Abstract
Helicobacter pylori requires urease activity in order to survive in the acid environment of the human stomach. Urease is regulated in part by nickelation, a process that requires the HypA protein, which is a putative nickel metallochaperone that is generally associated with hydrogenase maturation. However, in H. pylori, HypA plays a dual role. In addition to an N-terminal nickel binding site, HypA proteins also contain a structural zinc site that is coordinated by two rigorously conserved CXXC sequences, which in H. pylori are flanked by His residues. These structural Zn sites are known to be dynamic, converting from Zn(Cys)4 centers at pH 7.2 to Zn(Cys)2(His)2 centers at pH 6.3 in the presence of Ni(II) ions. In this study, mutant strains of H. pylori that express zinc site variants of the HypA protein are used to show that the structural changes in the zinc site are important for the acid viability of the bacterium, and that a reduction in acid viability in these variants can be traced in large measure to deficient urease activity. This in turn leads to a model that connects the Zn(Cys)4 coordination to urease maturation.
Introduction
Helicobacter pylori (H. pylori) is a Gram negative bacterium that colonizes the human gastric mucosa1. Infection causes chronic inflammation and is a major risk factor for development of peptic ulcers and cancer2. H. pylori is a prevalent pathogen that colonizes approximately one-third to one-half of the worldwide adult population3. Successful colonization of the acidic environment of the stomach requires the activity of urease4, a nickel-dependent enzyme that catalyzes the hydrolysis of urea to ammonia and carbon dioxide5. Urease is highly expressed in H. pylori, representing up to 10% of the total protein.6 However, the enzyme remains inactive without maturation through nickel insertion into the active site.5
H. pylori urease maturation is dependent on accessory proteins UreIEFGH.5 Additionally, HypA and HypB, accessory proteins normally involved in the maturation of NiFe-hydrogenases, also aid in urease maturation; deletion of hypA or hypB results in urease deficiency in H. pylori despite the presence of the full ureIEFGH cascade.7 Urease deficient phenotypes in hypAB deletion mutants can be compensated for by addition of Ni(II) to the media, implicating the role of HypA and HypB in nickel delivery to the urease maturation pathway.7a, 7c The traditionally hydrogenase-specific nickel metallochaperone, HypA, has been shown to interact directly with a urease-specific nickel metallochaperone, UreE, in vitro, providing a possible link for the role of HypA in urease maturation.7c Several studies have interrogated the interaction between HypA and UreE. Biophysical characterizations show that HypA can outcompete UreG for interaction with UreE.8 Additionally, nickel transfer from HypA to UreE has been shown to involve the interaction of the HypA nickel-binding domain and the UreE nickel-binding C-terminus.9
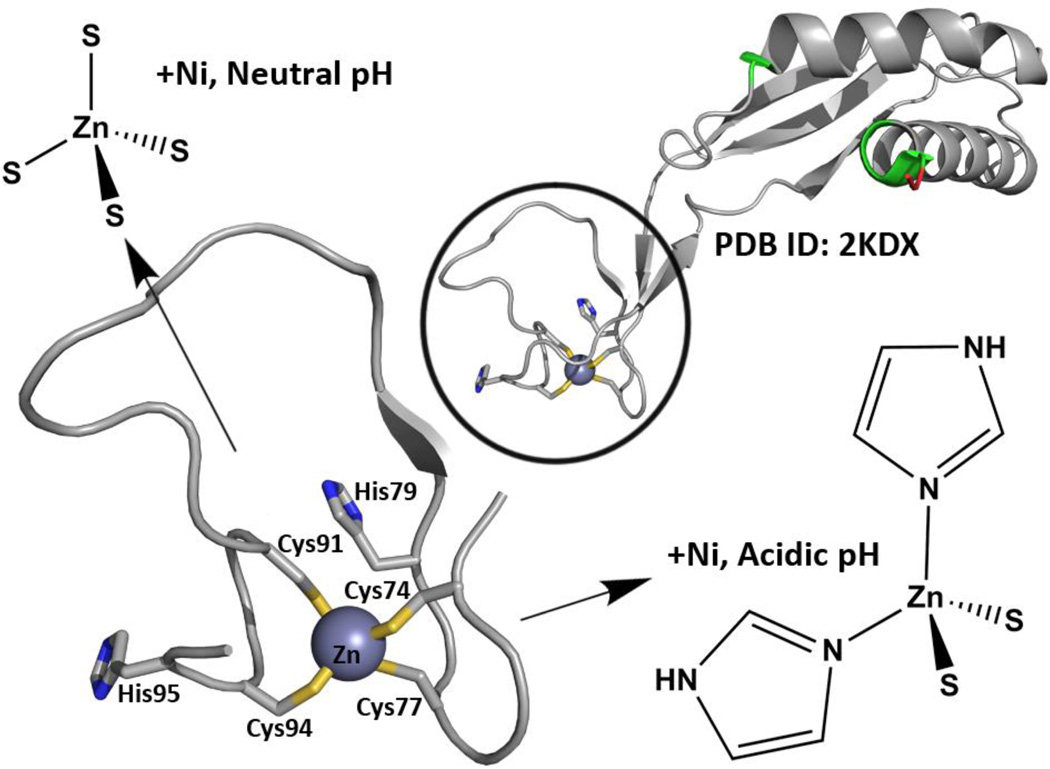
As shown in Fig.1, the overall protein structure of HypA has been shown by nuclear magnetic resonance (NMR) to have a distinct nickel-binding domain and structural zinc-binding domain [PDB: 2KDX].10 In addition to the two conserved CXXC motifs in the HypA structural zinc site, the H. pylori zinc binding motifs also have His residues closely flanking the conserved motifs. The structural zinc site has also been extensively characterized by X-ray absorption spectroscopy (XAS), demonstrating that the WT HypA protein structural zinc site is dynamic.11 The average Zn coordination at pH 7.2 is Zn(Cys)4, which changes to Zn(Cys)2(His)2 at pH 6.3 (the estimated internal pH of H. pylori under acid shock conditions) with nickel bound in the nickel site.11 The dynamic nature of the HypA zinc site is lost when any one of the four cysteine residues in the CXXC motif is mutated to alanine or aspartate, and results in locking of the zinc coordination in the Zn-Cys2His2 structure (acidic conformation).11b The dynamic nature of the HypA zinc site is also lost when either of the histidine residues flanking the CXXC motifs is mutated to alanine; this change results in locking of the zinc coordination in the Zn-Cys4 structure (neutral conformation).11b These structure-locking mutants of HypA are ideal for interrogation of the role of the HypA zinc site and corresponding protein conformations in urease maturation and acid resistance in H. pylori, and are the focus of this study. Herein we create a series of isogenic H. pylori strains that each express zinc site mutant hypA variants under the control of the endogenous promoter, and then utilize these mutants to define the contribution of the conserved HypA motifs to H. pylori acid viability and urease activity. These studies connect the point mutations in the structural zinc site of a nickel metallochaperone, HypA, to changes in acid viability and urease maturation in vivo.
Fig. 1.
The structure and dynamics of the HypA zinc site have been characterized. The NMR structure of HypA10 (PDB ID: 2KDX) shows distinct Ni- and Zn-binding domains, where known Ni-binding residues are colored in green (non-native residues leftover from affinity-tag processing shown in red). Close-up look of the Zn site reveals two CXXC motifs with flanking His residues (Zn shown as gray sphere). The Zn site has also been shown to alter its average structure depending on pH and Ni-binding; at acidic conditions with Ni-bound, two His imidazoles replace two of the sulphur donors from the Cys residues.
Materials and methods
Bacterial growth conditions
All H. pylori strains were maintained at −80°C in brain heart infusion broth (Becton Dickinson) supplemented with 10% fetal bovine serum (FBS) and 20% glycerol and were cultivated on horse blood agar (HBA) medium containing 4% columbia agar base (Neogen Corp), 5% defibrinated horse blood (HemoStat Laboratories, Dixon, CA), 0.2% β-cyclodextrin (Sigma), 10 µg/mL of vancomycin (Amresco), 2.5 U/mL of polymyxin B (Sigma), 5 µg/mL of trimethoprim (Sigma), and 5 µg/mL of amphotericin B (Amresco). Where required, 5% sucrose was added to HBA for selection of sucrose sensitive strains. Liquid growth of H. pylori was performed in brucella broth (Neogen Corp) with 10% FBS and 10µg/mL of vancomycin. All H. pylori cultures were grown under microaerobic conditions (5% O2, 10% CO2, and 85% N2) at 37°C with 100rpm shaking for liquid cultures. H. pylori strain G27 was used for all experiments12. Escherichia coli Top10 cells were either grown on LB agar or in LB liquid medium with shaking at 225rpm. Kanamycin (25µg/mL) and ampicillin (100µg/mL) were used for bacterial selection.
hypA mutant construction
H. pylori G27 genomic DNA was used as the template for mutant construct development. A hypA mutant strain containing a kan-sacB cassette insertion was constructed as follows. The HypA_Up_F and HypA_Up_R primers (Table 1) were used to amplify a 556 base pair segment of DNA that spanned 398 base pairs upstream of hypA (HPG27_832) and 158 base pairs into the hypA coding region. Additionally, the HypA_Dn_F and HypA_Dn_R primers (Table 1) were used to amplify a 532 base pair region that began at nucleotide position 160 of hypA and spanned 337 base pairs downstream of the hypA stop codon. The HypA_Up_R and HypA_Dn_F primers were designed to contain identical flanking sequences, which harbored XhoI and XbaI restriction enzyme sites. Thus, the upstream and downstream amplicons were next fused together utilizing splicing by overlap extension (SOE) PCR13. The resulting spliced product was cloned into the pGEM-T easy vector (Promega) and transformed into E. coli Top10 cells. The kan-sacB cassette14, which confers resistance to kanamycin and sensitivity to sucrose, was amplified from pKSF-II14 using primers Kan_SacB_F and Kan_SacB_R, which were designed to incorporate a XhoI and XbaI restriction site, respectively. The resulting fragment was digested with XhoI and XbaI and was then inserted between the spliced hypA fragments that had been similarly digested. The resulting construct was then transformed into H. pylori G27 and transformants were selected on HBA plates containing kanamycin. Successful insertion of the kan-sacB cassette into the hypA gene was confirmed by PCR and sequencing using the HypA_Confirm_F/R and HypA_seq_F/R primers, respectively. The resulting mutant strain was named DSM1283.
Table 1.
Strain, plasmids, and primers used in this study
| Strains | Description | Reference |
| DSM1 | G27 WT | 12 |
| DSM43 | G27 ΔureB KanR | 15, This study |
| DSM1283 | G27 hypA::kan-sacB KanR SucS | This study |
| DSM1295 | G27 hypA restorant | This study |
| DSM1296 | G27 hypA C74A | This study |
| DSM1297 | G27 hypA C94D | This study |
| DSM1298 | G27 hypA C91A | This study |
| DSM1299 | G27 hypA C91D | This study |
| DSM1300 | G27 hypA H95A | This study |
| DSM1301 | G27 hypA C74D | This study |
| DSM1363 | G27 hypA C77A | This study |
| DSM1364 | G27 hypA C77D | This study |
| DSM1365 | G27 hypA H79A | This study |
| DSM1366 | G27 hypA C94A | This study |
| Plasmids | Description | Reference |
| pDSM3 | pKSF-II | 14 |
| pDSM32 | pEJ22 | 15 |
| pJI110 | pET22b(+) vector with hypA WT coding sequence | 11, This study |
| pET22b-HypA(C74A) | pET22b(+) vector with hypA C74A coding sequence | This study |
| pET22b-HypA(C74D) | pET22b(+) vector with hypA C74D coding sequence | This study |
| pET22b-HypA(C77A) | pET22b(+) vector with hypA C77A coding sequence | This study |
| pET22b-HypA(C77D) | pET22b(+) vector with hypA C77D coding sequence | This study |
| pET22b-HypA(H79A) | pET22b(+) vector with hypA H95A coding sequence | This study |
| pET22b-HypA(C91A) | pET22b(+) vector with hypA C91A coding sequence | This study |
| pET22b-HypA(C91D) | pET22b(+) vector with hypA C91D coding sequence | This study |
| pET22b-HypA(C94A) | pET22b(+) vector with hypA C94A coding sequence | This study |
| pET22b-HypA(C94D) | pET22b(+) vector with hypA C94D coding sequence | This study |
| pET22b-HypA(H95A) | pET22b(+) vector with hypA H95A coding sequence | This study |
| Primers | Sequence (5’-3’) | Reference |
| HypA_Up_F | CCGCTTTGATTCAGATGGGGTG | This study |
| HypA_Up_R* | TCTAGAAGCTTGCGATCGCTCGAGACTCTAAAAGTCTCAAACGCGCTC | This study |
| HypA_Dn_F* | CTCGAGCGATCGCAAGCTTCTAGAGAATCTTTGGTGTGTAAAGACGC | This study |
| HypA_Dn_R | GCAAAACGCTGCGGTATTGC | This study |
| Kan_SacB_F* | GTGGGCTCGAGCCCGGGCGAACCATTTGAGGTGA | This study |
| Kan_SacB_R* | GCGCGTCTAGATATAAGCCCATTTTCATGC | This study |
| HypA_Confirm_F | GGCTAACGAGCGTGGATAAG | This study |
| HypA_Confirm_R | GCACTCACTAAAATCGTGGGC | This study |
| HypA_seq_F | CTAAAGCGGTAACCACATCCG | This study |
| HypA_C74A_F | GGTTGAATTAGAAGCCAAGGATTGTTCGCATGTTTTTAAGCCTAACGCG | 11b, This study |
| HypA_C74A_R | CGCGTTAGGCTTAAAAACATGCGAACAATCCTTGGCTTCTAATTCAACC | 11b, This study |
| HypA_C74D_F | GGTTGAATTAGAAGACAAGGATTGTTCGCATGTTTTTAAGCCTAACGCG | 11b, This study |
| HypA_C74D_R | CGCGTTAGGCTTAAAAACATGCGAACAATCCTTGTCTTCTAATTCAACC | 11b, This study |
| HypA_C77A_F | GAATTAGAATGCAAGGATGCTTCGCATGTTTTTAAGCCTAACGCGC | 11b, This study |
| HypA_C77A_R | GCGCGTTAGGCTTAAAAACATGCGAAGCATCCTTGCATTCTAATT | 11b, This study |
| HypA_C77D_F | GAATTAGAATGCAAGGATGATTCGCATGTTTTTAAGCCTAACGCGC | 11b, This study |
| HypA_C77D_R | GCGCGTAGGCTTAAAAACATGCGAATCATCCTTGCATTCTAATTC | 11b, This study |
| HypA_H79A_F | GCAAGGATTGTTCGGCTGTTTTTAAGCCTAACGCGCTAG | 11b, This study |
| HypA_H79A_R | GTTAGGCTTAAAAACAGCCGAACAC | This study |
| HypA_C91A_F | GCGCTAGATTATGGGGTGGCTGAGAAATGCCACAGC | 11b, This study |
| HypA_C91A_R | GCTGTGGCATTTCTCAGCCACCCCATAATCTAGCGC | 11b, This study |
| HypA_C91D_F | CGCCGTAGATTATGGGGTGGATGAGAAATGCCACAGC | 11b, This study |
| HypA_C91D_R | GCTGTGGCACTTTCTCATCCACCCCATAATCTAGCGCG | 11b, This study |
| HypA_C94A_F | GGTGTGTGAGAAAGCCCACAGCAAG | This study |
| HypA_C94A_R | AACATTCTTGCTGTGGGCTTTCTCAC | This study |
| HypA_C94D_F | GGGGTGTGTGAGAAAGACCACAGCAAGAATGTTATTATCAC | 11b, This study |
| HypA_C94D_R | GTGATAATAACATTCTTGCTGTGGTCTTTCTCACACACCCC | 11b, This study |
| HypA_H95A_F | GTGTGTGAGAAATGCGCCAGCAAGAATGTTATTATC | 11b, This study |
| HypA_H95A_R | GATAATAACATTCTTGCTGGCGCATTTCTCACACAC | 11b, This study |
Restriction enzyme sites are italicized (XhoI or XbaI); Underline denotes nucleotide changes made to the pJI110 (wild type hypA) plasmid
Site-directed mutagenesis of hypA Zn-binding site
Mutations of the cysteines in the HypA CXXC motifs (Cys71, Cys74, Cys91 and Cys94) to alanine or aspartic acid and the flanking histidines (His79 and His95) to alanine were constructed by polymerase chain reaction (PCR) using the PJI110 plasmid (wild type hypA sequence carried on the pET-22b(+) vector, Table 1) as a template.11b The PJI110 plasmid was transformed into NovaBlue (Novagen) competent cells and then re-isolated using the Axyprep Plasmid MiniPrep Kit (Axygen) and used as the DNA template in all subsequent PCR reactions. PCR primers were designed to incorporate the desired mutations and are listed in Table 1. Reactions were carried out in 50µL volumes using 1ng of template DNA and 2.5ng or 2µM of each primer per reaction. Successful PCR amplifications were confirmed by 0.8% agarose gel electrophoresis, and then methylated PJI110 template was digested with 20 units of DpnI (New England Biolabs) for 1 hour at 37°C. The resulting PCR mixture was transformed into NovaBlue competent cells. Single colonies were selected and grown to saturation in 5mL liquid cultures of LB-Miller (Fisher Scientific) media supplemented with ampicillin at 37°C. Cells were pelleted by centrifugation at 6,000 g for 5 minutes and plasmids were isolated using the Axyprep Plasmid MiniPrep Kit. Successful mutations were confirmed by plasmid sequencing (GENEWIZ, Inc.).
We utilized the wild-type or mutant HypA pET-22b(+) plasmids, which each contained the entire HypA coding region, to move the mutant constructs of interest into the H. pylori chromosome. Each of the constructs was individually transformed into DSM1283, and double crossover events in which the kan-sacB cassette was replaced by the mutagenized hypA gene carried on the pET-22b(+) vector were selected for based on sucrose resistance. Sucrose resistant transformants were screened for kanamycin sensitivity and proper integration of the hypA construct was confirmed by PCR and sequencing using the HypA_Confirm_F/R and HypA_seq_F/R primers, respectively. In total, H. pylori mutant strains were made that contained the following hypA mutations: C74A (DSM1296), C74D (DSM1301), C77A (DSM1363), C77D (DSM1364), H79A (DSM1365), C91A (DSM1298), C91D (DSM1299), C94A (DSM1366), C94D (DSM1297), and H95A (DSM1300). In addition, a hypA-restorant (hypA-R; DSM1295) in which the kan-sacB cassette was replaced by the wild type hypA gene was also created in order to control for any defects that may have resulted due to genetic manipulation.
Acid resistance testing
Each of the ten hypA Zn-site H. pylori mutants were tested for acid resistance. Additionally, the wild type H. pylori strain, the hypA::kan-sacB mutant, and the hypA restorant were included as controls. Furthermore, a urease-deficient mutant of H. pylori (DSM43) in which the kanamycin resistance cassette replaced the ureB subunit was used as a positive control for acid sensitivity; DSM43 was created by transforming wild type H. pylori with vector pEJ22 as previously described.15 Assays were conducted as follows: 20mL liquid cultures of H. pylori were inoculated to an optical density (600nm) of 0.05 from overnight liquid grown bacterial cells and then allowed to grow for approximately 19 hours. 1mL aliquots were removed from the culture and pelleted by centrifugation. The supernatants were removed and the bacterial pellet was re-suspended in 1mL of phosphate buffered saline (PBS) at pH 6 or 2.3, with or without supplementation with 5mM urea. Immediately after the bacterial pellets were re-suspended, an aliquot was removed, serially diluted in brucella broth and plated on HBA plates to determine colony forming units (CFU) per milliliter. The cultures were then incubated in 1.5mL capped tubes for 1 hour at 37°C. At this point, a second aliquot was removed from the cultures, immediately serially diluted, and plated to determine CFU/mL as described above. Percent survival after 1-hour incubation in the various PBS solutions was determined for each bacterial strain. At least three biological replicates were performed for each strain.
Urease activity assay
Urease activities were determined for each of the ten hypA Zn-site variants, the wild type H. pylori strain, the hypA::kan-sacB mutant, and the hypA restorant strain. For each strain, 8mL liquid cultures of H. pylori were inoculated to an optical density (600nm) of 0.05 from overnight liquid grown bacterial cells and then allowed to grow for approximately 22 hours. At that point, 1mL aliquots were removed from the culture and pelleted by centrifugation (~108 cells). The supernatants were removed and the bacterial pellets were stored at −20°C until ready for urease assays. The frozen cells were thawed and then re-suspended in 750µL of ice-cold HEPES buffer (pH 7.0), 1mM phenylmethanesulfonyl fluoride (PMSF) (MP Biomedicals, LLC), and 1× protease inhibitor cocktail (Sigma-Aldrich) and then lysed by sonication at 70% power for 6 pulses (2-second each) on ice. Lysate was centrifuged at 15,000-g for 10 minutes to remove insoluble fractions from soluble whole cell extracts. Soluble whole cell extracts were kept at 4°C for up to one month and insoluble fractions were stored at −20°C. Total protein concentration in soluble whole cell extract was assessed by Bradford Assay using the Coomassie Protein Assay Kit (Thermo Scientific).
Urease activities for each strain were determined using a modified phenol-hypochlorite method to assay the amount of ammonia released in the soluble whole cell extract of H. pylori lysate in the presence of urea16. For each strain, 5µL of whole cell extract was added to 245µL of urease reaction buffer (50mM HEPES, 25mM Urea, pH 7.0), and incubated at 37°C for 20 minutes to allow for ammonia production. The reaction was quenched with the sequential addition of 375µL of phenol-hypochlorite buffer A (100mM phenol, 167.8µM sodium nitroprusside) and then the addition of 375µL of phenol-hypochlorite buffer B (125mM NaOH, 0.044% NaClO); samples were mixed with quick vortexing after the addition of each buffer. The assay mixture was incubated at 37°C for 30 minutes to allow for color development (the conversion of ammonia to indophenol) and the absorbance was evaluated at 625nm. Assays were performed alongside a standard curve created using known amounts of ammonium chloride (0.24 – 500nmol) in place of whole cell extract. The urease activity of ΔureB strain was set as background and subtracted from the activity of all other strains. Urease activity for the various mutants was normalized to the hypA-restorant (hypA-R; DSM1295) H. pylori as 100%. All experiments were performed in triplicate with two independently grown cultures.
Results
HypA Zn-binding sites are important for acid survival
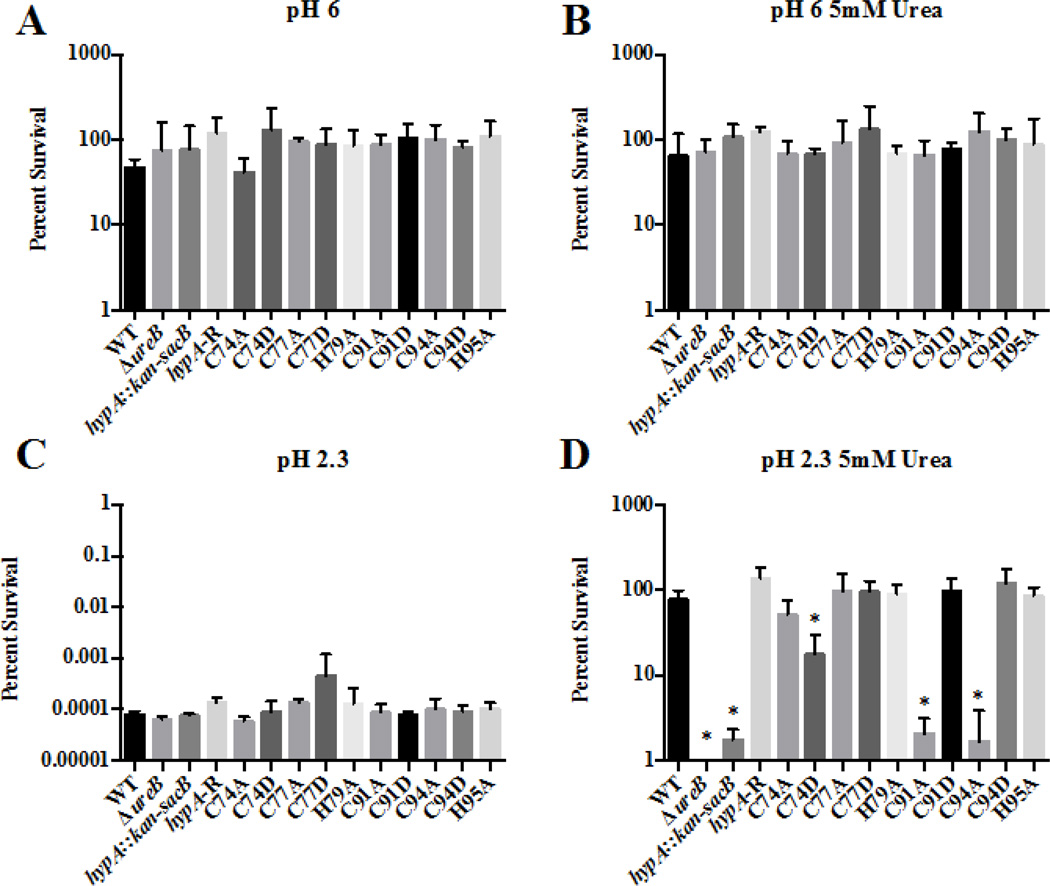
To determine whether changes in the zinc binding sites of HypA affected acid resistance of H. pylori, we created strains of H. pylori in which the Cys residues found in the two CXXC motifs associated with zinc binding (Cys74, Cys77, Cys91 and Cys94) and the two His residues (His79 and His95) that flank these two CXXC motifs were mutated. Cys residues were changed to Asp and Ala, while His residues were changed to Ala. We then utilized these 10 HypA mutant strains in combination with the wild type strain, a hypA mutant (hypA::kan-sacB), a hypA-restorant (hypA-R) and the ureB mutant (ΔureB) control strains to assess acid resistance. All 14 of these H. pylori strains showed similar robust survival profiles when exposed to pH 6 in the presence or absence of 5mM urea (Fig. 2A, B). Conversely, when exposed to pH 2.3, all of the H. pylori strains, including the wild type, showed a dramatic decrease in viability; less than 0.01% of the inoculum survived at pH 2.3 in the absence of urea (Fig. 2C). However, when urea was supplemented to the pH 2.3 buffer, we began to detect differences in acid resistance across the various strains (Fig. 2D). As expected, the wild type H. pylori strain and the hypA-restorant were able to efficiently utilize the supplemented urea as a substrate for the urease system and survive the acidic challenge. Also as expected, the urease deficient ΔureB strain was incredibly acid sensitive; no surviving bacteria were detected (limit of detection 500 CFU/mL equating to 0.0001% survival). The ΔhypA strain that carried an insertion in the hypA coding sequence was also deficient in its ability to resist acidic stress (< 3% survival). This result confirmed previous studies that indicated that HypA is necessary for efficient urease activity7. For the various Zn-binding site mutants, mutation of Cys77, His79, and His95 of HypA resulted in no changes in acid resistance. Conversely, significant decreases in acid resistance, as compared to the wild type strain, were observed for the C74D, C91A, and C94A mutant strains: 17.6%, 2.1% and 1.7% average survival, respectively. This finding suggests that these zinc-binding site residues play a critical role in acid viability, presumably by affecting the ability of HypA to provide nickel to the urease maturation pathway.
Fig. 2.
Specific amino acid mutations within the zinc-binding site of the HypA protein in Helicobacter pylori result in decreased acid resistance. The ten hypA mutants, as well as the wild type (WT), ureB knockout (ΔureB), hypA interrupted mutant (hypA::kan-sacB), and the hypA-restorant (hypA-R) were exposed to various environments for 1 hour: pH 6 (A), pH 6 containing 5mM urea (B), pH 2.3 (C), and pH 2.3 containing 5mM urea. Percent survival was calculated for each strain. Data represent mean ± standard deviation. * = Acid resistance was significantly reduced when compared to wild type (p < 0.01, one-way ANOVA followed by Dunnett’s test for multiple comparisons).
Mutation to HypA Zn-binding sites impacts urease activity
The acid survival data suggested that the C74, C91 and C94 residues play a critical role in the ability of HypA to facilitate urease maturation.7 We next directly investigated urease activity in each of the ten Zn-binding site mutants of hypA, as well as in the ΔureB, the hypA::kan-sacB, the hypA-restorant and the wild type strains. The hypA-restorant showed three-fold less urease activity than wild type H. pylori, which may be a consequence of genetic manipulation. Consequently, urease activities were normalized to the hypA-restorant strain. Since the ΔureB strain of H. pylori is missing the nickel-containing β subunit of the urease enzyme and should have no urease activity, the average urease activity of the ΔureB strain was subtracted from measurements of all strains to correct for any background levels of ammonia present in whole cell extract.
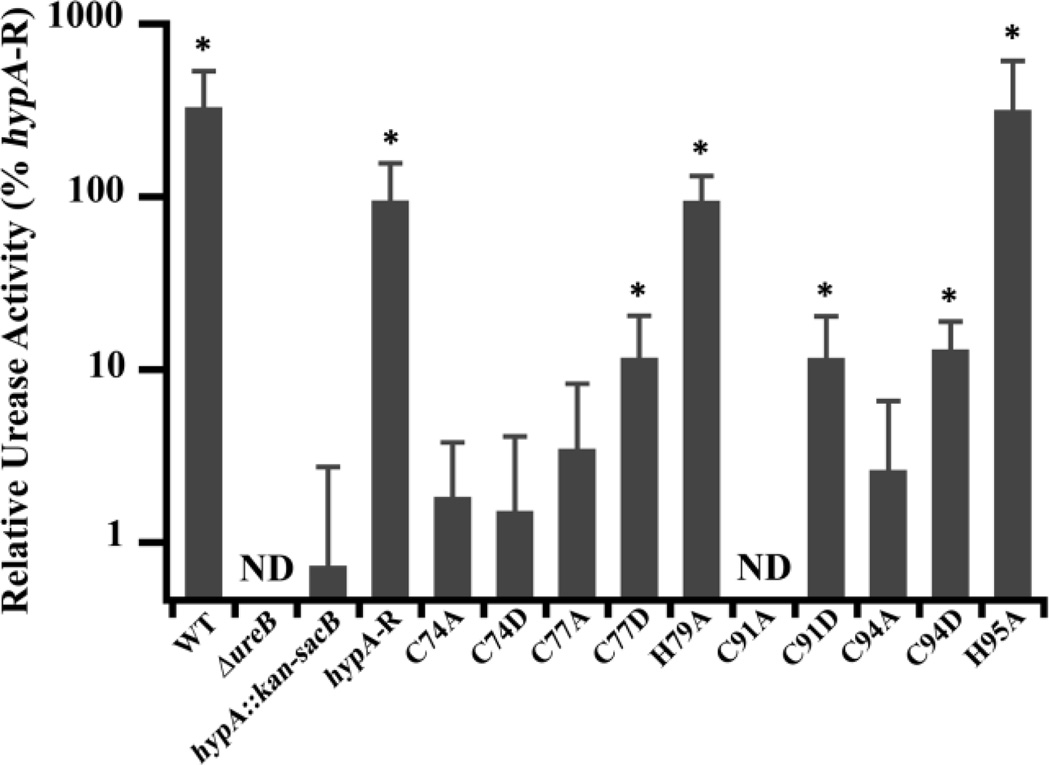
As shown in Fig. 3, we identified two classes of mutations of the HypA structural zinc site, those that had no effect on urease activity (WT-like), and those that decreased urease activity. This latter class can be subdivided into moderately deficient and severely deficient (hypA::kan-sacB-like) subclasses. Mutations of His residues (H79A and H95A) flanking the HypA CXXC motifs resulted in strains that retained WT-like activity, suggesting that these His residues are not important for urease maturation. In contrast, all of the mutations involving Cys residues in the HypA CXXC motifs were deficient in urease activity to some extent (less than 15% compared with the hypA-restorant strain), indicating the importance of the Cys residues in the proper function of HypA in the urease maturation pathway. Comparisons of the urease activities in the Cys mutant strains with the ΔhypA stain revealed more subtle phenotypes. The activity of C77D, C91D, and C94D were statistically different (p < 0.05) from the hypA::kan-sacB strain (12 – 13% compared with the hypA-restorant strain) constituting a moderately deficient phenotype (Fig. 3). The remaining Cys mutants, C74A, C74D, C77A, C91A, and C94A, showed activity levels similar to the ΔhypA strain (> 4% compared with hypA-restorant strain). These urease activity results corroborate the acid survival phenotypes, where C74D, C91A, and C94A mutants were found to be both acid-sensitive and severely deficient in urease activity. Contrary to the acid survival results, C74A and C77A mutants were not acid sensitive under our test conditions, but were found to be severely deficient in urease activity. These findings suggest that, although all of the Cys residues in the CXXC motifs are important for proper HypA function in the urease maturation pathway, there is a position-dependent gradient of importance based on the residue position as well as the type of mutation.
Fig. 3.
Zn-site mutations of hypA in H. pylori lead to deficiencies in urease activity. Soluble whole cell extracts were obtained from lysis of ten hypA Zn-site mutant strains, as well as the wild type (WT), ureB knockout (ΔureB), hypA interrupted mutant (hypA::kan-sacB), and the hypA-restorant (hypA-R) strains. The phenol-hypochlorite method was used to assay the ammonia production (urease activity) of the strains. Amount of ammonia produced was quantified by comparison with a standard curve constructed with known amount of ammonium chloride. The ΔureB strain activity was subtracted from each strain as background ammonia in cell extracts and all activity was normalized to hypA-R strain. * = Urease activity was significantly different from the hypA::kan-sacB strain (p < 0.05). ND = none detected.
Discussion
The H. pylori HypA protein is a putative nickel metallochaperone that is involved in both hydrogenase maturation (as are all HypA proteins) and urease maturation, a function that is unique to H. pylori. NMR structural studies of a monomeric construct of H. pylori HypA revealed that it contains a structural zinc site associated with two rigorously conserved CXXC motifs in addition to a nickel-binding site associated with the N-terminus.10 The amino acid sequence of H. pylori reveals that the zinc-binding CXXC motifs are unique among other HypA proteins in that each is flanked by a His residue.11b The structural Zn-sites of HypA were previously shown to change coordination in a pH and nickel-dependent manner, adopting a Zn(Cys)4 conformation at neutral pH (7.2), and a Zn(Cys)2(His)2 conformation at more acidic pH (6.3) (Fig. 1) in the presence of Ni(II).11 These two structures were correlated with other physical properties of dimeric HypA, including nickel binding stoichiometry and thermal stability, with the lower pH form associated with greater thermal stability.11b Mutagenesis studies revealed that altering any Cys residue in the two CXXC motifs (to Ala or Asp) resulted in adoption of the acidic conformation (Zn(Cys)2(His2)), and any mutation of the flanking His residues resulted in the neutral conformation (Zn(Cys)4), in the presence of Ni(II) at both pH values, as determined by EXAFS analysis of data collected at ~ 50K.11b These pH-insensitive and structure-locking mutants of HypA were important tools employed in the present study to probe the role of HypA in urease maturation and acid viability in vivo.
In vivo urease activities of H. pylori hypA Zn site variant strains correlate well with previous in vitro studies employing addition of purified HypA proteins to whole cell extract from a ΔhypA mutant of H. pylori.11b Results of the in vitro studies show a general decrease in urease activity upon addition of the zinc site variants relative to addition of wild type HypA protein, and a generally greater effect associated with Cys mutants as compared to His mutants.11b The in vivo studies presented here reveal two distinct phenotypes: WT-like urease activity, represented by His mutants, and urease-deficient strains associated with Cys-mutants. These two classes of mutants correspond to the Zn(Cys)4 or Zn(Cys)2(His)2 conformations, respectively. Our data suggest that the all the Cys residues in the HypA Zn site are important in the maturation of urease in H. pylori, a result that is consistent with a model in which the conformation associated with Zn(Cys)2(His)2 sites, or in which access to the Zn(Cys)4 structure is inhibited in a dynamic sense, leads to lower levels of urease activation. Among the Cys mutations, there were two levels of urease activity observed, a moderate decrease associated with C77D, C91D, and C94D, all Cys→ Asp mutations, and a severely deficient level associated primarily with Cys→ Ala mutations, which have activity levels similar to the hypA::kan-sacB strain (C74A, C74D, C77A, C91A, and C94A). With the exception of C74D, Cys→ Asp mutations give rise to more active variants than Cys→ Ala mutations, suggesting that at least part of the difference lies in coordination of the CXXC motifs. A Cys→ Ala mutation results in a loss of a ligand in the CXXC motifs, while a Cys→ Asp mutation results in the potential substitution of a Cys thiolate ligand by a carboxylate ligand. Thus, the Cys→ Asp substitution provides a mechanism to access a Zn(Cys)3Asp site that favors the wild type-like Zn(Cys)4 conformation, which appears to preserve greater HypA function in moderately affected mutants-- C77D, C91D, and C94D, but not in C74D (vide infra).
A similar trend can be discerned in the study of acid viability: His mutations have no effect, whereas C91A and C94A mutations are associated with low acid survival (comparable to the hypA::kan-sacB strain), and C91D and C94D have wild type survival under the assay conditions employed. Interestingly, mutations in the more N-terminal CXXC motif lead to strains that are significantly less acid sensitive. Taken together with the urease activity assays, this suggests that the ability of HypA to deliver Ni to the urease maturation pathway may lie with the dynamic nature of the structural zinc sites. While no clear reason is apparent for the functional differences observed for the two CXXC motifs, it is noteworthy that two Cys residues are substituted by His at low pH, whereas two are retained and anchor the Zn site. This pair-wise difference in function is one possible explanation for the differences between the more N- and C- terminal CXXC sequences. Further elucidation of this difference awaits structural studies of the low pH form of HypA.
It is clear from both the urease assay data and acid viability data that Cys74 is a special position among the Cys variants. It is the only position where the Cys→ Asp mutation results in a less acid viable strain than the Cys→ Ala mutation, and the only position where urease activity is not higher for the Asp variant than for the Ala variant. Examination of the NMR structure (Fig 1) shows that the Zn site is formed by a loop that begins with Cys74 and ends with Cys94. However, the terminal Cys residues in the loop differ in that Cys94 is flanked by a His residue and Cys74 is not. In the HypA dimer crystal structure from Thermococcus kodakarensis KOD1 (PDB ID: 3A44),17 Cys74 and 94 have a similar relationship, though these residues are now derived from different monomers in the strand-swap dimer.17 The lack of an alternative ligand could make Cys74 a more vulnerable position, particularly if it plays an anchoring role, the loss of which may result in a more severe phenotype due to destabilization of the entire structural zinc site.
HypA and HypB are accessory proteins that are generally associated with hydrogenase maturation, but in H. pylori have been shown to be important for the maturation of both NiFe-hydrogenase7a, b and urease7. In addition to interacting with HypB7b, 18, HypA has also been shown to interact directly with the urease-specific Ni metallochaperone, UreE7c, 8–9. Interaction of HypA with both HypB18 and UreE9 has been demonstrated to involve the N-terminal Ni-binding domain of HypA and Ni-transfer between proteins. In this study we demonstrated that altering the HypA structural zinc site can also severely affect acid viability and urease maturation in vivo. These findings suggest that protein structural dynamics, reflected in the HypA zinc site structure, may result in subtle alterations of the overall HypA fold and differential interactions between HypA and partnering proteins, ultimately leading to a gradient in urease and hydrogenase enzymatic activities that is a function of cellular pH and nickel availability. Since mutations of the His residues do not lead to a large effect on urease activity, it is possible that the conformation associated with Zn(Cys)2(His)2 coordination is involved in supporting Ni trafficking to hydrogenase under acidic conditions. This possibility is the focus of ongoing studies. In this regard, it is worth noting that the stoichiometry of Ni binding associated with Zn(Cys)4 coordination is two per protein, while Zn(Cys)2(His)2 coordination supports binding of only a single Ni ion,11b and that the active sites of urease and hydrogenase require incorporation of two and one Ni ion, respectively.
Acknowledgments
Funding
This work was supported by NIH Grant R01-GM069696 to M.J.M, a Henry M. Jackson Fellowship to R.C.J and NIH R56 AI065529 to D.S.M.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/b000000x/
Footnotes should appear here. These might include comments relevant to but not central to the matter under discussion, limited experimental and spectral data, and crystallographic data.
Notes and references
- 1.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.(a) Uemura N, Okamoto S, Yamamoto S, Matsumura N, Yamaguchi S, Yamakido M, Taniyama K, Sasaki N, Schlemper RJ. Helicobacter pylori infection and the development of gastric cancer. The New England journal of medicine. 2001;345:784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]; (b) Wroblewski LE, Peek RM, Jr, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clinical microbiology reviews. 2010;23:713–739. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Salama NR, Hartung ML, Muller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nature reviews. Microbiology. 2013;11:385–399. doi: 10.1038/nrmicro3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eusebi LH, Zagari RM, Bazzoli F. Epidemiology of Helicobacter pylori infection. Helicobacter. 2014;19(Suppl 1):1–5. doi: 10.1111/hel.12165. [DOI] [PubMed] [Google Scholar]
- 4.(a) Eaton KA, Brooks CL, Morgan DR, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infection and immunity. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Eaton KA, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infection and immunity. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Eaton KA, Krakowka S. Avirulent, urease-deficient Helicobacter pylori colonizes gastric epithelial explants ex vivo. Scandinavian journal of gastroenterology. 1995;30:434–437. doi: 10.3109/00365529509093303. [DOI] [PubMed] [Google Scholar]
- 5.(a) Zambelli B, Musiani F, Benini S, Ciurli S. Chemistry of Ni2+ in urease: sensing, trafficking, and catalysis. Accounts of chemical research. 2011;44:520–530. doi: 10.1021/ar200041k. [DOI] [PubMed] [Google Scholar]; (b) Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Interplay of metal ions and urease. Metallomics : integrated biometal science. 2009;1:207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauerfeind P, Garner R, Dunn BE, Mobley HL. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut. 1997;40:25–30. doi: 10.1136/gut.40.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Molecular microbiology. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]; (b) Mehta N, Olson JW, Maier RJ. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. Journal of bacteriology. 2003;185:726–734. doi: 10.1128/JB.185.3.726-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Benoit SL, Mehta N, Weinberg MV, Maier C, Maier RJ. Interaction between the Helicobacter pylori accessory proteins HypA and UreE is needed for urease maturation. Microbiology. 2007;153:1474–1482. doi: 10.1099/mic.0.2006/003228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benoit SL, McMurry JL, Hill SA, Maier RJ. Helicobacter pylori hydrogenase accessory protein HypA and urease accessory protein UreG compete with each other for UreE recognition. Biochimica et biophysica acta. 2012;1820:1519–1525. doi: 10.1016/j.bbagen.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Li H, Cheng T, Xia W, Lai YT, Sun H. Nickel translocation between metallochaperones HypA and UreE in Helicobacter pylori. Metallomics : integrated biometal science. 2014;6:1731–1736. doi: 10.1039/c4mt00134f. [DOI] [PubMed] [Google Scholar]
- 10.Xia W, Li H, Sze KH, Sun H. Structure of a nickel chaperone, HypA, from Helicobacter pylori reveals two distinct metal binding sites. Journal of the American Chemical Society. 2009;131:10031–10040. doi: 10.1021/ja900543y. [DOI] [PubMed] [Google Scholar]
- 11.(a) Kennedy DC, Herbst RW, Iwig JS, Chivers PT, Maroney MJ. A dynamic Zn site in Helicobacter pylori HypA: a potential mechanism for metal-specific protein activity. Journal of the American Chemical Society. 2007;129:16–17. doi: 10.1021/ja066958x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Herbst RW, Perovic I, Martin-Diaconescu V, O'Brien K, Chivers PT, Pochapsky SS, Pochapsky TC, Maroney MJ. Communication between the zinc and nickel sites in dimeric HypA: metal recognition and pH sensing. Journal of the American Chemical Society. 2010;132:10338–10351. doi: 10.1021/ja1005724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. The complete genome sequence of Helicobacter pylori strain G27. Journal of bacteriology. 2009;191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horton RM, Ho SN, Pullen JK, Hunt HD, Cai Z, Pease LR. Gene splicing by overlap extension. Methods in enzymology. 1993;217:270–279. doi: 10.1016/0076-6879(93)17067-f. [DOI] [PubMed] [Google Scholar]
- 14.Copass M, Grandi G, Rappuoli R. Introduction of unmarked mutations in the Helicobacter pylori vacA gene with a sucrose sensitivity marker. Infection and immunity. 1997;65:1949–1952. doi: 10.1128/iai.65.5.1949-1952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joyce EA, Gilbert JV, Eaton KA, Plaut A, Wright A. Differential gene expression from two transcriptional units in the cag pathogenicity island of Helicobacter pylori. Infection and immunity. 2001;69:4202–4209. doi: 10.1128/IAI.69.7.4202-4209.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) McGee DJ, May CA, Garner RM, Himpsl JM, Mobley HL. Isolation of Helicobacter pylori genes that modulate urease activity. Journal of bacteriology. 1999;181:2477–2484. doi: 10.1128/jb.181.8.2477-2484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Weatherb.Mw, Phenol-Hypochlorite Reaction for Determination of Ammonia. Anal Chem. 1967;39 971-&, DOI: [Google Scholar]
- 17.Watanabe S, Arai T, Matsumi R, Atomi H, Imanaka T, Miki K. Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation. Journal of molecular biology. 2009;394:448–459. doi: 10.1016/j.jmb.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 18.Xia W, Li H, Yang X, Wong KB, Sun H. Metallo-GTPase HypB from Helicobacter pylori and its interaction with nickel chaperone protein HypA. The Journal of biological chemistry. 2012;287:6753–6763. doi: 10.1074/jbc.M111.287581. [DOI] [PMC free article] [PubMed] [Google Scholar]