Abstract
Understanding how eukaryotic enhancers are bound and regulated by specific combinations of transcription factors is still a major challenge. To better map transcription factor binding genome-wide at nucleotide resolution in vivo, we have developed a robust ChIP-exo protocol called ChIP experiments with nucleotide resolution through exonuclease, unique barcode and single ligation (ChIP-nexus), which utilizes an efficient DNA self-circularization step during library preparation. Application of ChIP-nexus to four proteins—human TBP and Drosophila NFkB, Twist and Max— demonstrates that it outperforms existing ChIP protocols in resolution and specificity, pinpoints relevant binding sites within enhancers containing multiple binding motifs and allows the analysis of in vivo binding specificities. Notably, we show that Max frequently interacts with DNA sequences next to its motif, and that this binding pattern correlates with local DNA sequence features such as DNA shape. ChIP-nexus will be broadly applicable to studying in vivo transcription factor binding specificity and its relationship to cis-regulatory changes in humans and model organisms.
The ability to precisely map transcription factor binding footprints in vivo at a single-nucleotide resolution is essential to understand the mechanisms of combinatorial control by transcription factors 1. The occupancy of specific transcription factors can be mapped by chromatin immunoprecipitation (ChIP) coupled to deep sequencing (ChIP-seq), but the resolution of this technique is limited by the minimal DNA fragment size required for unique alignment to the genome (e.g. see Bardet et al. 2). In an improvement to ChIP-seq called ChIP-exo, the immunoprecipitated chromatin fragments are treated with lambda exonuclease, which digests one strand of the double-stranded DNA in a 5′-to-3′ direction and stops when it encounters a cross-linked protein 3, 4. In this manner the exact bases bordering a DNA-bound protein (the ‘stop bases’) can be mapped at essentially nucleotide resolution, enabling new biological insights 3, 5, 6. However, we found significant technical hurdles in applying ChIP-exo. The additional wash and digestion steps reduce the amount of DNA that can be recovered compared to conventional ChIP-seq experiments, which is critical for the quality of a ChIP library. For amplification during library preparation, DNA fragments must complete two inefficient ligation steps to acquire adapters on both ends. Low amounts of starting DNA often lead to over-amplification artifacts during PCR, producing noisy data that are not reproducible 7, 8. Another hurdle is that the original ChIP-exo protocol is designed for the SOLiD platform, although Illumina versions have recently become available 9, 10.
Here we describe a more robust and reproducible Illumina-based ChIP-exo protocol. As lambda exonuclease digestion of ChIP DNA mostly yields single-stranded DNA and requires retention of strand information, we combined the standard ChIP-exo protocol with the library preparation protocol from the iCLIP method for mapping RNA-protein interactions 11 to improve the efficiency by which DNA fragments are incorporated into the library. In addition, we added a unique randomized barcode to the adapter, which enables monitoring of over-amplification 7, 8. This combined protocol, called ChIP-nexus, is more efficient because it requires only one successful ligation per DNA fragment. Although ChIP-nexus adapters were designed to be ligated to both DNA ends as in conventional ChIP-seq and ChIP-exo protocols, a library product will still be generated even if the adapter is only ligated to one end. This is because lambda exonuclease digests the 5′ end of each strand independently of whether an adapter is present, thus a single ChIP-nexus adapter on the 3′ end is sufficient. The fragment is then circularized, which brings Illumina library primers to the digested end. Because intramolecular circularization is far more efficient than intermolecular ligation, library generation is more efficient compared to a classical library preparation protocol where two independent ligations are required to generate a library product. As a result, ChIP-nexus produces high-quality libraries without requiring more starting material than conventional ChIP-seq experiments. The protocol is outlined in Figure 1a and in the Online Methods. A detailed protocol is available as Supplementary Protocol 1 or from our web page (http://research.stowers.org/zeitlingerlab).
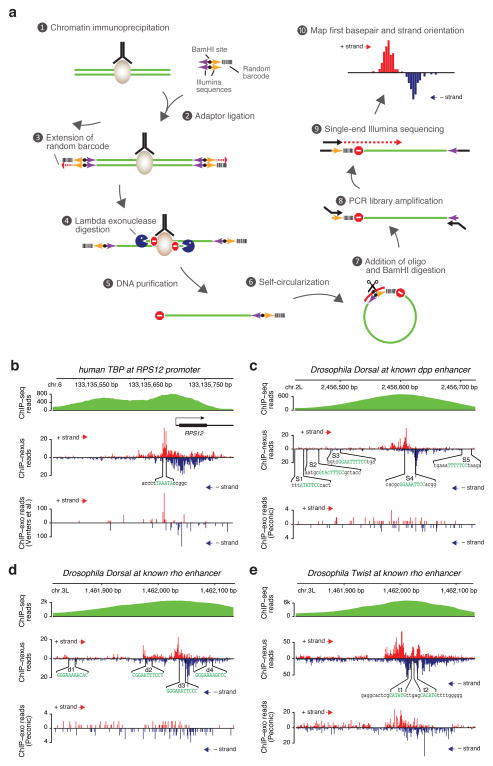
Figure 1. Superior performance of ChIP-nexus in discovering relevant binding footprints for transcription factors.
(a) Outline of ChIP-nexus 1) The transcription factor of interest (brown) is immunoprecipitated from chromatin fragments with antibodies in the same way as during conventional ChIP-seq experiments. 2) While still bound to the antibodies, the DNA ends are repaired, dA-tailed and then ligated to a special adaptor that contains a pair of sequences for library amplification (arrows indicate the correct orientation for them to be functional), a BamHI site (black dot) for linearization, and a 9-nucleotide barcode containing 5 random bases and 4 fixed bases to remove reads resulting from over-amplification of library DNA. The barcode is part of a 5′ overhang, which reduces adaptor-adaptor ligation. 3) After the adaptor ligation step, the 5′ overhang is filled, copying the random barcode and generating blunt ends for lambda exonuclease digestion. 4) Lambda exonuclease (blue Pacman) digests until it encounters a physical barrier such as a cross-linked protein-DNA complex (‘Do not enter’ sign = ‘stop base’). 5) Single-stranded DNA is eluted and purified. 6) Self-circularization places the barcode next to the ‘stop base’. 7) An oligonucleotide (red arc) is paired with the region around the BamHI site for BamHI digestion (black scissors). 8) The digestion results in re-linearized DNA fragments with suitable Illumina sequences on both ends, ready for PCR library amplification. 9) Using single-end sequencing with the standard Illumina primer, each fragment is sequenced: first the barcode, then the genomic sequence starting with the ‘stop base’. 10) After alignment of the genomic sequences, reads with identical start positions and identical barcodes are removed. The final output is the position, number and strand orientation of the ‘stop’ bases. The frequencies of ‘stop’ bases on the positive strand are shown in red, while those on the negative strand are shown in blue. (b–e) Comparison of conventional ChIP-seq data (extended reads), ChIP-nexus data (raw stop base reads) and data generated using the original ChIP-exo protocol (raw stop base reads). (b) TBP profiles in human K562 cells at the RPS12 promoter. Although ChIP-nexus and ChIP-exo generally agree on TBP binding footprints, ChIP-nexus provides better coverage and richer details than ChIP-exo, which shows signs of over-amplification as large numbers of reads accumulate at a few discreet bases. (c) Dorsal profiles at the D. melanogaster decapentaplegic (dpp) enhancer. Five “Strong” dorsal binding sites (S1–S5) were previously mapped by in vitro DNase footprinting 12. Note that ChIP-nexus identifies S4 as the only site with significant Dorsal binding in vivo. At the same time, ChIP-exo performed by Peconic did not detect any clear Dorsal footprint within the enhancer, in part due to the low read counts obtained. (d) Dorsal profiles at the rhomboid (rho) NEE enhancer. Four Dorsal binding sites (d1–d4) were previously mapped by in vitro DNase footprinting 14. Note that ChIP-nexus identifies d3 as the strongest dorsal binding site in vivo, consistent with its close proximity to two Twist binding sites. Again, the original ChIP-exo protocol did not detect any clear Dorsal footprint within the enhancer. (e) Twist profiles at the same rho enhancer. Note that ChIP-nexus shows strong Twist footprints surrounding the two Twist binding sites (t1, t2) 14. In this case, ChIP-exo performed by Peconic identified a similar Twist footprint. This shows that the Peconic experiments, which were performed with the same chromatin extracts as the Dorsal experiments, worked in principle but were less robust than our ChIP-nexus experiments.
We compared the results from the ChIP-nexus protocol to published results on human TBP obtained with the original ChIP-exo protocol adapted to the Illumina sequencing platform9, Our ChIP-nexus experiments were performed using the same number of K562 cells and the same TBP antibody as in the previous study and the locations of the stop bases on each strand were plotted. As exemplified by the RPS12 locus9, ChIP-nexus produced visibly better results (Fig. 1b). When the previous ChIP-exo data were plotted in the same way, they show signs of over-amplification, i.e. the reads often occur in extremely high numbers at the same position without reads detected at neighboring positions. In contrast, ChIP-nexus produces a signal across the entire promoter region in a pattern that can only be observed with regular ChIP-exo data after averaging across many genes. Thus, while the overall readout is comparable to the original ChIP-exo protocol, ChIP-nexus produces higher quality data that can be analyzed at the single-gene level.
Next, we studied transcription factors in the early Drosophila embryo, where many well-characterized enhancers allow us to assess the performance of ChIP-nexus compared to other techniques. One of the best-studied transcriptional regulatory networks is dorso-ventral patterning, which is controlled by an activity gradient of Dorsal, the homologue of the vertebrate transcription factor NFκB. One well-characterized enhancer is located in an intron of decapentaplegic (dpp) and is ventrally repressed by Dorsal 12. Based on in vitro footprinting, Dorsal binds to multiple binding sites in the enhancer but simultaneous mutation of two specific sites (S3 and S4) almost completely abolished ventral repression 12. Our ChIP-nexus data showed a clear footprint of Dorsal at the previously mapped S4 binding site, but not at S3 or other previously mapped Dorsal sites (Fig. 1c), suggesting that S4 is the most critical site for dpp repression. We also note that the boundaries of the ChIP-nexus footprint are similar to the DNase footprints in vitro 12, extending beyond the NFkB consensus motif by a similar number of nucleotides.
To further test whether ChIP-nexus footprints are preferentially found at critical binding sites, we also analyzed Dorsal interactions at an extensively characterized rhomboid (rho) enhancer, which drives expression in the neurectoderm (rho NEE) 13–15. In vitro footprinting revealed four Dorsal sites in the rho NEE enhancer (d1–d4), and simultaneous mutation of d2, d3, and d4 almost completely abolishes the enhancer activity14. ChIP-nexus showed a strong Dorsal footprint directly over the d3 binding site, while weaker footprints were found at the other Dorsal binding sites (Fig. 1d). Indeed, d3 is likely to be the most important Dorsal binding site due to its proximity to two E-box motifs 13, 16. Both E-boxes can be bound by the basic helix-loop-helix (bHLH) transcription factor Twist in vitro 14 and are important for enhancer activity in vivo 14, 17. We therefore tested whether ChIP-nexus with Twist would identify these two binding sites. Indeed, prominent ChIP-nexus footprints of Twist were found exactly over the two known binding sites next to the d3 Dorsal site (Fig. 1e).
To compare these results to ChIP-exo, we had Peconic LLC perform ChIP-exo experiments for Dorsal and Twist using the original ChIP-exo protocol. Although both experiments were performed in biological replicates from the same chromatin extracts, Twist showed a footprint at the rho NEE enhancer, while Dorsal did not show footprints at known target sites (Fig. 1c–e). The reduced quality of the Dorsal experiment is in part because of the lower read number obtained (Fig. 1c, d). But even the Twist ChIP-exo experiment, which has comparable read counts to our ChIP-nexus data, shows a less precise footprint (Fig. 1e, Supplementary Fig. 1), supporting our conclusion that ChIP-nexus produces better results at the single-gene level.
Taken together, the strong concordance between ChIP-nexus binding and previously characterized sites suggests that ChIP-nexus is an effective approach that can pinpoint critical binding sites within an enhancer. The analyses of Dorsal also suggest that its in vivo binding sites may differ from those bound in vitro, consistent with studies on other transcription factors 18, 19.
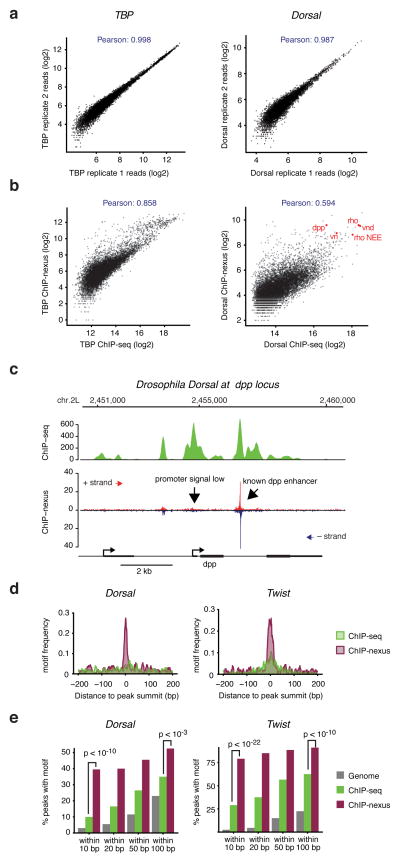
To test the robustness of the ChIP-nexus protocol, we analyzed the correlation between replicates at bound regions. Although peak finding algorithms are not designed for ChIP-nexus data 2, 3, 9, we found that MACS 20 (version 2) and Peakzilla 2 identified thousands of binding peaks in all cases. Using a maximum of 10,000 peaks, the ChIP-nexus reads highly correlated between replicates (Fig. 2a, Pearson correlations for TBP 0.998, Dorsal 0.986, Twist 0.993), showing that our ChIP-nexus data are highly reproducible.
Figure 2. High reproducibility, resolution and specificity of ChIP-nexus as compared to ChIP-seq.
(a) Comparisons between biological ChIP-nexus replicates were performed by calling peaks using MACS 2 20 in replicate 1 (200 bp centered on the peak summit, up to 10,000 peaks as arbitrary cutoff) and by plotting the average raw reads for each peak in both replicates. A tight line is observed for all factors, corresponding to Pearson correlations of 0.98–0.99. TBP, which has the highest correlation, is shown on the left, whereas Dorsal, which has the lowest correlation, is shown on the right. (b) Comparison between ChIP-seq and ChIP-nexus. Peaks were called in the ChIP-seq data as in (a) and reads in these peaks from ChIP-seq and ChIP-nexus data are shown as a scatter plot. As can be seen for both TBP and Twist, there is an overall good correlation between the bulk data (Pearson correlations between 0.5–0.9). However, the ChIP-nexus data show an increased signal for a fraction of peaks. (c) Examination of individual examples shows that the ChIP-nexus signal is indeed highly specific. For example, the known dpp enhancer as shown in Figure 2 has a strong ChIP-nexus footprint (arrow), whereas the signal at the dpp promoter, which is equally high in the ChIP-seq data, has much lower and more distributed ChIP-nexus reads without any typical footprint (arrow). (d) Frequency distribution of consensus motifs in peaks identified by ChIP-seq (green) and ChIP-nexus (purple). Shown are the examples of Dorsal (left), for which ChIP-nexus shows a dramatic increase in motifs directly at the summit of the peaks, as well as for Twist (right), for which ChIP-nexus shows a more moderate improvement in motif frequency over ChIP-seq. (e) Quantification of the motif frequency in random genomic regions, in ChIP-seq peaks and in ChIP-nexus peaks within increasing windows from the peaks’ summits for Dorsal and Twist. ChIP-nexus performs much better at a close interval to the peak summit (within 10 bp on either side, Chi2 test, Dorsal p<10−11, Twist p<10−14), underscoring the increased specificity of ChIP-nexus. But even at wider intervals (within 100 bp on either side of the summit), ChIP-nexus peaks contain more motifs (Chi2 test, Dorsal p<2×10−3, Twist p<10−5), suggesting that ChIP-nexus has higher specificity as compared to ChIP-seq.
We next analyzed the relationship between ChIP-nexus and ChIP-seq signal. The Pearson correlation of the reads was lower than between replicates but still very high (Fig. 2b, TBP 0.85, Dorsal 0.59). Scatterplots confirm that the bulk signal is similar between ChIP-nexus and ChIP-seq signal but that many bound regions have higher signal in the ChIP-nexus data (Fig. 2b). Regions with higher ChIP-nexus/ChIP-seq ratio include many known Dorsal enhancers (e.g. rho NEE, dpp, zen, vnd, vn), while regions with lower ChIP-nexus/ChIP-seq signal often lack a specific footprint, indicating that they may be enriched through unspecific binding to open chromatin. For instance, the dpp promoter shows high Dorsal ChIP-seq enrichments comparable to the known dpp enhancer, but has no specific footprint in the ChIP-nexus data (Fig. 2c).
To test more systematically whether ChIP-nexus indeed has increased specificity and resolution compared to ChIP-seq, we analyzed the presence and location of consensus binding motifs within peaks (Fig. 2c, d). Among the top 200 Dorsal and Twist ChIP-nexus binding peaks, the corresponding consensus motif was found directly at the center of the ChIP-nexus binding peaks much more frequently than at the ChIP-seq binding peaks (Fig. 2d), underscoring the increased resolution. Indeed, within 10 bp of the peak summit, there was a significant improvement in motif enrichment in the ChIP-nexus data compared to the ChIP-seq data (Chi2 test, Dorsal p<10−10, Twist p<10−22, Fig. 2e). Yet even at 100 bp from the summit, ChIP-nexus still had significantly higher motif enrichment than ChIP-seq (Chi2 test, Dorsal p<10−3, Twist p<10−10, Fig. 3e), supporting the notion that ChIP-nexus not only has improved resolution but also improved specificity.
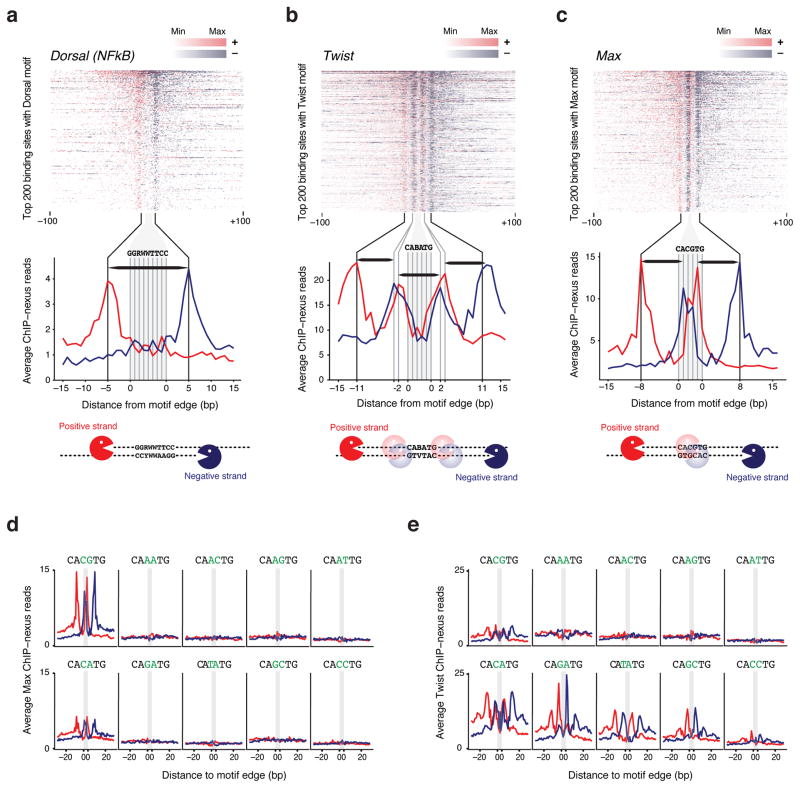
Figure 3. Analysis of the Dorsal, Twist and Max in vivo footprint.
(a–c) For each factor, the top 200 motifs with the highest ChIP-nexus read counts were selected and are shown in descending order as heat map. The footprints show a consistent boundary on the positive strand (red) and negative strand (blue) around each motif. The zoomed-in average profile below reveals that the footprints are wider than the motif. A schematic representation of the digestion pattern is shown below using Pacman symbols for lambda exonuclease. (a) The ChIP-nexus footprint for Dorsal (NFkB) on its canonical motif (GGRWWTTCC with up to one mismatch) extends on average 5 bp away from the motif edge. Thus, the average dorsal footprint is 18 bp long (horizontal black bar). (b) The Twist ChIP-nexus footprint on the E-box motif CABATG (no mismatch) has two outside boundaries, one at 11 bp, and one at 2 bp away from the motif edge, suggesting interactions with flanking DNA sequences. Each portion of the footprint is around 8–9bp long (horizontal black bar). (c) The Max ChIP-nexus footprint on its canonical E-box motif (CACGTG, no mismatch) has an outside boundary at 8 bp away from the motif edge, as well as a boundary inside the motif (at the A/T base), suggesting two partial footprints (horizontal black bars). (d, e) Average Max and Twist ChIP-nexus footprints at the top 200 sites for all possible E-box variants (CANNTG). Each variant profile includes its reverse complement. (d) Max binds specifically to the canonical CACGTG motif and to a lesser extent to the CACATG motif. Note that the Max footprint shape looks identical between the two motifs. (e) In contrast, the Twist binding specificity and the footprint shape is more complex. Notably, the outer boundary at -11bp is stronger at the CATATG and CACATG motif, whereas the inner boundary at -2 bp is stronger at the CAGATG motif.
We next examined the binding profile of the Dorsal footprint when bound to a Dorsal consensus binding motif (GGRWWTTCC). Using the 200 motifs with the highest ChIP-nexus counts, we generated the average Dorsal footprint (Fig. 3a). It is very similar to the footprints on known Dorsal targets, with the boundaries located five nucleotides upstream of the motif. This distance is consistent with the crystal structure of NFkB, which also suggests that the footprint is wider than the binding sequence16. Whether lambda exonuclease stops exactly at the protein-DNA boundary or perhaps a few nucleotides before also remains unclear.
Next we analyzed the ChIP-nexus footprint of Twist over the known binding motifs (CABATG, thus CATATG, CACATG or CAGATG). We found that Twist has two boundaries, one located 11 nucleotides and another one two nucleotides upstream of the motif (Fig. 3b), indicating interactions between Twist and the DNA flanking sequences outside the binding motif. To obtain further insights into the binding of bHLH transcription factors in general, we then analyzed Max, which binds to the palindromic E-box CACGTG either as a homodimer or as a heterodimer with other bHLH proteins such as Myc 21, 22. The average ChIP-nexus footprint of Max had a second set of boundaries located 8 bp upstream of the motif (Fig. 3c), again indicating interactions with flanking DNA sequences. The crystal structure of Max-Max, Max-Myc and Max-Mad only included 6 base pairs flanking either side of the E-box motif and did not use full-length Max or Myc 23, 24. However, in vitro footprinting assays of Max and Myc show protection of 4–6 bases beyond the motif 25, 26, consistent with our results.
We next tested whether the binding footprint of Max and Twist varies across E-box variants of the pattern CANNTG (Fig. 3d, e). For each possible middle sequence, we selected the 200 motifs with the highest ChIP-nexus read counts. As expected, Max binding was strongest at the canonical CACGTG motif. A weaker but similar pattern was detected at the CACATG motif (Fig. 3d), consistent with its binding specificity measured by a bacterial one-hybrid system 27. Consistent with previous data 17, 27, Twist binding occurred at multiple E-boxes (Fig. 3e). But the shapes of these footprints varied in that the outer boundary (at 11 bp from the motif) was dominant at the CATATG motif and to a lesser extent the CACATG motif, the two motifs with the highest evolutionary conservation across Drosophila species 17. In contrast, the inner boundary (at 2 bp from the motif) was more prominent at the CAGATG motif. Although the basis for these differences in footprints is unknown, the results may indicate an unappreciated specificity in the way transcription factors are detected in vivo.
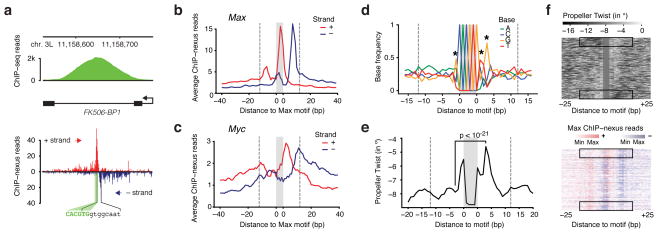
The average footprint of Max suggested interactions with flanking DNA sequences on both sides of the motif. Inspection of the footprints at individual genes, however, suggested that Max often has a favored interaction to one side of the motif (Fig. 4a). A favored interaction side was also found for Twist, especially at the CATATG motif (Supplementary Fig. 1), but we will focus here on the analysis of Max.
Figure 4. Favored interaction side of Max at E-Box motifs correlates with DNA features in the flanking sequences.
(a) Single-gene examples of the ChIP-nexus footprints show that the Max profile indeed consists of two separate footprints, one of which is frequently dominant. For example, in the Fk506-BP1 intron, the Max footprint (black brackets) is found to the right of the E-box motif (green). (b) Average Max ChIP-nexus profile at the top 200 CACGTG motifs after orienting each footprint such that the higher signal is to the right. The area of the motif is shaded in grey and the extended area of the footprint is demarcated with dotted lines from the motif (at 12 bp away from the motif to include most reads from the footprint). (c) Average Myc ChIP-nexus profile at the same motifs shown in (b) shows that Myc’s footprint is generally localized to the same side of the motif as Max. (d) Average base composition of the oriented E-box motifs from (b). Significant differences in nucleotides within the area of the footprint are marked with a star (Chi2 test, p < 10−24 for the G to the right and p < 10−12 for all others). The consensus sequence for orientation to the right is RCACGTGYTG. (e) The oriented sequences also show a marked difference in predicted DNA shape, notably the propeller twist score between a base pair (measured in degrees of rotation). At the third position from the motif, the difference is the highest (paired t-test, p<10−21). Note that on the favored interaction side, the predicted propeller twist is more neutral (seen as peak due to the negative scale). (f) Differences in DNA propeller twist in regions flanking the E-box motif correlate with Max ChIP-nexus footprint level. In the upper panel, the top 200 motifs were ordered by the difference in the mean DNA propeller twist measurements within the 6 bp flanking the E-box on both sides. The Max ChIP-nexus heatmap with the same order of motifs (lower panel) shows that the favored interaction side is most pronounced when there is an asymmetry in the DNA propeller twist around the motif (black boxes).
To analyze the basis for the Max binding asymmetry, we determined the dominant side for each of the top 200 Max binding footprints (based on the difference in read counts observed between the right and left sides of each motif). Because the CACGTG motif is palindromic and thus not strand-specific, we then oriented the binding footprints such that the dominant side is to the right of the motif. The average footprint after orientating the motifs is shown in Fig. 4b. We then searched for differences between the left and the right side.
To test whether binding to a half site might reflect the binding of Max as a heterodimer with its partner Myc, we performed ChIP-nexus with Myc. If the Myc-Max heterodimer determines the orientation, we would expect the Myc footprint to follow the opposite trend as Max at the orientated binding sites, i.e. the higher signal would be found to the left of the motif. Although there are differences between the binding footprints, the Myc profile, like the Max profile, was also oriented to the right (Fig. 4c), suggesting that the favored interaction side is not determined by heterodimer orientation.
Next, we searched for differences in the DNA sequences surrounding the Max motif that could explain the favored interaction side (Fig. 4d, e). We found that the base composition shows significant biases next to the E-box (indicated as stars in Fig. 4d), which creates a directional motif of the consensus RCACGTGYTG. The nucleotide biases outside the motif could either mediate direct contacts with the Mac-Myc dimer or could indirectly affect the protein interactions through the overall DNA shape 28. Indeed, the specificity of bHLH factors has previously been shown to correlate with parameters of DNA shape in flanking sequences 19, 29. We therefore examined predicted DNA shape parameters 30 for all 200 sequences and found that the propeller twist, a measurement for the relative rotation between two paired bases, is on average significantly stronger at the less favored interaction side (Fig. 4e, paired t-test p<10−21). To visualize the correlation between propeller twist and favored interaction side, we sorted our 200 Max footprints based on the difference in propeller twist between the two sides and then plotted the Max footprint in the same order (Fig. 4f). This shows that a strong asymmetry with regard to the propeller twist is highly correlated with the favored interaction side.
In summary, ChIP-nexus achieves increased resolution compared to conventional ChIP-seq and enhanced robustness compared to ChIP-exo to provide a detailed view of the in vivo binding landscape of transcription factors. ChIP-nexus uses a similar amount of cells, as ChIP-seq yet pinpoints binding sites within individual enhancers more precisely and provides new information on how different motif variants are bound in vivo. Although high-resolution in vivo binding data can also be obtained by digital genomic footprinting 31, this method requires substantially more sequencing depth and does not reveal the identity of the bound transcription factors.
The increased resolution suggests that the Max binding footprint is influenced by DNA sequences flanking the motif and that this interaction is often stronger at one side of the motif. The favored interaction side correlates with differences in specific nucleotides as well as parameters of DNA shape, and might explain why the reads from conventional ChIP-seq experiments often do not peak directly over the binding motif (e.g. Twist at the CATATG motif 17). While we cannot exclude the possibility that the favored side is the preferred side of cross-linking by formaldehyde, it is unlikely that this is the only explanation. It is becoming more and more evident that local DNA features around a motif contribute to the specificity of protein-DNA interactions, whether measured in vitro without formaldehyde cross-linking 19 or in vivo using reporter assays 32. Thus, it is possible that Max indeed has a favored interaction side in vivo, but whether this preference has a functional consequence is not known.
The high resolution and robustness of the protocol opens the possibility for a more extended analysis of the in vivo binding site specificity of transcription factors. For example, ChIP-nexus is ideally suited for identification of single nucleotide polymorphisms (SNPs) that alter transcription factor binding, either across species or between individuals within a population. Furthermore, as it precisely identifies which binding motif is bound in vivo, it will help in identifying the influence of nucleosomes, other transcription factors or DNA methylation on the in vivo binding of transcription factors. Therefore, ChIP-nexus could become a useful tool for untangling the mechanisms of combinatorial regulation.
Accession number, availability of analysis code and experimental protocol
All ChIP-nexus, ChIP-exo and ChIP-seq samples analyzed in this study are available from the NCBI Gene Expression Omnibus (GEO) under accession number GSE55306. All analysis code used for data processing and figure generation is available via GitHub at https://github.com/zeitlingerlab. In addition, we have prepared a Linux virtual machine containing all software tools, analysis code, raw data and processed data used in this study. Instructions for accessing the virtual machine via Amazon Web Services, as well as a detailed ChIP-nexus protocol, can be found at our website (http://research.stowers.org/zeitlingerlab).
Contributions
Q.H. and J.Z. conceived and designed the ChIP-nexus protocol. Q.H. performed all experiments. J.J. developed all computational analysis tools. Q.H., J.J. and J.Z. analyzed and interpreted the data and wrote the manuscript.
Competing financial interests
A patent for ChIP-nexus has been submitted.
Online Methods
Preparation of K562 cells
K562 cells from ATTC were grown at 37 °C, 5% CO2 w/ humidity in Iscove’s DMEM media with 10% fetal bovine serum. Ten million cells were harvested for each ChIP-seq or ChIP-nexus experiment, respectively. Cells were cross-linked in 1% formaldehyde (in 50 mM HEPES-KOH, pH 7.5; 100 mM NaCl; 1 mM EDTA; 0.5 mM EGTA) and rotated for 10 minutes at room temperature. Cross-linking was quenched by adding glycine to 0.125 M and cells and rotating for 5 minutes at room temperature. Cells were spun down, washed with PBS and re-suspended in A1 buffer (15 mM HEPES pH 7.5; 15 mM NaCl; 60 mM KCl; 4 mM MgCl2; 0.5% Triton X-100; 0.5 mM DTT), transferred to a Wheaton Dounce homogenizer, and broken down by twenty strokes with each pestle. Homogenates were spun down at 3000 g and washed three times with A1 buffer and once with A2 buffer (15 mM HEPES pH 7.5; 140 mM NaCl; 1 mM EDTA; .5 mM EGTA; 1% Triton X-100; 0.1% sodium deoxycholate; 1% SDS; 0.5% N-lauroylsarcosine sodium). Nuclei were re-suspended in 0.7 ml A2 buffer. The chromatin was fragmented with a Bioruptor by two rounds of 15 minutes sonication at high power. Chromatin was cleared by centrifugation and the supernatant was used for ChIP.
Preparation of Drosophila embryos
D. melanogaster embryos were collected on apple plates from Oregon-R flies raised and kept at 25 °C and 60% humidity. The apple plates were placed into fly cages for 2 h and then incubated for another 2 h outside the cage such that the embryos were aged 2–4 h after egg laying (AEL). Embryo collections and whole cell extract (WCE) preparations were performed as previously described 33, 34. About 0.1 g of fixed embryos was used per ChIP-seq or ChIP-nexus.
Preparation of Drosophila S2 cells
S2 cells from Invitrogen were grown at 25°C in HyClone SFX-Insect Cell Culture Media with 1x penicillin and streptomycin (Sigma-Aldrich). About 20 million cells were harvested for each ChIP-seq or ChIP-nexus experiment. S2 sells were cross-linked with 1% formaldehyde for 10 minutes at room temperature. Formaldehyde was quenched by 0.125 M glycine for 5 minutes. Cells were washed with PBS, re-suspended in Orlando and Paro’s Buffer A (0.25% triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM Tris-HCl, pH 8.0) and rotated for 10 minutes at room temperature. Nuclei were spun down and re-suspended in RIPA buffer (10 mM Tris-HCl, pH 8.0; 140 mM NaCl; 0.1% SDS; 0.1% sodium deoxycholate; 0.5% sarkosyl; 1% Triton X-100). The chromatin was fragmented with a Bioruptor by two rounds of 15 min sonication at high power. Chromatin was cleared by centrifugation and the supernatant was used for ChIP.
Chromatin immunoprecipitation
Chromatin immunoprecipitations were performed in biological duplicates as previously described 35 with rabbit polyclonal antibodies against TBP (sc-204X, 3 μg/ChIP), Dorsal (20 μg/ChIP), Twist (10 μg/ChIP), Max (sc-28209, 8 μg/ChIP) and Myc (sc-28207, 8 μg/ChIP). The rabbit polyclonal antibodies against Dorsal protein (a.a. 39–346) and Twist protein (C-terminal a.a. 340–490) were produced by GenScript. The ChIP-seq pattern with these antibodies matched those obtained previously 34. Enrichments for each transcription factor of interest were confirmed at known target sites by real-time PCR (StepOnePlus, Applied Biosystem) before library preparation. Primers are available upon request.
ChIP-nexus oligonucleotides
Nex_adapter_UBamHI:
/5Phos/GATCGGAAGAGCACACGTCTGGATCCACGACGCTCTTCC
Nex_adapter_BN5BamHI:
/5Phos/TCAGNNNNNAGATCGGAAGAGCGTCGTGGATCCAGACGTGTGCTCTTCCGATCT
To anneal the two Nex_adapter oligonucleotides, 50 μM of each are mixed in 1x TE and 50 mM NaCl and placed in a thermocycler: 95 °C for 5 minutes, then the temperature is ramped down to 25 °C at a rate of ~ 3.5 °C/minute, and held at 25 °C for 30 minutes.
Nex_cut_BamHI (cut oligo):
GAAGAGCGTCGTGGATCCAGACGTG
Nex_primer_U (universal PCR primer with 3′ phosphoro-thioate bond):
AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T
Nex_primer_B01 (barcoded PCR primer with 3′ phosphoro-thioate bond, other barcodes may be used):
CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATC*T
ChIP-nexus digestion steps
The digestion with lambda exonuclease was carried out using a modified version of the published ChIP-exo protocol 3, 4, while the chromatin is immunoprecipitated on Dynabeads.
The chromatin was first washed five times with the following buffers: Wash Buffer A (10 mM Tris-EDTA, 0.1% Triton X-100), Wash Buffer B (150 mM NaCl, 20 mM Tris-HCl [pH 8.0], 5 mM EDTA, 5.2% sucrose, 1.0% Triton X-100, 0.2% SDS), Wash Buffer C (250 mM NaCl, 5 mM Tris-HCl [pH 8.0], 25 mM HEPES, 0.5% Triton X-100, 0.05% sodium deoxycholate, 0.5 mM EDTA), Wash Buffer D (250 mM LiCl, 0.5% IGEPAL CA-630, 10 mM Tris-HCl [pH 8.0], 0.5% sodium deoxycholate, 10 mM EDTA), Tris Buffer (10 mM Tris pH 7.5, 10 mM Tris pH 8.0 or 10 mM Tris pH 9.5 depending on the next enzymatic step).
After the last wash, residual buffer was drained before the next enzymatic reaction was added. These washing steps were repeated between all following steps.
To repair the DNA ends, each sample was incubated with 0.05 u/μl DNA polymerase I, large fragment (New England Biolabs, M0210), 0.15 u/μl T4 DNA polymerase (New England Biolabs, M0203), 0.5 u/μl T4 polynucleotide kinase (New England Biolabs, M0201) and 0.4 mM/μl dNTPs in 30–40 μl 1x NEB T4 ligase buffer (New England Biolabs, B0202) at 12 °C for 30 min, followed by washing steps as above.
For dA tailing, each sample was incubated with 0.3 u/μl klenow fragment (3′ → 5′ exo−) (New England Biolabs, M0212) and 0.2 mM/μl ATP in 50 μl 1x NEBuffer 2 at 37 °C for 30 min, followed by washing steps as above.
The adapters were then ligated by incubation in 200 u/μl Quick T4 DNA ligase (New England Biolabs, M2200) and 60 nM/μl Nex_adapter in 50 μl 1x Quick Ligation Reaction Buffer at 25 °C for 60 min, followed by washing steps as above.
To fill the ends of the adapters, each sample was incubated with 0.1 u/μl klenow fragment (3′ → 5′ exo−) (New England Biolabs, M0212) and 0.1 mM/μl dNTPs in 50 μl 1x NEBuffer 2 at 37 °C for 30 min, followed by washing steps as above.
The ends were then trimmed by incubation in 0.09 u/μl T4 DNA polymerase (New England Biolabs, M0203) and 0.1 mM/μl dNTPs in 50 μl 1x NEBuffer 2 at 12 °C for 5 min, followed by washing steps as above.
For lambda exonuclease digestion, each sample was incubated in 0.2 u/μl lambda exonuclease (New England Biolabs, M0262), 5% DMSO and 0.1% triton X-100 in 100 μl 1x NEB Lambda exonuclease reaction buffer at 37 °C for 60 min with constant agitation, followed by washing steps as above.
Finally, RecJf exonuclease digestion occurred in 0.75 u/μl RecJf exonuclease (New England Biolabs, M0264), 5% DMSO and 0.1% triton X-100 in 100 μl 1x NEBuffer 2 at 37 °C for 60 min with constant agitation. After RecJf digestion, the Dynabeads were washed three times with RIPA buffer (50 mM HEPES, pH7.5, 1 mM EDTA, 0.7% sodium deoxycholate, 1% IGEPAL CA-630, 0.5 M LiCl). DNA elution, reverse cross-linking, DNA purification and precipitation were performed as previously described 34, 35.
ChIP-nexus library preparation
The library preparation protocol is based on the iCLIP protocol 11. After the DNA is purified and precipitated, each sample is dissolved in 11.25 μl H2O, 1.5 μl 10× CircLigase buffer, 0.75 μl 1 mM ATP, 0.75 μl 50 mM MnCl2, 0.75 μl CircLigase (Epicentre) and incubated at 60 °C for 60 min for self-circularization. To anneal the oligonucleotide complementary to the BamHI restriction site (cut oligo Nex_cut_BamHI), 26 μl H2O, 5 μl FastDigest buffer (Fermentas) and 1 μl 10 μM cut oligo were added to each sample. The mixture was incubated with the following program on a thermocycler: 95 °C for 5 min, ramp down to 25 °C at a rate of ~ 3.5 °C/minute, and hold at 25 °C for 30 min. For BamHI digestion, 3 μl Fastdigest BamHI (Fermentas) were added and the sample was incubated at 37 °C for 30 min. The samples were then precipitated by adding 150 μl TE buffer, 30 μg glycogen, 20 μl 3 M/l sodium acetate (pH 5.5) and 500 μl 100% ethanol and incubated at −80 °C for 2.5 h. After centrifugation at 4 °C for 30 min at 16,100 g, the samples were washed with 500 μl 80 % ethanol, dried overnight at room temperature and resuspended in 36 μl H2O.
For PCR amplification, 10 μl 5x Phusion buffer, 1.5 μl 10 mM dNTP, 1 μl each of 10 μM universal and barcoded PCR primers (Nex_primer_U and Nex_primer_B01), and 0.5 μl Phusion Polymerase (New England Biolabs, M0530) were added to each sample in a total volume of 50 μl. The DNA was amplified with the following program: 98 °C for 30 s; 18x (98 °C for 10 s, 65 °C for 30 s, 72 °C for 30 s); 72 °C for 5 min. To remove contaminating adapter dimers, the PCR products are run on a 2% agarose gel. The adapter dimers usually form a thin bright band migrating at the front edge of the library DNA, which forms a smear. The library DNA is carefully sliced out, purified by MinElute kit (Qiagen, 28006) and eluted into 12 μl elution buffer. After Bioanalyzer analysis, libraries were sequenced on an Illumina HiSeq platform with the single-end sequencing primer over 50 cycles of extension according to manufacturer’s instructions.
Data processing for ChIP-nexus samples
Sequencing reads passing the default Illumina quality filter (CASAVA v1.8.2) were further filtered for the presence of the fixed barcode CTGA starting at read position 6. The random and fixed barcode sequences were then removed (read positions 1 through 9), while retaining the 5-bp random barcode sequence for each read separately. Adapter sequences from the right end were then trimmed using the cutadapt tool 36. All reads of at least 22 bp in length after adapter trimming were then aligned to the appropriate reference genome (dm3 for Drosophila melanogaster and hg19 for Homo sapiens) using bowtie v1.0.0 37. Only uniquely aligning reads with a maximum of 2 mismatches were kept. To remove duplicates, reads with identical alignment coordinates (chromosome, start position and strand) and identical random barcode were removed using R 38 and Bioconductor 39. All reads were then split by strand orientation and a genome-wide count of the start positions (lambda exonuclease’s stop position) was calculated for each strand.
Data processing for ChIP-exo samples
The published ChIP-exo TBP samples in human K562 cells 9 were downloaded from the Sequence Read Archive (accession numbers SRR770743 and SRR770744) and aligned to the UCSC hg19 reference genome using the same parameters as for ChIP-nexus samples. Peconic provided aligned BAM files for both Dorsal and Twist ChIP-exo replicates. Aligned reads for all ChIP-exo experiments were separated by strand and reduced to the first sequenced base (lambda exonuclease’s stop position), and genome-wide counts for read start positions were calculated.
Data processing for ChIP-seq samples
ChIP-seq reads were aligned to the appropriate reference genome (dm3 or hg19) using the same parameters as for the ChIP-nexus samples. After alignment, reads were extended in the 5′ to 3′ direction to each sample’s estimated library insert size as determined by a Bioanalyzer. These extensions were 136 bp for Dorsal, 124 bp for Twist, 83 bp for Max and 74 bp for TBP. After extension, genome-wide coverage values were calculated.
Reference genome modification for Drosophila Oregon-R embryos
Multiple SNPs in our Oregon-R strain resulted in gaps in read coverage at a number of regions of interest (including the rho enhancer used as an example). To correct this, the Dorsal and Twist ChIP-seq samples were combined and re-aligned to the reference genome while allowing up to 3 mismatches. Samtools 40 was then used to identify variants genome-wide using the following parameters:
samtools mpileup -uD -f dm3.fasta embryo_combined_chipseq.bam | bcftools view -vcg
The identified single-allele variants were then used to create a modified reference genome matching the sequence of our Oregon-R strain. ChIP-seq and ChIP-nexus samples for Dorsal and Twist were aligned to this modified reference genome. As Peconic did not provide the unaligned reads for the Dorsal ChIP-exo data, we could only perform this read recovery procedure on our ChIP-seq and ChIP-nexus data.
Peak calling
MACS v.2.0.10 20 was run on the ChIP-nexus replicate #1 samples and the ChIP-seq samples for TBP, Dorsal, Twist and Max using the following parameters:
macs2 callpeak –g dm –keep-dup=all –call-summits
Resulting peak summits were sorted by score and a maximum of 10,000 were retained per sample.
Comparison scatterplots
For each scatterplot, the peaks detected in the sample on the x axis were resized to 201 bp centered at the summit. Each peak was scored using the genome-wide coverage values for the two samples. For ChIP-seq, these coverage values were calculated using the entire extended fragment size. For ChIP-nexus and ChIP-exo, coverage values were calculating using only the first base pair of each aligned fragment. Pearson correlations were calculated using the raw values before log transformation.
ChIP-nexus and ChIP-seq motif presence
For Dorsal, Twist and Max, the top 200 peaks by MACS score were used. Motif frequency plots were generated by scoring each position in the genome as either 1 or 0 based on the presence of a consensus motif for each factor. These consensus motifs were GGRWWTTCC with up to one mismatch for Dorsal, CABATG with no mismatches for Twist and CACGTG with no mismatches for Max. The average motif presence around the top 200 peak summits was then calculated and plotted for both ChIP-seq and ChIP-nexus (replicate 1) samples.
For each peak, the distance from the peak summit to the nearest consensus motif was calculated. For distance thresholds of 10, 20, 50 and 100 bp, a two-sided Chi-squared test was used to test for a significant difference in proportion of peaks near a consensus motif between ChIP-nexus and ChIP-seq.
Motif average profiles and heatmaps
For each factor, all non-overlapping instances of its motif with up to one mismatch were scored for ChIP-nexus signal (replicate 1) by summing the total reads from both strands in a fixed region centered on the motif (29 bp for Dorsal, 15 bp for Max and 51 bp for Twist).. The heatmaps of the top 200 motifs were oriented such that the motif is on the positive strand and sorted by total reads in a 50 bp window centered on the motif. Positive and negative strand reads (relative to the strand of the motif) were normalized from zero reads (minimum) to the read value at the 98th percentile or higher (maximum) for display.
The E-box specificity plots shown in Figure 3 were constructed by separately averaging the positive and negative strand ChIP-nexus signal among the top scoring 200 non-overlapping instances of each unique E-box motif CANNTG. Each motif was scored by summing the ChIP-nexus reads in a window 50 bp centered on the motif.
To analyze the favored interaction side of Max in Figure 4, the same top 200 Max motifs described above were scored for ChIP-nexus signal on the left and right side based on the observed average pattern. The left side signal was calculated by summing the positive strand reads in a region 9-bp wide centered 8-bp upstream of the motif and the negative strand reads in a region 9-bp wide centered on the motif +1 position. The right side signal was calculated by summing the positive strand reads in a region 9-bp wide centered on the motif +4 position and the negative strand reads in a region 9-bp wide centered 8-bp downstream of the motif. Each motif was then oriented so that the side with higher signal was to the right of the motif.
Analysis of DNA shape
Genome-wide DNA shape parameters were collected for the positive strand of the Drosophila melanogaster UCSC dm3 reference genome. First, all 1,024 DNA pentamers were uploaded to the DNA Shape web service 30 to obtain predictions for minor groove width and propeller twist. For both DNA shape parameters, a single value was provided for the center base of each pentamer. These values were applied genome-wide by aligning the pentamers to the positive strand of the reference genome.
To order the top 200 Max-bound E-box motifs by the difference in DNA propeller twist (Figure 4F), we calculated the mean propeller twist for the six base pairs immediately to the left and right of the motif. The motifs were then ordered by the difference between right and left mean propeller twist.
Supplementary Material
Acknowledgments
We thank Marco Blanchette and Alex Stark for discussions, as well as Robb Krumlauf and Ryan Mohan for critical comments on the manuscript. This work was funded by the NIH New Innovator Award 1DP2 OD004561-01 to J.Z. and the Stowers Institute for Medical Research.
References
- 1.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 2.Bardet AF, et al. Identification of transcription factor binding sites from ChIP-seq data at high resolution. Bioinformatics. 2013;29:2705–2713. doi: 10.1093/bioinformatics/btt470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee HS, Pugh BF. ChIP-exo method for identifying genomic location of DNA-binding proteins with near-single-nucleotide accuracy. Curr Protoc Mol Biol. 2012;Chapter 21(Unit 21):24. doi: 10.1002/0471142727.mb2124s100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic preinitiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kivioja T, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods. 2012;9:72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 8.Casbon JA, Osborne RJ, Brenner S, Lichtenstein CP. A method for counting PCR template molecules with application to next-generation sequencing. Nucleic Acids Res. 2011;39:e81. doi: 10.1093/nar/gkr217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venters BJ, Pugh BF. Genomic organization of human transcription initiation complexes. Nature. 2013;502:53–58. doi: 10.1038/nature12535. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Serandour AA, Brown GD, Cohen JD, Carroll JS. Development of an Illumina-based ChIP-exonuclease method provides insight into FoxA1-DNA binding properties. Genome Biol. 2013;14:R147. doi: 10.1186/gb-2013-14-12-r147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang JD, Schwyter DH, Shirokawa JM, Courey AJ. The interplay between multiple enhancer and silencer elements defines the pattern of decapentaplegic expression. Genes Dev. 1993;7:694–704. doi: 10.1101/gad.7.4.694. [DOI] [PubMed] [Google Scholar]
- 13.Fakhouri WD, et al. Deciphering a transcriptional regulatory code: modeling short-range repression in the Drosophila embryo. Mol Syst Biol. 2010;6:341. doi: 10.1038/msb.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1992;6:1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 15.Zinzen RP, Senger K, Levine M, Papatsenko D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol. 2006;16:1358–1365. doi: 10.1016/j.cub.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 16.Szymanski P, Levine M. Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. Embo J. 1995;14:2229–2238. doi: 10.1002/j.1460-2075.1995.tb07217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozdemir A, et al. High resolution mapping of Twist to DNA in Drosophila embryos: Efficient functional analysis and evolutionary conservation. Genome Res. 2011;21:566–577. doi: 10.1101/gr.104018.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Lee CK, Granek JA, Clarke ND, Lieb JD. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 2006;16:1517–1528. doi: 10.1101/gr.5655606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordan R, et al. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 2013;3:1093–1104. doi: 10.1016/j.celrep.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 22.Prendergast GC, Lawe D, Ziff EB. Association of Myn, the murine homolog of max, with c-Myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 23.Ferre-D’Amare AR, Prendergast GC, Ziff EB, Burley SK. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 24.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 25.Walhout AJ, Gubbels JM, Bernards R, van der Vliet PC, Timmers HT. c-Myc/Max heterodimers bind cooperatively to the E-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene. Nucleic Acids Res. 1997;25:1493–1501. doi: 10.1093/nar/25.8.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wechsler DS, Papoulas O, Dang CV, Kingston RE. Differential binding of c-Myc and Max to nucleosomal DNA. Mol Cell Biol. 1994;14:4097–4107. doi: 10.1128/mcb.14.6.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu LJ, et al. FlyFactorSurvey: a database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 2011;39:D111–117. doi: 10.1093/nar/gkq858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohs R, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, et al. TFBSshape: a motif database for DNA shape features of transcription factor binding sites. Nucleic Acids Res. 2014;42:D148–155. doi: 10.1093/nar/gkt1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou T, et al. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 2013;41:W56–62. doi: 10.1093/nar/gkt437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White MA, Myers CA, Corbo JC, Cohen BA. Massively parallel in vivo enhancer assay reveals that highly local features determine the cis-regulatory function of ChIP-seq peaks. Proc Natl Acad Sci U S A. 2013;110:11952–11957. doi: 10.1073/pnas.1307449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandmann T, et al. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21:436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeitlinger J, et al. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Q, et al. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet. 2011;43:414–420. doi: 10.1038/ng.808. [DOI] [PubMed] [Google Scholar]
- 36.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17:10–12. [Google Scholar]
- 37.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Team, R.D.C. R: a language and environment for statistical computing. 2013 Available online at http://www.R-project.org/
- 39.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.