) to qm(v|t) is to assume proportionality, namely pm(v|t) ∝ qm(v|t). However, other assumptions are plausible and can substantially affect the resulting estimates of the overall impact of wMel on dengue transmission. We undertake some sensitivity analysis therefore, by assuming pm(v|t) is determined by a similar functional form to equation 4, but with modified parameters. We examine the impact of varying the infectious dose parameters by a fixed multiplier to mimic the effect of the infectious dose from mosquitoes to humans being either larger or smaller than that seen with the assay we used to assess infectious virus in saliva.
Inferential framework
Model fitting was undertaken in a Bayesian framework using Markov Chain Monte Carlo (MCMC) methods (20). To account for the over-dispersion of the data (Figures 1 and 2), a Beta-binomial likelihood function was used rather than a simple binomial likelihood. The Beta-binomial was parameterized in terms of the mean binomial proportion, Θ, and its over-dispersion ρ, defined such that the mean and variance of a sample of n draws is given by nΘ and nΘ(1−Θ)[1+(n−1)ρ], respectively. The over-dispersion parameter ρ was fitted separately for the abdominal and saliva data. Uninformative uniform priors were assumed for all parameters, with an upper bound of 200 for all parameters, and a lower bound of 0 for all parameters other than δ, for which a lower bound of −200 was used. Sensitivity to changing the upper and (where appropriate) lower bounds on priors was tested and none found so long as upper and lower bounds lay outside the 99.9th percentile of the posterior distribution. Parameters were updated individually, with a single update sweep defined as a sequence of proposed updates to each parameter in turn. For computational efficiency, a uniform proposal distribution was used for each parameter, centered around the current parameter value and with width manually tuned to give 20–40% acceptance rates (proposal acceptance rates were monitored separately for each parameter). MCMC chains were equilibriated with 100,000 update sweeps and posterior distributions estimated from the following 500,000 update sweeps, sampling once every 500 sweeps. Convergence was checked visually and by running multiple chains from different starting points. Analyses were undertaken in Microsoft Excel and the statistical language R.
In exploratory but non-exhaustive analyses, a variety of functional forms were explored for both pi(v) and qm(v|t): in particular, we examined how model fit could be significantly improved by making a parameter vary by serotype or by Wolbachia infection status while retaining parameter identifiability. We found little evidence for any serotype-dependence beyond the overall scaling of the dose-response relationships expressed in the functional forms used above. Similarly, significant differences (assessed by non-overlapping 95% credible intervals and the DIC) between estimates for WT and wMel-infected mosquitoes were only seen for the parameters δ, ε and, to a lesser extent (see discussion above), β.
Supplementary Material
Table S1: Study population characteristics
Table S2: List of primers and probes used
Figure S1: Human DENV viremia kinetics
Supplementary data: Mosquito biting study data
Accessible Summary.
How infecting mosquitoes with the bacteria Wolbachia can help control dengue
Dengue is the most common mosquito borne viral infection in humans. Here we report experiments which assessed the extent to which infecting mosquitoes with a bacterium called Wolbachia was able to prevent those mosquitoes from being infected with dengue after they were fed on blood collected from dengue patients. One Wolbachia strain (wMelPop) almost completely prevented dengue infection. A second strain (wMel) partially blocked dengue infection. A mathematical model fitted to the data collected on the wMel strain suggested that wMel could reduce the transmissibility of dengue by 66–75%, sufficient to eliminate dengue in low or moderate transmission settings.
Acknowledgments
Funding: Supported by the Wellcome Trust; the Bill and Melinda Gates Foundation (BMGF); the Foundation for the National Institutes of Health as part of the Grand Challenges in Global Health Initiative of BMGF; the National Health and Medical Research Council, Australia; the UK Medical Research Council; the NIGMS MIDAS initiative; the European Union FP7 EMPERIE consortium.
Footnotes
Author contributions: CPS, NMF, SLO designed the study; DTHK, VTT, TNBC, VTL, LTD, HLN, NVVC performed mosquito biting experiments; JP, PAR, SLO, EAM developed Wolbachia infected A. aegypti; NMF, HC, RA performed the analysis; NMF, CPS drafted the manuscript.
Competing interests: NF is an informal and unpaid advisor on dengue control measures (including Wolbachia and vaccines) and dengue modeling for the Bill and Melinda Gates Foundation and Sanofi Pasteur Inc. CS has a paid consulting position with Sanofi Pasteur who have a business interest in developing dengue vaccines. PR is named as a coinventor on a patent for Wolbachia mosquito strains. The other authors declare no competing interests.
Data and materials availability: The data collected in this study are provided in the Supplementary information.
References and Notes
- 1.Simmons CP, Farrar JJ, Nguyen V, Wills vB. Dengue. The New England journal of medicine. 2012;366:1423–1432. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- 2.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McMeniman CJ, Lane RV, Cass BN, Fong AW, Sidhu M, Wang YF, O’Neill SL. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323:141–144. doi: 10.1126/science.1165326. [DOI] [PubMed] [Google Scholar]
- 4.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, Leong YS, Dong Y, Axford J, Kriesner P, Lloyd AL, Ritchie SA, O’Neill SL, Hoffmann AA. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–453. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- 5.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O’Neill SL. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell. 2009;139:1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 6.McMeniman CJ, O’Neill SL. A virulent Wolbachia infection decreases the viability of the dengue vector Aedes aegypti during periods of embryonic quiescence. PLoS Neglected Tropical Diseases. 2010;4:e748. doi: 10.1371/journal.pntd.0000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turley AP, Moreira LA, O’Neill SL, McGraw EA. Wolbachia infection reduces blood-feeding success in the dengue fever mosquito, Aedes aegypti. PLoS Neglected Tropical Diseases. 2009;3:e516. doi: 10.1371/journal.pntd.0000516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang G, Hussain M, O’Neill SL, Asgari S. Wolbachia uses a host microRNA to regulate transcripts of a methyltransferase, contributing to dengue virus inhibition in Aedes aegypti. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10276–10281. doi: 10.1073/pnas.1303603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caragata EP, Rances E, Hedges LM, Gofton AW, Johnson KN, O’Neill SL, McGraw EA. Dietary cholesterol modulates pathogen blocking by Wolbachia. PLoS Pathogens. 2013;9:e1003459. doi: 10.1371/journal.ppat.1003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan X, Zhou G, Wu J, Bian G, Lu P, Raikhel AS, Xi Z. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E23–31. doi: 10.1073/pnas.1116932108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, Greenfield M, Durkan M, Leong YS, Dong Y, Cook H, Axford J, Callahan AG, Kenny N, Omodei C, McGraw EA, Ryan PA, Ritchie SA, Turelli M, O’Neill SL. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476:454–457. doi: 10.1038/nature10356. [DOI] [PubMed] [Google Scholar]
- 12.Johansson MA, Hombach J, Cummings DA. Models of the impact of dengue vaccines: a review of current research and potential approaches. Vaccine. 2011;29:5860–5868. doi: 10.1016/j.vaccine.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen MN, Thi Hue Kien D, Tuan TV, Quyen NT, Tran CN, Vo Thi L, Thi DL, Nguyen HL, Farrar JJ, Holmes EC, Rabaa MA, Bryant JE, Nguyen TT, Nguyen HT, Nguyen LT, Pham MP, Luong TT, Wills B, Nguyen CV, Wolbers M, Simmons CP. Host and viral features of human dengue cases shape the population of infected and infectious Aedes aegypti mosquitoes. Proceedings of the National Academy of Sciences of the United States of America. 2013;11:9072–9077. doi: 10.1073/pnas.1303395110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet. 2012;380:1559–1567. doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- 15.Xi Z, Khoo CC, Dobson SL. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science. 2005;310:326–328. doi: 10.1126/science.1117607. [DOI] [PubMed] [Google Scholar]
- 16.Hue KD, Tuan TV, Thi HT, Bich CT, Anh HH, Wills BA, Simmons CP. Validation of an internally controlled one-step real-time multiplex RT-PCR assay for the detection and quantitation of dengue virus RNA in plasma. Journal of Virological Methods. 2011;177:168–173. doi: 10.1016/j.jviromet.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrington LC, Buonaccorsi JP, Edman JD, Costero A, Kittayapong P, Clark GG, Scott TW. Analysis of survival of young and old Aedes aegypti (Diptera : Culicidae) from Puerto Rico and Thailand. J Med Entomol. 2001;38:537–547. doi: 10.1603/0022-2585-38.4.537. [DOI] [PubMed] [Google Scholar]
- 18.Maciel-De-Freitas R, Codeco CT, Lourenco-De-Oliveira R. Daily survival rates and dispersal of Aedes aegypti females in Rio de Janeiro, Brazil. Am J Trop Med Hyg. 2007;76:659–665. [PubMed] [Google Scholar]
- 19.Sheppard PM, Macdonald WW, Tonn RJ, Grab B. The Dynamics of an Adult Population of Aedes aegypti in Relation to Dengue Haemorrhagic Fever in Bangkok. Journal of Animal Ecology. 1969;38:661–702. [Google Scholar]
- 20.Gilks RSWR, Spiegelhalter DJ. Markov chain Monte Carlo in practice. Chapman & Hall; London: 1996. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Study population characteristics
Table S2: List of primers and probes used
Figure S1: Human DENV viremia kinetics
Supplementary data: Mosquito biting study data