Abstract
Biosynthesis of ubiquinones requires the intramembrane UbiA enzyme, an archetypal member of a superfamily of prenyltransferases that generates lipophilic aromatic compounds. Mutations in eukaryotic superfamily members have been linked to cardiovascular degeneration and Parkinson's disease. To understand how quinones are produced within membranes, we report the crystal structures of an archaeal UbiA in its apo and substrate-bound states at 3.3 and 3.6Å resolution, respectively. The structures reveal nine transmembrane helices and an extra-membrane cap domain that surround a large central cavity containing the active site. To facilitate the catalysis inside membranes, UbiA has an unusual active site that opens laterally to the lipid bilayer. Our studies illuminate general mechanisms for substrate recognition and catalysis in the UbiA superfamily and rationalize disease-related mutations in humans.
The UbiA superfamily of intramembrane prenyltranferases catalyzes a key step in the synthesis of ubiquinones, menaquinones, chlorophylls, hemes, and vitamin E, which are released into membranes to serve as electron and proton carriers for cellular respiration and photosynthesis and as antioxidants to reduce cell damage. The UbiA superfamily (fig. S1) includes the UbiA and MenA enzymes in bacteria and archaea, chlorophyll synthases and homogentisate prenyltransferases in photosynthetic organisms, and the COQ2 (1) and UBIAD1 (2) enzymes that play important physiological roles in eukaryotes (3). COQ2 is involved in the biosynthesis of ubiquinones, which function as electron carriers for the mitochondria respiration. The UBIAD1 enzyme is involved in maintaining vascular homeostasis (4), preventing oxidative damage in cardiovascular tissues (5), and sustaining mitochondrial function (6).
Members in the UbiA superfamily share considerable sequence similarity (fig. S2) and catalyze a common reaction of fusing a phytyl- or isoprenyl-chain to an aromatic ring. As the prototype of the superfamily, the UbiA enzyme catalyzes the condensation of isoprenylpyrophosphate (IPP) with the aromatic p-hydroxybenzoate (PHB). UbiA cleaves the pyrophosphate from the IPP substrate to generate a carbocation intermediate at the end of the isoprenyl chain, which reacts at the meta-position of the aromatic PHB substrate to form a C-C bond (Fig. 1A). Although the prenylation of PHB is regiospecific, UbiA promiscuously recognizes IPPs of various chain lengths to generate the ubiquinones CoQ6-10 in different species (7–9). Short-chain substrates such as geranylpyrophosphate (GPP) can be used by UbiA in vitro (10, 11). UbiA is a transmembrane protein that contains two conserved Asp-rich motifs (fig. S2) and requires magnesium (Mg) for catalysis (10). UbiA may be evolutionarily related to trans-prenyltransferases that catalyze the elongation of isoprenyl chains (12), but shares no sequence similarity with soluble aromatic prenyltransferases that synthesize secondary metabolites (3). Unlike these soluble enzymes, UbiA recognizes long isoprenyl chains and releases a prenylated quinone product directly into the membrane. Structural knowledge of UbiA is essential to understand how prenyl transfer is catalyzed within lipid bilayers.
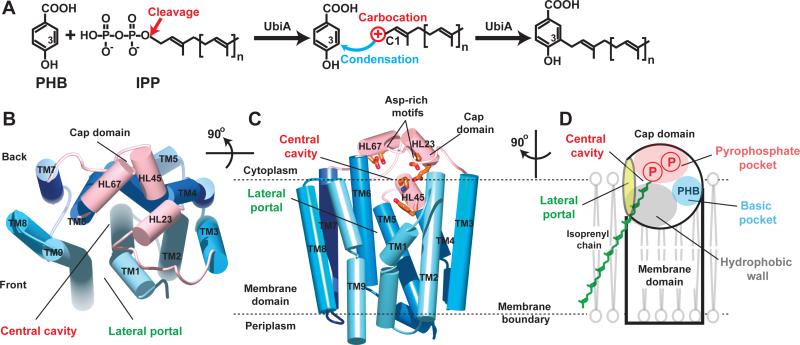
Figure 1. Scheme of UbiA catalysis and structure of ApUbiA.
(A) UbiA catalysis. Cleavage of the pyrophosphate group from the IPP substrate generates a carbocation intermediate, which reacts at the meta-position of the PHB substrate to complete the condensation reaction. (B) Apo-structure of ApUbiA viewed from the cytoplasmic side. The cap domain is shown in pink and the TMs are shown in different blue colors. (C) A side view of the same structure. Conserved residues in the central cavity are shown in orange. (D) Cartoon representation of the structure, which has a unique lateral portal that opens to membrane. The large central cavity contains a polar pocket (pink) for pyrophosphate binding, a hydrophobic wall (grey) for the binding of isoprenyl chains, and a small basic pocket (blue) for PHB binding (see fig. S5).
Here we report the 3.3Å crystal structure of a UbiA homolog from Aeropyrum pernix (ApUbiA). The overall structure of ApUbiA contains nine transmembrane helices (TM) that are arranged counterclockwise in a U-shape with a large central cavity (Fig. 1B and fig. S3). When viewed from the cytoplasmic side, the central cavity is surrounded by TM1, TM2, TM9 at the front and TM5, TM6 from the back. The C-terminal extensions of TM2, TM4, and TM6 are kinked to create a short extramembrane helix followed by a C-terminal loop. These helix/loop regions are termed HL2-3, HL4-5, and HL6-7, respectively, and they form a cap over the central cavity of the transmembrane domain. HL2-3 and HL6-7 each contain an Asp-rich motif, D54XXXD58 and D182XXXD186, respectively (13, 14), and HL4-5 harbors another conserved motif, Y115XXXK119 (fig. S2). All these conserved residues protrude into the central cavity (Fig. 1C) where they are likely involved in substrate binding or catalysis. One side of the central cavity has an opening that we term the lateral portal that is largely buried in the membrane (Fig. 1C). The lateral portal is delineated by TM1 and TM9, both of which are kinked helices with a proline in the middle. The central cavity has a hydrophobic portion leading to the lateral portal (fig. S5) that could accommodate the isoprenyl chain of the IPP substrate (Fig. 1D).
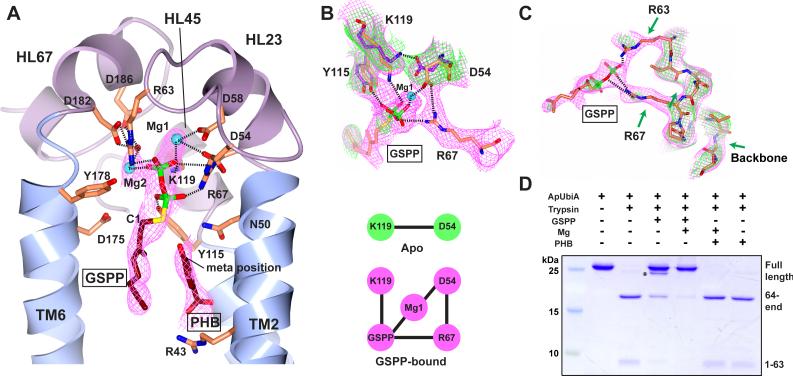
To capture a substrate-bound state, we soaked ApUbiA crystals in a mixture of PHB, Mg, and an uncleavable IPP analog, geranyl thiolopyrophosphate (GSPP). This 3.6Å structure shows GSPP binding in the central cavity (Fig. 2A and fig. S3C). A cluster of conserved residues around GSPP implies the formation of an extensive interaction network that recognizes the pyrophosphate group of GSPP. Since Mg2+ ions are required to mediate the interaction between Asp residues and the pyrophosphate group (10, 15), we modeled (16) two Mg into the electron density (Fig. 2A): Mg1 may bridge the O1 atom of the pyrophosphate to Asp54 and Asp58, and Mg2 may bridge the O2 atom to Asp182 and Asp186 through a water. Asp54 and Asp182 seem to have an additional role of positioning the side chains of Arg67 and Arg63, respectively. These two arginine residues, together with Tyr115 and Lys119, may in turn interact with other oxygens of the pyrophosphate group.
Figure 2. Substrate binding of ApUbiA.
(A) The ApUbiA structure in complex with the substrates. The B-factor sharpened experimental map (in purple) is contoured at 1σ. Due to the limited resolution, the interactions (dashed lines) between individual atoms are putative. (B) Conformational changes induced by GSPP binding. Top: the experimental maps (1σ) of the apo (green) and GSPP-bound (purple) structures suggest the change of interactions. Bottom: a cartoon shows that an interaction network is established upon GSPP binding. (C) Comparison ofthe loop region near Arg63 and Arg67 (same maps as in B). The green arrows indicate a few disordered regions in the apo-structure. (D) Trypsin digestion of the ApUbiA protein in presence of GSPP, PHB, and Mg. The primary digestion site is at Arg63 and a minor digestion site (asterisk) is in a linker region of the N-terminal His-tag.
Comparison of the electron density maps (Fig. 2B) in the apo and substrate-bound states suggests that binding of GSPP induces conformational changes in several active site residues. The interaction between Lys119 and Asp54 in the apo structure is “unlocked” and a new interaction network is established, with Lys119 recognizing the pyrophosphate and Asp54 interacting with Arg67. Moreover, the loop region containing Arg63 and Arg67 becomes ordered (Fig. 2C), consistent with protection of this loop region from trypsin digestion when GSPP binds in a Mg-dependent manner (Fig. 2D). GSPP binding seems to only induce local changes in the active site of ApUbiA, whereas the overall architecture is retained (fig. S4A).
The large central cavity of UbiA contains a small basic pocket near the GSPP binding site (Fig. 1D and fig. S5). The electron density contained in this pocket is suggestive of PHB binding (fig. S5). With PHB modeled so that its carboxyl group contacts Arg43 (Fig. 2A and fig. S5), the meta-position of PHB can be close to the C1 atom of the IPP, consistent with C-C bond formation during the condensation reaction.
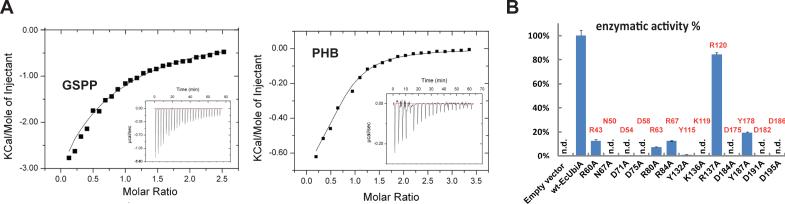
The residues lining the central cavity are highly conserved in the superfamily (fig. S2) and are essential for ApUbiA's binding of GSPP and PHB in vitro. Binding constants of wild-type ApUbiA for GSPP and PHB were determined by isothermal titration calorimetry to be about 0.3 mM and 0.1 mM, respectively (Fig. 3A). These ApUbiA affinities are comparable to the KM values of GPP (0.255 mM) and PHB (0.188 mM) for the condensation reaction catalyzed by E. coli UbiA (10). Consistent with the structure (Fig. 2A), Ala mutations of residues that bind GSPP significantly lowered its binding affinity for ApUbiA (fig. S6). In addition, Arg43Ala and Asn50Ala mutations in the small basic pocket interfered with PHB binding (fig. S7).
Figure 3. Binding and activity assays of UbiA enzymes.
(A) ITC measurements of the GSPP (left) and PHB (right) binding to ApUbiA in presence of Mg. (B) Prenyltransferase activities of EcUbiA mutants. Conserved residues at the central cavity are mutated (n.d.: not detected; error bars: s.e.m. of duplicated assays). Corresponding residues in the ApUbiA structure (Fig. 2A) are labeled in red. R137 is a positive control that is not at the active site in our structure, yet was predicted important in an in silico model (19).
To identify residues essential for UbiA catalysis, we went on to test the enzymatic activities of mutant UbiA enzymes. We were unable to establish an activity assay for the hyperthermophilic ApUbiA at elevated temperature (16) probably because this enzyme was expressed in E. coli membranes (17, 18). Therefore, our subsequent enzymatic assays used E. coli UbiA (EcUbiA), a model enzyme (10, 14) that shares 52% sequence similarity to ApUbiA and has all the conserved active site residues (fig. S2). Among the residues that bound GSPP in the ApUbiA structure (Fig. 2A), several single mutations abolished EcUbiA catalysis (Fig. 3B). As a more stringent test for activity, most of these EcUbiA mutants failed to rescue the growth defect of a quinone-deficient strain of E. coli (fig. S8). The Asp residues in the two DXXXD motifs are essential, likely because they chelate the Mg2+ ions required for UbiA catalysis (10, 15). In addition, the Asn67Ala and Asp184Ala mutations, but not Tyr187Ala, completely abolished the activity. These residues correspond to those (Asn50, Asp175 and Tyr178 in ApUbiA) close to the C1 atom of GSPP (Fig. 2A) and therefore are candidates for stabilizing the carbocation intermediate. For those mutants with partial activity (Fig. 3B), we were able to investigate the enzyme kinetics. We found that the Arg60Ala mutation increased the KM of PHB to about 3mM (ten times of the wild-type enzyme), whereas mutating residues involved in pyrophosphate binding did not generate this dramatic effect (fig. S9). The Arg60 residue in EcUbiA corresponds to Arg43 in the ApUbiA structure (Fig. 2A), which is predicted to bind PHB's carboxyl group. Because Arg60Ala did not completely abolish the EcUbiA activity, the PHB binding could involve other residues around the basic pocket.
Previous models postulated that one of the Asp-rich motifs binds to PHB (13, 14, 19), but the ApUbiA structure suggests that both Asp motifs in UbiA are likely to engage the pyrophosphate group of IPP through Mg-dependent interactions (Fig. 2A). As for trans-prenyltransferases (12), the Mg2+ ions may trigger the ionization of the carbocation (20), which could be stabilized by nearby residues such as Asn67 or Asp184 in EcUbiA (Figs. 2A and 3B). Although the active site of UbiA opens laterally to the bilayer, the highly reactive carbocation intermediate would be protected from water by the surrounding lipids (Fig. 1D). In contrast to previous models (13, 14, 19), PHB is probably bound in a basic pocket (fig. S5) adjacent to the carbocation. Consistent with this proposal, mutating residues in the basic pocket affects PHB binding (figs S7 and S9). Our results for EcUbiA and ApUbiA are also consistent with the enzymatic characterization of LePGT1, a UbiA homolog in plants. LePGT1 mutations corresponding to Arg43Ala and Asn50Ala (Fig. 2A and fig. S5) increased the KM for PHB by 10-fold and 50-fold, respectively (13). At this binding site, the covalent bond formation between the meta-position of PHB and the C1 atom of IPP may proceed without a significant conformational change.
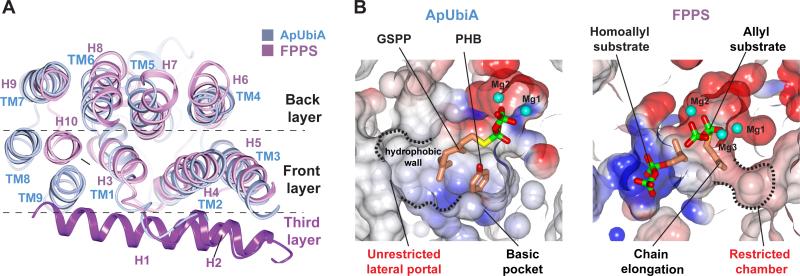
Ubiquinones incorporate long-chain, highly lipophilic tails of six to ten isoprenyl units (7–9). The promiscuous use of long-chain IPP substrates by UbiA can now be explained by structural comparisons (Fig. 4). UbiA is an all α-helical structure that is completely different from the α/β barrel structure of soluble aromatic prenyltransferases (21–24). But interestingly, the membrane-bound UbiA shares some structural features with a soluble trans-prenyltransferase (Fig. 4A), farnesyl pyrophosphate synthase (20) (FPPS), which catalyzes isoprenyl chain elongation and adopts a typical isoprenyl synthase fold (12, 20, 25) with three layers of helices. It appears that part of the allylic binding pocket in FPPS corresponds to the PHB binding pocket of UbiA (Fig. 4B). Conversely, the IPP substrate in UbiA is positioned similarly to the homoallylic substrate in FPPS and also extended to the pyrophosphate pocket of FPPS's allylic substrate. Unlike FPPS, which has two defined chambers for substrate binding (Fig. 4B), the absence of the third layer of helices in UbiA (Fig. 4A) leaves an unrestricted opening (the lateral portal) to the central cavity. Without a defined chamber to restrict the chain length, UbiA can promiscuously recognize a variety of long-chain IPP substrates and generate the precursors of coenzyme Q6-10 in different species (7–9). During the condensation reaction, these long isoprenyl chains may extend through the lateral portal and directly contact lipid molecules (Fig. 1D). Thus, the lateral portal can facilitate the binding of long-chain IPP substrates and the prenylated products can be directly released into membranes through this portal. This is an interesting strategy that intramembrane enzymes can use to catalyze reactions in lipid bilayers.
Figure 4. Structural comparison of ApUbiA and a soluble trans-prenyltransferase.
The FPPS (1RQI (20)) structure is a top DALI (29) hit of the ApUbiA structure. (A) The superimposed structures of ApUbiA (blue) and FPPS (purple). (B) Electrostatic representation of the binding pockets in UbiA (left) and FPPS (right). The two pockets are generated from superimposed structures and shown in the same orientation.
Mutations in eukaryotic CoQ2 and UBIAD1 have been linked to various diseases (4–6, 26–28). Several of these mutations correspond to conserved residues (fig. S2) located in the active site of ApUbiA (Fig. 2A). We have shown that mutating these residues in EcUbiA lowers enzymatic activity (Fig. 3B) or substrate binding affinity (figs. S6, S7) and therefore is likely to have similar effects in the human enzymes (fig. S10).
In summary, the structures of ApUbiA reveal mechanisms of substrate recognition and catalysis that are likely generally applicable to all superfamily members catalyzing the synthesis of lipid-soluble aromatic compounds. Our studies build a framework to understand other important UbiA superfamily members and to design chemoenzymatic synthesis of new aromatic compounds (11, 14).
Supplementary Material
Acknowledgements
We thank the staff at Advanced Photon Source beamline ID-24C (GM103403) for support with data collection, T. Lohman and A. Kozlov for assistance with ITC experiments, J. Janetka and Z. Han for aid with the HPLC analysis, J. Wang, F. Murphy and J. Schuermann for help with structure analysis, S. Chen for mass spectrometry analysis of protein samples, N. Ke and J. Beckwith for generating the quinone-deficient strains, R. Zhang for help with the figures, and D. Fremont, J. Chai, T. Lohman, T. Ellenberger for critical reading of the manuscript. W. L. is supported by a R00 grant 5R00HL097083 from the National Heart, Lung, and Blood Institute and a scholar award from the American Society of Hematology. The structure factors and coordinates were deposited in the protein data bank (accession code 4OD4 for the apo structure and accession code 4OD5 for the substrate-bound structure). The authors declare no conflicts of interest.
Footnotes
Crystal structures of a UbiA superfamily prenyltransferase reveal a catalytic strategy in membranes.
References and notes
- 1.Forsgren M, et al. Biochem. J. 2004;382:519–26. doi: 10.1042/BJ20040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakagawa K, et al. Nature. 2010;468:117–21. doi: 10.1038/nature09464. [DOI] [PubMed] [Google Scholar]
- 3.Bonitz T, Alva V, Saleh O, Lupas AN, Heide L. PLoS One. 2011;6:e27336. doi: 10.1371/journal.pone.0027336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hegarty JJM, Yang H, Chi NCN. Development. 2013;140:1713–9. doi: 10.1242/dev.093112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mugoni V, et al. Cell. 2013;152:504–18. doi: 10.1016/j.cell.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vos M, et al. Science. 2012;336:1306–10. doi: 10.1126/science.1218632. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki K, et al. Biosci. Biotechnol. Biochem. 1994;58:1814–9. doi: 10.1271/bbb.58.1814. [DOI] [PubMed] [Google Scholar]
- 8.Swiezewska E, Dallner G, Andersson B, Ernster L. J. Biol. Chem. 1993;268:1494–9. [PubMed] [Google Scholar]
- 9.Kalén, Appelkvist EL, Chojnacki T, Dallner G. J. Biol. Chem. 1990;265:1158–64. [PubMed] [Google Scholar]
- 10.Melzer M, Heide L. Biochim. Biophys. Acta. 1994;1212:93–102. doi: 10.1016/0005-2760(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 11.Wessjohann L, Sontag B. Angew. Chemie Int. Ed. English. 1996;35:1697–1699. [Google Scholar]
- 12.Wallrapp FH, et al. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1196–202. doi: 10.1073/pnas.1300632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohara K, Muroya A, Fukushima N, Yazaki K. Biochem. J. 2009;421:231–41. doi: 10.1042/BJ20081968. [DOI] [PubMed] [Google Scholar]
- 14.Bräuer L, Brandt W, Schulze D, Zakharova S, Wessjohann L. Chembiochem. 2008;9:982–92. doi: 10.1002/cbic.200700575. [DOI] [PubMed] [Google Scholar]
- 15.Young IG, Leppik RA, Hamilton JA, Gibson F, Bacteriol J. 1972;110:18–25. doi: 10.1128/jb.110.1.18-25.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Material and methods are available as Supplementary Material on Science Online.
- 17.Yernool D, Boudker O, Jin Y, Gouaux E. Nature. 2004;431:811–818. doi: 10.1038/nature03018. [DOI] [PubMed] [Google Scholar]
- 18.Jormakka M, et al. Nat. Struct. Mol. Biol. 2008;15:730–737. doi: 10.1038/nsmb.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bräuer L, Brandt W, Wessjohann L. a. J. Mol. Model. 2004;10:317–27. doi: 10.1007/s00894-004-0197-6. [DOI] [PubMed] [Google Scholar]
- 20.Hosfield DJ, et al. J. Biol. Chem. 2004;279:8526–9. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 21.Kuzuyama T, Noel JP, Richard SB. Nature. 2005;435:983–7. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metzger U, Keller S, Stevenson CEM, Heide L, Lawson DM. J. Mol. Biol. 2010;404:611–26. doi: 10.1016/j.jmb.2010.09.067. [DOI] [PubMed] [Google Scholar]
- 23.Metzger U, et al. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14309–14. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Miao Y, Wang B, Cui G, Merz KM. Biochemistry. 2012;51:2606–18. doi: 10.1021/bi201800m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kellogg B, Poulter C. Curr. Opin. Chem. Biol. 1997:570–578. doi: 10.1016/s1367-5931(97)80054-3. [DOI] [PubMed] [Google Scholar]
- 26.Weiss JS, et al. Invest. Ophthalmol. Vis. Sci. 2007;48:5007–12. doi: 10.1167/iovs.07-0845. [DOI] [PubMed] [Google Scholar]
- 27.Nickerson ML, et al. PLoS One. 2010;5:e10760. doi: 10.1371/journal.pone.0010760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fredericks WJ, et al. J. Cell. Biochem. 2013;114:2170–87. doi: 10.1002/jcb.24567. [DOI] [PubMed] [Google Scholar]
- 29.Holm L, Rosenström P. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.