Abstract
Objective
To determine normative ranges for fetal ocular biometrics between 19 and 38 weeks gestational age (GA) using volumetric MRI reconstruction.
Method
3D images of 114 healthy fetuses between 19 and 38 weeks GA were created using super-resolution volume reconstructions from MRI slice acquisitions. These 3D images were semi-automatically segmented to measure fetal orbit volume, binocular distance (BOD), interocular distance (IOD), and ocular diameter (OD).
Results
All biometry correlated with GA (Volume, CC = 0.9680; BOD, CC = 0.9552; OD, CC = 0.9445; and IOD, CC = 0.8429), and growth curves were plotted against linear and quadratic growth models. Regression analysis showed quadratic models to best fit BOD, IOD and OD, and a linear model to best fit volume.
Conclusion
Orbital volume had the greatest correlation with GA, though BOD and OD also showed strong correlation. The normative data found in this study may be helpful for the detection of congenital fetal anomalies with more consistent measurements than are currently available.
Keywords: MRI, fetal development, orbit, ocular, eyes, biometrics, volume, gestational age
INTRODUCTION
Fetal ocular biometry is a valuable resource for the evaluation of fetal growth and as a clue to the detection of congenital abnormalities during pregnancy1. Conditions such as hypotelorism, hypertelorism, micro- or anophthalmia and cataracts can be indicative of many congenital anomalies and syndromes2, 3, and the orbits, being conspicuous and easily measured, provide an accessible tool for imagers and clinicians to use as they attempt to determine a proper course of action as early as possible4-6. Timely and accurate diagnosis can be crucial to improving prognosis, prenatal consultation, and management of pregnancy and birth. Several published studies have characterized normal fetal eye development using both ultrasound (US)6-13 and, more recently, fetal magnetic resonance imaging (MRI)5, 14-16. However, these studies have lacked consistent methods for measurement, data interpretation and image acquisition. Clinically, fetal growth can be quantified and compared with normative data from corresponding gestational age (GA) weeks to identify and describe the condition of the fetus Some studies have investigated the relationship between disease and fetal ocular growth17, 18.
Due to the differences of tissue visualization in MRI and US, different anatomical landmarks are sometimes used for biometric measurements and in result data may have biases depending on the image modality. Thus, it is important to reestablish normal values for fetal ocular growth when using MRI, which has several advantages over US in the detection and visualization of craniofacial defects, including: sharper image quality, improved soft-tissue contrast, the capability to collect useful images regardless of fetal position, and the ability to accurately measure non-oblique images3.
When the fetal head is fully engaged in the maternal pelvis it is often possible to better visualize fetal craniofacial features on MRI than on US because MRI provides better soft-tissue contrast and can better show fetal anatomic structures which cannot be seen by sonography as they are made of or are surrounded by bone3, 19. Engagement of the fetal head into the maternal pelvis also has the benefit of reducing fetal head motion. Fetuses at more advanced gestational ages are even more vulnerable to limitations of US because fetal crowding limits the amount of amniotic fluid surrounding the fetal head acting as a contrasting background to the structure of interest7.
One shortcoming of many previous US and MRI studies is the use of 2D images for measurements. Two-dimensional (2D) images may be oblique, which can cause the selection of false anatomical landmarks. Due to the nature of fetal MRI, which involves relatively slow acquisitions that are susceptible to fetal motion, acquiring perfectly orthogonal images can be difficult and, due to the small size of the region of interest in fetal ocular biometry, there may not be an optimal orthogonal slice showing both orbits for accurate distance measurements. Volumetric or 3D images, on the other hand, ensure that the appropriate voxels are chosen for the desired landmark, as orthogonal reconstructions can be created from oblique, suboptimal images. In addition, 3D methodology enables volume measurements for the orbits and lens. Odeh et al.(2009)7 measured orbit volume using 3D ultrasound technology (3DUS). However, the creation of the 3D model was based on a 2D image and assumed the shape to be a perfect sphere, whereas it has been shown that the fetal orbit may be better characterized as an ellipsoid15, 20 which undergoes changes in shape throughout embryonic, fetal, and neonatal periods of human development. Using the same 3DUS technology, Bojikian et al. (2013)21 accounted for this by tracing the eye in six planes as well as reporting biometrics assuming a perfect spheroid shape.
Three-dimensional MRI is not easily achieved because intermittent fetal and maternal motion disrupts the spatial encoding necessary for 3D MRI acquisitions. Clinical fetal MRI is thus performed using high-quality, fast 2D acquisitions in order to freeze the motion of the fetus. Unfortunately, significant inter-slice motion artifacts appear in out-of-plane views when 3D images are rendered from the stacks of fast 2D slice acquisitions. These artifacts hinder volumetric 3D analysis of fetal ocular growth. However, recent advances in image analysis have allowed retrospective inter-slice motion correction and reconstruction of high-resolution volumetric images from multiple 2D MRI acquisitions22.
In this study, we sought to determine normal values for fetal ocular biometry and to characterize the pattern of ocular development between 19 and 38 weeks gestation using 3D super-resolution reconstructions22 created from MRI scans of 114 healthy fetuses. The GA range in this study, which covers the late second and the third trimesters, is one of rapid change and critical fetal development. The use of advanced image analysis allowed us, for the first time, to systematically compute four measurements of ocular growth in the fetus based on volumetric fetal MRI: binocular distance (BOD), interocular distance (IOD), average ocular diameter (OD), and average orbit volume. We used linear and polynomial regression analysis to determine the best fit model of growth for each biometric value in the relevant GA range.
MATERIALS AND METHODS
Data
This was a retrospective analysis of the MRI images of 114 fetuses between 19 and 38 weeks gestation imaged in the Department of Radiology at Boston Children’s Hospital. These exams included MRI scans performed between 2007 and September, 2014 involving 87 pregnant research volunteers whose fetuses had no known anomalies, and 27 clinical fetal MRI scans in which there were no significant fetal findings. The duration of MRI scans used for the measurements in this study was between 15 and 30 minutes.
The GA at the time of each MRI scan was calculated from the reported “expected date of delivery” (EDD) and the date of service of the MRI scans. All recruited research subjects gave informed consent prior to imaging and retrospective use of data was approved by the Boston Children’s Hospital Committee on Clinical Investigation.
The following inclusion criteria were applied to the selection of healthy fetuses used in our analysis: 1) documented neuroradiologist and pediatric radiologist reports of an anatomically and developmentally normal fetus, 2) absence of a documented fetal syndromic or chromosomal anomaly, and 3) singleton pregnancy. Exclusion criteria were: 1) Any reported congenital abnormality of the fetal brain, face or body and 2) fetuses whose images were of insufficient quality to perform volumetric reconstruction. Poor image quality was caused by a number of factors, including fetal motion, image artifacts, low signal-to-noise (SNR) ratio, and scans with a limited field of view that did not cover the entire fetal head.
Fetal MRI was performed on state of the art equipment from several vendors. Image acquisition settings are detailed in Table 1 and the frequency of use for each magnet and sequence is detailed in Table 2.
Table 1.
Magnet hardware and acquisition settings.
| Magnet | Sequence Name | TE (ms) | TR (ms) | Slice thickness (mm) | Matrix |
|---|---|---|---|---|---|
| 1.5 T Phillips | SShTSE | 120 | 1200 – 2500 | 2.0 – 4.0 | 204 × 256 |
| 1.5 T GE | SSFSE | 80 – 102 | 1357 – 4500 | 3.0 – 4.0 | 384 × 256 |
| 1.5 T GE | FIESTA | 1.34 – 2.04 | 3.18 – 4.56 | 5.0 – 6.0 | 340 × 192 |
| 1.5 T Siemens | HASTE | 60 | 2000 | 3.0 | 256 × 256 |
| 3.0 T Siemens | HASTE | 100 – 120 | 1400 – 1600 | 2.0 | 256 × 256 |
| 3.0 T Siemens | TRUFISP | 1.85 | 4.31 | 2.0 | 320 × 320 |
TE, echo time; TR, repetition time; SShTSE, Single-Shot Turbo Spin Echo; SSFSE, Single-Shot Fast Spin Echo; HASTE, Half-Fourier Acquired Single-shot Turbo spin Echo; FIESTA, Fast Imaging Employing Steady sTate Acquisition; TRUFISP, True Fast Imaging with Steady-state Precession.
Table 2.
Magnet use detailed. “Research” refers to normal recruited research subjects. “Clinical” refers to hospital scans that were performed for a clinical ultrasound indication or as a screening due to family history of an anomaly. FIESTA refers to the number of volumetric reconstructions created from FIESTA or TRUFISP acquisitions.
| Magnet | n | Research | Clinical | FIESTA | Dates acquired |
|---|---|---|---|---|---|
| 1.5 T Phillips | 60 | 59 | 1 | 0 | Jan 2007 – July 2011 |
| 1.5 T GE | 16 | 0 | 16 | 5 | Feb 2011 – Sep 2013 |
| 1.5 T Siemens | 2 | 0 | 2 | 0 | Dec 2012 – Feb 2013 |
| 3.0 T Siemens | 36 | 28 | 8 | 1 | Aug 2013 – Sep 2014 |
Image Reconstruction
After the scan was completed, a volumetric (3D) image was reconstructed from multiple sets of fast 2D slice acquisitions, including SShTSE, SSFSE, HASTE, or TRUFISP, depending on the magnet and the type of MR imaging. This volumetric image was obtained using a previously validated algorithm22 which iteratively corrects for inter-slice motion and reconstructs a 3D image by combining information from motion-corrected 2D slices. In this algorithm, fetal head motion is estimated per slice acquisition as a 6 degree-of-freedom rigid transformation in 3D space that aligns each slice within an estimated volumetric image of the fetal head. Motion-corrected slices are then combined within a robust error minimization framework and an improved estimation of the fetal head volumetric image is obtained. This image is then used as reference for slice motion correction in the next iteration, and the iterations of motion correction and volume reconstruction continue until they converge and a high-resolution motion-corrected volumetric image of the fetal head (including brain and orbits) is obtained. The reconstruction technique was successfully applied in other published work on fetal brain image analysis23, 24.
Measurements
Orbit segmentations from the 3D reconstructed images were made using the semi-automatic segmentation tool in ITK-SNAP25 (Figure 1). A small “seed” label was placed within each orbit manually, and the “Image edges” option for automatic snake segmentation was used to complete the segmentation of both orbits. Thus, the border of the segmentation was defined by the contrast in image intensity between the vitreous body (high signal) and the surrounding tissue of the sclera, choroid and ciliary body (low signal). A single operator executed all of the semi-automatic segmentations, and manually refined the label in the event of obvious program error, such as the label “spilling” out of the orbit into surrounding tissue, or the presence of a gap of labeled voxels within the shape of the orbit.
Figure 1.
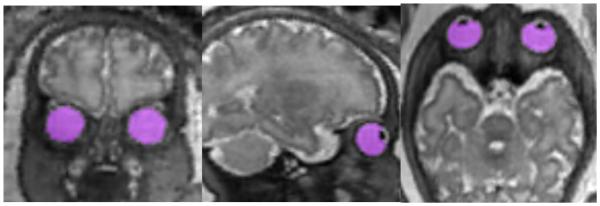
Super-resolution 3D reconstruction from 2D MRI acquisitions displayed in coronal, sagittal and axial directions with semi-automatic orbit segmentations (magenta) created in ITK-SNAP25.
The snake segmentation did not include the partial-volume voxels representing the border of the eye, where the vitreous meets the tissue of the sclera. In order to comply with the routine practice in manual measurements, in which calipers are placed manually on the outside border of the fluid-filled vitreous, the images were resampled from 1.0 mm3 to 0.5 mm3 resolution and the labels were dilated by 0.5 mm (half of the original voxel size) using a morphological dilation process to include the outside boundary of the orbit.
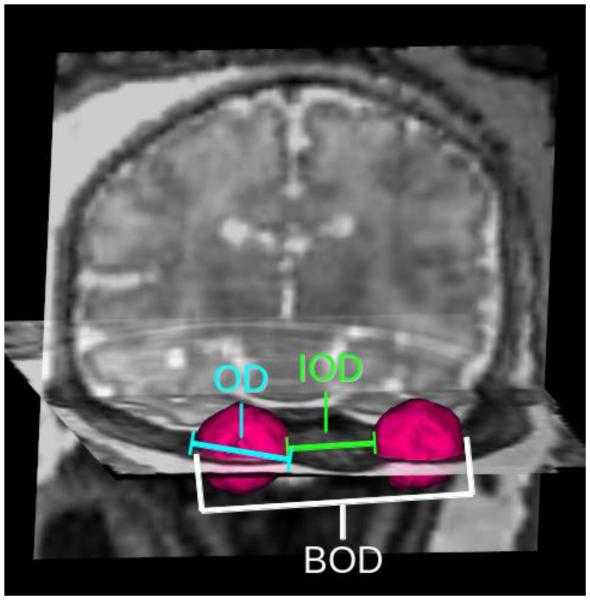
All measurements were computed by an automated algorithm (Figure 2). BOD was calculated as the maximum distance found between any two voxels (one from each orbit). IOD was calculated as the minimum distance found between two voxels (one from each orbit). OD was calculated as the maximum distance between any two voxels in a single orbit. The physical distance between two voxels was calculated based on the physical coordinates of the voxel boundaries. For the purposes of this study, OD for each case was reported as the averaged diameters of the two orbits. Orbital volume was calculated based on the number of voxels contained within each orbit multiplied by the volume of each voxel in cubic millimeters. Volume was reported as the averaged volume of the two orbits in each fetus.
Figure 2.
Visual representation of ocular biometrics. The 3D model of the orbit segmentation (pink) is overlaid on coronal and sagittal slices taken from a 3D reconstruction of the fetal head. (a) Binocular distance (BOD), the greatest distance between two voxels from opposite orbits; (b) interocular distance (IOD), the smallest distance between two voxels from opposite orbits; (c) ocular diameter* (OD), the greatest distance between two voxels from a single orbit. Volume* is calculated as the total number of voxels in the orbit segmentation multiplied by voxel volume.
*Average of left and right measurements.
Statistical analysis
MATLAB (Mathworks, Natick, MA), and the STATA software (Release 13. StataCorp LP, College Station, TX), were used for processing the data and for statistical analysis. All ocular biometric measurements were correlated with gestational age, and were analyzed against gestational age using linear and polynomial regression models, where mean, standard deviation, 5%, 50% and 95% confidence intervals, and goodness-of-fit were calculated.
RESULTS
Data summary including mean and standard deviation (SD) for BOD, IOD, OD and volume from 19 to 38 weeks gestation are displayed in Table 3.
Table 3.
Data summary. Mean and standard deviation (SD) per gestational age (GA) week for normative binocular distance (BOD), interocular distance (IOD), ocular diameter* (OD) and volume*, as well as the number of fetuses imaged at each GA are displayed.
| Gestational age (weeks) |
n | BOD |
IOD |
OD |
Volume |
||||
|---|---|---|---|---|---|---|---|---|---|
| mean (mm) |
SD | mean (mm) |
SD | mean (mm) |
SD | mean (mL) |
SD | ||
| 19 | 1 | 29.51 | - | 13.00 | - | 9.36 | - | 0.32 | - |
| 20 | 2 | 31.34 | 1.77 | 13.02 | 0.03 | 9.76 | 0.64 | 0.34 | 0.06 |
| 21 | 5 | 32.28 | 1.33 | 13.84 | 1.28 | 10.17 | 0.38 | 0.39 | 0.05 |
| 22 | 3 | 35.77 | 1.88 | 14.03 | 0.98 | 11.56 | 0.48 | 0.56 | 0.06 |
| 23 | 5 | 36.08 | 1.39 | 15.43 | 0.84 | 11.72 | 0.92 | 0.54 | 0.08 |
| 24 | 4 | 37.52 | 0.65 | 16.00 | 1.15 | 12.18 | 0.53 | 0.67 | 0.07 |
| 25 | 1 | 42.88 | - | 17.22 | - | 13.37 | - | 0.80 | - |
| 26 | 7 | 42.08 | 1.09 | 16.60 | 0.77 | 13.81 | 0.52 | 0.98 | 0.10 |
| 27 | 8 | 42.97 | 1.40 | 17.13 | 0.83 | 13.79 | 0.68 | 1.02 | 0.16 |
| 28 | 16 | 45.27 | 1.36 | 17.73 | 0.63 | 14.77 | 0.63 | 1.20 | 0.09 |
| 29 | 11 | 46.19 | 1.48 | 17.69 | 0.96 | 15.07 | 0.50 | 1.36 | 0.19 |
| 30 | 7 | 46.65 | 1.12 | 17.58 | 0.54 | 15.40 | 0.62 | 1.38 | 0.10 |
| 31 | 9 | 47.37 | 1.46 | 18.44 | 1.56 | 15.76 | 0.96 | 1.38 | 0.09 |
| 32 | 5 | 48.31 | 2.95 | 18.80 | 2.18 | 15.58 | 0.46 | 1.47 | 0.09 |
| 33 | 8 | 50.75 | 2.32 | 18.84 | 2.03 | 17.05 | 0.67 | 1.78 | 0.12 |
| 34 | 3 | 51.54 | 2.26 | 20.10 | 0.18 | 16.64 | 0.81 | 1.80 | 0.12 |
| 35 | 6 | 53.61 | 1.95 | 20.33 | 1.02 | 17.37 | 0.70 | 2.07 | 0.23 |
| 36 | 4 | 53.17 | 1.14 | 20.50 | 1.29 | 17.18 | 0.39 | 2.05 | 0.07 |
| 37 | 7 | 52.80 | 1.28 | 19.61 | 1.23 | 17.66 | 0.45 | 2.12 | 0.14 |
| 38 | 2 | 55.47 | 0.18 | 21.51 | 0.69 | 18.22 | 1.00 | 2.15 | 0.23 |
Average of left and right measurements.
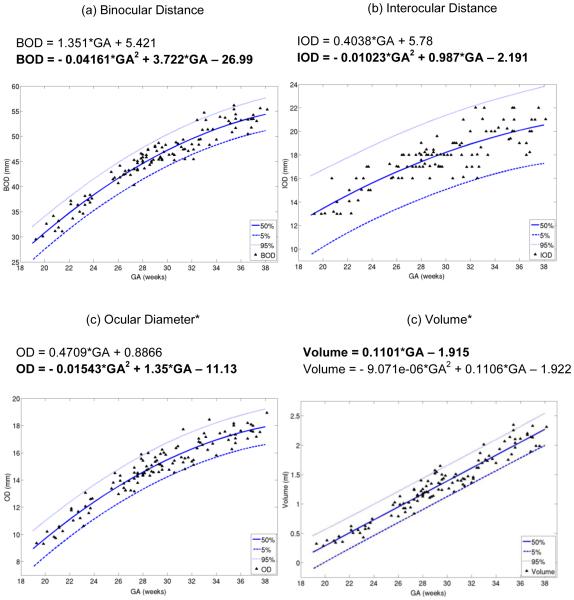
The goodness-of-fit measures, adjusted R-squared and Root Mean Square Error (RMSE), values for linear and quadratic models of growth for BOD, IOD, OD and volume are displayed in Table 4. All measurements exhibited high correlation between GA and BOD, IOD, OD, and volume. For the GA range in this study of 19-38 weeks, a quadratic model of growth was the best fit for BOD, IOD and OD (P < .001), but the quadratic model appeared to over fit orbit volume growth with a P value of 0.986 for the quadratic term. Volume did, however, fit a linear model of growth (P < .001). The order of Pearson’s correlation coefficient with GA for each biometric, from greatest correlation to least, was volume (CC = 0.9680), BOD (CC = 0.9552), OD (CC = 0.9445) and IOD (CC = 0.8429).
Table 4.
Pearson’s correlation coefficient and regression analysis between biometry and gestational age for linear and quadratic models of growth.
| Measure | CC | Linear Adj R2 |
Linear RMSE |
Quadratic Adj R2 |
Quadratic RMSE |
|---|---|---|---|---|---|
| BOD (mm) | 0.9552 | 0.9117 | 1.8767 | 0.9376 | 1.5771 |
| IOD (mm) | 0.8429 | 0.7079 | 1.1634 | 0.7192 | 1.1407 |
| OD* (mm) | 0.9445 | 0.8912 | 0.7329 | 0.9199 | 0.6288 |
| Volume* (mL) | 0.9680 | 0.9365 | 0.1323 | 0.9360 | 0.1329 |
CC, Pearson’s correlation coefficient; Adj R2, adjusted r-squared; RMSE, root mean square error; BOD, binocular distance; IOD, interocular distance; OD, ocular diameter*.
Average of left and right measurements.
The 5%, 50% and 95% confidence intervals based on the regression analysis for each biometric are displayed in Table 5. Each biometric was plotted against their respective models of growth in Figure 3.
Table 5.
Normative biometric values in relation to gestational age fitted to the mean. 5%, 50% and 95% confidence intervals for binocular distance (BOD), interocular distance (IOD), ocular diameter* (OD) and volume* by gestational age.
| Gestational age (weeks) |
BOD |
IOD |
OD |
Volume |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5% | 50% | 95% | 5% | 50% | 95% | 5% | 50% | 95% | 5% | 50% | 95% | |
| 19 | 25.37 | 28.71 | 32.05 | 10.45 | 12.87 | 15.29 | 7.62 | 8.95 | 10.29 | −0.11 | 0.18 | 0.46 |
| 20 | 27.54 | 30.81 | 34.08 | 11.09 | 13.46 | 15.82 | 8.40 | 9.70 | 11.01 | 0.01 | 0.29 | 0.56 |
| 21 | 29.60 | 32.83 | 36.05 | 11.69 | 14.02 | 16.36 | 9.13 | 10.42 | 11.71 | 0.13 | 0.40 | 0.67 |
| 22 | 31.57 | 34.76 | 37.95 | 12.26 | 14.57 | 16.88 | 9.84 | 11.11 | 12.38 | 0.24 | 0.51 | 0.78 |
| 23 | 33.44 | 36.61 | 39.78 | 12.81 | 15.10 | 17.39 | 10.50 | 11.76 | 13.03 | 0.35 | 0.62 | 0.89 |
| 24 | 35.22 | 38.38 | 41.53 | 13.32 | 15.60 | 17.89 | 11.13 | 12.39 | 13.65 | 0.46 | 0.73 | 0.99 |
| 25 | 36.91 | 40.06 | 43.21 | 13.81 | 16.09 | 18.37 | 11.73 | 12.98 | 14.24 | 0.57 | 0.84 | 1.10 |
| 26 | 38.51 | 41.66 | 44.81 | 14.28 | 16.55 | 18.83 | 12.29 | 13.55 | 14.80 | 0.68 | 0.95 | 1.21 |
| 27 | 40.03 | 43.18 | 46.32 | 14.72 | 17.00 | 19.28 | 12.82 | 14.08 | 15.33 | 0.79 | 1.06 | 1.32 |
| 28 | 41.46 | 44.61 | 47.76 | 15.15 | 17.42 | 19.70 | 13.33 | 14.58 | 15.84 | 0.90 | 1.17 | 1.43 |
| 29 | 42.81 | 45.96 | 49.11 | 15.55 | 17.83 | 20.10 | 13.80 | 15.05 | 16.31 | 1.01 | 1.28 | 1.54 |
| 30 | 44.08 | 47.23 | 50.38 | 15.93 | 18.21 | 20.49 | 14.24 | 15.49 | 16.75 | 1.12 | 1.39 | 1.65 |
| 31 | 45.26 | 48.41 | 51.56 | 16.30 | 18.57 | 20.85 | 14.65 | 15.90 | 17.16 | 1.23 | 1.50 | 1.76 |
| 32 | 46.37 | 49.51 | 52.66 | 16.64 | 18.92 | 21.19 | 15.02 | 16.28 | 17.53 | 1.34 | 1.61 | 1.87 |
| 33 | 47.38 | 50.53 | 53.68 | 16.96 | 19.24 | 21.51 | 15.37 | 16.63 | 17.88 | 1.45 | 1.72 | 1.98 |
| 34 | 48.31 | 51.46 | 54.62 | 17.26 | 19.54 | 21.82 | 15.69 | 16.94 | 18.20 | 1.56 | 1.83 | 2.10 |
| 35 | 49.15 | 52.32 | 55.48 | 17.53 | 19.82 | 22.11 | 15.97 | 17.23 | 18.49 | 1.67 | 1.94 | 2.21 |
| 36 | 49.90 | 53.08 | 56.27 | 17.78 | 20.08 | 22.38 | 16.21 | 17.48 | 18.75 | 1.78 | 2.05 | 2.32 |
| 37 | 50.55 | 53.77 | 56.99 | 17.99 | 20.32 | 22.65 | 16.42 | 17.71 | 18.99 | 1.89 | 2.16 | 2.43 |
| 38 | 51.11 | 54.37 | 57.64 | 18.18 | 20.54 | 22.90 | 16.60 | 17.90 | 19.20 | 1.99 | 2.27 | 2.54 |
Average of left and right measurements.
Figure 3.
Biometric data and 5%, 50% and 95% confidence intervals. Quadratic and linear regression equations are displayed above each plot. BOD, IOD and OD best fit a quadratic model of growth, and volume best fits a linear model of growth for 19-38 weeks GA. (a) Binocular distance (BOD), (b) interocular distance (IOD), (c) ocular diameter* (OD) and (d) volume* plotted against gestational age (GA)
*Average of left and right measurements.
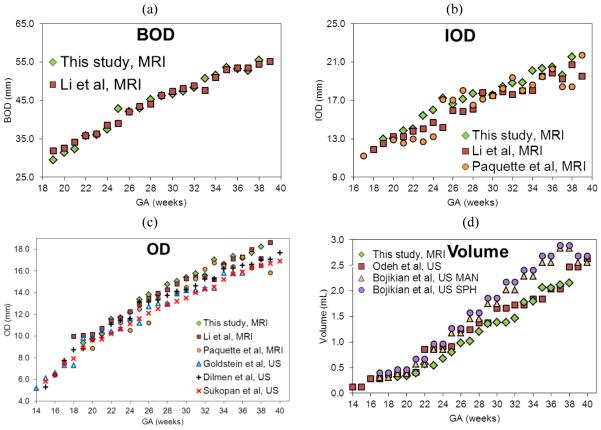
The data from this study were compared and plotted with data reported in other MRI and US studies in Figure 4. BOD, IOD and OD measurements were comparable to those found in other MRI studies 14, 15, as well as US studies which only shared OD as a biometric8, 9, 11. Our volume measurements showed a slight decrease when compared to previous US studies7, 21.
Figure 4.
Comparison of biometric values reported in fetal ocular research studies. (a) BOD compared with Li et al (2010).15, (b) IOD compared with Li et al.15 and Paquette et al. (2009)14, (c) OD* data compared with MRI studies14, 15 and US studies8, 9, 11, (d) volume* compared with Odeh et al. (2009)7 and two measurements from Bojikian et al. (2013)21; slice by slice tracing (MANUAL) and a perfect sphere with diameter set according to the internal limits of the eye (SPHERE). All biometrics are plotted against GA. BOD, binocular distance; IOD, interocular distance; OD, ocular diameter*; GA, gestational age; MRI, MRI-based measurements; US, ultrasound-based measurements.
*Average of left and right measurements.
DISCUSSION
This study demonstrates that MRI 3D reconstruction is an effective method for obtaining fetal ocular measurements and for establishing normative data for fetal ocular growth from 19-38 weeks gestation. The data in this study supports a quadratic longitudinal model of growth for BOD, IOD and OD in the 19 to 38 week GA range, and a linear model of growth for orbit volume. Previous studies have reported on various ocular biometrics during gestation. However, the bulk of the previous work used US data. To our knowledge, this is the first MRI study to report orbital volume, as well as the first study to use 3D MRI reconstructions, semi-automatic segmentation and automatic biometry to compute BOD, IOD, OD and volume measurements.
The quadratic growth model for BOD, IOD and OD found in this research is supported by other recent findings in Paquette et al. (2009)14 and Li et al. (2010)15, both MRI studies. The linear model of growth for volume in this study differed from the exponential model reported by Odeh et al. (2009)7. Earlier studies of ocular biometry, for the most part, reported linear growth patterns. These include US studies8, 9, 11, 12 and one early MRI study16. Birnholz et al. (1985)6 reported a pattern with a piece-wise linear fit. Goodall et al. (2009)26 reported a linear growth rate with two distinct phases in an MRI study of chick embryo growth. Most recently, Kivilevitch et al. (2010)10 reported a linear growth rate as measured by interlens distance using US. Robinson et al. (2008)5 reported a logarithmic growth pattern in an MRI study. Bojikian et al. (2013)21 assumed no growth model and implied that more complex models of growth may be at work for fetal ocular development.
Differences in visualization between US and MRI result in the selection of different landmarks as the orbit boundary. US ocular measurements are typically made from the bony medial and lateral orbital walls which surround the orbit, while MRI measurements use the edge of the brightly visualized vitreous body. Thus, biometric measurements from MRI may differ from their gestational age counterparts in US studies5, 14, 15, but more truly represent the 3D anatomic and surgical anatomy of the human eye by utilizing landmarks from the vitreous body and/or scleral tissue, i.e. the eye itself.
To our knowledge this is the first MRI study to collect volume measurements of in-vivo fetal eyes, as well as the first application of 3D images reconstructed from MRI acquisitions for fetal ocular biometry. Two previous studies reported volume measurements for ocular growth during gestation using a 3DUS method, Virtual Organ Computer-aided AnaLysis (VOCAL). Odeh et al. (2009)7 created orbital measurements using sphere mode, which fits a perfect sphere into position after an operator positions image calibrators on the edge of the anteroposterior (AP) diameter of the orbit. Bojikian et al. (2013)21 reported volume data using a manual mode, in which the contours of the orbit are manually traced in six consecutive planes, as well as the sphere mode. They reported that measurements from manually traced orbits exhibited a fixed bias of being smaller than sphere shaped measurements. The volume data reported in our study was based on voxel counting on the actual shape of the orbits. Therefore, we could expect the measurement to not only display a bias towards smaller volumes due to border selection, but also because of the ellipsoid shape of the orbit.
Another possible source of bias in our study was that the lens, which appears dark on T2-weighted images, was not included as part of the orbit label because the semi-automatic snake segmentation selects voxels based on image intensity. To examine the effect of lens omission, we manually segmented the lens in 10 cases spread across 19-38 weeks GA and calculated average lens volume as a fraction of orbit volume (mean = 0.038, SD = 0.018). This indicates that including the lens as part of the orbit, as did authors Odeh et al. (2009)7 and Bojikian et al. (2013)21, can result in an average of about 4% increase in orbit volume. We did indeed find that our results were consistently smaller than their 3DUS counterparts.
This study had several strengths. First, the reconstruction algorithm uses multiple MRI scan inputs to correct for fetal motion and up-sample the image in order to create a consistent, smoothed 3D shape that accurately represents the true contours of the orbit. This enabled the measurement of volume as a biometric. Volume may best correlate with GA due to its robustness as a measure. A small mistake, such as the omission or addition of a handful of voxels, in a 3D segmentation of an orbit will not cause a large change in the resulting volume.
Second, the use of a 3D image removes the risk of a suboptimal plane of measurement. When using a 2D methodology, an oblique MRI or US acquisition can make measurement impossible or inaccurate. Due to the nature of fetal MRI, perfect orthogonal alignment is difficult to achieve because the fetus cannot be positioned for the scan. In practice, orthogonal images are obtained by adjusting the direction of the scan based on the previous acquisition, however this is subject to operator error and the fetus may move between or during acquisitions. Obliquity is not an issue for a 3D image that can be manipulated to proper orientation or measured automatically.
For example, Li et al. (2010)15, Paquette et al. (2009)14 and Robinson et al. (2008)5, by necessity, measured BOD, IOD and OD in the plane of 2D MRI acquisitions which may have been suboptimal, i.e. not contained the true diameter or edges of the ellipsoid eye. Furthermore, Robinson et al. (2008) calculated OD as the difference between BOD and IOD. Fetal eyes are often rotated and do not point in the same direction, in which case OD cannot be accurately measured in the same plane as BOD and IOD (an example can be seen in their figure 3a). These biases were observed in comparison with our data.
Third, the presence of a 3D representation of the orbit enabled automated measurement by a computer algorithm. This improved reproducibility by removing human inconsistency and allowed us to present unambiguous and easily reproducible definitions for each biometric. The use of a computer algorithm also reduced operator effort and time commitment. The computer measurement only took several seconds for each image with no human input.
Fourth, we utilized a wide range of MRI hardware and image sequences. This included the use of 1.5T and 3T magnets provided by different manufacturers. We were successful in using both FIESTA and HASTE acquisitions to measure orbital volume. Our approach is practical in the clinical setting, as it is not strongly dependent on the acquisition settings. To test whether the field strength and acquisition type had any statistically significant effect on our biometric measurements, we performed multiple regression analysis tests with the magnetic field strength (1.5T vs 3.0T) and acquisition type (SShTSE, SSFSE, FIESTA, and HASTE) as possible predictors of biometric measurements in addition to GA, and found no significant effect for any of those parameters and their interaction terms with GA in regression models.
The data presented in this study is helpful as a reference point for diagnosis after fetal imaging. Structural conditions such as microphthalmia6, hypo- and hypertelorism are often indicative of an underlying developmental disorder2. For example, small vitreous body and lens measurements have been associated with microcephaly and agenesis of the corpus callosum13. Early detection and diagnosis of a congenital condition may help parents and healthcare providers to decide upon a thoughtful course of action. In addition normative biometrics like those found in this study, when used in conjunction with other measurements and data, can be helpful in estimating gestational age and for identifying ocular anomalies. Future work will involve the use of these reference biometrics to detect and diagnose anomalies.
The vast majority of MRI scans are done after 16-20 weeks gestational age. This is partly due to the lack of adequate signal from small and moving fetal structures early in pregnancy. MRI resolution is limited by partial volume averaging and the slice-thickness of the acquisition making it difficult to obtain images during the first trimester19. Future work may also involve collecting data from fetuses at lower gestation age, which would help complete the record of normal fetal development throughout gestation.
This study had some limitations. First, the accuracy of all measurements was limited by the effective resolution of T2-weighted MRI, which was about 1 mm, and caused partial voluming that did not allow measuring the thickness of the scleral tissue. Consequently the measurements in this study have an error margin of ±0.5 mm. Second, the exclusion of the lens due to contrast with the vitreous may have caused our volume measurements to be smaller as compared to previously reported values, which included the lens. Third, the number of subjects was limited for some gestational ages. Future work will involve adding more data points to corroborate our biometrics, leading to increased statistical power for the entire GA range.
CONCLUSION
In conclusion, we have presented normative fetal ocular biometry using 3D super-resolution reconstructions of MRI acquisitions as an accurate tool for measurement. Between 19 and 38 weeks, BOD, IOD and OD followed a quadratic model of growth, indicating a slowing rate of growth throughout gestation, while orbit volume followed a linear growth model. Orbital volume had the greatest correlation with GA. Thus, volumetric analysis is potentially helpful as a marker for detecting ocular and orbital anomalies and, when used with other fetal biometric measurements, may aid in establishing an accurate gestational age due to more consistent and accurate measurements than are currently available.
What’s already known about this topic?
Fetal ocular measurements are known to correspond with gestational age
Abnormal ocular development can suggest the presence of congenital anomalies. What does this study add?
Gestational age-based fetal ocular measurements, including volume, based on the 3D shape of the orbits using volumetric MRI reconstruction.
Supports a quadratic growth model for BOD, IOD and OD, and linear growth model for orbit volume for the examined gestational age.
Semi-automatic procedure for orbit segmentation and automatic biometric measurements.
Acknowledgements
This research was supported in part by NIH grants R03 DE22109 and R01 EB013248, the Thrasher Research Fund, the Harvard Catalyst - Harvard Clinical and Translational Science Center for Research Resources and the National Center for Advancing Translational Sciences award UL1TR001102, and by a faculty career development award from the Office of Faculty Development at Boston Children’s Hospital.
Footnotes
There are no potential conflicts of interest to report in regards to this study.
References
- 1.Burns NS, Iyer RS, Robinson AJ, Chapman T. Diagnostic imaging of fetal and pediatric orbital abnormalities. American Journal of Roentgenology. 2013;201(6):W797–W808. doi: 10.2214/AJR.13.10949. [DOI] [PubMed] [Google Scholar]
- 2.Nyberg DA. Diagnostic imaging of fetal anomalies. Lippincott Williams & Wilkins; 2003. [Google Scholar]
- 3.Twining P, McHugo JM, Pilling DW. Textbook of fetal abnormalities. Elsevier Health Sciences; 2007. [Google Scholar]
- 4.Paquette L, Randolph L, Incerpi M, Panigrahy A. Fetal microphthalmia diagnosed by magnetic resonance imaging. Fetal Diagnosis and Therapy. 2008;24(3):182–5. doi: 10.1159/000151335. [DOI] [PubMed] [Google Scholar]
- 5.Robinson AJ, Blaser S, Toi A, et al. MRI of the fetal eyes: morphologic and biometric assessment for abnormal development with ultrasonographic and clinicopathologic correlation. Pediatric Radiology. 2008;38(9):971–81. doi: 10.1007/s00247-008-0929-3. [DOI] [PubMed] [Google Scholar]
- 6.Birnholz JC. Ultrasonic fetal ophthalmology. Early Human Development. 1985;12(2):199–209. doi: 10.1016/0378-3782(85)90182-3. [DOI] [PubMed] [Google Scholar]
- 7.Odeh M, Feldman Y, Degani S, et al. Fetal eyeball volume: relationship to gestational age and biparietal diameter. Prenatal Diagnosis. 2009;29(8):749–52. doi: 10.1002/pd.2274. [DOI] [PubMed] [Google Scholar]
- 8.Dilmen G, Köktener A, Turhan NÖ, Tez S. Growth of the fetal lens and orbit. International Journal of Gynecology & Obstetrics. 2002;76(3):267–71. doi: 10.1016/s0020-7292(01)00581-1. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein I, Tamir A, Zimmer E, Itskovitz-Eldor J. Growth of the fetal orbit and lens in normal pregnancies. Ultrasound in Obstetrics & Gynecology. 1998;12(3):175–9. doi: 10.1046/j.1469-0705.1998.12030175.x. [DOI] [PubMed] [Google Scholar]
- 10.Kivilevitch Z, Salomon L, Benoit B, Achiron R. Fetal interlens distance: normal values during pregnancy. Ultrasound in Obstetrics & Gynecology. 2010;36(2):186–90. doi: 10.1002/uog.7531. [DOI] [PubMed] [Google Scholar]
- 11.Sukonpan K, Phupong V. A biometric study of the fetal orbit and lens in normal pregnancies. Journal of Clinical Ultrasound. 2009;37(2):69–74. doi: 10.1002/jcu.20537. [DOI] [PubMed] [Google Scholar]
- 12.Achiron R, Kreiser D, Achiron A. Axial growth of the fetal eye and evaluation of the hyaloid artery: in utero ultrasonographic study. Prenatal Diagnosis. 2000;20(11):894–9. doi: 10.1002/1097-0223(200011)20:11<894::aid-pd949>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 13.Achiron R, Gottlieb Z, Yaron Y, et al. The development of the fetal eye: in utero ultrasonographic measurements of the vitreous and lens. Prenatal Diagnosis. 1995;15(2):155–60. doi: 10.1002/pd.1970150208. [DOI] [PubMed] [Google Scholar]
- 14.Paquette L, Jackson H, Tavare C, et al. In utero eye development documented by fetal MR imaging. American Journal of Neuroradiology. 2009;30(9):1787–91. doi: 10.3174/ajnr.A1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li XB, Kasprian G, Hodge JC, et al. Fetal ocular measurements by MRI. Prenatal Diagnosis. 2010;30(11):1064–71. doi: 10.1002/pd.2612. [DOI] [PubMed] [Google Scholar]
- 16.Bremond-Gignac D, Benali K, Deplus S, et al. In utero eyeball development study by magnetic resonance imaging. Surgical and Radiologic Anatomy. 1997;19(5):319–22. doi: 10.1007/BF01637602. [DOI] [PubMed] [Google Scholar]
- 17.Blazer S, Zimmer EZ, Mezer E, Bronshtein M. Early and late onset fetal microphthalmia. American Journal of Obstetrics and Gynecology. 2006;194(5):1354–9. doi: 10.1016/j.ajog.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Meenken C, Assies J, Van Nieuwenhuizen O, et al. Long term ocular and neurological involvement in severe congenital toxoplasmosis. British journal of Ophthalmology. 1995;79(6):581–4. doi: 10.1136/bjo.79.6.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pugash D, Brugger PC, Bettelheim D, Prayer D. Prenatal ultrasound and fetal MRI: the comparative value of each modality in prenatal diagnosis. European Journal of Radiology. 2008;68(2):214–26. doi: 10.1016/j.ejrad.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 20.Ishii K, Iwata H, Oshika T. Quantitative evaluation of changes in eyeball shape in emmetropization and myopic changes based on elliptic Fourier descriptors. Investigative Ophthalmology & Visual Science. 2011;52(12):8585–91. doi: 10.1167/iovs.11-7221. [DOI] [PubMed] [Google Scholar]
- 21.Bojikian KD, de Moura CR, Tavares IM, et al. Fetal ocular measurements by three-dimensional ultrasound. Journal of American Association for Pediatric Ophthalmology and Strabismus. 2013;17(3):276–81. doi: 10.1016/j.jaapos.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Gholipour A, Estroff JA, Warfield SK. Robust super-resolution volume reconstruction from slice acquisitions: application to fetal brain MRI. Medical Imaging, IEEE Transactions on. 2010;29(10):1739–58. doi: 10.1109/TMI.2010.2051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clouchoux C, Kudelski D, Gholipour A, et al. Quantitative in vivo MRI measurement of cortical development in the fetus. Brain Structure and Function. 2012;217(1):127–39. doi: 10.1007/s00429-011-0325-x. [DOI] [PubMed] [Google Scholar]
- 24.Gholipour A, Estroff JA, Barnewolt CE, et al. Fetal brain volumetry through MRI volumetric reconstruction and segmentation. International Journal of Computer Assisted Radiology and Surgery. 2011;6(3):329–39. doi: 10.1007/s11548-010-0512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–28. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Goodall N, Kisiswa L, Prashar A, et al. 3-Dimensional modelling of chick embryo eye development and growth using high resolution magnetic resonance imaging. Experimental Eye Research. 2009;89(4):511–21. doi: 10.1016/j.exer.2009.05.014. [DOI] [PubMed] [Google Scholar]