Abstract
Vitrification is commonly used in the cryopreservation of mammalian blastocysts to overcome the temporal and spatial limitations of embryo transfer. Previous studies have shown that the implantation ability of vitrified blastocysts is impaired and that microRNAs (miRNAs) regulate the critical genes for embryo implantation. However, little information is available about the effect of vitrification on the miRNA transcriptome in blastocysts. In the present study, the miRNA transcriptomes in fresh and vitrified mouse blastocysts were analyzed by miRNA Taqman assay based method, and the results were validated using quantitative real-time PCR (qRT-PCR). Then, the differentially expressed miRNAs were assessed using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. Overall, 760 known mouse miRNAs were detected in the vitrified and fresh mouse blastocysts. Of these, the expression levels of five miRNAs differed significantly: in the vitrified blastocysts, four miRNAs (mmu-miR-199a-5p, mmu-miR-329-3p, mmu-miR-136-5p and mmu-miR-16-1-3p) were upregulated, and one (mmu-miR-212-3p) was downregulated. The expression levels of all miRNAs measured by the miRNA Taqman assay based method and qRT-PCR were consistent. The four upregulated miRNAs were predicted to regulate 877 candidate target genes, and the downregulated miRNA was predicted to regulate 231 genes. The biological analysis further showed that the differentially expressed miRNAs mainly regulated the implantation of embryos. In conclusion, the results of our study showed that vitrification significantly altered the miRNA transcriptome in mouse blastocysts, which may decrease the implantation potential of vitrified blastocysts.
Introduction
Blastocyst transfer has been shown to be an effective approach to improve implantation and pregnancy rates during the transfer of embryos produced in vitro [1]. To overcome the temporal and spatial limitations of embryo transfer, several types of cryopreservation methods have been developed, including controlled freezing [2] and vitrification [3]. Many experiments have shown that vitrification has a higher efficiency of cryopreservation than controlled freezing [4–7], mainly due to the higher rapid cooling rate of vitrification (>20,000°C/min) [8].
Vitrification is commonly applied in bovine [8], mouse [9], and human [10] blastocysts. However, the implantation of vitrified blastocysts is impaired. Therefore, researchers are now focusing on the possible influence of blastocyst vitrification on factors such as the inner cell mass number [11], spindle formation [12], fragmented DNA in nuclei [7,13], the expression levels of important development-related genes [14], and the sex ratio of offspring after transfer [15]. Recently, our research group reported the effect of vitrification on the promoter methylation levels and the mRNA expression levels of octamer-binding transcription factor 4 (OCT4), Nanog homeobox (NANOG), and caudal-type homeobox 2 (CDX2) in mouse blastocysts [16].
MicroRNAs (miRNAs) are a family of small RNAs that are 21–25 nucleotides in length and are involved in the negative regulation of gene expression at the post-transcriptional level [17,18]. Many miRNA knockout experiments have illustrated the physiological importance of individual miRNAs in mice [19–22]. Several studies have reported that miRNAs are important regulators of pluripotency and differentiation and, consequently, of early lineage segregation in embryonic development [23,24] and maternal-to-embryonic transition [25]. MiRNAs also regulate the critical genes for embryo implantation [26], and the disruption of blastocyst miRNA expression is associated with human infertility [27]. Nuclear transfer by microinjection has been found to alter the miRNA profile of enucleated mouse oocytes [28], indicating that external stimuli may affect the miRNA profiles of mammalian oocytes. Currently, however, little information is available on the effect of vitrification on the miRNA transcriptome profile in oocytes and embryos.
In the present study, the TaqMan Array Rodent MicroRNA A+B Cards Set v3.0 with an Applied Biosystems 7900 HT Fast Real-time PCR system was utilized to detect the miRNA transcriptome in fresh and vitrified mouse blastocysts. Five of 760 known mouse miRNAs were found to be differentially expressed between the vitrified and fresh mouse blastocysts. The biological analysis further showed that the differentially expressed miRNAs mainly regulated the implantation of embryos, which may partially explain the impaired implantation ability of vitrified blastocysts.
Results
Of the 2416 blastocysts that were vitrified in this experiment, 2188 (90.56%) survived after thawing.
miRNA expression in vitrified and fresh mouse blastocysts
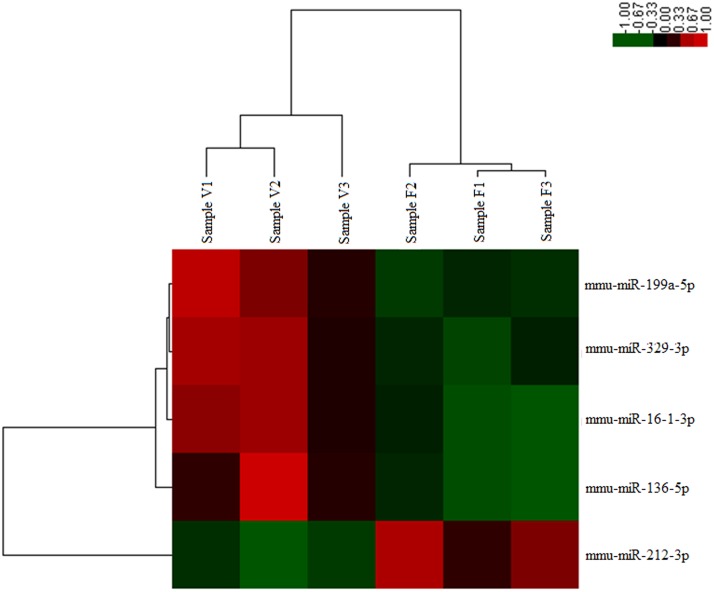
S1 Table presented the fold change of the miRNAs in vitrified blastocysts. Compared to the expression levels in the fresh blastocysts, five miRNAs showed significantly different expression in the vitrified blastocysts (Fig 1). Of these miRNAs, four were upregulated (mmu-miR-199a-5p, mmu-miR-329-3p, mmu-miR-136-5p, and mmu-miR-16-1-3p), and one was downregulated (mmu-miR-212-3p).
Fig 1. Heat map of differentially expressed miRNAs in the vitrification and control groups.
F1, F2, and F3 are the samples from the fresh blastocyst group. V1, V2, and V3 are the samples from the vitrified blastocyst group. Red indicates upregulated expression, and green indicates downregulated expression, with respect to a reference expression level.
Validation of miRNA array results by qRT-PCR
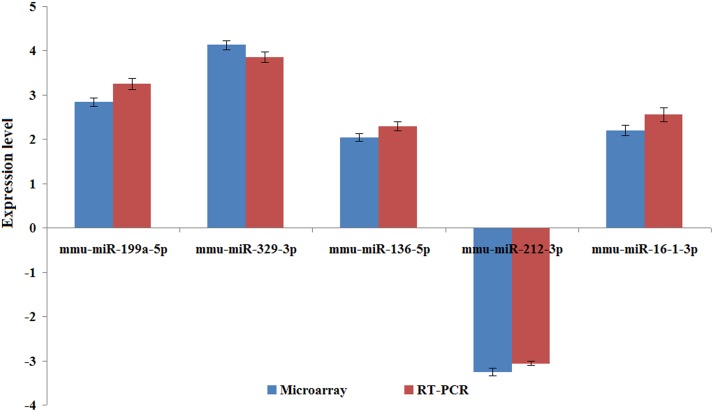
A miRNA assay (No. 4427975) targeting the five differentially expressed miRNAs was used to confirm the results obtained from the miRNA Taqman assay based method (Fig 1). The miRNA expression levels quantified using qRT-PCR and the microarray were compared (Fig 2), and the results of both the methods were found to be consistent.
Fig 2. Validation of miRNA array results by qRT-PCR.
Three biological replicates were used, and U6 snRNA was used as an internal control. The Y-axis denotes the fold change between the vitrified and fresh blastocyst groups.
Prediction of target genes and functional annotation
Among the upregulated miRNAs, 292, 178, 164, and 243 candidate target genes were predicted for mmu-miR-199a-5p, mmu-miR-329-3p, mmu-miR-136-5p, and mmu-miR-16-1-3p, respectively (S2 Table). For the downregulated miRNA, mmu-miR-212-3p, 231 candidate target genes were predicted (S3 Table).
GO was performed to determine the functions of the target genes regulated by these differentially expressed miRNAs. Many of the target genes were found to be involved in the regulation of gene transcription, DNA methylation, histone acetylation, and embryo implantation (Table 1 and Table 2).
Table 1. GO analysis of candidate target genes regulated by the upregulated miRNAs.
| GO_ID | GO_name | P-value |
|---|---|---|
| GO:0006355 | Regulation of transcription, DNA-dependent | 4.33E-60 |
| GO:0008152 | Metabolic process | 2.56E-09 |
| GO:0001701 | In utero embryonic development | 7.14E-07 |
| GO:0006917 | Induction of apoptosis | 4.05E-05 |
| GO:0050896 | Response to stimulus | 0.0001535 |
| GO:0060136 | Embryonic process involved in female pregnancy | 0.0002918 |
| GO:0072593 | Reactive oxygen species metabolic process | 0.0007675 |
| GO:0042771 | Intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 0.0010413 |
| GO:0080111 | DNA demethylation | 0.0010992 |
| GO:0044027 | Hypermethylation of CpG island | 0.0023122 |
| GO:0071470 | Cellular response to osmotic stress | 0.0023122 |
| GO:0043407 | Negative regulation of MAP kinase activity | 0.0025463 |
| GO:0010424 | DNA methylation on cytosine within a CG sequence | 0.0038178 |
| GO:0032873 | Negative regulation of stress-activated MAPK cascade | 0.0038178 |
| GO:0000188 | Inactivation of MAPK activity | 0.0038292 |
| GO:0006309 | Apoptotic DNA fragmentation | 0.0038292 |
| GO:0010629 | Negative regulation of gene expression | 0.0049572 |
| GO:0006979 | Response to oxidative stress | 0.0052717 |
| GO:0023019 | Signal transduction involved in regulation of gene expression | 0.006932 |
| GO:0048864 | Stem cell development | 0.0078691 |
Table 2. GO analysis of candidate target genes regulated by the downregulated miRNA.
| GO_ ID | GO_name | P-value |
|---|---|---|
| GO:0006355 | Regulation of transcription, DNA-dependent | 1.10E-27 |
| GO:0006915 | Apoptotic process | 1.21E-08 |
| GO:0009791 | Post-embryonic development | 4.24E-06 |
| GO:0043066 | Negative regulation of apoptotic process | 5.24E-05 |
| GO:0006349 | Regulation of gene expression by genetic imprinting | 8.11E-05 |
| GO:0010468 | Regulation of gene expression | 0.0001458 |
| GO:0035067 | Negative regulation of histone acetylation | 0.0011141 |
| GO:2000036 | Regulation of stem cell maintenance | 0.0011141 |
| GO:0016573 | Histone acetylation | 0.0062976 |
| GO:0032259 | Methylation | 0.0069064 |
| GO:0034721 | Histone H3-K4 demethylation, trimethyl-H3-K4-specific | 0.0079573 |
| GO:0070512 | Positive regulation of histone H4-K20 methylation | 0.0079573 |
| GO:2000617 | Positive regulation of histone H3-K9 acetylation | 0.0079573 |
| GO:2000620 | Positive regulation of histone H4-K16 acetylation | 0.0079573 |
| GO:0016571 | Histone methylation | 0.008903 |
KEGG pathway annotations of the candidate target genes regulated by differentially expressed miRNAs were also performed. A number of KEGG pathways were identified as relevant to embryo implantation (Table 3 and Table 4).
Table 3. Significant pathways of candidate target genes regulated by the upregulated miRNAs.
| Path_ID | Path_name | Path_gene_count | P-value |
|---|---|---|---|
| 4310 | Wnt signaling pathway | 148 | 8.22E-12 |
| 4510 | Focal adhesion | 205 | 3.28E-11 |
| 4520 | Adherens junction | 75 | 1.24E-09 |
| 4910 | Insulin signaling pathway | 141 | 2.85E-08 |
| 4010 | MAPK signaling pathway | 259 | 7.37E-08 |
| 4530 | Tight junction | 138 | 8.83E-06 |
| 4115 | p53 signaling pathway | 69 | 0.0001053 |
| 4150 | mTOR signaling pathway | 62 | 0.0004703 |
| 4630 | Jak-STAT signaling pathway | 156 | 0.0007667 |
Table 4. Significant pathways of candidate target genes regulated by the downregulated miRNA.
| Path_ID | Path_name | Path_gene_count | P-value |
|---|---|---|---|
| 4010 | MAPK signaling pathway | 259 | 2.17E-07 |
| 4310 | Wnt signaling pathway | 148 | 3.25E-07 |
| 4510 | Focal adhesion | 205 | 3.82E-06 |
| 4915 | Estrogen signaling pathway | 99 | 0.000100728 |
| 4370 | VEGF signaling pathway | 66 | 0.000289995 |
| 4520 | Adherens junction | 75 | 0.000475568 |
| 4910 | Insulin signaling pathway | 141 | 0.00053183 |
| 4150 | mTOR signaling pathway | 62 | 0.003954979 |
| 4530 | Tight junction | 138 | 0.004656746 |
Validation of changes in mRNA expression of target genes
From candidate target genes of these differentially expressed miRNAs, 14 genes were randomly selected to quantify their mRNA expression levels (S4 Table). The results showed that vitrification significantly altered the mRNA expression level of all these selected genes (Fig 3).
Fig 3. Validation of changes in mRNA expression of target genes.
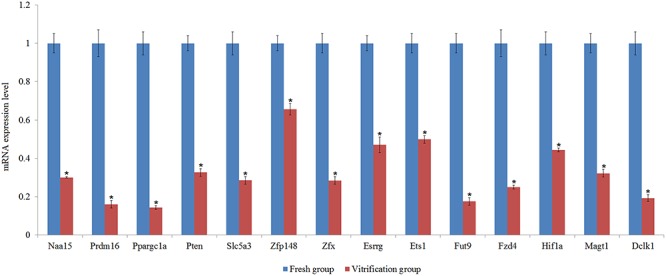
Means of vitrification group with star on the columns differed significantly with those of the fresh group (P<0.05).
Discussion
In this study, 5 of the 760 identified miRNAs, namely, mmu-miR-199a-5p, mmu-miR-329-3p, mmu-miR-136-5p, mmu-miR-16-1-3p, and mmu-miR-212-3p, showed significantly different expression between the vitrified and fresh mouse blastocysts.
Functions of the differentially expressed miRNAs
Of the five differentially expressed miRNAs, four miRNAs, namely, mmu-miR-199a-5p, mmu-miR-329-3p, mmu-miR-136-5p, and mmu-miR-16-1-3p, were significantly upregulated in the vitrified blastocysts.
The mmu-miR-199a-5p miRNA showed the highest expression level at the embryonic stage at 12.5 days postcoitus [29]. A previous study reported that mmu-miR-199a-5p could silence the self-renewal of mouse embryonic stem cells [30]. Mmu-miR-199a-5p is upregulated during the fibrogenic response to tissue injury and mediates TGFbeta-induced lung fibroblast activation by targeting caveolin-1 [31]. Mmu-miR-199a* was found to be differentially expressed in the mouse uterus during implantation in coordination with the expression of cyclooxygenase-2 (COX-2), a gene critical for implantation [26]. In humans, miR-329 may inhibit cell proliferation in human glioma cells by regulating E2F1-mediated suppression of the Akt pathway [32]. Mmu-mir-16 is reported to be related to apoptosis in mouse embryos [33], and miRNA profiling suggested that genes targeted by this miRNA are important in spindle organization in mouse oocytes [34]. In Drosophila, miR-316 is expressed in the mesoderm [35].
In our study, mmu-miR-212-3p was downregulated in vitrified mouse blastocysts. Previously, mmu-miR-212 was found to post-transcriptionally regulate C-terminal-binding protein 1 [36], a protein that was recently shown to repress the expression of steroidogenic factor 1 [37], a nuclear receptor involved in ovarian, adrenal, and testis development and function [38]. In mouse periovulatory granulosa cells, mmu-miR-212 was found to be highly upregulated following luteinizing hormone (LH)/hCG induction [36].
Thus, the miRNAs that were found to be differentially expressed in the vitrified blastocysts are mainly involved in embryo implantation and cell development.
GO analysis of candidate genes regulated by differentially expressed miRNAs in vitrified blastocysts
The GO analysis of the candidate target genes regulated by the differentially expressed miRNAs illustrated a high enrichment of genes involved in the regulation of transcription, implantation, metabolism, DNA methylation, and histone acetylation.
The results of the present study indicated that the candidate target genes regulated by significantly altered miRNAs induced by vitrification (such as GO:0006355, GO:0010629, GO:0006349, and GO:0010468) are involved in the regulation of transcription, which may partially explain the mechanisms through which vitrification alters the expression levels of pluripotent genes in mouse blastocysts [16], development- [14] and stress-related [39] genes in bovine blastocysts and the gene expression profiles of in vivo-derived 8-cell stage mouse embryos [40].
Previous experiments have reported that rabbit embryo vitrification reduced early fetal growth [41], increased losses throughout gestation [41,42], and affected gene and protein expression in the placenta at day 14 after the transfer of vitrified morulae [43]. A higher rate of major postpartum hemorrhage was observed in humans receiving vitrified blastocyst transfers [10]. The present study showed that the candidate target genes of miRNAs whose expression levels were significantly altered by vitrification (e.g., GO:0001701, GO:0060136, and GO:0009791) were involved in the regulation of implantation; these results explained the impaired implantation ability of vitrified blastocysts.
The target genes regulated by miRNAs whose expression levels were significantly altered by vitrification (e.g., GO: 0008152 and GO: 0072593) are also involved in the regulation of metabolism: this may explain the altered energy metabolism reported in vitrified blastocysts [44] and oocytes [45–47], as well as the altered oxygen metabolism in blastocysts [44] and oocytes [45,46].
In oocytes, vitrification has been reported to significantly alter the overall methylation levels in the OCT4 and sex determining region y-box 2 (SOX2) promoters in mice [48] and the H4 acetylation and H3K9 methylation levels in mice [49] and pigs [50]. Moreover, vitrification has been shown to significantly alter the imprinted cortical region methylation of H19 in bovine embryos [51], the H19 differentially methylated domain in mouse fetuses [52], and the methylation levels of the OCT4, NANOG, and CDX2 promoters in mouse blastocysts [16]. The results of the present study indicated that candidate target genes regulated by miRNAs whose expression levels were significantly altered by vitrification (e.g., GO:0080111, GO:0044027, GO:0016573, and GO:0032259) are involved in the regulation of methylation and acetylation; these findings may explain the altered methylation and acetylation levels in vitrified embryos or oocytes.
Pathway analysis of candidate genes regulated by the differentially expressed miRNAs
The results of the present study indicated that the significant pathways of the candidate target genes are mainly involved in embryo implantation, such as the Wnt, adherens, and tight junction genes (Table 3 and Table 4), which partially explains the reduced implantation ability and post-implantation development potential of vitrified embryos [41,42].
Wnt signaling plays an important role in mouse embryonic development and is involved in processes such as axis patterning, cell fate specification, cell proliferation, and cell migration [53], as well as in the coordination of mouse uterus—embryo interactions required for implantation [54]. Both adherens and tight junction proteins are tightly regulated at implantation sites [55]; therefore, trophectodermal cells must become mobile and penetrate between the endometrial luminal epithelial cells [56]. Focal adhesions play an important role in promoting embryo invasion; in particular, they disassemble at the time of implantation in rats, facilitating the detachment of the uterine luminal epithelium to allow the embryo to invade the endometrium [57]. The insulin signaling pathway is involved in the pregnancy of murine blastocysts [58]. The Jak/STAT pathway is activated by the actions of multiple cytokines such as IL11 and LIF, which are known for their roles in implantation [59]. VEGF can act on human blastocysts, enhancing their outgrowth and adhesive capacity [60]. mTOR signaling leads to the development of mouse trophoblast cell motility and the initiation of implantation [61]. Maternal estrogen signaling induces adhesion complex assembly in the trophectoderm [62]. The MAP kinase pathway has been implicated in stem cells [63].
Validation of changes in mRNA expression of target genes
In the present study, the mRNA expression levels of these selected genes regulated by the differentially expressed miRNAs were all decreased by the vitrification. It has been reported that the predominant regulatory effect of miRNAs is to repress their target mRNAs; mechanisms for this include translational repression, mRNA cleavage, mRNA deadenylation or alteration of mRNA stability [64], which contributes to explain the mRNA expression changes of these selected genes in the present study.
Conclusions
By using the TaqMan Array Rodent MicroRNA A+B Cards Set v3.0 with an Applied Biosystems 7900 HT Fast Real-time PCR system, the expression levels of 760 known miRNAs were compared between fresh and vitrified mouse blastocysts, and five of these mRNAs were found to differ significantly between the two blastocyst groups. The expression levels of all miRNAs measured by the miRNA Taqman assay based method and qRT-PCR were consistent. Bioinformatic analysis of the differentially expressed miRNAs showed that major affected pathways are known to be involved in implantation [41,42], which partially explain the impaired implantation ability of vitrified blastocysts.
Materials and Methods
Ethics Statement
All animal procedures were performed according to the guidelines developed by the China Council on Animal Care, and all protocols were approved by the Animal Care and Use Committee of Beijing, China. The approval ID or permit numbers were SYXK (Beijing) 2008–007 and SYXK (Beijing) 2008–008.
Chemicals
All chemicals and media were purchased from Sigma Chemical Co. (St. Louis, MO, USA), and all plasticware was obtained from Nunc-ware (Nunc; Nalge Nunc International, Roskilde, Denmark), unless indicated otherwise.
Open-pulled straw (OPS) preparation
OPSs were prepared according to the method described previously [8]. Briefly, 0.25-mL plastic straws (IMV, L’Aigle, France) were heat-softened and manually pulled to a length of 2–3 cm to attain an approximate outer diameter of 0.23 mm and a wall thickness of 0.02 mm.
Vitrification solutions
The pretreatment solution consisted of 10% ethylene glycol (EG) and 10% dimethyl sulfoxide (DMSO) in Dulbecco’s phosphate-buffered saline (DPBS) containing 3 mg/mL bovine serum albumin (BSA). EDFS30 solution, which was used for vitrification, contained 15% EG and 15% DMSO in Ficoll-sucrose (FS) solution (DPBS containing 300 mg/mL Ficoll, 171.2 g/L sucrose, and 3 mg/mL BSA).
Collection of blastocysts
Five week-old Kunming white mice were superovulated with an intraperitoneal (ip) injection of 10 IU equine chorionic gonadotropin (eCG; Ningbo Hormone Products Co., Ltd, Zhejiang, China) followed by an ip injection of 10 IU human chorionic gonadotropin (hCG; Ningbo Hormone Products Co., Ltd.) 48 hours later. The mice were then mated with fertile males, and the successfully mated females were identified the next day by detecting a vaginal plug. Blastocysts were collected from the excised oviducts of mated females within 92–94 hours after the hCG injection.
Vitrification and thawing of blastocysts
The embryos were equilibrated in pretreatment solution for 30 seconds, loaded into an OPS with EDFS30 for 25 seconds, and plunged into liquid nitrogen (LN2).
The blastocysts were thawed by rinsing in 0.5 M sucrose under mineral oil for 5 minutes and washed three times in CZB medium.
Blastocysts with morphologically intact inner cell mass, trophoderm, and re-expanding blastocoele at 18–20 hours after warming were considered to have survived, according to the method described by Son et al. [65].
Total RNA isolation
Total RNA was extracted from each pool of 300 (n = 3) vitrified/fresh blastocysts using an RNeasy Mini Kit (Qiagen, CA, USA) according to the manufacturer’s instructions. Genomic DNA contamination was removed by treating the isolated RNA with DNase using a DNA-free kit (Ambion, TX, USA). TriPure Isolation Reagent (Roche, NSW, Australia) was used to purify the extracted RNA. A Nanodrop spectrophotometer (Thermo-Fisher) was utilized to measure the concentrations of total RNA.
MiRNA array
MiRNAs were measured in triplicate using the TaqMan Array Rodent MicroRNA A+B Cards Set v3.0 with an Applied Biosystems 7900 HT Fast Real-time PCR system according to the method described by Granjon et al. [66].
Reverse transcription of 30 ng of total RNA from fresh and vitrified blastocysts was carried out using a TaqMan MicroRNA Reverse Transcription Kit. TaqMan PreAmp Master Mix was used to pre-amplify small amounts of cDNA without introducing amplification bias to the sample. The pre-amplified products were diluted in the ratio of 1:5 and then used as a template for the PCR reaction and loaded into the corresponding fill port. Individual singleplex PCR reactions were carried out in 384-well plates, and the miRNA expression level was measured using Ct (threshold cycle) determined by RQ Manager software. U6 snRNA was used as an endogenous control to normalize the data, as described in previous studies [67–69]. Microarray data have been deposited in the NCBI’s Gene Expression Omnibus database (accession number GSE62581).
The Ct value for each miRNA was calculated using an ABI 7900HT Fast Real-time PCR system. Raw Ct values considered “undetermined” by the software or at a level of ≥40 cycles were excluded from the analysis. Relative quantification was calculated using the comparative Ct (2–ΔΔCt) method [70].
The qRT-PCR data were analyzed by Student's t-test and calculated using Statistical Analysis System (SAS) software (SAS Institute; Cary, NC, USA). Each experiment was repeated three times, and a P-value of <. 05 was considered statistically significant.
Validation of miRNA array results by qRT-PCR
The MiRNA assay (ABI) was used to confirm the results of the miRNA Taqman assay based method. qRT-PCR was performed using a TaqMan Universal PCR Master Mix II and 7500 Fast system (Applied Biosystems, CA, USA) with the comparative Ct (2–ΔΔCt) method [70]. The isolation and reverse transcription of total RNA and preamplification of cDNA were performed as described previously.
Gene target prediction and functional annotation
MiRanda (http://www.microrna.org/microrna/) and TargetScan (http://www.targetscan.org/) were utilized to scan targets for the differentially expressed miRNAs obtained above. The intersection of these two prediction programs was selected in the present study. The thresholds of the candidate target sites were an exact match to positions 2–7 of the mature miRNA with a downstream ‘A’ across from position 1 of the miRNA, to positions 2–8 of the mature miRNA, or to positions 2–8 of the mature miRNA with a downstream ‘A’ across from position 1 of the miRNA. The 3’-untranslated region (UTR) sequences of the target genes in mice were obtained from the National Center for Biotechnology Information (NCBI) database.
The main functions of the candidate target genes regulated by differentially expressed miRNAs were determined using the Gene Ontology (GO) analysis. GO analysis, which was provided by the NCBI, was used to organize genes into hierarchical categories and uncover the gene regulatory network on the basis of biological processes and molecular functions [71]. Significance was set at P <. 01.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/) was used to perform pathway annotations of the candidate target genes regulated by differentially expressed miRNAs. Significance was set at P <. 01.
Validation of changes in mRNA expression of target genes
Total RNA was isolated from each pool (n = 3) containing 100 fresh or vitrified blastocysts according to the method as mentioned above and dissolved in sterile water at -80°C until use. First-strand cDNA was synthesized using 0.05 μg RNA with random hexamers. qRT-PCR was performed using an ABI 7500 Fast system with the comparative Ct (2–ΔΔCt) method [70]. The β-actin gene was used as a reference gene, and the primers for each gene were listed in S4 Table.
Supporting Information
(XLS)
(XLS)
(XLS)
(DOCX)
Data Availability
All Microarray data files are available from the NCBI’s Gene Expression Omnibus database (accession number GSE62581).
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (31472100), the Basic Research Fund of IAS, CAAS (2014ywf-yb-2) and The Agricultural Science and Technology Innovation Program (ASTIP-IAS06-2014, ASTIP-IAS-TS-5).
References
- 1. Cruz JR, Dubey AK, Patel J, Peak D, Hartog B, Gindoff PR (1999) Is blastocyst transfer useful as an alterative treatment for patients with multiple in vitro fertilization failures? Fertil Steril 72: 218– 220. [DOI] [PubMed] [Google Scholar]
- 2. Whittingham DG, Leibo SP, Mazur P (1972) Survival of mouse embryos frozen to -196°C and -269°C. Science 178:411–414. [PubMed] [Google Scholar]
- 3. Rall WF, Fahy GM (1985) Ice-free cryopreservation of mouse embryos at -196°C by vitrification. Nature 313:573–575. [DOI] [PubMed] [Google Scholar]
- 4. Sommerfeld V, Niemann H (1999) Cryopreservation of bovine in vitro produced embryos using ethylene glycol in controlled freezing or vitrification. Cryobiology 38:95–105. [DOI] [PubMed] [Google Scholar]
- 5. Nedambale TL, Dinnyés A, Groen W, Dobrinsky JR, Tian XC, Yang X (2004) Comparison on in vitro fertilized bovine embryos cultured in KSOM or SOF and cryopreserved by slow freezing or vitrification. Theriogenology 62:437–449. [DOI] [PubMed] [Google Scholar]
- 6. Zhao XM, Quan GB, Zhou GB, Hou YP, Zhu SE (2007) Conventional freezing, straw and open-pulled straw vitrification of moue two pronuclear (2-PN) embryos. Anim Biotech 18: 203– 212. [DOI] [PubMed] [Google Scholar]
- 7. Li L, Zhang X, Zhao L, Xia X, Wang W (2012) Comparison of DNA apoptosis in mouse and human blastocysts after vitrification and slow freezing. Mol Reprod Dev 79:229–236. 10.1002/mrd.22018 [DOI] [PubMed] [Google Scholar]
- 8. Vajta G, Holm P, Kuwayama M, Booth PJ, Jacobsen H, Greve T, et al. (1998) Open pulled straw (OPS) vitrification: a new way to reduce cryoinjuries of bovine ova and embryos. Mol Reprod Dev 51: 53–58. [DOI] [PubMed] [Google Scholar]
- 9. Huang CC, Lee TH, Chen SU, Chen HH, Cheng TC, Liu CH, et al. (2005) Successful pregnancy following blastocyst cryopreservation using super-cooling ultra-rapid vitrification. Hum Reprod 20:122–128. [DOI] [PubMed] [Google Scholar]
- 10. Wikland M, Hardarson T, Hillensjö T, Westin C, Westlander G, Wood M, et al. (2010) Obstetric outcomes after transfer of vitrified blastocysts. Hum Reprod 25:1699–1707. 10.1093/humrep/deq117 [DOI] [PubMed] [Google Scholar]
- 11. Gómez E, Muñoz M, Rodríguez A, Caamaño JN, Facal N, Díez C, et al. (2009) Vitrification of bovine blastocysts produced in vitro inflicts selective damage to the inner cell mass. Reprod Domest Anim 44:194–199. 10.1111/j.1439-0531.2007.01026.x [DOI] [PubMed] [Google Scholar]
- 12. Chatzimeletiou K, Morrison EE, Panagiotidis Y, Vanderzwalmen P, Prapas N, Prapas Y, et al. (2012) Cytoskeletal analysis of human blastocysts by confocal laser scanning microscopy following vitrification. Hum Reprod 27:106–113. 10.1093/humrep/der344 [DOI] [PubMed] [Google Scholar]
- 13. Fabian D, Gjørret JO, Berthelot F, Martinat-Botté F, Maddox-Hyttel P (2005) Ultrastructure and cell death of in vivo derived and vitrified porcine blastocysts. Mol Reprod Dev 70:155–65. [DOI] [PubMed] [Google Scholar]
- 14. Stinshoff H, Wilkening S, Hanstedt A, Brüning K, Wrenzycki C (2011) Cryopreservation affects the quality of in vitro produced bovine embryos at the molecular level. Theriogenology 76:1433–1441. 10.1016/j.theriogenology.2011.06.013 [DOI] [PubMed] [Google Scholar]
- 15. Lin PY, Huang FJ, Kung FT, Wang LJ, Chang SY, Lan KC (2009) Comparison of the offspring sex ratio between fresh and vitrification-thawed blastocyst transfer. Fertil Steril 92:1764–1766. 10.1016/j.fertnstert.2009.05.011 [DOI] [PubMed] [Google Scholar]
- 16. Zhao XM, Du WH, Hao HS,Wang D, Qin T, Liu Y, et al. (2012) Effect of vitrification on promoter methylation and the expression of pluripotency and differentiation genes in mouse blastocysts. Mol Reprod Dev 79:445–450. 10.1002/mrd.22052 [DOI] [PubMed] [Google Scholar]
- 17. Ambros V (2003) MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113:673–676. [DOI] [PubMed] [Google Scholar]
- 18. Lai EC (2003) microRNAs: runts of the genome assert themselves. Curr Biol 13:R925–936. [DOI] [PubMed] [Google Scholar]
- 19. Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. (2008) Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132: 875–86. 10.1016/j.cell.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, et al. (2008) Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development 135: 3989–3993. 10.1242/dev.029736 [DOI] [PubMed] [Google Scholar]
- 21. Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, et al. (2009) The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ 16:1590–1598. 10.1038/cdd.2009.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, et al. (2010) The miR-144/451 locus is required for erythroid homeostasis. J Exp Med 207: 1351–1358. 10.1084/jem.20100458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I (2008) MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455:1124–1128. 10.1038/nature07299 [DOI] [PubMed] [Google Scholar]
- 24. Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R (2008) Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40:1478–1483. 10.1038/ng.250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mondou E, Dufort I, Gohin M, Fournier E, Sirard MA (2012) Analysis of microRNAs and their precursors in bovine early embryonic development. Mol Hum Reprod 18:425–434. 10.1093/molehr/gas015 [DOI] [PubMed] [Google Scholar]
- 26. Chakrabarty A, Tranguch S, Daikoku T, Jensen K, Furneaux H, Dey SK (2007) MicroRNA regulation of cyclooxygenase-2 during embryo implantation. Proc Natl Acad Sci USA 104: 15144–15149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCallie B, Schoolcraft WB, Katz-Jaffe MG (2010) Aberration of blastocyst microRNA expression is associated with human infertility. Fertil Steril 93:2374–2382. 10.1016/j.fertnstert.2009.01.069 [DOI] [PubMed] [Google Scholar]
- 28. Amanai M, Brahmajosyula M, Perry AC (2006) A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol Reprod 75:877–884. [DOI] [PubMed] [Google Scholar]
- 29. Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB (2009) Twist-1 regulates the miR- 199a/214 cluster during development. Nucleic Acids Res 37:123–128. 10.1093/nar/gkn920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Y, Melton C, Li YP, Shenoy A, Zhang XX, Subramanyam D, et al. (2013) miR-294/miR-302 promotes proliferation, suppresses G1-S restriction point, and inhibits ESC differentiation through separable mechanisms. Cell Rep 4:99–109. 10.1016/j.celrep.2013.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lino Cardenas CL, Henaoui IS, Courcot E, Roderburg C, Cauffiez C, Copin MC, et al. (2013) miR-199a- 5p is upregulated during fibrogenic response to tissue injury and mediates TGFbeta-induced lung fibroblast activation by targeting caveolin-1. PLoS Genet 9: e1003291 10.1371/journal.pgen.1003291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiao B, Tan L, He B, Liu Z, Xu R (2013) MiRNA-329 targeting E2F1 inhibits cell proliferation in glioma cells. J Transl Med 11:172 10.1186/1479-5876-11-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen XH, Han YJ, Yang BC, Cui XS, Kim NH (2009) Hyperglycemia reduces mitochondrial content and glucose transporter expression in mouse embryos developing in vitro. J Reprod Dev 55:534–541. [DOI] [PubMed] [Google Scholar]
- 34. Hawkins SM, Matzuk MM (2010) Oocyte-somatic cell communication and microRNA function in the ovary. Ann Endocrinol (Paris) 71:144–148. 10.1016/j.ando.2010.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC (2005) Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci USA 102:18017–18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fiedler SD, Carletti MZ, Hong X, Christenson LK (2008) Hormonal regulation of MicroRNA expression in periovulatory mouse mural granulosa cells. Biol Reprod 79:1030–1037. 10.1095/biolreprod.108.069690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dammer EB, Sewer MB (2008) Phosphorylation of CtBP1 by cAMP-dependent protein kinase modulates induction of CYP17 by stimulating partnering of CtBP1 and 2. J Biol Chem 283: 6925–6934. 10.1074/jbc.M708432200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lala DS, Rice DA, Parker KL (1992) Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol Endocrinol 6:1249– 1258. [DOI] [PubMed] [Google Scholar]
- 39. Aksu DA, Agca C, Aksu S, Bagis H, Akkoc T, Caputcu AT, et al. (2012) Gene expression profiles of vitrified in vitro- and in vivo-derived bovine blastocysts. Mol Reprod Dev 79: 613–625. 10.1002/mrd.22068 [DOI] [PubMed] [Google Scholar]
- 40. Mamo S, Bodo S, Kobolak J, Polgar Z, Tolgyesi G, Dinnyes A (2006) Gene expression profiles of vitrified in vivo derived 8-cell stage mouse embryos detected by high density oligonucleotide microarrays. Mol Reprod Dev 73:1380–1392. [DOI] [PubMed] [Google Scholar]
- 41. Vicente JS, Saenz-de-Juano MD, Jiménez-Trigos E, Viudes-de-Castro MP, Peñaranda DS, Marco-Jiménez F (2013) Rabbit morula vitrification reduces early foetal growth and increases losses throughout gestation. Cryobiology 67:321–326. 10.1016/j.cryobiol.2013.09.165 [DOI] [PubMed] [Google Scholar]
- 42. Mocé ML, Blasco A, Santacreu MA (2010) In vivo development of vitrified rabbit embryos: effects on prenatal survival and placental development. Theriogenology 73:704–710. 10.1016/j.theriogenology.2009.11.010 [DOI] [PubMed] [Google Scholar]
- 43. Saenz-de-Juano MD, Marco-Jiménez F, Schmaltz-Panneau B, Jiménez-Trigos E, Viudes-de- Castro MP, Peñaranda DS, et al. (2014) Vitrification alters rabbit foetal placenta at transcriptomic and proteomic level. Reproduction 147:789–801. 10.1530/REP-14-0019 [DOI] [PubMed] [Google Scholar]
- 44. Zhao XM, Du WH, Wang D, Hao HS, Qin T, Liu Y, et al. (2012) Controlled freezing and open-pulled straw (OPS) vitrification of in vitro produced bovine blastocysts following analysis of ATP content and reactive oxygen species (ROS) level. J Integ Agric 11: 446–455. [Google Scholar]
- 45. Zhao XM, Du WH, Wang D, Hao HS, Liu Y, Qin T, et al. (2011) Recovery of mitochondrial function and endogenous antioxidant systems in vitrified bovine oocytes during extended in vitro culture. Mol Reprod Dev 78:942–950. 10.1002/mrd.21389 [DOI] [PubMed] [Google Scholar]
- 46. Zhao XM, Du WH, Wang D, Hao HS, Liu Y, Qin T, et al. (2011) Effect of cyclosporine pretreatment on mitochondrial function in vitrified bovine mature oocytes. Fertil Steril 95: 2786–2788. 10.1016/j.fertnstert.2011.04.089 [DOI] [PubMed] [Google Scholar]
- 47. Salvetti P, Buff S, Afanassieff M, Daniel N, Guérin P, Joly T (2010) Structural, metabolic and developmental evaluation of ovulated rabbit oocytes before and after cryopreservation by vitrification and slow freezing. Theriogenology 74:847–855. 10.1016/j.theriogenology.2010.04.009 [DOI] [PubMed] [Google Scholar]
- 48. Milroy C, Liu L, Hammoud S, Hammoud A, Peterson CM, Carrell DT (2011) Differential methylation of pluripotency gene promoters in in vitro matured and vitrified, in vivo-matured mouse oocytes. Fertil Steril 95:2094–2099. 10.1016/j.fertnstert.2011.02.011 [DOI] [PubMed] [Google Scholar]
- 49. Yan LY, Yan J, Qiao J, Zhao PL, Liu P (2010) Effects of oocyte vitrification on histone modifications. Reprod Fertil Dev 22:920–925. 10.1071/RD09312 [DOI] [PubMed] [Google Scholar]
- 50. Spinaci M, Vallorani C, Bucci D, Tamanini C, Porcu E, Galeati G (2012) Vitrification of pig oocytes induces changes in histone H4 acetylation and histone H3 lysine 9 methylation(H3K9).Vet Res Commun 36:165–171. 10.1007/s11259-012-9527-9 [DOI] [PubMed] [Google Scholar]
- 51. Zhao XM, Ren JJ, Du WH, Hao HS, Wang D, Liu Y, et al. (2012) Effect of 5-aza-2′-deoxycytidine on methylation of the putative imprinted control region of H19 during the in vitro development of vitrified bovine two-cell embryos. Fertil Steril 98:222–227. 10.1016/j.fertnstert.2012.04.014 [DOI] [PubMed] [Google Scholar]
- 52. Wang Z, Xu L, He F (2010) Embryo vitrification affects the methylation of the H19/Igf2 differentially methylated domain and the expression of H19 and Igf2. Fertil Steril 93: 2729– 2733. 10.1016/j.fertnstert.2010.03.025 [DOI] [PubMed] [Google Scholar]
- 53. Kemp CR, Hendrickx M, Willems E, Wawrzak D, Métioui M, Leyns L (2007) The roles of Wnt signaling in early mouse development and embryonic stem cells. Func Dev Embryol 1: 1–13. [Google Scholar]
- 54. Mohamed OA, Jonnaert M, Labelle-Dumais C, Kuroda K, Clarke HJ, Dufort D (2005) Uterin Wnt/ beta-catenin signaling is required for implantation. Proc Natl Acad Sci USA 102: 8579–8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Murphy CR (2000) Junctional barrier complexes undergo major alterations during the plasma membrane transformation of uterine epithelial cells. Hum Reprod 15 (Suppl 3): 182–188. [DOI] [PubMed] [Google Scholar]
- 56. Achache H, Revel A (2006) Endometrial receptivity markers, the journey to successful embryo implantation. Hum Reprod Update 12:731–746. [DOI] [PubMed] [Google Scholar]
- 57. Kaneko Y, Lecce L, Day ML, Murphy CR (2012) Focal adhesion kinase localizes to sites of cell-to-cell contact in vivo and increases apically in rat uterine luminal epithelium and the blastocyst at the time of implantation. J Morphol 273:639–650. 10.1002/jmor.20010 [DOI] [PubMed] [Google Scholar]
- 58. Louden E, Chi MM, Moley KH (2008) Crosstalk between the AMP-activated kinase and insulin signaling pathways rescues murine blastocyst cells from insulin resistance. Reproduction 136:335–344. 10.1530/REP-08-0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Paiva P, Menkhorst E, Salamonsen L, Dimitriadis E (2009) Leukemia inhibitory factor and interleukin-11: critical regulators in the establishment of pregnancy. Cytokine Growth Factor Rev 20: 319–328. 10.1016/j.cytogfr.2009.07.001 [DOI] [PubMed] [Google Scholar]
- 60. Hannan NJ, Paiva P, Meehan KL, Rombauts LJ, Gardner DK, Salamonsen LA (2011) Analysis of fertility-related soluble mediators in human uterine fluid identifies VEGF as a key regulator of embryo implantation. Endocrinology 152: 4948–4956. 10.1210/en.2011-1248 [DOI] [PubMed] [Google Scholar]
- 61. Martin PM, Sutherland AE, Van Winkle LJ (2003) Amino acid transport regulates blastocyst implantation. Biol Reprod 69:1101–1108. [DOI] [PubMed] [Google Scholar]
- 62. Chaen T, Konno T, Egashira M, Bai R, Nomura N, Nomura S (2012) Estrogen-dependent uterine secretion of osteopontin activates blastocyst adhesion competence. PLoS One 7: e48933 10.1371/journal.pone.0048933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brivanlou AH, Darnell JE Jr (2002) Signal transduction and the control of gene expression. Science 295:813–818. [DOI] [PubMed] [Google Scholar]
- 64. Pillai RS, Bhattacharyya SN, Filipowicz W (2007) Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 17:118–126. [DOI] [PubMed] [Google Scholar]
- 65. Son WY, Yoon SH, Yoon HJ, Lee SM, Lim JH (2003) Pregnancy outcome following transfer of human blastocysts vitrified on electron microscopy grids after induced collapse of the blastocoele. Hum Reprod 18:137–139. [DOI] [PubMed] [Google Scholar]
- 66. Granjon A, Gustin MP, Rieusset J, Lefai E, Meugnier E, Güller I, et al. (2009) The microRNA signature in response to insulin reveals its implication in the transcriptional action of insulin in human skeletal muscle and the role of a sterol regulatory element-binding protein-1c/myocyte enhancer factor 2C pathway. Diabetes 58:2555–2564. 10.2337/db09-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li J, Yao B, Huang H, Wang Z, Sun C, Fan Y, et al. (2009) Real-time polymerase chain reaction microRNA detection based on enzymatic stem-loop probes ligation. Anal Chem 81:5446– 5451. 10.1021/ac900598d [DOI] [PubMed] [Google Scholar]
- 68. Tesfaye D, Worku D, Rings F, Phatsara C, Tholen E, Schellander K, et al. (2009) Identification and expression profiling of microRNAs during bovine oocyte maturation using heterologous approach. Mol Reprod Dev 76:665–677. 10.1002/mrd.21005 [DOI] [PubMed] [Google Scholar]
- 69. Roa W, Brunet B, Guo L, Amanie J, Fairchild A, Gabos Z, et al. (2010) Identification of a new microRNA expression profile as a potential cancer screening tool. Clin Invest Med 33: E124 [DOI] [PubMed] [Google Scholar]
- 70. Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C (T) method. Nat Protoc 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 71. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
(DOCX)
Data Availability Statement
All Microarray data files are available from the NCBI’s Gene Expression Omnibus database (accession number GSE62581).