Abstract
Repeated measurement designs have been widely used in various randomized controlled trials for evaluating long term intervention efficacies. For some clinical trials, the primary research question is to compare two treatments at a fixed time, using a t-test. Though simple, robust, and convenient, this type of analysis fails to utilize a large amount of collected information. Alternatively, the mixed effects model is commonly used for repeated measurement data. It models all available data jointly and allows explicit assessment of the overall treatment effects across the entire time spectrum. In this paper, we propose an analytic strategy for longitudinal clinical trial data where the mixed effects model is coupled with a model selection scheme. The proposed test statistics not only make full use of all available data but also utilize the information from the optimal model deemed for the data. The performance of the proposed method under various setups, including different data missing mechanisms, is evaluated via extensive Monte Carlo simulations. Our numerical results demonstrate that the proposed analytic procedure is more powerful than the t-test when the primary interest is to test for the treatment effect at the last time point. Simulations also reveal that the proposed method outperforms the usual mixed effects model for testing the overall treatment effects across time. In addition, the proposed framework is more robust and flexible in dealing with missing data compared to several competing methods. The utility of the proposed method is demonstrated by analyzing a clinical trial on the cognitive effect of testosterone in geriatric men with low baseline testosterone levels.
Keywords: Repeated measurement, Mixed effects model, t-test, Model selection, Missing data, Bootstrap
1. Introduction
In many clinical trials, multiple responses (or longitudinal measurements) at various time points including baseline and after a randomized treatment assignment are often collected for each subject. The repeated measurement design is useful in investigating intervention efficacies, risk factors of chronic diseases, long term treatment effects, etc. For some studies, researchers are interested in intervention efficacy at a fixed point in time and thus restrict their analysis to the response differences from the last time point to baseline via a t-test. For properly randomized trials, the systematic differences in baseline covariates are eliminated. Thus, this t-test is valid for evaluating the last time point intervention efficacy even under a repeated measurement design. Another commonly used analysis strategy is analysis of covariance (ANCOVA) [1] in which the baseline values are included as covariates. For instance, for a longitudinal clinical trial study in evaluating the effect of intracoronary transfer of autologous bone-marrow cells on improving the global left-ventricular ejection fraction [2], ANCOVA was applied for comparing the response differences from the last follow up time to baseline between the two treatment groups, i.e. optimum postinfarction medical treatment versus bone-marrow-cell. It has been shown that the ANCOVA model can reduce potential bias in the treatment effect estimate due to randomly imbalanced baseline covariates [3].
Though simple and convenient, these approaches only make use of the information at baseline and the last time point and ignore a large amount of potentially valuable information. To utilize complete data, the mixed effects model [4] is often used to simultaneously model the longitudinal responses and the covariance structure of repeated measurements flexibly. In addition to assessing the last time point treatment effect, the mixed effects model allows one to evaluate and test the overall effects across every time point. Furthermore, the mixed effects model, if correctly specified, can provide more accurate and efficient treatment effect estimates. Another advantage of mixed effects model is its flexibility in dealing with missing data, which occur frequently in clinical trials. As an example, in the DURATION-2 trial [5], each patient with type 2 diabetes was randomly assigned to one of three intervention groups, i.e. exenatide, sitagliptin, or pioglitazone. The efficacy of exenatide was compared to that of sitagliptin or pioglitazone in terms of glycosylated haemoglobin (HbA(1c)) changes from baseline to week 26 post treatment start. In this study, the discontinuation rates at the final time point were 20.6%, 13.3% and 20.6% for the three treatment arms respectively. By taking the longitudinal information and correlations among the repeated measurements into account using mixed effects model, the information from the non-missing observations can help partially recover the lost information in the missing data. However, in practice, the optimal mixed effects model and the valid covariance structure deemed for the data are unknown. For well designed randomized trials, it is common to employ the grand full model by including all possible fixed effect terms along with an unstructured covariance matrix. This strategy is valid in general, but not necessarily efficient. The full model can be very conservative under certain situations (see Section 3 for simulation results). In searching for an efficient analysis strategy, researchers may first identify the optimal model deemed for the data via a given model selection procedure, and then perform statistical analysis on the selected model. Model selection criteria, AIC [6], BIC [7], corrected AIC [8] and others are available and often used in practice. However, the stochastic nature associated with the model selection procedure [9] makes the post-model selection statistical inference challenging.
In this paper, we propose an analytic procedure for longitudinal randomized controlled trial data. We first select the optimal model over a set of mixed effects models based on BIC criteria. BIC has been shown to be consistent and asymptotically selects the correct model if the correct model is among the candidate model set [10–12]. Based on the selected optimal model, we propose two test statistics, one for testing the overall treatment effects across time and the other for testing the treatment effect at the last time point. The proposed statistics not only make full use of the available data but also utilize the information from the optimal model deemed for the data. Further, we develop a restricted cluster bootstrap resampling scheme to take the stochastic variation associated with the model selection procedure into account in the proposed test statistics. The organization of the paper is as follows: In Section 2, the details of the proposed method are illustrated; In Section 3, simulation studies under different settings and missing data scenarios are conducted to investigate the performance of the proposed method; The practical usage of the proposed method is demonstrated in Section 4 with a real clinical trial; Final remarks and a discussion are given in Section 5.
2. Methods
To fix the notation, we first define xijk as the (continuous) response variable of jth subject (j = 1, · · · , ni) from the ith treatment group (where i = 1 & 0 refers the treated and untreated group, respectively) measured at the kth time point (k = 0, · · · , T) with xij0 being the baseline measurement. Further, let yijk ≡ xijk − xij0 (k = 1, · · · , T) represent the response difference at time point k from baseline.
For well designed longitudinal clinical trials, the following mixed effects model is commonly used to analyze all available observations jointly, allowing one to test for overall treatment effects and for effects at any single time point, including the last time point, T.
| (1) |
where I is the indicator function (1 if true, 0 otherwise); α0 is the intercept and αTrt is the treatment effect corresponding to the last measurement time point; the αks′ (k = 1, · · · , T − 1) are the main time effects and the αk+T−1 s′ (k = 1, · · · , T − 1) are the corresponding time × treatment interactions; the rijks′ are the subject specific random effects and rij ~ N(0, Ω) where rij = (rij1, · · · , rijT) and the covariance matrix Ωdescribes the correlation structure among the repeated measurements; the random measurement error . The hypothesis corresponding to the overall treatment effects is thus
| (2) |
To test the hypothesis, we employ the likelihood ratio statistics for testing between the full model (1) and the following null model:
| (3) |
with an unstructured covariance matrix Ω. We denote this likelihood ratio test statistics as ζMBP, with MBP referring to the mixed effects model based power analysis. In addition, we may similarly test the last time point treatment effect by simply testing the hypothesis H0 : αTrt = 0 vs Ha : αTrt ≠ 0.
The full mixed effects model is valid but not necessarily efficient when there exists no time × treatment interaction. When no such interaction exists, a more efficient main effect only model is the following:
| (4) |
and the hypothesis for testing the overall treatment effects now becomes H0 : αTrt = 0 vs Ha : αTrt ≠ 0.
However, for a given dataset, it is not known a priori if time × treatment interaction exists. A common practice is to first identify an optimal model deemed for the data via either a model selection procedure or testing procedure. Based on the selected model, a formal statistical analysis follows. One issue with the two-step approach is that due to the stochastic nature associated with the selected model, the post-model selection inference without adjusting for the selection variation will no longer be valid. Below we describe a valid post-model selection testing procedure under the mixed effects model framework.
Post-Model Selection Statistical Inference:
We first select the optimal model using the BIC criteria from a set of J candidate models including the full model (1) and the main effects model (4) with different covariance structures. In particular, the covariance structures that we consider include compound symmetry (CS), autoregressive (AR), and unstructured (UN) covariance matrices that are commonly used in practice. However, it is straightforward to include other candidate covariance structures. Let M̂ be the selected optimal model, based on which we obtain the maximum likelihood estimates (MLE) of the parameters and make the statistical inference.
Overall Treatment Effects Testing:
To test the overall treatment effects, we propose the following test statistics where MSA stands for model selection test of overall treatment effects:
where is the MLE of α′ = (αTrt, αT , · · · , α2T−2) from the optimal model M̂ if it happens to be the full model (1), while is the MLE of αTrt from the optimal model M̂ when it happens to be the main effects model (4). The and are the corresponding estimated covariance matrix and standard error of and , respectively. Details on obtaining and will be discussed in the next subsection. Under H0, if the selected optimal model M̂ belongs to the full model set (1), ζMSA ~ χ2 (T). Otherwise, ζMSA ~ χ2 (1). Note that, with the built in model selection procedure, testing the overall treatment effects reduces to testing the last time point effect when the optimal model belongs to the main effects model set, leading to improved power, as will be shown by simulation studies in Section 3.
Last Time Point Treatment Effect Testing
In practice, it is common to test the last time point treatment effect by the following simple t-test:
where is the sample mean difference between the two treatment groups. , , and n1 & n0 are the number of subjects in the treated and untreated groups, respectively. A well randomized procedure guarantees that there exists no systematic difference for baseline covariates between the two comparison groups, which in turn ensures the validity of the t-test above.
In addition to the t-test, the full mixed effects model (1) along with an unstructured covariance matrix can be used to fit repeated measurement data and the following test statistics ζFUN (referring full mixed effects model with unstructured covariance) can be constructed for testing the last time point treatment effect:
where and are the last time point treatment effect maximum likelihood estimate from the full mixed effects model (1) using unstructured covariance matrix and its standard error, respectively.
Post-model selection analysis can be similarly restricted to test for the last time point treatment effect αTrt in model (1) or (4) with the following statistics
where is the MLE of αTrt from the optimal model M̂ and is the post-model selection standard error of . Under H0, ζMSL ~ N(0, 1).
Restricted Cluster Bootstrapping Covariance Estimations
The covariance and variance estimation post-model selection is rather complex owing to the stochastic nature associated with the selected optimal model. Shao [9] proposed a bootstrapping procedure for simple linear regression models which we adopt here for mixed effects models. However, the empirical procedure of Shao [9] cannot be applied directly here, since observations within each subject are correlated and hence treating each observation as a resampling unit is no longer applicable. Instead, the cluster (or block) bootstrapping scheme [13, 14] can be used to preserve the correlation structure within each subject. For simplicity of presentation, we describe the estimation procedure for , given the fact that the optimal model is from the full model set (1). The variance estimation of can be similarly constructed. The process is defined as follows:
For a given dataset D, conduct model selection using the BIC criteria and denote the selected optimal model as M̂. Let B be the total number of bootstrapping iterations. For the bth bootstrapped dataset, Db (b = 1, · · · ,B), let M̂b be the optimal model selected. If M̂b = M̂ , let be the MLE of α from M̂b. The bootstrapping covariance estimate, of is then calculated as:
where , and ⨂ represents the outer product. In summary, the restricted cluster bootstrapping procedure works as follows:
Step 1: Perform model selection on the observed dataset, D, to get the optimal model M̂.
Step 2: Conduct cluster level resampling with replacement to get a resampled dataset and perform model selection on the resampled dataset Db. Obtain the selected optimal model M̂b. If M̂b = M̂, i.e. the same model is chosen in the original model selection and the resampling, then the MLE of α, i.e. , is used for the calculation of .
Step 3: Repeat Step 2 B times and calculate , i.e. the post-model selection covariance matrix estimate of .
3. Simulation Studies
To evaluate the performance of the proposed model selection test statistics ζMSL and ζMSA, we compared them with ζt, ζFUN and ζMBP via extensive simulations which are also used to evaluate the proposed restricted cluster bootstrapping procedure. Specifically, for testing the last time point treatment effect, we compare the proposed model selection test statistics ζMSL with the t-test ζt and full mixed effects model with unstructured covariance test statistics ζFUN . For overall treatment effects testing, we compare the proposed model selection test statistics ζMSA with the mixed effects model based likelihood ratio test statistics ζMBP. Numerical studies are conducted under two categories:
Case I (No Missing): Outcomes are generated according to the following mixed effects model with different time × treatment interactions to mimic real world clinical trial data:
| (5) |
Specifically, 4 sets of data are generated with 1) strong interaction (α1 = 0.45, α2 = 0.42, α3 = 0.39, α4 = 0.36); 2) moderate interaction (α1 = 0.24, α2 = 0.27, α3 = 0.33, α4 = 0.36); 3) weak interaction (α1 = 0.1, α2 = 0.075, α3 = 0.05, α4 = 0.025); and 4) no interaction where all the interaction terms are set to 0. The last time point treatment effect is set to 0.3 with total 5 post randomization repeated measurements. The random measurement error εijk ~ N(0, 1) and the random cluster effect rij ~ N(0, ). We simulated data with various forms of Ω, including CS, AR, and UN. Simulations were also conducted for different sample sizes and correlation strength scenarios. However, similar conclusions are obtained and for ease of presentation, only the results based on the simulated data with the following compound symmetry covariance structure Ω are presented:
For each setting, the total number of simulations is set to 1000. Results for the last time point treatment effect and the overall treatment effects testing with 100 subjects in each treatment arm are given in Tables 1 and 2, respectively.
Table 1.
Last Time Point Treatment Effect Testing without Missinga
| Data Model | Interaction | Statisticsb | Power | Type I Error |
|---|---|---|---|---|
| Full (CS) | Strong | ζ t | 0.336 | 0.044 |
| ζ FUN | 0.343 | 0.046 | ||
| ζ MSL | 0.981 | 0.048 | ||
| Moderate | ζ t | 0.336 | 0.044 | |
| ζ FUN | 0.343 | 0.046 | ||
| ζ MSL | 0.987 | 0.048 | ||
| Weak | ζ t | 0.336 | 0.044 | |
| ζ FUN | 0.343 | 0.046 | ||
| ζ MSL | 0.834 | 0.048 | ||
| Main Effects (CS) | None | ζ t | 0.336 | 0.044 |
| ζ FUN | 0.343 | 0.046 | ||
| ζ MSL | 0.710 | 0.048 | ||
results based on 1000 simulations with 100 subjects at each treatment arm
model selection via BIC with 200 bootstrap resampling
ζt: t-test for last time point effect
ζFUN: test last time point effect via full mixed effect model with UN covariance
ζMSL: model selection test statistics for testing last time point effect
Table 2.
Overall Treatment Effects Testing without Missinga
| Data Model | Interaction | Statisticsb | Power | Type I Error |
|---|---|---|---|---|
| Full (CS) | Strong | ζ MBP | 0.996 | 0.044 |
| ζ MSA | 1.000 | 0.048 | ||
| Moderate | ζ MBP | 0.967 | 0.044 | |
| ζ MSA | 0.995 | 0.048 | ||
| Weak | ζ MBP | 0.621 | 0.044 | |
| ζ MSA | 0.834 | 0.048 | ||
| Main Effects (CS) | None | ζ MBP | 0.479 | 0.044 |
| ζ MSA | 0.710 | 0.048 | ||
results based on 1000 simulations with 100 subjects at each treatment arm
model selection via BIC with 200 bootstrap resampling
ζMBP: mixed effects model based likelihood ratio test for overall effects
ζMBP: model selection statistics for testing overall treatment effects
From the “Type I Error” column of Table 1, it is clear that the type I errors of the three tests (i.e. ζt, ζFUN , and ζMSL) are well controlled near the nominal level 0.05. Examining the column of “Power” reveals that the proposed model selection test statistics ζMSL is consistently more powerful than the t-test ζt or the full mixed effects model test statistic ζFUN. This observation holds whether the data are generated from the full model or main effects model, or when time × treatment interaction is strong or weak. For example, for data generated from the main effects model, the power of ζt is 0.336 and 0.343 for ζFUN while the power of ζMSL is 0.71, more than doubled. Similarly, for data generated from the full model (5) with weak time × treatment interaction, the power gain of ζMSL is even more evident: 0.834 for ζMSL versus 0.336 for ζt and 0.343 for ζFUN . This simulation result also demonstrates that there is almost no gain for using ζFUN over ζt to test the last time point treatment effect. Note, the term of “strong”, “moderate”, and “weak” that describe the interaction are all in a relative sense. Even for the “strong” interaction setting, we purposely set the interaction to be strong but not in an overwhelming way such that the full mixed effects model is not uniformly selected. Otherwise, ζMSL and ζFUN will end up with the same or similar power.
Examining Table 2 for the overall treatment effects testing, the type I error rates for both test statistics, i.e. ζMBP and ζMSA, are controlled near the nominal level 0.05. Checking the “Power” column in Table 2 again further reveals that the proposed test statistics ζMSA is much more powerful than ζMBP when the data are generated from the main effects model. A clear pattern is that the weaker time × treatment interaction is, the more power ζMSA gains over ζMBP is. For instance, the power of ζMBP is 0.479 and 0.71 for ζMSA when data are from the main effects model. When time × treatment interactions are relatively strong, the two tests are comparable.
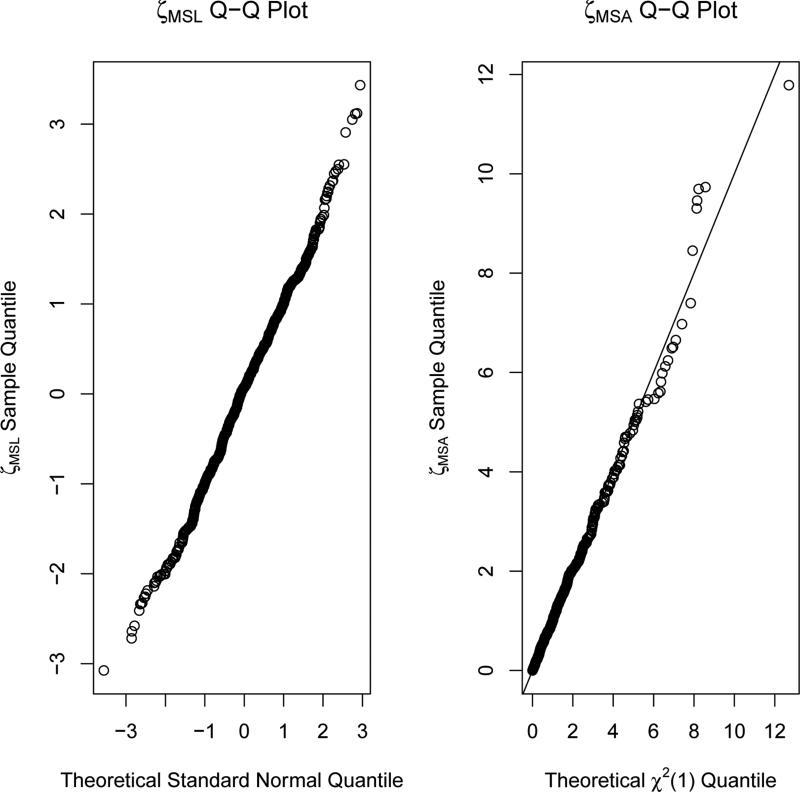
To investigate the performance in controlling the type I error rate for the proposed test statistics, we generated Q-Q plots of the proposed test statistics ζMSL and ζMSA under the null. These plots are presented in Figure 1 and they follow the theoretical distributions quite well, demonstrating their good performance for controlling the type I error rate in testing the last time point and overall treatment effects.
Figure 1.
Model Selection Test Statistics Distributions Under the Null
In practice, researchers could test for interaction first. If the interaction term comes out non-significant, the reduced main effects model is then used for assessing the treatment effect. However, this two-stage procedure is essentially another variable/model selection-based procedure and again has the stochastic nature associated with the selected model. Due to the stochastic nature of determining whether the full or main effects model is used, this procedure could fail to control the type I error rate. In the above simulation settings for Table 1 and 2, the type I error rate of this two-stage procedure is 0.085 while it is 0.048 for our proposed test statistics.
To further compare the proposed strategy with the ANCOVA method for testing the treatment effect at the last time point as suggested by one of the reviewers, we design the following simulation setting with the baseline measurement as a covariate and the response as the raw measurement at each post-baseline time point, instead of the change from the baseline.
where xij0 ~ N(0, 1). All other specifications are the same as that in model (5). When testing the last time point effect, we consider the ANCOVA model test statistics where is the MLE from the following ANCOVA model: xijT = α0 + αTrtI(i = 1) + αxxij0 + εij. Correspondingly, for all mixed effects models, the baseline response measurement xij0 is included as a fixed effect covariate. The model selection procedures proceed exactly the same as mentioned previously. Simulation results are presented in Tables 3 and 4 for testing the last time point and overall treatment effects.
Table 3.
Last Time Point Treatment Effect Testing with Baseline as a Covariatea
| Data Model | Interaction | Statisticsb | Power | Type I Error |
|---|---|---|---|---|
| Full (CS) | Strong | ζ t | 0.531 | 0.059 |
| ζ ANCOVA | 0.573 | 0.053 | ||
| ζ FUN | 0.579 | 0.053 | ||
| ζ MSL | 0.951 | 0.042 | ||
| Moderate | ζ t | 0.531 | 0.059 | |
| ζ ANCOVA | 0.573 | 0.053 | ||
| ζ FUN | 0.579 | 0.053 | ||
| ζ MSL | 0.988 | 0.042 | ||
| Weak | ζ t | 0.531 | 0.059 | |
| ζ ANCOVA | 0.573 | 0.053 | ||
| ζ FUN | 0.579 | 0.053 | ||
| ζ MSL | 0.988 | 0.042 | ||
| Main Effects (CS) | None | ζ t | 0.531 | 0.059 |
| ζ ANCOVA | 0.573 | 0.053 | ||
| ζ FUN | 0.579 | 0.053 | ||
| ζ MSL | 0.942 | 0.042 | ||
results based on 1000 simulations with 100 subjects at each treatment arm
model selection via BIC with 200 bootstrap resampling
ζt: t-test for last time point effect
ζANCOVA: test last time point effect via ANCOVA model
ζFUN: test last time point effect via full mixed effect model with UN covariance
ζMSL: model selection test statistics for testing last time point effect
Table 4.
Overall Treatment Effects Testing with Baseline as a Covariatea
| Data Model | Interaction | Statisticsb | Power | Type I Error |
|---|---|---|---|---|
| Full (CS) | Strong | ζ MBP | 1.000 | 0.047 |
| ζ MSA | 1.000 | 0.042 | ||
| Moderate | ζ MBP | 1.000 | 0.047 | |
| ζ MSA | 1.000 | 0.042 | ||
| Weak | ζ MBP | 0.917 | 0.047 | |
| ζ MSA | 0.988 | 0.042 | ||
| Main Effects (CS) | None | ζ MBP | 0.775 | 0.047 |
| ζ MSA | 0.842 | 0.042 | ||
results based on 1000 simulations with 100 subjects at each treatment arm
model selection via BIC with 200 bootstrap resampling
ζMBP: mixed effects model based likelihood ratio test for overall effects
ζMBP: model selection statistics for testing overall treatment effects
Inspecting Table 3, we see that the ANCOVA model is more powerful than the t-test method. The proposed test statistic, ζMSL, still has the best performance among the four test statistics while controlling the type I error rate at the same level. Results in Table 4 show that the proposed test statistic ζMSA outperforms ζMBP for testing the overall treatment effects when the raw score is used as the outcome instead of the change from the baseline while the baseline measurement is treated as a fixed effects covariate.
Case II (Missing Data Scenarios): Under the repeated measurement design, missing data are frequently observed due to, for example, patient withdrawal related or unrelated to their treatment or response sequence. Statistical analysis with missing data becomes more complicated and challenging. We next investigate the impact of missing data on the proposed tests. The first scenario is missing completely at random (MCAR) where the numbers of missing observations per treatment arm are (5, 5, 5, 5, 20) for the five measurement time points, respectively, and missing data are randomly selected across all samples. Thus the missing probability does not depend on any observed or unobserved variables. In this simulation, data are generated in the same manner as those in Tables 1 and 2 except that we randomly mark some observations as missing. The results for data with moderate time × treatment interaction or no interaction are presented in Table 5.
Table 5.
Missing Completely At Random (MCAR)a
| Data Model | Interaction | Statisticsb | Hypothesis Test | Powerc | Type I Errorc |
|---|---|---|---|---|---|
| Full (CS) | Moderate | ζ t | Last Time Point | 0.267(0.336) | 0.042(0.044) |
| ζ FUN | 0.288(0.343) | 0.046(0.046) | |||
| ζ MSL | 0.989(0.987) | 0.050(0.048) | |||
| ζ MBP | Overall Effect | 0.961(0.967) | 0.038(0.044) | ||
| ζ MSA | 0.995(0.995) | 0.050(0.048) | |||
| Main Effects (CS) | None | ζ t | Last Time Point | 0.267(0.336) | 0.042(0.044) |
| ζ FUN | 0.288(0.343) | 0.046(0.046) | |||
| ζ MSL | 0.699(0.710) | 0.050(0.048) | |||
| ζ MBP | Overall Effect | 0.459(0.479) | 0.038(0.044) | ||
| ζ MSA | 0.699(0.710) | 0.050(0.048) | |||
results based on 1000 simulations with 100 subjects at each treatment arm
model selection via BIC with 200 bootstrap resampling
values inside parentheses are based on no missing scenario
ζt: t-test for last time point effect
ζFUN: test last time point effect via full mixed effect model with UN covariance
ζMSL: model selection test statistics for testing last time point effect
ζMBP: mixed effects model based likelihood ratio test for overall effects
ζMSA: model selection statistics for testing overall effects
Comparing the numbers in Table 5 outside and inside the parentheses, which correspond to the analysis results from data with and without missing observations, it shows that MCAR had minimal impact on either type I error or power for both the mixed effects model based and the proposed model selection based test statistics. This is probably due to the relatively small proportion of missing observations. However, the changes in type I error and power are noticeable for the simple t-test, i.e. type I error is slightly inflated and power drops noticeably with missing data. The power of ζFUN in testing the last time point effect suffers noticeably when there exist data missing completely at random. This demonstrates the robustness of the proposed methods in dealing with MCAR. Furthermore, the power gains as we observed previously for ζMSA over ζMBP and ζMSL over ζt & ζFUN still hold which further reveal the efficiency and robustness of the proposed methods.
For many clinical studies, missing mechanisms may not be completely at random and depend on some observed baseline covariates, or missing at random (MAR). Next we investigate the performance of the proposed framework under MAR. In this simulation, data are generated similarly to those in Table 5 except that an additional covariate uij, associated with the response variable, is introduced and used for inducing missing data:
| (6) |
where uij ~ Unif(−1, 1). The missing data are generated as: for the treatment arm 0 and one of the 1st four time points k (k = 1, 2, 3, 4), five subjects with uij > 0 are randomly selected to have their observations set to missing, while for the treatment arm 1, 5 subjects with uij < 0 are randomly selected to create missing observations. For the last time point, we randomly select 20 subjects in the treatment arm 0 with uij > 0.5 and another 20 subjects from the treatment arm 1 with uij < −0.5 and set their observations to missing. We set α1, · · · , α4 to be moderate or 0.
Instead of imputing missing data, we analyze the simulated data with models (1), (3) and (4) except that we add baseline uij as a covariate into these models. Under this setting, the baseline covariate uij is imbalanced and becomes a confounding variable. As a further comparison, when testing the last time point effect, we include the ANCOVA model test statistics where is the MLE from the following ANCOVA model: yijT = α0 + αTrtI(i = 1) + αuuij + εij. The simulation results are presented in Table 6.
Table 6.
Missing At Random (MAR)a
| Data Model | Interaction | Statisticsb | Hypothesis Test | Powerc | Type I Errorc |
|---|---|---|---|---|---|
| Full (CS) | Moderate | ζ t | Last Time Point | 0.193(0.328) | 0.060(0.047) |
| ζ ANCOVA | 0.285(0.327) | 0.064(0.044) | |||
| ζ FUN | 0.310(0.343) | 0.049(0.049) | |||
| ζ MSL | 0.989(0.987) | 0.054(0.046) | |||
| ζ MBP | Overall Effect | 0.968(0.965) | 0.054(0.044) | ||
| ζ MSA | 0.995(0.995) | 0.054(0.046) | |||
| Main Effects (CS) | None | ζ t | Last Time Point | 0.176(0.325) | 0.060(0.047) |
| ζ ANCOVA | 0.265(0.331) | 0.064(0.044) | |||
| ζ FUN | 0.303(0.338) | 0.049(0.049) | |||
| ζ MSL | 0.689(0.710) | 0.054(0.046) | |||
| ζ MBP | Overall Effect | 0.463(0.486) | 0.054(0.044) | ||
| ζ MSA | 0.689(0.710) | 0.054(0.046) | |||
results based on 1000 simulations with 100 subjects at each treatment arm
model selection via BIC with 200 bootstrap resampling
values inside parentheses are based on no missing scenario
ζt: t-test for last time point effect
ζANCOVA: test last time point effect via ANCOVA model
ζFUN: test last time point effect via full mixed effect model with UN covariance
ζMSL: model selection test statistics for testing last time point effect
ζMBP: mixed effects model based likelihood ratio test for overall effects
ζMSA: model selection statistics for testing overall effects
Comparing the numbers in Table 6 outside and inside the parentheses which correspond to the analysis results from the data with and without missing observations, it demonstrates that MAR appears to have an effect on the type I error rate for the t-test. For example, for the full data without missing observations, the type I error is 0.047 but this number is inflated to 0.06 with missing data for the situation with moderate interaction. By contrast, the type I errors for the proposed methods are well controlled. Further, the impact of missing data on the power is larger for the t-test and ζFUN than that for the proposed methods. For instance, the power, 0.325 for the data without missing, is sharply reduced to 0.176 with missing data for the scenario of no interaction. In this scenario, the power change using ζFUN for testing the last time point effect is minor, i.e. from 0.338 to 0.303. Also the power gain of ζMSA over ζMBP is consistent with the results reported in the previous tables. The power gain of ζMSL over t-test and ζFUN is even more dramatic, further indicating the efficiency and robustness of the proposed methods. Due to the missing data, the baseline covariates become imbalanced and confounded with the response values. Compared with the t-test, ANCOVA statistics are more robust to missing data by adjusting for imbalanced baseline covariates as confounding factors [3]. Clearly, the t-test is invalid under this scenario since it violates the randomized controlled trial assumption that there exists no systematic differences for the baseline covariates.
In practice, missing mechanisms can be more complicated than MAR for repeated measurement studies where missing mechanism depends on unobserved variables, resulting in the so called missing not at random (MNAR) data. Our next simulation considers MNAR data. Specifically, we analyze the same data as simulated under MAR, except that the baseline covariate uij is now assumed to be unobserved, but instead we have another covariate vij which is observed and correlated with uij as follows:
Similar to the analysis of MAR data, we analyze the simulated data with models (1), (3) and (4) but we add the observed baseline vij instead of uij as a covariate into these models. The simulation results are presented in Table 7.
Table 7.
Missing Not At Random (MNAR)a
| Data Model | Interaction | Statisticsb | Hypothesis Test | Powerc | Type I Errorc |
|---|---|---|---|---|---|
| Full (CS) | Moderate | ζ t | Last Time Point | 0.171(0.327) | 0.061(0.043) |
| ζ FUN | 0.251(0.342) | 0.062(0.051) | |||
| ζ MSL | 0.989(0.985) | 0.051(0.050) | |||
| ζ MBP | Overall Effect | 0.955(0.963) | 0.052(0.048) | ||
| ζ MSA | 0.994(0.994) | 0.051(0.050) | |||
| Main Effects (CS) | None | ζ t | Last Time Point | 0.168(0.338) | 0.061(0.043) |
| ζ FUN | 0.262(0.347) | 0.062(0.051) | |||
| ζ MSL | 0.671(0.690) | 0.051(0.050) | |||
| ζ MBP | Overall Effect | 0.430(0.476) | 0.052(0.048) | ||
| ζ MSA | 0.671(0.690) | 0.051(0.050) | |||
results based on 1000 simulations with 100 subjects at each treatment arm
model selection via BIC with 200 bootstrap resampling
values inside parentheses are based on no missing scenario
ζt: t-test for last time point effect
ζFUN: test last time point effect via full mixed effect model with UN covariance
ζMSL: model selection test statistics for testing last time point effect
ζMBP: mixed effects model based likelihood ratio test for overall effects
ζMSA: model selection statistics for testing overall effects
From Table 7, it is evident that similar conclusions hold here. The t-test is sensitive to MNAR data for both the type I error and power. However, the proposed methods are robust to missing data and have well controlled type I error. Again, the power gain for ζMSA over ζMBP and ζMSL over t-test and ζFUN under MNAR persists as seen previously. In practice, we can adjust for these baseline values by including them as covariates in the proposed analysis procedures. Provided they are good surrogate variables of the unobserved covariates related to the missing data, they can help to reduce potential bias in the treatment effect analysis and boost the testing power.
4. Real Data Analysis
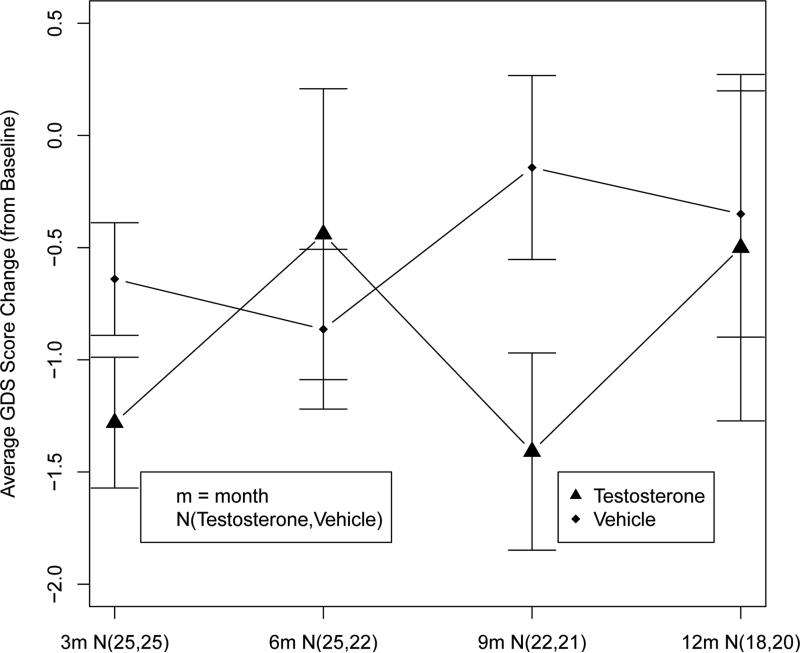
To illustrate a practical application of the proposed methods, we apply them to a clinical trial [15] regarding the cognitive effect of testosterone on geriatric men (age ≥ 65). In this study, 60 subjects were recruited and randomly assigned to two treatment groups, testosterone (Trt = 1) and vehicle (Trt = 0). The primary outcome of the study is the geriatric depression scale (GDS) which is continuous and ranges from 0 to 30. Higher GDS scores indicate more severe depression. GDSs for the subjects were repeatedly measured at baseline, 3, 6, 9, and 12 months. Our primary interest is to compare the overall treatment effects between the two treatment groups across time.
The summary of the data is depicted in Figure 2 where the average GDS scores change from baseline for each of the time by treatment group is presented. It seems that there exists some time by treatment interactions although the interaction terms are not significantly strong enough to be detected by the BIC model selection procedure or the likelihood ratio test statistics (p-value=0.10).
Figure 2.
Average GDS Score Change (from Baseline) Comparison
Based on BIC model selection criteria, the main effects only model with unstructured covariance structure best fits this dataset. Therefore, the overall treatment effects testing reduces to testing the hypothesis H0 : αTrt = 0 vs Ha : αTrt ≠ 0 by using the proposed test statistic . With 200 rounds restricted cluster bootstrapping for variance estimates, we conclude that there exists an overall significant (p-value=0.04) difference between the two treatment groups. For a further comparison, we also tested the overall treatment effects using ζMBP with the grand full mixed effects model, and the p-value is 0.04. This result is consistent with what we observed in the simulation study, i.e. the performance of ζMBP is comparable to that of ζMSA in testing the overall treatment effects when the time by treatment interaction is not weak. However, if the t-test was used to evaluate the treatment effect with only the last time point data, we would obtain an insignificant treatment effect (p-value=0.87).
5. Discussion
In this paper, we proposed a new analytic framework for repeated measurement randomized trial data based on the mixed effects model. The proposed analysis scheme combines a model selection procedure with a testing procedure. The model selection procedure makes use of the information from the optimal model deemed for the data for testing treatment effect(s) to increase power. To ensure the validity of the post-model selection inference, we further developed a restricted cluster bootstrapping variance estimation strategy to estimate the (co)variance of the treatment effect estimate(s). Our proposed test statistics take into account the stochastic nature of the model selection process such that the proposed test statistics asymptotically follow χ2 distributions under the null. This provides us the validity to control the type I error rate. As shown by our simulations, the proposed method outperforms the simple t-test, the grand full mixed effects model and the likelihood ratio test (LRT) between the grand full model and null model. The power gain for our proposed method over the t-test and the grand full mixed effects model for testing the last time point effect is substantial across all of our simulation scenarios. Compared to the LRT from the full mixed effects model, our method is superior under most of the simulation scenarios, except that the two methods are comparable when the data are generated from the full model with strong time by treatment interaction. In practice, sometimes the clinical trial protocol may specify the use of the full mixed effects model to test the last time point treatment effect regardless of whether the full mixed effects model best fits the data or not. For protocol integrity purposes, the proposed methodologies no longer apply. It is worth mentioning that in the repeated measurement clinical trial setting, an alternative to the mixed effects model approach considered in this paper, is a (weighted) summary measures method, which uses the (weighted) post-baseline measurements mean difference as the estimator of the treatment effects, and it can be efficient under some scenarios. In this regard, more details about these schemes could be found in [16, 17]. Furthermore, the performance of ANCOVA model on the mean of all post-baseline treatment values for evaluating the overall treatment effects deserves further investigation.
The simulations also reveal that the substantial power gain of the proposed methods over the t-test and the LRT from the mixed effects model remains for data simulated under various missing mechanisms. The proposed analytic procedure is more robust to missing data than the simple t-test because of its flexibility in adding covariates related to missing data directly or indirectly (i.e. surrogate variables) to reduce the bias due to missing data. However, this paper does not intend to comprehensively resolve the missing data problem and our simulations on missing data are simple and used strictly to demonstrate the performance of the proposed method over the other existing methods. How the proposed methods behave for data under more complicated missing mechanisms deserves further attention and is beyond the scope of this paper. Note, the proposed analysis framework is based on continuous data. Extending the proposed framework to other response data types, such as binary responses or censored survival data should be investigated.
Acknowledgement
Zou and Shuster were partially supported by NIH grant 1UL1TR000064 from the National Center for Advancing Translational Sciences and Zhou was partially supported by NIH grants R01ES021900 and P01CA142538. Authors would like to thank the editor and two anonymous reviewers for their constructive comments.
References
- 1.Frison L, Pocock SJ. Repeated measures in clinical trials: analysis using mean summary statistics and its implications for design. Statistics in Medicine. 1992;11:1685–1704. doi: 10.1002/sim.4780111304. [DOI] [PubMed] [Google Scholar]
- 2.Wollert KC, Meyer GP, Lotz J, Ringes-Lichtenberg S, Lippolt P, Breidenbach C, Fichtner S, Korte T, Hornig B, Messinger D, Arseniev L, Hertenstein B, Ganser A, Drexler H. Intracoronary autologous bone-marrow cell transfer after myocardial infarction: the BOOST randomised controlled clinical trial. Lancet. 2004;364:141–148. doi: 10.1016/S0140-6736(04)16626-9. [DOI] [PubMed] [Google Scholar]
- 3.Senn SJ. Covariate imbalance and random allocation in clinical trials. Statistics in Medicine. 1989;8(4):467–475. doi: 10.1002/sim.4780080410. [DOI] [PubMed] [Google Scholar]
- 4.Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38(4):963–974. [PubMed] [Google Scholar]
- 5.Bergenstal RM, Wysham C, Macconell L, Malloy J, Walsh B, Yan P, Wilhelm K, Malone J, Porter LE. Efficacy and safety of exenatide once weekly versus sitagliptin or pioglitazone as an adjunct to metformin for treatment of type 2 diabetes (DURATION-2): a randomised trial. Lancet. 2010;376(9739):431–439. doi: 10.1016/S0140-6736(10)60590-9. [DOI] [PubMed] [Google Scholar]
- 6.Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19(6):716–723. [Google Scholar]
- 7.Schwarz GE. Estimating the dimension of a model. Annals of Statistics. 1978;6(2):461–464. [Google Scholar]
- 8.Hurvich CM, Tsai CL. Regression and time series model selection in small samples. Biometrika. 1989;76:297–307. [Google Scholar]
- 9.Shao J. Bootstrap model selection. Journal of the American Statistical Association. 1996;91(434):655–665. [Google Scholar]
- 10.Shao J. An asymptotic theory for linear model selection. Statistica Sinica. 1997;7:221–264. [Google Scholar]
- 11.Yang Y. Can the strengths of AIC and BIC be shared? A conflict between model identification and regression estimation. Biometrika. 2005;92:937–950. [Google Scholar]
- 12.Niu F, Pu P. Selecting mixed-effects models based on generalized information criterion. Journal of Multivariate Analysis. 2006;97:733–758. [Google Scholar]
- 13.Carlstein E. The use of subseries methods for estimating the variance of a general statistic from a stationary time series. Annal of Statistics. 1986;14:1171–1179. [Google Scholar]
- 14.Politis DN, Romano JP. The Stationary Bootstrap. Journal of American Statistical Association. 1994;89:1303–1313. [Google Scholar]
- 15.Borst SE, Yarrow JF, Fernandez C, Conover CF, Ye F, Meuleman JR, Morrow M, Zou B, Shuster JJ. Cognitive effects of testosterone and finasteride administration in older hypogonadal men. Clinical Interventions in Aging. 2014;2014(9):1327–1333. doi: 10.2147/CIA.S61760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senn S, Stevens L, Chaturvedi N. Repeated measures in clinical trials: simple strategies for analysis using summary measures. Statistics in Medicine. 2000;19(6):861–877. doi: 10.1002/(sici)1097-0258(20000330)19:6<861::aid-sim407>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 17.Bamia C, White IR, Kenward MG. Some consequences of assuming simple patterns for the treatment effect over time in a linear mixed model. Statistics in Medicine. 2013;32(15):1171–1179. doi: 10.1002/sim.5707. [DOI] [PubMed] [Google Scholar]