Abstract
As the key cellular effectors of adaptive immunity, T and B lymphocytes utilize specialized receptors to recognize, respond to, and neutralize a diverse array of extrinsic threats. These receptors (immunoglobulins in B lymphocytes, T cell receptors in T lymphocytes) are incredibly variable, the products of specialized genetic diversification mechanisms that generate complex lymphocyte repertoires with extensive collections of antigen specificities. Recent advances in high throughput sequencing (HTS) technologies have transformed our ability to examine antigen receptor repertoires at single nucleotide, and more recently, single cell, resolution. Here we review current approaches to examining antigen receptor repertoires by HTS, and discuss inherent biological and technical challenges. We further describe emerging applications of this powerful methodology for exploring the adaptive immune system.
Keywords: immune repertoires, immunoglobulin diversity, antibody diversity, T cell receptor diversity, high throughput sequencing, immune profiling
Lymphocyte antigen receptors: diverse sequences and specificities
Adaptive immunity can provide acute and long-term protection against a virtually limitless array of pathogenic hazards. To contend with the broad variety and unpredictability of potential threats, the adaptive immune system relies on somatic diversification processes that generate immense sequence variation in B cell immunoglobulin (herein referred to as B cell receptor, BCR) and T cell receptor (TCR) genes to create massive repertoires of lymphocytes with distinct immune receptors and antigen specificities. Upon recognition of their specific antigens, lymphocytes can undergo clonal expansion with appropriate pathogen-targeted effector and subsequent memory functions.
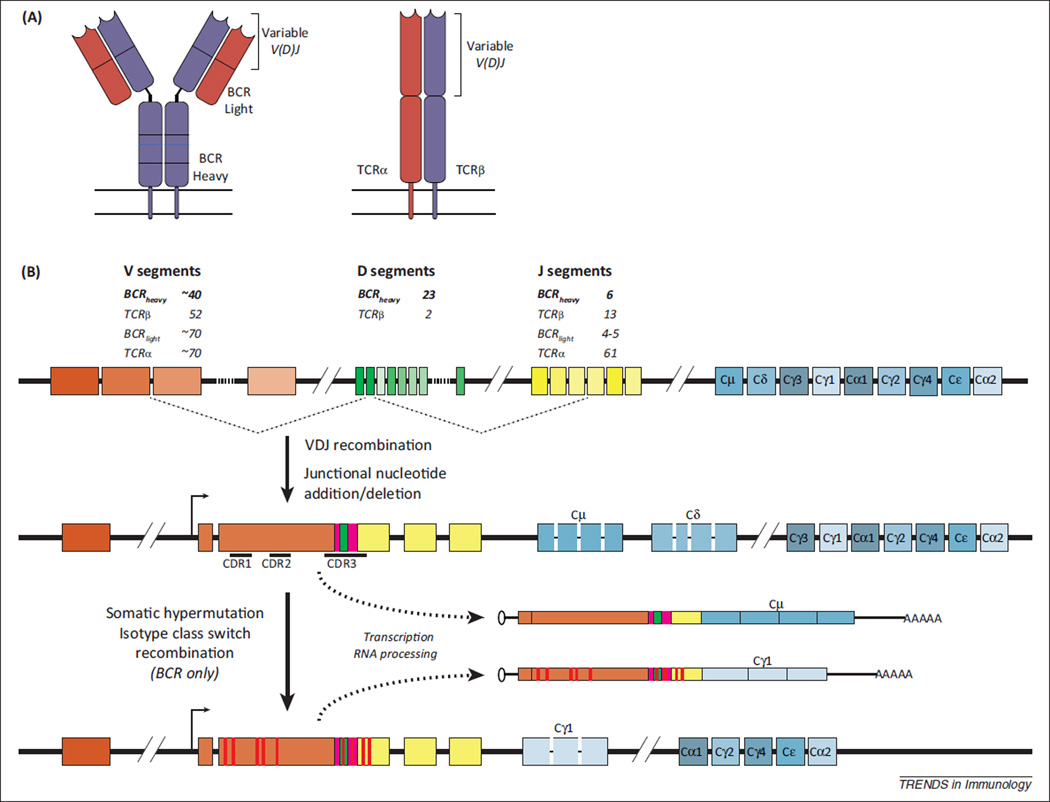
Although functionally distinct, BCRs and TCRs are similarly organized and correspondingly diverse (Figure 1A). Both are composed of two distinct subunit chains, each chain containing a variable domain that contributes to the antigen binding surface of the heterodimeric receptor. Primary diversification of the genes encoding these variable domains proceeds by analogous mechanisms for BCRs and TCRs. On account of these similarities, hereafter we refer to BCRs and TCRs collectively as antigen receptors, with specific distinction where appropriate. During lymphocyte development, variable antigen receptor gene segments (Variable, Joining, Diversity: V, J, D) are rearranged through targeted DNA recombination events (Figure 1B, reviewed in [1]). Substantial sequence complexity is also introduced by the addition or removal of nucleotides at the junctions of these segments. While the entire variable region shapes receptor function, sequence within several complementarity determining regions (CDRs), and CDR3 in particular, contribute most to BCR and TCR specificities [2]. As this recombination process occurs separately for both sub-unit chains, subsequent heterodimeric pairing brings forth still greater combinatorial diversity. Taken together, the diversity established through these molecular mechanisms is staggering, with the theoretical number of distinct BCRs and αβ TCRs estimated to exceed 1013 and 1018 [2], respectively. In addition, upon antigen recognition, mature B lymphocytes may also undergo secondary diversification processes in lymphoid germinal centers. Here, activation-induced cytidine deaminase (AID) and error-prone repair mechanisms introduce somatic hypermutation (SHM) in BCR variable region sequences, enabling selection of lymphocytes with superior BCR properties (a process known as affinity maturation) [3]. BCRs may also undergo class-switch recombination (CSR), in which gene segments encoding immunoglobulin constant regions are recombined to ‘switch the isotype of the expressed antibody, thereby altering its effector properties [4].
Figure 1.
Diversification of antigen receptor repertoires. (A) BCRs and TCRs are similarly organized. Each receptor is composed of two distinct subunit chains (BCR: light chain and heavy chain, TCR: α chain and β chain). The antigen binding surface is formed by the variable region of each chain, which is encoded by recombined V, J, and D (BCR heavy and TCRβ) gene segments. (B) Antigen receptor diversification. A schematic of the BCR heavy locus is shown; with the exception of somatic hypermutation and class-switch recombination, analogous mechanisms proceed at the TCRβ locus (with differences in segment organization). Antigen receptor repertoire diversity is primarily established during lymphocyte development, during which V (orange), D (green), and J (yellow) gene segments are rearranged through the process of V(D)J recombination. Numbers of distinct V, D, and J segments are shown for each antigen receptor locus [2]. During the recombination process, nucleotides may be added or deleted at segment junctions (magenta), contributing to additional sequence diversity. Complementarity determining regions are indicated. BCR-specific secondary diversification may occur following antigen recognition. In somatic hypermutation processes, mutations (red) are introduced throughout the variable region such that modified BCRs may be selected through affinity maturation. In class-switch recombination, gene segments encoding constant regions (blue) are rearranged resulting in the production of antibodies with different isotypes and corresponding effector functions. Abbreviations: BCR, B cell receptor; TCR, T cell receptor; V, J, and D, Variable, Joining, and Diversity gene segments.
As the principal sites for antigen recognition, BCRs and TCRs are fundamental in lymphocyte development, effector function, and immune memory. As such, immunologists have developed a variety of techniques in attempts to measure diversity and/or perturbations of antigen receptor repertoires. Traditional molecular cloning techniques coupled with Sanger sequencing provided early perspectives on TCR [5] and BCR variation (reviewed in [6]), but were relatively limited in their capacity to assess repertoire diversity by their inherently low throughput. Spectratyping strategies, in which CDR3 length distributions in lymphocyte pools are measured from PCR-amplified VDJ segments, offer a more general view of repertoire diversity (reviewed in [6]). However, despite its effectiveness in estimating overall heterogeneity and assessing monoclonal expansions, spectratyping is relatively limited in resolution and interrogates only a single property of receptor diversity (CDR3 length), without providing the underlying sequence information that encodes specificity.
The exquisite specificity and memory capacity of the adaptive immune system operates at the level of individual receptor sequences of single lymphocytes. Therefore, antigen receptor repertoires are ideally examined at the nucleotide level with individual receptor resolution. Moreover, very large numbers of individual sequences are needed to capture even a moderate degree of the diversity present in a typical lymphocyte repertoire. These demands make BCRs and TCRs attractive targets for high throughput sequencing (HTS) technologies. Yet, while HTS has become nearly routine for genome and transcriptome sequencing, its application to lymphocyte repertoires poses unique technical challenges, due in large part to the excessive diversity it aims to examine. However, with careful methodological consideration and experimental design, antigen receptor repertoire HTS techniques are powerful tools with which to explore lymphocyte biology. Although not without limitations, these new methods have already provided unprecedented new insights into lymphocyte development, repertoire diversity, infectious disease, autoimmunity, and beyond.
HTS of antigen receptor repertoires: technical strategies and considerations
HTS and analysis of antigen receptor repertoires can be used to characterize key features of adaptive immunity, such as repertoire diversity, clonal expansion, and receptor properties, in a variety of immunological contexts (including steady-state, infection, vaccination, lymphocyte development, autoimmune disease, among others). However, despite the standardization of ‘traditional’ HTS applications such as RNA-Seq or genome resequencing, there is no ‘one size fits all’ approach to immune repertoire HTS; at present, technical limitations and evolving methodologies demand that investigators carefully consider different approaches to define an HTS strategy appropriate for each particular immunological problem.
'What to sequence?'
When designing an HTS experiment directed at BCRs or TCRs, an important first question is ‘what to sequence?’ Although antigen receptors are heterodimeric, until recently, technical obstacles have limited HTS analyses to individual receptor subunit chains. While ‘paired chain’ techniques are now becoming available (discussed later), most methods target single subunits (typically BCR heavy chains, TCRα or TCRβ chains). These approaches are more straightforward and provide sufficient information to measure many immunological parameters. Additionally, although some questions may require sequencing of complete variable regions, many HTS approaches target only CDR3. With its tremendous potential for diversity in both BCRs and TCRs, CDR3 sequence can be used as an identifier of lymphocyte clonal origin, and can be effectively sequenced with shorter reads and higher throughput. Nevertheless, CDR3-targeted strategies may not be appropriate for certain applications, such as examining complete SHM profiles and/or heavily-mutated BCRs, as found in some broadly-neutralizing HIV antibodies [7–9].
Another nontrivial technical consideration of ‘what to sequence’ is the type of material used as input for HTS library construction: genomic DNA or transcript RNA. In genomic DNA, recombined gene segments are present within the context of ‘unused’ segments and introns, which can create PCR amplification challenges. In addition, genomic DNA libraries typically contain all VDJ receptor recombinants, whether they contribute to a productive coding sequence and expressed receptor or a nonproductive segment arrangement. The presence of nonproductive sequences in HTS datasets can be advantageous for studying receptor diversification and selection in lymphocyte development, but can also supply unwanted reads and obfuscate analysis of the expressed receptor repertoire. When lymphocyte RNA is used as input material, nonproductive receptor transcripts are dramatically underrepresented [10]. In addition, the close proximity of variable and constant regions following mRNA splicing can facilitate simplified PCR amplification strategies (discussed later). Highly expressed TCR and BCR RNA transcripts are far more abundant than their genomic DNA templates, which in the case of antigen receptor recombinants are limited to one copy per cell. Although a potential obstacle when input material is limited, the consistent copy number of genomic DNA offers considerable advantages in quantifying lymphocyte clonal expansions within diverse repertoires. Assuming no bias in genomic DNA amplification and library preparation, the number of HTS reads representing a particular CDR3 sequence should be directly proportional to the number of clonal lymphocytes bearing a certain receptor. One strategy for quantifying clonal frequencies employs replicate library preparations, in which a lymphocyte DNA sample is partitioned into multiple amplification reactions and sequenced in parallel [11–14]. As genomic DNA contains discrete counts of template molecules (one productive V(D)J segment per cell), CDR3 sequences observed in more than one PCR library must therefore be derived from different cells. The number of replicates in which each CDR3 sequence is observed can be used to estimate clonal frequencies. RNA-derived libraries may also be used to approximate clone sizes, but quantification can be affected by irregular BCR [15] and TCR [16] transcript abundance among different cells.
HTS library strategies for variable antigen receptor sequences
Whether DNA or RNA is selected as input material, selectively incorporating antigen receptor sequences into sequencing libraries is typically achieved by targeted PCR amplification. However, as BCR and TCR variable region sequences are extraordinarily diverse by definition, the principal challenge is the design of PCR strategies that allow for comprehensive and unbiased amplification of exceedingly variable template mixtures. Minimizing amplification bias is particularly critical for experiments in which receptors and/or lymphocyte clones are to be quantified by HTS, as uneven amplification efficiencies can skew clonal frequency measurements.
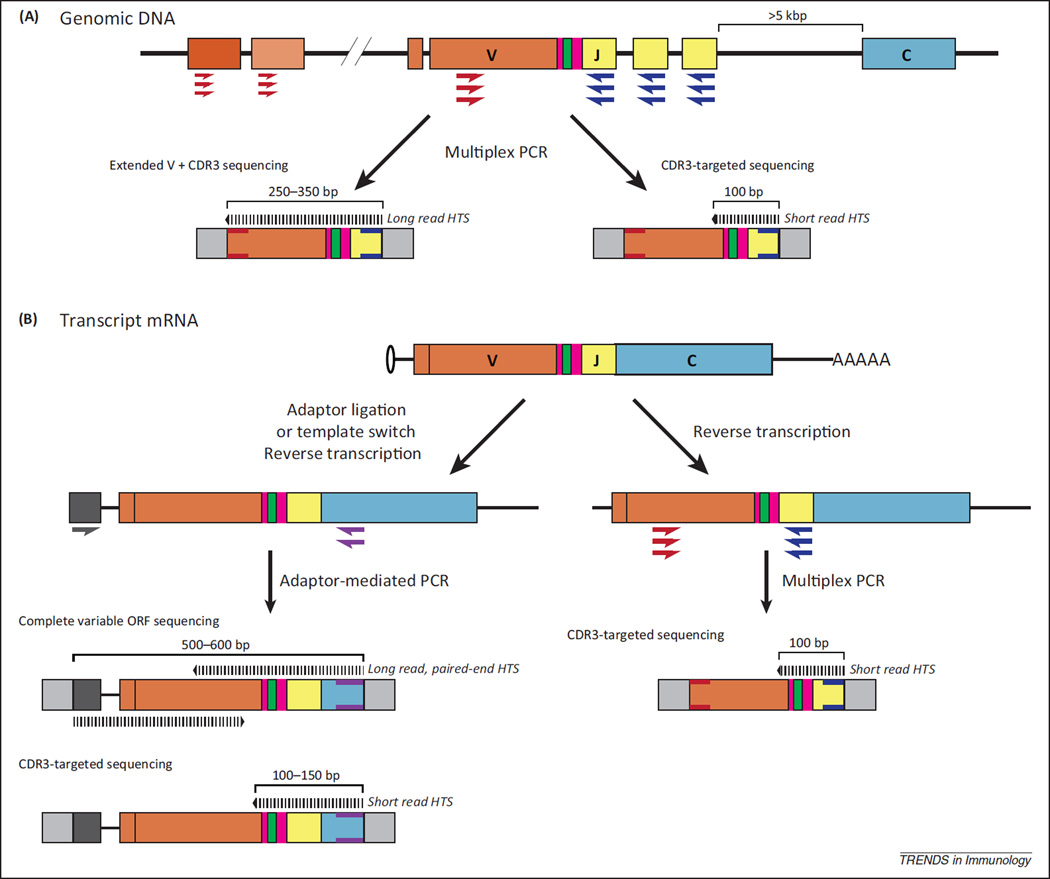
These challenges have been addressed with a variety of amplification strategies (Figure 2). Several approaches employ multiplex PCR with complex mixtures of forward primers complementary to many or all possible V segments [12,17–24]. J segments or constant region exons (for mRNA transcripts) are used for reverse priming. However, achieving unbiased amplification of the multitude of different antigen receptors in a typical repertoire is extremely challenging [25]. Furthermore, in the case of BCRs, SHM has the potential to alter primer-binding sequences within V and J segments. Bias can be minimized through multistep PCR [21,22] or adjusted for using synthetic template controls [25]. However, there are also alternative strategies that circumvent multiplex amplification by priming PCR from engineered adaptor sites [26–28]. In these methods, used with RNA input material, an invariable adaptor sequence is introduced upstream of transcript V regions prior to (by RNA ligation [29]) or concordant with (by template switch [26,28,30,31]) reverse transcription. PCR amplification is performed with a single forward primer (complementary to the adaptor sequence), and reverse primers targeting J segments or constant regions. As the priming sites are invariable, amplification bias is effectively minimized. Furthermore, the resulting library contains the complete variable region open reading frame (ORF). Effectively captured by long read HTS platforms, these sequences can be used in engineering and expressing TCRs or immunoglobulins for functional studies.
Figure 2.
Library preparation strategies for antigen receptor HTS. The extraordinary diversity of antigen receptor sequences poses challenges for targeted amplification and library preparation; although not comprehensive, a general overview of select PCR priming strategies is presented here. A generic antigen receptor schematic representative of BCR heavy or TCRβ is shown. (A) PCR amplification and library preparation from genomic DNA. Multiplex PCR strategies, in which complex mixtures of primers complementary to many or all possible V segment sequences, can be used to amplify portions of variable region sequences from genomic DNA. Multiplex primers targeting portions of V segments (red) can be used in conjunction with J segment primers (navy blue) for amplification. Upon incorporation of HTS adaptors (light gray boxes), long read HTS can be used to capture a majority of variable region sequence. Alternatively, short read HTS can be used to sequence only the CDR3 region. (B) Reverse transcription PCR amplification and library preparation from mRNA. In antigen receptor transcript mRNA, the juxtaposition of constant region exons adjacent to the variable region offers a reverse priming site with minimal diversity. In invariant adaptor strategies, the complexities and potential biases of V segment multiplex PCR are bypassed by incorporating a defined adaptor sequence (dark gray box) upstream of the variable region by oligonucleotide ligation or template switch methods during reverse transcription. A single primer to the adaptor sequence (dark gray) can then be used with constant region primers (violet) to generate amplicons that contain complete variable region open reading frames (ORFs). These can be sequenced in entirety by long read paired-end HTS or sequenced with short read HTS for CDR3-targeted studies. Similar adaptor-mediated PCR strategies using multiplex J primers for CDR3 sequencing are also available (not shown). Alternatively, following reverse transcription without invariant adaptors, multiplex primers to V regions and J regions (as for genomic DNA amplification) or constant regions (not shown), can be used for CDR3-targeted short read HTS. Hashed lines, HTS reads. V, D, J, and constant segment colors as in Figure 1. Abbreviations: BCR, B cell receptor; TCR, T cell receptor; V, J, and D, Variable, Joining, and Diversity gene segments; HTS, high throughput sequencing; CDR3, complementarity determining region 3.
HTS platforms
Several different HTS platforms are suitable for sequencing antigen receptor repertoires; each differs in read lengths, sequencing depth, error frequency, and error type, all of which impact BCR and TCR analysis [32]. Reads must be of sufficient length to include regions of interest (CDR3 or complete variable region), and greater read depth permits the examination of larger and more complex repertoires. Depth is also important for managing the relatively high error rates of HTS by enabling consensus assemblies from multiple reads per sequence. Not accounting for ‘real’ errors introduced during library preparation (e.g., low-fidelity reverse transcription, introduction and perpetuation of mutations during PCR), rates and types of nucleotide errors vary among different HTS platforms. For example, Roche-454, Ion Torrent, and Pacific Biosciences instruments are prone to insertions or deletions (indels), which are relatively rare on Illumina technology [33–36]. Indel errors can be particularly problematic in antigen receptor HTS, specifically in differentiating them from the ‘true indels’ present in CDR3 sequences, the result of nucleotide addition and deletion during recombination. At present, Illumina instruments (MiSeq for longer reads, HiSeq for CDR3-targeted short reads) provide a reasonable read length, depth, and error profile for most studies, although other platforms are preferred for certain applications. As HTS technology advances at a rapid pace, new and superior options are expected in the coming years [37].
Data processing and analysis
Given the complexity of both the biological input and digital output, antigen receptor HTS data requires specialized computational tools and workflows. Interpretation of experimental results requires two independent but related tasks: data processing, in which raw sequence reads are filtered, assembled, and corrected for errors; and data analysis, which will vary considerably by project.
Data processing workflows focus on managing the relatively high error rates of HTS. Although not major obstacles to identifying germline V and J segments, sequence errors can be confounding for measuring repertoire diversity; without proper correction, it can be impossible to differentiate real sequence differences in the receptor repertoire from artifactual differences caused by sequencing errors. A typical strategy for identifying and correcting sequencing and late-cycle PCR errors involves generating consensus assemblies from highly similar reads. However, depending on the algorithm used, grouping by sequence can result in erroneous clustering (and consensus building) of reads derived from highly similar but distinct antigen receptor sequences, thereby eliminating ‘real’ diversity from subsequent analysis. Related strategies address these issues by coupling error management demands with library construction, in which random sequence ‘barcodes’ are appended to input templates prior to amplification. During data processing, which can be performed with software tools specialized for antigen receptor HTS [38,39], reads are organized by barcode, effectively grouping those sequences that originated from the same template molecule for consensus assembly [22,39]. These approaches also enable the direct quantification of input template molecules, which can aid in normalization and comparative analyses [40]. While effective, barcode strategies cannot entirely correct for errors introduced prior to, or during early cycles of, PCR amplification. Managing such errors remains a significant challenge, although recently developed tools designed to address erroneous amplification in TCR sequences by detecting and correcting for PCR error ‘hotspots’ show promise [39].
Making ‘immunological sense’ of antigen receptor HTS data can be challenging. While particular questions may require specialized methods, most analysis workflows involve sequence annotation and some combination of measurements for diversity, clonal expansion, selection, and/or SHM (for BCRs). Select strategies and computational tools for analysis are summarized in Box 1. Although beyond the scope of this review to describe these tools in detail, it should be noted that it is often the details that matter most in appropriately informative data analysis. We advise researchers to familiarize themselves with a given approach and assess how outcomes might be influenced by factors such as repertoire size, clonality, sequencing errors, or undersampling.
Box 1. Selected analysis strategies for antigen receptor HTS.
V(D)J germline segment classification
Identifying the V, D, and J segments present in antigen receptor sequences is a common first step in many analysis workflows. VDJ assignment (and CDR3 annotation) is useful for general classification, diversity measures, and repertoire comparisons. Moreover, discrepancies between germline reference sequences and HTS data may be used to identify SHM or sequencing errors. In general, VDJ classification tools identify segments by alignment to reference databases from NCBI [86] or IMGT [87], although alignment methods vary. A noncomprehensive list of select tools and strategies appears as follows, although not all are specifically tailored to HTS data:
Repertoire diversity analytics
Antigen receptor repertoire diversity can be described by size (i.e., number of unique CDR3 sequences) or by measures that account for frequencies of different receptors. In the latter case, Simpson's diversity index can be used to estimate the probability that different sequences can be randomly picked from the repertoire [96]. Alternatively, 'unseen species' analytics, drawn from the ecology field, can be used to estimate repertoire diversity while accounting for undersampling [12].
Repertoire comparisons
A variety of strategies can be employed to compare repertoires, such as between different individuals [27,48,50], or across different lymphocyte subsets [21]. One straightforward approach entails quantifying the number of shared CDR3 nucleotide [27] or amino acid [27,48,50] sequences between repertoires. Comparisons of variable segment usage frequencies in HTS data have been performed using metrics such as Jensen–Shannon divergence [50]. Many additional diversity and distance tools for antigen receptor repertoires exist and have been comprehensively reviewed by Mehr et al. [53].
SHM and affinity maturation
BCR sequences that have undergone SHM and affinity maturation pose special challenges, such as identifying B lymphocyte clonal lineages, measuring selection, and characterizing SHM patterns.
BCR lineages. B lymphocyte clonal lineages can be identified and visualized by specialized clustering methods that incorporate CDR3 sequence and VJ similarity [97], or by forming genealogical trees rooted with germline sequences [98,99]. A recently developed tool, ImmuniTree [100], models and accounts for sequencing noise common to certain HTS data in constructing cell lineage trees.
Detecting selection in affinity maturation. During affinity maturation, BCR residues with greater contributions to antigen recognition undergo stronger positive selection. Therefore, selection processes can be assessed by tracking the association of specific mutations with expanded lineages using tools such as BASELINe [101] and other specialized lineage tree analytics [102,103].
Immunological applications of antigen receptor HTS
The advent of HTS technology and its application to antigen receptors has enabled new perspectives on adaptive immunity. In just the past several years, these approaches have led to notable advances in both fundamental immunology and clinical applications.
Antigen receptor repertoire analysis
Determining the size and complexity of lymphocyte repertoires has been a long-standing challenge in immunology. Previously, estimates of repertoire complexity were extrapolated from relatively low throughput Sanger sequencing data (reviewed in [6]). HTS approaches have enabled more direct measurements of repertoire diversity. One of the first antigen receptor repertoires to be analyzed by HTS, the zebrafish BCR heavy chain complement was estimated at approximately 5000–6000 unique sequences per fish [41]. Considerably more sequence complexity has been assessed in human lymphocyte populations: tens of thousands of distinct BCR heavy chains [12,22,42] and up to 2 million TCRβ chains [18,21,27,43,44] have been independently and directly observed in single individuals. These data have been used to estimate the total BCR heavy chain repertoire between 0.5 × 106 and 5 × 106 per individual [12,22]. The TCRβ repertoire has been estimated to contain at least 0.5 × 106 to 3 × 106 unique sequences per individual [18,21]. These values are still estimates, as truly comprehensive sequencing of a complete repertoire is limited not by HTS capabilities but by practical restraints: lymphocyte numbers in even large volumes of blood undersample total diversity [27] and only a fraction of the complete lymphocyte repertoire may be present in peripheral blood [45,46].
Antigen receptor HTS has also been used to compare lymphocyte repertoires between individuals, and thereby explore factors that shape differences and similarities. HTS analyses have demonstrated that although the frequencies of BCR segment (V, J, and/or D) usage are often correlated between unrelated individuals, significant deviations of individual segments are not uncommon [47,48]. Measurements of similar segment usage patterns in monozygotic twins suggest that segment preferences are determined genetically [48]. Indeed, Boyd et al. took advantage of the single nucleotide resolution of BCR HTS data to detect sequence polymorphisms and infer genomic structural variations at the heavy chain locus, and suggest that these differences can affect the frequency of segment representation in the BCR repertoire [47]. At the level of CDR3 amino acid sequences, steady-state repertoires have been described as largely unique to each individual [48,49], although some minimal overlap has been observed [49].
In similar HTS analyses, TCRβ repertoires exhibit common segment usage patterns [43]. Furthermore, considerable numbers of identical TCRβ CDR3 nucleotide [27,44] and amino acid [27,43,44,50] sequences have been observed in different individuals. Of note, the degree of observed overlap between TCRβ repertoires has been shown to be directly related to the depth at which the repertoires are sampled [51]. The surprising amount of TCRβ repertoire overlap observed by HTS methods may be explained, at least in part, by convergent recombination models, in which certain CDR3 clonotypes are consistently generated at higher efficiencies during V(D)J recombination [52], and therefore more likely to be shared in different individuals. Furthermore, although at least one study has inferred that HLA-matched individuals display increased TCRβ CDR3 repertoire overlap [27], other deep profiling studies of unrelated subjects [43,50] and/or monozygotic twins [50] suggest that repertoire overlap between individuals is generally independent of HLA type. These results raise questions about the impact of environmental exposure, genetics, and thymic selection in shaping the mature T lymphocyte repertoire. Although these types of comparisons can suffer from undersampling [27] and present significant statistical challenges [53], more conclusive answers may become available as additional repertoires are surveyed at greater depth.
Comparative analysis of HTS receptor data has also been used to study lymphocyte development and lineage commitment. Exploring long-standing questions about the stochastic nature of V(D)J recombination, Callan and colleagues exploited the large, high-resolution datasets produced by TCR HTS to generate a probabilistic model for the generation of CDR3 sequences [54]. Using genomic DNA sequences from nonproductive recombinants to exclude effects of selection, these analyses provide insight into the degree to which different molecular processes of TCR diversification contribute to repertoire composition, and will likely prove useful in future TCR HTS studies. Analysis of V(D)J recombinants can also be used to trace lymphocyte lineage history. For example, using a genomic DNA library construction and HTS strategy, Sherwood et al. observed that only 4% of γδT cells contain rearranged TCRβ loci, while all αβ T cells contain rearranged TCRγ loci [19]. These data support a model in which developing T lymphocytes commit to the αβ lineage only after diverging from the γδ program.
Additional comparative studies have explored how a variety of factors shape immune repertoires. Adaptive immune diversity and function are known to decline with age, but the mechanistic underpinnings of this deterioration are incompletely understood [55,56]. HTS methods enabling direct assessment and comparison of antigen receptor diversity have demonstrated diminished TCR diversity [40] and perturbed BCR CDR3 patterns [13] in aged versus young individuals. Moreover, HTS methods have been particularly useful in detecting pronounced differences in BCR hypermutation patterns and CDR3 lengths in elderly vaccine recipients [57–59]. A related analysis revealed significant B lymphocyte repertoire perturbations associated with CMV seropositivity [13]. These initial studies not only demonstrate the utility of comparing repertoires at the sequence level but also hint that similar tools might one day be useful in measuring immune function based on lymphocyte diversity. Indeed, several clinical studies have already demonstrated the utility of antigen receptor HTS for assessing diversity in lymphocyte repertoires derived from hematopoietic stem cell transplants [60–62].
Characterizing antigen-specific immune responses
In the steady state, BCR and TCR sequences in memory lymphocyte populations offer tractable targets for assessing the broad immune history of an individual by HTS. For example, BCR heavy [42], TCRα [21], and TCRβ [18,21] sequences in memory repertoires have been observed as more oligoclonal than their naïve counterparts.
Moving beyond steady-state measurements, HTS techniques are also capable of tracking lymphocytes that respond to a specific antigenic challenge at exquisite sensitivity and resolution. Recent studies are just beginning to apply HTS techniques to characterize immune responses to vaccination. Using BCR heavy chain HTS to evaluate responses to influenza vaccination, Quake and colleagues were able to characterize pre- and post-vaccination repertoires, quantify their diversity, and reconstruct clonal lineages based on SHM patterns [57]. Single cell plasmablast cloning experiments demonstrated that influenza-specific BCR heavy chain sequences were represented in some of the identified lineages. In another study profiling B cell responses to seasonal and H1N1 pandemic influenza vaccination, vaccine-induced clonal expansions were quantified from replicate genomic DNA BCR heavy chain HTS libraries [11]. The magnitude of B lymphocyte clonal expansion post-vaccination (day 7) was shown to correlate with serum antibody titers 2 weeks later (day 21) [11]. Furthermore, BCR heavy chain sequences expanded in response to H1N1 vaccination shared similar characteristics (including CDR3 properties) across several individuals, demonstrating apparent convergent immunological evolution [11]. HTS data have also proved useful in detecting and tracking B lymphocyte memory recall responses to repeated annual influenza vaccination [22]. These influenza studies exemplify a new level of detail for assessing vaccine response.
Antigen receptor HTS has also enabled the characterization of immune responses to infectious agents including HIV [7,63] and Dengue virus [14]. In the Dengue study, BCR sequence data were used to track expanding clones during acute primary and secondary infections in comparison to non-Dengue fever patients and healthy controls. Remarkably, convergent and apparently Dengue-specific similar CDR3 signatures were detectable in many individuals. Taken together with the observation of interindividual convergent response to H1N1 influenza vaccine described above, these results suggest that in the future, BCR HTS may have clinical utility in evaluating vaccination efficacy and/or immune function during ongoing infection. Outside the context of infectious disease, TCR HTS has demonstrated patterns suggestive of antigen-specific expansions among tumor-infiltrating lymphocytes for a variety of malignancies, including ovarian [64], clear cell renal [65], and colorectal [66] cancers.
Assessing immune dysfunction
Antigen receptor HTS strategies have considerable potential for characterizing lymphocyte repertoires in dysfunctional contexts, such as autoimmunity and lymphoid malignancies. From both basic research and clinical perspectives, examining antigen receptor sequences can serve multiple goals: (i) to identify potentially pathogenic lymphocyte clones, and (ii) to monitor such pathogenic clones prior to and following therapeutic intervention.
At present, these goals are best exemplified by the application of receptor HTS to diagnose and monitor lymphoid malignancies. As the product of unchecked proliferation originating from a single lymphoid cell, malignant cells can share a dominant V(D)J rearrangement and antigen receptor, or somatically hypermutated versions thereof [23]. Therefore, for many lymphoid malignancies, the clonal receptor sequence can be used as a characteristic biomarker for malignant cells and to monitor treatment response. Recently, several groups have demonstrated the efficacy of antigen receptor HTS to track minimal residual disease in a variety of lymphoid malignancies, including chronic lymphocytic leukemia [12,23,67], B lymphoblastic leukemia [68], T lymphoblastic leukemia [69], cutaneous T cell lymphoma [70], and others. Logan et al. reported superior specificity (greater than 99.9%) and sensitivity (1:100 000) as compared with traditional techniques [23]. Furthermore, HTS approaches have proved useful in characterizing molecular features of certain B lymphoblastic leukemias difficult to assess by traditional methods, such as ongoing intraclonal V(D)J recombination and heterogeneity [68,71]. Although further development, standardization, and validation are needed, antigen receptor HTS is fast becoming an important tool in clinical oncology.
Autoimmunity is characterized by the inappropriate recognition of, and response to, self antigens. In many autoimmune diseases pathogenesis is directly linked to specific autoantibodies. However, in other diseases the significance of a specific immune response to pathogenesis is unclear. In these cases, it may be that disease is triggered by the inappropriate activation of autoreactive lymphocytes normally present in the steady-state repertoire of susceptible individuals. Alternatively, autoimmune pathogenesis may be initiated by the generalized dysregulation of lymphocytes bearing irrelevant and/or polyreactive antigen receptors. Antigen receptor HTS offers a new approach with which to address these questions. Identifying potentially pathogenic antigen receptors may be challenging, as autoreactive lymphocyte clones may be relatively rare in the peripheral repertoire and/or sequestered in relatively inaccessible tissues. A recent HTS-based study identified oligoclonal BCR heavy chain sequences in the affected joints of multiple rheumatoid arthritis patients, suggestive of potential autoreactivity [72]. These clones were not detected in the peripheral blood of these patients; it remains unclear whether this is a consequence of complete tissue sequestration, undersampling, or both. Although additional reports describing HTS characterization of lymphocyte repertoires in autoimmunity have been somewhat limited, the approach shows great promise, and we anticipate that this will become a more active area of research as additional investigators incorporate antigen receptor HTS into clinical study designs.
Recent advances
‘Single chain’ sequencing techniques have been instrumental in broadening our understanding of the adaptive immune system and will continue to be extremely important experimental tools. However, they do not describe the combinatorial heterodimeric pairing of receptor subunits, which is an important determinant of both BCR [73] and TCR [74] antigen specificity. This pairing information is necessary for a comprehensive characterization of repertoire diversity and for downstream experiments that aim to produce complete antigen receptors and/or characterize their function. Thus, there has been considerable interest in exploiting the advantages of HTS in the analysis of paired antigen receptor sequences. One strategy using an abundance ranking algorithm to predict most likely pairing frequencies from single chain HTS in antigen-specific B cell expansions has shown efficacy in identifying high affinity antibodies [75], but this approach is limited to samples pre-enriched for a particular specificity.
Broadly applicable paired analysis essentially requires HTS at single lymphocyte resolution for large cell numbers. Although they vary in implementation, a series of recently developed methodologies has been successful in ‘linking’ the sequence information of paired chains from individual lymphocytes, either molecularly or bioinformatically. DeKosky et al. used specially designed microwell arrays to segregate B lymphocytes during lysis and bead-based RNA capture [76]. Following bead encapsulation in emulsion droplets, reverse transcription proceeds separately for each bead, and subsequent PCR is used to assemble a heavy chain–light chain fusion amplicon. This fusion is then sequenced as a single unit. A related strategy for TCRs, in which assembly PCR couples TCRα and TCRβ sequences of droplet-encapsulated T lymphocytes, was recently described [77]. Wardemann and colleagues, taking advantage of HTS readouts, developed a sequence barcode indexing strategy to link BCR heavy and light chain sequence data informatically [78]. After single cells are sorted to individual wells, RT-PCR amplification of heavy and light chain transcripts is performed using primers appended with well-specific barcode sequences. HTS output from pooled amplicons contains BCR heavy chain, BCR light chain, and associated barcode sequences, which are used to connect data to the initial well/lymphocyte. These first examples of paired antigen receptor sequencing have shown tremendous promise, but remain technically demanding and process far fewer cells than single chain techniques. We anticipate that additional improvements such as increased throughput and integrated, single-lymphocyte functional analyses will enable characterization of adaptive immunity at even greater detail.
Concluding remarks, future directions and challenges
While antigen receptor HTS techniques continue to advance at a rapid pace, numerous technical, scientific, and clinical problems remain to be addressed. In the technical category, additional, broadly applicable strategies for managing errors in antigen receptor sequences are needed. In addition, standardization of techniques and data analysis will be important for sharing and comparing results across experiments and between laboratories [79]. Furthermore, emerging HTS technologies may offer opportunities to circumvent some of the current difficulties in library preparation. Aside from these technical improvements, new methods for augmenting antigen receptor HTS datasets with phenotypic gene expression data would be an important advance. This might be achieved by integrating flow cytometry approaches with single-cell resolution library preparations [78], or more directly by simultaneously assessing transcript expression and receptor sequences with HTS. Indeed, a recent report described a medium throughput, single-cell RT-PCR approach, in which TCR transcripts and a panel of lymphocyte phenotypic marker transcripts are amplified and barcoded by microplate well, with each well containing a single lymphocyte [80]. Following HTS, these ‘cellular barcodes’ are used to link expression of phenotypic markers to corresponding TCR sequences. Improving the throughput and sensitivity of these and similar methods will be necessary to assess functionally larger portions of the lymphocyte repertoire.
Clinical applications for antigen receptor HTS are likely to increase in the coming years. For some applications, such as monitoring lymphoid malignancies and hematopoietic transplants, progress has been rapid and additional data and methodological validation may soon establish these techniques as widely used tools in oncology. Autoimmune disease represents a key area for future research with antigen receptor HTS; comparative repertoire analyses are expected to be useful in better understanding pathogenesis, and perhaps also in characterizing clinical isolates. Lastly, as more repertoire data become available, it may be possible to begin establishing the parameters that define a ‘normal’ or ‘healthy’ immune repertoire; such metrics would have research and clinical utility.
One of the most demanding research challenges for high throughput immune repertoire analysis is effectively linking large collections of BCR and TCR sequences to antigen specificities. Emerging methodologies integrating BCR HTS with mass spectrometry have demonstrated feasibility in identifying antibody sequences with specificities to antigens of interest [81–83]. Conversely, recently described peptide–MHC screening strategies can be used to identify antigenic peptides recognized by TCRs of interest [84,85]. Additional development of these and related techniques, along with corresponding computational methods, is needed for the high throughput integration of sequence data, antigen specificity, and immune function. These and other advances will help ensure that antigen receptor HTS continues to contribute to important advances in our understanding of adaptive immunity.
Acknowledgments
We thank F.N. Papavasiliou and G. Livshits for helpful discussions, comments, and critical reading of the manuscript. This work was supported by the Netherlands Organization for Scientific Research (NWO Rubicon, J.J.A.C.), the John C. Whitehead Presidential Fellowship (B.R.R.), and the National Institutes of Health under award number DP5OD012142 (B.R.R.).
References
- 1.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 2.Murphy K. Janeway’s Immunobiology. Garland Science; 2012. [Google Scholar]
- 3.Teng G, Papavasiliou FN. Immunoglobulin somatic hypermutation. Annu. Rev. Genet. 2007;41:107–120. doi: 10.1146/annurev.genet.41.110306.130340. [DOI] [PubMed] [Google Scholar]
- 4.Matthews AJ, et al. Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair. Adv. Immunol. 2014;122:1–57. doi: 10.1016/B978-0-12-800267-4.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arstila TP, et al. A direct estimate of the human T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 6.Six A, et al. The past, present, and future of immune repertoire biology - the rise of next-generation repertoire analysis. Front. Immunol. 2013;4:413. doi: 10.3389/fimmu.2013.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheid JF, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- 9.Mouquet H, et al. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS ONE. 2011;6:e24078. doi: 10.1371/journal.pone.0024078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S, Wilkinson M. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/s1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- 11.Jackson KJL, et al. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe. 2014;16:105–114. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyd SD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel V-D-J pyrosequencing. Sci. Transl. Med. 2009;1:1–8. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, et al. Effects of aging, cytomegalovirus infection, and EBV infection on human B cell repertoires. J. Immunol. 2014;192:603–611. doi: 10.4049/jimmunol.1301384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parameswaran P, et al. Convergent antibody signatures in human dengue. Cell Host Microbe. 2013;13:691–700. doi: 10.1016/j.chom.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jack H, Wabi M. mRNA stability varies during B lymphocyte differentiation. EMBO J. 1988;7:1041–1046. doi: 10.1002/j.1460-2075.1988.tb02911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paillard F, et al. Lymphokine mRNA and T cell multireceptor mRNA of the Ig super gene family are reciprocally modulated during human T cell activation. Eur. J. Immunol. 1988;18:1643–1646. doi: 10.1002/eji.1830181028. [DOI] [PubMed] [Google Scholar]
- 17.Van Dongen JJM, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 18.Robins HS, et al. Comprehensive assessment of T-cell receptor β-chain diversity in αβ T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherwood AM, et al. Deep sequencing of the human TCRγ and TCRβ repertoires suggests that TCRβ rearranges after αβ and γδ T cell commitment. Sci. Transl. Med. 2011;3:1–7. doi: 10.1126/scitranslmed.3002536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larimore K, et al. Shaping of human germline IgH repertoires revealed by deep sequencing. J. Immunol. 2012;189:3221–3230. doi: 10.4049/jimmunol.1201303. [DOI] [PubMed] [Google Scholar]
- 21.Wang C, et al. High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proc. Natl. Acad. Sci. U.S.A. 2010;107:1518–1523. doi: 10.1073/pnas.0913939107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vollmers C, et al. Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13463–13468. doi: 10.1073/pnas.1312146110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Logan AC, et al. High-throughput VDJ sequencing for quantification of minimal residual disease in chronic lymphocytic leukemia and immune reconstitution assessment. Proc. Natl. Acad. Sci. U.S.A. 2011;108:21194–21199. doi: 10.1073/pnas.1118357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiller T, et al. Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods. 2009;350:183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Carlson CS, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat. Commun. 2013;4:1–9. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- 26.Freeman JD, et al. Profiling the T-cell receptor β-chain repertoire by massively parallel sequencing. Genome Res. 2009;19:1817–1824. doi: 10.1101/gr.092924.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warren RL, et al. Exhaustive T-cell repertoire sequencing of human peripheral blood samples reveals signatures of antigen selection and a directly measured repertoire size of at least 1 million clonotypes. Genome Res. 2011;21:790–797. doi: 10.1101/gr.115428.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mamedov IZ, et al. Preparing unbiased T-cell receptor and antibody cDNA libraries for the deep next generation sequencing profiling. Front. Immunol. 2013;4:1–10. doi: 10.3389/fimmu.2013.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee E-C, et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nat. Biotechnol. 2014;32:356–363. doi: 10.1038/nbt.2825. [DOI] [PubMed] [Google Scholar]
- 30.Matz M, et al. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558–1560. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Douek DC, et al. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 2002;168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- 32.Bolotin D, et al. Next generation sequencing for TCR repertoire profiling: platform-specific features and correction algorithms. Eur. J. Immunol. 2012;42:3073–3083. doi: 10.1002/eji.201242517. [DOI] [PubMed] [Google Scholar]
- 33.Metzker ML. Sequencing technologies - the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 34.Bragg LM, et al. Shining a light on dark sequencing: characterising errors in Ion Torrent PGM data. PLoS Comput. Biol. 2013;9:e1003031. doi: 10.1371/journal.pcbi.1003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carneiro MO, et al. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics. 2012;13:1–7. doi: 10.1186/1471-2164-13-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loman NJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 37.McGinn S, Gut IG. DNA sequencing – spanning the generations. N. Biotechnol. 2013;30:366–372. doi: 10.1016/j.nbt.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 38.Vander Heiden JA, et al. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014;30:1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shugay M, et al. Towards error-free profiling of immune repertoires. Nat. Methods. 2014;11:653–655. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 40.Britanova OV, et al. Age-related decrease in TCR repertoire diversity measured with deep and normalized sequence profiling. J. Immunol. 2014;192:2689–2698. doi: 10.4049/jimmunol.1302064. [DOI] [PubMed] [Google Scholar]
- 41.Weinstein J, et al. High-throughput sequencing of the zebrafish antibody repertoire. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Briney BS, et al. High-throughput antibody sequencing reveals genetic evidence of global regulation of the naïve and memory repertoires that extends across individuals. Genes Immun. 2012;13:469–473. doi: 10.1038/gene.2012.20. [DOI] [PubMed] [Google Scholar]
- 43.Robins HS, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci. Transl. Med. 2010;2:1–9. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Putintseva EV, et al. Mother and child T cell receptor repertoires: deep profiling study. Front. Immunol. 2013;4:1–13. doi: 10.3389/fimmu.2013.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farber DL, et al. Human memory T cells: generation, compartmentalization and homeostasis. Nat. Rev. Immunol. 2014;14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Briney BS, et al. Tissue-specific expressed antibody variable gene repertoires. PLoS ONE. 2014;9:e100839. doi: 10.1371/journal.pone.0100839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyd SD, et al. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J. Immunol. 2010;184:6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glanville J, et al. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:20066–20071. doi: 10.1073/pnas.1107498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnaout R, et al. High-resolution description of antibody heavy-chain repertoires in humans. PLoS ONE. 2011;6:e22365. doi: 10.1371/journal.pone.0022365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zvyagin IV, et al. Distinctive properties of identical twins’ TCR repertoires revealed by high-throughput sequencing. Proc. Natl. Acad. Sci. U.S.A. 2014;111:5980–5985. doi: 10.1073/pnas.1319389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shugay M, et al. Huge overlap of individual TCRβ repertoires. Front. Immunol. 2013;4:1–3. doi: 10.3389/fimmu.2013.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venturi V, et al. A mechanism for TCR sharing between T cell subsets and individuals revealed by pyrosequencing. J. Immunol. 2011;186:4285–4294. doi: 10.4049/jimmunol.1003898. [DOI] [PubMed] [Google Scholar]
- 53.Mehr R, et al. Models and methods for analysis of lymphocyte repertoire generation, development, selection and evolution. Immunol. Lett. 2012;148:11–22. doi: 10.1016/j.imlet.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 54.Murugan A, et al. Statistical inference of the generation probability of T-cell receptors from sequence repertoires. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16161–16166. doi: 10.1073/pnas.1212755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boraschi D, et al. The gracefully aging immune system. Sci. Transl. Med. 2013;5:1–9. doi: 10.1126/scitranslmed.3005624. [DOI] [PubMed] [Google Scholar]
- 56.Boyd SD. Diagnostic applications of high-throughput DNA sequencing. Annu. Rev. Pathol. 2013;8:381–410. doi: 10.1146/annurev-pathol-020712-164026. [DOI] [PubMed] [Google Scholar]
- 57.Jiang N, et al. Lineage structure of the human antibody repertoire in response to influenza vaccination. Sci. Transl. Med. 2013;5:1–9. doi: 10.1126/scitranslmed.3004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Y-CB, et al. Age-related changes in human peripheral blood IGH repertoire following vaccination. Front. Immunol. 2012;3:1–12. doi: 10.3389/fimmu.2012.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ademokun A, et al. Vaccination-induced changes in human B-cell repertoire and pneumococcal IgM and IgA antibody at different ages. Aging Cell. 2011;10:922–930. doi: 10.1111/j.1474-9726.2011.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Heijst JWJ, et al. Quantitative assessment of T cell repertoire recovery after hematopoietic stem cell transplantation. Nat. Med. 2013;19:372–377. doi: 10.1038/nm.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muraro PA, et al. T cell repertoire following autologous stem cell transplantation for multiple sclerosis. J. Clin. Invest. 2014;124:1168–1172. doi: 10.1172/JCI71691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mamedov IZ, et al. Quantitative tracking of T cell clones after haematopoietic stem cell transplantation. EMBO Mol. Med. 2011;3:201–207. doi: 10.1002/emmm.201100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao H-X, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J. Exp. Med. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Emerson RO, et al. High-throughput sequencing of T-cell receptors reveals a homogeneous repertoire of tumour-infiltrating lymphocytes in ovarian cancer. J. Pathol. 2013;231:433–440. doi: 10.1002/path.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerlinger M, et al. Ultra-deep T cell receptor sequencing reveals the complexity and intratumour heterogeneity of T cell clones in renal cell carcinomas. J. Pathol. 2013;231:424–432. doi: 10.1002/path.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sherwood AM, et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol. Immunother. 2013;62:1453–1461. doi: 10.1007/s00262-013-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grupp S, et al. Chimeric antigen receptor-modified T cells for acute lymphoid leukemia. N. Engl. J. Med. 2013;368:1509–1518. doi: 10.1056/NEJMoa1215134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu D, et al. Detection of minimal residual disease in patients with B lymphoblastic leukemia by high-throughput sequencing of IGH. Clin. Cancer Res. 2014;20:4540–4548. doi: 10.1158/1078-0432.CCR-13-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu D, et al. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci. Transl. Med. 2012;4:1–7. doi: 10.1126/scitranslmed.3003656. [DOI] [PubMed] [Google Scholar]
- 70.Weng W-K, et al. Minimal residual disease monitoring with high-throughput sequencing of T cell receptors in cutaneous T cell lymphoma. Sci. Transl. Med. 2013;5:1–9. doi: 10.1126/scitranslmed.3007420. [DOI] [PubMed] [Google Scholar]
- 71.Gawad C, et al. Massive evolution of the immunoglobulin heavy chain locus in children with B precursor acute lymphoblastic leukemia. Blood. 2012;120:4407–4417. doi: 10.1182/blood-2012-05-429811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doorenspleet ME, et al. Rheumatoid arthritis synovial tissue harbours dominant B-cell and plasma-cell clones associated with autoreactivity. Ann. Rheum. Dis. 2014;73:756–762. doi: 10.1136/annrheumdis-2012-202861. [DOI] [PubMed] [Google Scholar]
- 73.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 74.Jorgensen J, et al. Mapping T-cell receptor-peptide contacts by variant peptide immunization of single-chain transgenics. Nature. 1992;355:224–230. doi: 10.1038/355224a0. [DOI] [PubMed] [Google Scholar]
- 75.Reddy ST, et al. Monoclonal antibodies isolated without screening by analyzing the variable-gene repertoire of plasma cells. Nat. Biotechnol. 2010;28:965–969. doi: 10.1038/nbt.1673. [DOI] [PubMed] [Google Scholar]
- 76.DeKosky BJ, et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat. Biotechnol. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Turchaninova M, et al. Pairing of T-cell receptor chains via emulsion PCR. Eur. J. Immunol. 2013;43:2507–2515. doi: 10.1002/eji.201343453. [DOI] [PubMed] [Google Scholar]
- 78.Busse CE, et al. Single-cell based high-throughput sequencing of full-length immunoglobulin heavy and light chain genes. Eur. J. Immunol. 2014;44:597–603. doi: 10.1002/eji.201343917. [DOI] [PubMed] [Google Scholar]
- 79.Georgiou G, et al. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han A, et al. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cheung WC, et al. A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat. Biotechnol. 2012;30:447–452. doi: 10.1038/nbt.2167. [DOI] [PubMed] [Google Scholar]
- 82.Sato S, et al. Proteomics-directed cloning of circulating antiviral human monoclonal antibodies. Nat. Biotechnol. 2012;30:1039–1043. doi: 10.1038/nbt.2406. [DOI] [PubMed] [Google Scholar]
- 83.Lavinder JJ, et al. Identification and characterization of the constituent human serum antibodies elicited by vaccination. Proc. Natl. Acad. Sci. U.S.A. 2014;111:2259–2264. doi: 10.1073/pnas.1317793111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Birnbaum ME, et al. Deconstructing the peptide–MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pan X, et al. Combinatorial HLA–peptide bead libraries for high throughput identification of CD8+ T cell specificity. J. Immunol. Methods. 2013;403:72–78. doi: 10.1016/j.jim.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 86.Ye J, et al. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alamyar E, et al. IMGT/HIGHV-QUEST: the IMGT® web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis for NGS high throughput and deep sequencing. Immunome Res. 2012;8:1–15. [Google Scholar]
- 88.Brochet X, et al. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gaëta B, et al. iHMMune-align: hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. 2007;23:1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 90.Munshaw S, Kepler TB. SoDA2: a hidden Markov model approach for identification of immunoglobulin rearrangements. Bioinformatics. 2010;26:867–872. doi: 10.1093/bioinformatics/btq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohm-Laursen L, et al. No evidence for the use of DIR, D-D fusions, chromosome 15 open reading frames or VH replacement in the peripheral repertoire was found on application of an improved algorithm, JointML, to 6329 human immunoglobulin H rearrangements. Immunology. 2006;119:265–277. doi: 10.1111/j.1365-2567.2006.02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X, et al. Ab-origin: an enhanced tool to identify the sourcing gene segments in germline for rearranged antibodies. BMC Bioinformatics. 2008;9:1–9. doi: 10.1186/1471-2105-9-S12-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Souto-Carneiro MM, et al. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J. Immunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 94.Retter I, et al. VBASE2, an integrative V gene database. Nucleic Acids Res. 2005;33:D671–D674. doi: 10.1093/nar/gki088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bolotin D, et al. MiTCR: software for T-cell receptor sequencing data analysis. Nat. Methods. 2013;10:813–814. doi: 10.1038/nmeth.2555. [DOI] [PubMed] [Google Scholar]
- 96.Venturi V, et al. Methods for comparing the diversity of samples of the T cell receptor repertoire. J. Immunol. Methods. 2007;321:182–195. doi: 10.1016/j.jim.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 97.Chen Z, et al. Clustering-based identification of clonally-related immunoglobulin gene sequence sets. Immunome Res. 2010;6:1–7. doi: 10.1186/1745-7580-6-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kleinstein SH, et al. Estimating hypermutation rates from clonal tree data. J. Immunol. 2003;171:4639–4649. doi: 10.4049/jimmunol.171.9.4639. [DOI] [PubMed] [Google Scholar]
- 99.Barak M, et al. IgTree: creating immunoglobulin variable region gene lineage trees. J. Immunol. Methods. 2008;338:67–74. doi: 10.1016/j.jim.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 100.Sok D, et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog. 2013;9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yaari G, et al. Quantifying selection in high-throughput immunoglobulin sequencing data sets. Nucleic Acids Res. 2012;40:e134. doi: 10.1093/nar/gks457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Anderson SM, et al. Taking advantage: high-affinity B cells in the germinal center have lower death rates, but similar rates of division, compared to low-affinity cells. J. Immunol. 2009;183:7314–7325. doi: 10.4049/jimmunol.0902452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Uduman M, et al. Integrating B cell lineage information into statistical tests for detecting selection in Ig sequences. J. Immunol. 2014;192:867–874. doi: 10.4049/jimmunol.1301551. [DOI] [PMC free article] [PubMed] [Google Scholar]