Abstract
BRAF mutations occur in ~10% of colorectal cancer (CRC). While RAF inhibitor monotherapy is highly effective in BRAF-mutant melanoma, response rates in BRAF-mutant CRC are poor. Recent clinical trials of combined RAF/EGFR or RAF/MEK inhibition have produced improved efficacy, but patients ultimately develop resistance. To identify molecular alterations driving clinical acquired resistance, we performed whole-exome sequencing on paired pre-treatment and post-progression tumor biopsies from BRAF-mutant CRC patients treated with RAF inhibitor combinations. We identified alterations in MAPK pathway genes in resistant tumors not present in matched pre-treatment tumors, including KRAS amplification, BRAF amplification, and a MEK1 mutation. These alterations conferred resistance to RAF/EGFR or RAF/MEK combinations through sustained MAPK pathway activity, but an ERK inhibitor could suppress MAPK activity and overcome resistance. Identification of MAPK pathway reactivating alterations upon clinical acquired resistance underscores the MAPK pathway as a critical target in BRAF-mutant CRC and suggests therapeutic options to overcome resistance.
Keywords: BRAF, colorectal cancer, drug resistance
INTRODUCTION
BRAF valine 600 (V600) mutations occur in ~10% of CRC and confer poor prognosis in the metastatic setting(1, 2). BRAF, along with ARAF and CRAF, belongs to the RAF family of kinases, which are normally activated by RAS proteins(3). BRAF V600 mutations lead to constitutive activation of BRAF kinase activity, resulting in phosphorylation and activation of the MEK kinases (MEK1 and MEK2). Once activated, MEK kinases phosphorylate and activate ERK kinases, which phosphorylate a multitude of key cellular substrates involved in cell proliferation and survival.
RAF inhibitors, such as vemurafenib and dabrafenib, have produced response rates of ~50-80% in the roughly half of melanomas that also harbor BRAF V600 mutations, revolutionizing the treatment of these cancers and leading to the FDA approval of vemurafenib and dabrafenib for this disease(4, 5). However, in CRCs harboring the identical BRAF V600 mutation, RAF inhibitor monotherapy has proven disappointingly ineffective, with response rates of only ~5%(6). This striking disparity in sensitivity between these two tumor types represents a critical challenge to the development of effective therapies for BRAF-mutant CRC. Previous work by our group and others suggested that RAF inhibitor insensitivity in BRAF-mutant CRC is driven by feedback reactivation of MAPK signaling following RAF inhibitor treatment(7-9). In many, but not all, BRAF-mutant CRCs the feedback reactivation of MAPK signaling is driven by EGFR-mediated activation of RAS and CRAF(8, 10). Preclinical studies have suggested that RAF inhibitor combination strategies, including co-targeting RAF and EGFR or RAF and MEK, can suppress feedback reactivation of MAPK signaling, leading to more robust and sustained inhibition of the pathway and to improved efficacy in BRAF-mutant CRC.
Based on these data, RAF inhibitor combinations have been evaluated in clinical trials for BRAF-mutant CRC patients in recent years and are showing signs of improved efficacy compared to RAF inhibition alone. A clinical trial of combined RAF/MEK inhibition with dabrafenib and the MEK inhibitor trametinib in 43 BRAF V600 CRC patients produced a response rate of 12%, including one patient with a durable complete response that remains ongoing over three years. Additionally 51% of patients achieved disease stabilization on study(11). More recently, clinical trials of combined RAF/EGFR inhibition have been initiated, including the combination of vemurafenib and the anti-EGFR antibody cetuximab, the combination of the RAF inhibitor encorafenib (LGX-818) and cetuximab, and the combination of dabrafenib and the anti-EGFR antibody panitumumab(12-14). Some of these trials have produced preliminary response rates of as high as 29%. A trial of combined RAF/EGFR/MEK inhibition with the triple combination of dabrafenib, panitumumab, and trametinib is also underway and has shown an initial response rate of 40%(12). However, despite the promising signs of efficacy demonstrated by early clinical trials of RAF inhibitor combinations in BRAF-mutant CRC patients, those patients deriving initial benefit from therapy ultimately develop resistance to treatment and disease progression. An understanding of the mechanisms of clinical acquired resistance that arise to RAF inhibitor combinations in BRAF-mutant CRC patients may lead to valuable opportunities to overcome resistance and prolong clinical response.
Here, we find that MAPK pathway alterations leading to reactivation of MAPK signaling are important drivers of acquired resistance in BRAF-mutant CRC. In vitro modeling of acquired resistance to RAF/EGFR or RAF/MEK inhibitor combinations in BRAF-mutant CRC cell lines revealed activating KRAS exon 2 mutations as drivers of acquired resistance to either therapy through sustained MAPK pathway activation. Whole-exome sequencing of paired pre-treatment and post-progression biopsies from patients with initial clinical response or prolonged stable disease on RAF/EGFR or RAF/MEK combination therapy identified MAPK pathway alterations unique to the resistant tumor, including KRAS amplification, BRAF amplification, and MEK1 mutation. Each of these alterations led to resistance to RAF/EGFR or RAF/MEK combinations and to sustained MAPK pathway activation in vitro, but retained sensitivity to an ERK inhibitor. These findings highlight the critical importance of the MAPK pathway as a therapeutic target in BRAF-mutant CRC and point to possible therapeutic strategies to overcome clinical resistance.
RESULTS
In vitro modeling identifies RAS activation as a potential resistance mechanism
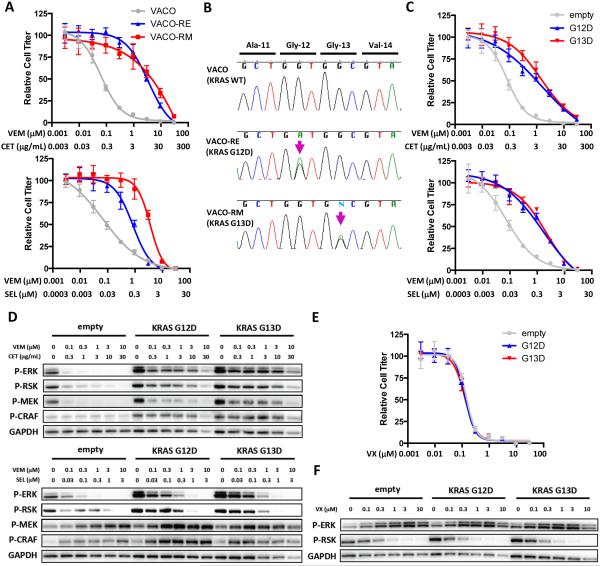
To begin to understand the spectrum of molecular alterations that can drive acquired resistance to RAF inhibitor combinations, we generated resistant variants of the sensitive BRAF-mutant CRC cell line VACO432 by culturing cells in the presence of a RAF/EGFR inhibitor combination using vemurafenib and cetuximab (VACO-RE cells) or a RAF/MEK combination using vemurafenib and selumetinib (VACO-RM cells). Interestingly, resistant cells derived using one RAF inhibitor combination were cross-resistant to the other RAF inhibitor combination (Fig. 1A). Sequence analysis of VACO-RE and VACO-RM cells revealed that each resistant variant retained the original BRAF V600E mutation, but that each acquired a distinct exon 2 mutation in KRAS (G12D and G13D, respectively) that was not present in the parental cell line, with no change in KRAS copy number (Figs. 1B, S1). Identification of KRAS mutations as a potential mechanism of acquired resistance is consistent with previous findings that NRAS mutations are a common cause of clinical acquired resistance in BRAF-mutant melanoma(15, 16). Exogenous expression of KRAS G12D or G13D in VACO432 cells recapitulated resistance to RAF/EGFR and RAF/MEK inhibitor combinations, including combinations currently in clinical trials (Fig. 1C, S2A). Expression of mutant KRAS led to increased basal levels of phosphorylated CRAF (P-CRAF), P-MEK, and P-ERK (Fig. 1D). Importantly, mutant KRAS expression also abrogated the ability of RAF/EGFR or RAF/MEK combinations to inhibit MAPK signaling, as evidenced by sustained phosphorylation of ERK and of the ERK target RSK (P-RSK). These findings suggest that activation of RAS can maintain MAPK pathway signaling and drive resistance to either combined RAF/EGFR or RAF/MEK inhibition.
Figure 1. KRAS mutation leads to resistance to combined RAF/EGFR and RAF/MEK inhibition.
(A) Parental VACO432 cells (VACO) and derivatives made resistant to combined RAF/EGFR inhibition (VACO-RE) or RAF/MEK inhibition (VACO-RM) were treated for 3d with the indicated concentrations of vemurafenib (VEM) and cetuximab (CET) or vemurafenib and selumetinib (SEL). Relative cell titer was determined by Cell TiterGlo assay. (B) Sanger sequencing of exon 2 of KRAS from genomic DNA isolated from VACO, VACO-RE, and VACO-RM cells. (C) VACO432 cells expressing exogenous KRAS G12D, G13D, or empty vector control were treated as in (A) and relative cell titer was determined. (D) VACO432 cells expressing KRAS G12D, G13D, or empty vector control were treated for 24h with the indicated concentrations of drugs, and western blotting was performed with the indicated antibodies. (E) VACO432 cells expressing exogenous KRAS G12D, G13D, or empty vector control were treated with the ERK inhibitor VX-11e (VX) as in (A) and relative cell titer was determined. (F) Cells were treated as in (D) for 24h with the indicated concentrations of VX-11e and western blotting was performed with the indicated antibodies.
Interestingly, an ERK inhibitor retained the ability to suppress MAPK despite expression of KRAS G12D or G13D, as measured by its ability to inhibit P-RSK levels (since certain ERK inhibitors like VX-11e lead to a feedback induction of P-ERK despite inhibition of ERK kinase activity(17, 18)), and was able to overcome resistance (Figs. 1E,F; S2B-D). These results emphasize the importance of sustained MAPK signaling in driving resistance to RAF inhibitor combinations in BRAF-mutant CRC cells and suggest that therapies which restore MAPK pathway suppression may be promising strategies to overcome resistance.
Clinical acquired resistance to combined RAF/EGFR inhibition
To identify acquired resistance mechanisms to RAF inhibitor combinations that occur clinically, we evaluated paired tumor biopsies taken before treatment and after eventual disease progression from BRAF-mutant CRC patients who achieved an initial response or prolonged stable disease on a RAF/EGFR or RAF/MEK inhibitor combination (Table S1). Pre-treatment and post-progression tumor biopsies were evaluated by whole-exome sequencing (WES) to identify novel molecular alterations arising in the drug-resistant post-progression tumor that were absent in the pre-treatment tumor. Paired biopsies from two patients with clinical acquired resistance to combined RAF/EGFR inhibition were evaluated.
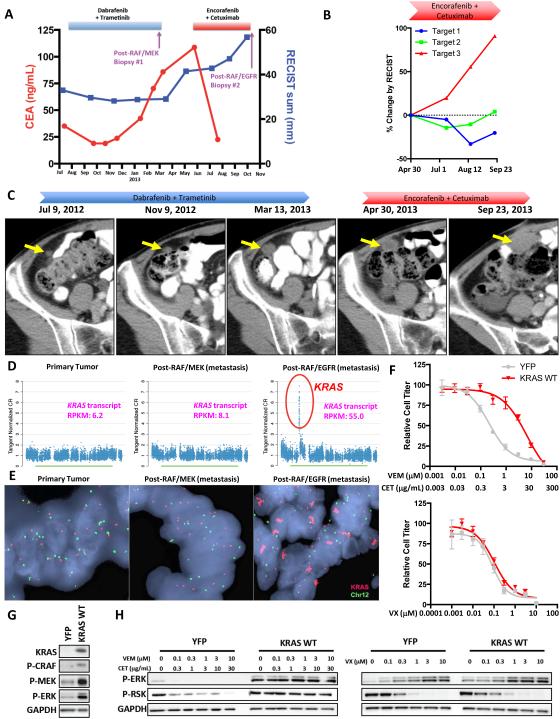
The first BRAF-mutant CRC patient was initially treated with the RAF/MEK combination of dabrafenib and trametinib and achieved a prolonged minor response lasting over seven months, as evidenced by the decrease in tumor burden as measured by Response Evaluation Criteria in Solid Tumors (RECIST) and the decrease in the patient’s serum carcinoembryonic antigen (CEA) tumor marker (Fig. 2A). Ultimately, the patient progressed on RAF/MEK combination therapy, and at that time the patient underwent complete excision of a rapidly progressing subcutaneous lesion near the umbilicus (post-RAF/MEK biopsy). The patient was then treated with the RAF/EGFR inhibitor combination of encorafenib and cetuximab and experienced a dramatic drop in the CEA tumor marker levels after initiating therapy, suggesting an overall decrease in tumor burden (Fig. 2A). The patient also experienced a decrease in two of three target lesions by RECIST (Fig. 2B). However, one target lesion progressed steadily through therapy, eventually causing the patient to meet RECIST criteria for progressive disease. Interestingly, this lesion was present while the patient was receiving RAF/MEK combination therapy, and although the lesion remained stable during the first few months of RAF/MEK therapy, the lesion began to progress during the latter months of RAF/MEK treatment (Fig. 2C). This suggests that perhaps the molecular alteration driving resistance in this lesion actually may have arisen as a mechanism of acquired resistance during RAF/MEK therapy and then caused this specific lesion to be immediately refractory to subsequent RAF/EGFR therapy, even though the remainder of the patient’s disease appeared to respond to combined RAF/EGFR inhibition.
Figure 2. KRAS amplification can drive clinical acquired resistance to combined RAF/EGFR or RAF/MEK inhibition.
(A) Clinical time course of therapy for BRAF-mutant CRC patient #1 showing dates of therapy and timing of biopsies. Biopsies #1 (post-RAF/MEK) and #2 (post-RAF/EGFR) were taken from distinct metastatic lesions. Serum CEA tumor marker levels and cumulative tumor diameter as measured by RECIST are shown throughout the treatment course. (B) RECIST measurements of individual target lesions during therapy with encorafenib plus cetuximab from April 30 to September 23, 2013. Target lesion #3 was biopsied upon progression. (C) CT images of target lesion #3 (indicated by yellow arrow) throughout therapy with dabrafenib plus trametinib and encorafenib plus cetuximab. (D) DNA copy number traces for chromosome 12 showing focal amplification of KRAS in the post-RAF/EGFR biopsy. KRAS transcript abundance as determined by RNA-seq are also shown for each sample (RPKM = reads per kilobase of transcript per million mapped reads). (E) FISH was performed on biopsy specimens using probes for KRAS (red) and chromosome 12 (Chr12; green). (F) VACO432 cells overexpressing YFP control or wild-type KRAS (KRAS WT) were treated for 72h with the indicated concentrations of drug, and relative cell titer was determined. (G) VACO432 cells overexpressing YFP or KRAS WT were lysed, and western blotting was performed with the indicated antibodies. (H) Western blot of VACO432 cells overexpressing YFP or KRAS WT were treated with the indicated concentrations of drug for 24h.
This progressing lesion (post-RAF/EGFR) was biopsied and was evaluated by WES and RNA sequencing compared to both the patient’s primary tumor and the separate metastatic lesion excised after progression on combined RAF/MEK therapy (post-RAF/MEK). The post-RAF/MEK biopsy retained the original BRAF V600E mutation, but harbored no new mutations compared to the primary tumor, and a definitive mechanism of resistance was not identified (Fig. S3). The post-RAF/EGFR progression biopsy retained the original BRAF V600E mutation, but no new candidate resistance mutations arising specifically in the post-RAF/EGFR biopsy were identified (Table S2). However, copy number analysis revealed focal amplification of KRAS on chromosome 12 in this resistant lesion that was not present in either of the two prior biopsy specimens (Fig. 2D). Amplification of wild-type KRAS has previously been implicated as a mechanism of resistance to targeted therapies, including anti-EGFR antibodies like cetuximab(19). RNA sequencing (RNA-seq) confirmed ~6-8 fold overexpression of KRAS transcript in the post-RAF/EGFR biopsy relative to each of the prior biopsies. Fluorescence in situ hybridization (FISH) confirmed ~25-fold amplification of KRAS in the post-RAF/EGFR biopsy (Fig. 2E), suggesting KRAS amplification as the likely driver of acquired resistance in this lesion.
Overexpression of wild-type KRAS conferred resistance to multiple RAF/EGFR inhibitor combinations (Fig. 2F, S4A). Notably, KRAS overexpression also conferred resistance to RAF/MEK inhibitor combinations (Fig. S4B), supporting the possibility that this alteration may have initially arisen as an acquired resistance mechanism to the patient’s original RAF/MEK therapy and then promoted upfront resistance to subsequent RAF/EGFR therapy. Similar to our observations with mutant KRAS expression, overexpression of wild-type KRAS led to increased basal levels of P-CRAF, P-MEK, and P-ERK, and abrogated the ability of combined RAF/EGFR or RAF/MEK inhibition to inhibit the MAPK pathway (Fig. 2G,H, S4C).
Importantly, an ERK inhibitor again was able to suppress the MAPK pathway in cells overexpressing wild-type KRAS and was able to overcome resistance (Figs. 2F,H). Collectively, our in vitro and clinical findings suggest that activation of RAS, either by mutation of KRAS or amplification of wild-type KRAS, can promote resistance to either RAF/EGFR or RAF/MEK inhibitor combinations by promoting sustained MAPK pathway signaling. However, in either case, an ERK inhibitor retains its ability to block MAPK signaling and can suppress resistance conferred by either KRAS alteration.
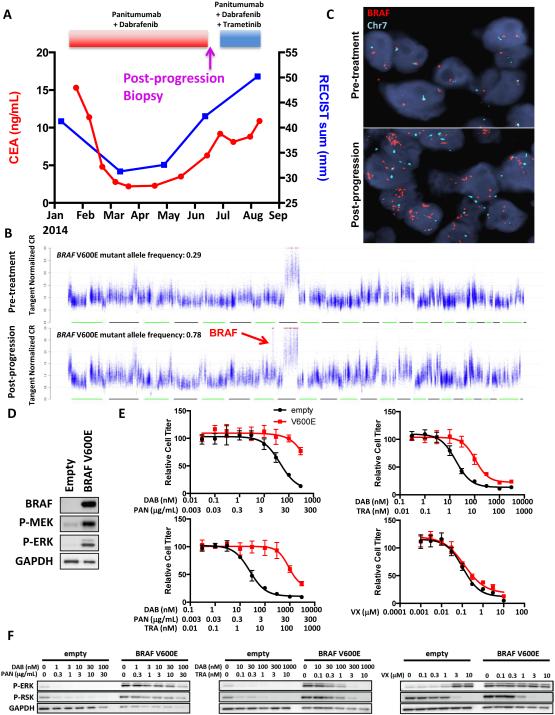
A second BRAF-mutant CRC patient achieved a minor response to combined RAF/EGFR inhibition with dabrafenib and panitumumab, lasting almost five months before eventual disease progression (Fig. 3A). A progressing retroperitoneal lymph node tumor was biopsied upon disease progression and evaluated by WES in comparison to a biopsy of the same lesion obtained immediately prior to the start of therapy. The patient was subsequently treated with the triple RAF/EGFR/MEK inhibitor combination of dabrafenib, panitumumab, and trametinib, but progressed rapidly through therapy.
Figure 3. BRAF amplification causes clinical acquired resistance to combined RAF/EGFR inhibition.
(A) Clinical time course of therapy for BRAF-mutant CRC patient #2 showing dates of therapy and timing of post-progression biopsy. CEA tumor marker levels and cumulative tumor diameter as measured by RECIST are shown throughout the treatment course. (B) DNA copy number traces are shown for the post-progression biopsy compared to a pre-treatment biopsy taken from the same lesion immediately prior to the start of panitumumab + dabrafenib therapy. Focal amplification of BRAF on chromosome 7 is shown in the post-progression biopsy. BRAF V600E mutant allele frequencies are shown for each sample. (C) FISH was performed on the pre-treatment and post-progression biopsies using probes for BRAF (red) and chromosome 7 (Chr7; green). (D) VACO432 cells overexpressing BRAF V600E compared to empty vector were lysed, and western blotting was performed with the indicated antibodies. (E) Cells from (D) were treated for 72h with the indicated concentrations of dabrafenib (DAB), panitumumab (PAN), trametinib (TRA), or VX-11e (VX) for 72h and relative cell titer was determined. (F) Cells from (D) were treated for 24h with the indicated concentrations of drugs, and western blotting was performed with the indicated antibodies.
WES revealed that the post-progression specimen retained the original BRAF V600E mutation, but did not identify any new mutations arising in the post-progression biopsy likely to explain acquired resistance (Table S2). However, copy number analysis identified a focal BRAF amplification on chromosome 7, and BRAF amplification was confirmed by FISH (Figs. 3B,C). The high allele frequency of the BRAF V600E mutation in the post-progression biopsy suggests predominant amplification of the mutant BRAF allele (Fig. 3B). Previously, our group identified amplification of mutant BRAF as a mechanism of resistance to RAF or MEK inhibition in BRAF-mutant CRC, and subsequently, BRAF amplification has been implicated as an important acquired resistance mechanism to RAF inhibitor monotherapy or combined RAF/MEK inhibition in BRAF-mutant melanoma(7, 20, 21). Overexpression of BRAF V600E led to increased basal levels of P-MEK and P-ERK in BRAF-mutant CRC cells and conferred resistance to combined RAF/EGFR inhibition (Fig. 3D,E). Notably, BRAF V600E overexpression also led to cross-resistance to combined BRAF/MEK and to combined BRAF/EGFR/MEK inhibition, perhaps explaining why the patient did not respond to the triple combination after initial disease progression on RAF/EGFR therapy (Fig 3E). BRAF V600E overexpression abrogated the ability of combined RAF/EGFR or RAF/MEK inhibition to suppress MAPK signaling (Fig. 3F). However, an ERK inhibitor retained the ability to suppress the MAPK pathway and to overcome resistance caused by BRAF V600E overexpression (Figs. 3E,F).
Clinical acquired resistance to combined RAF/MEK inhibition
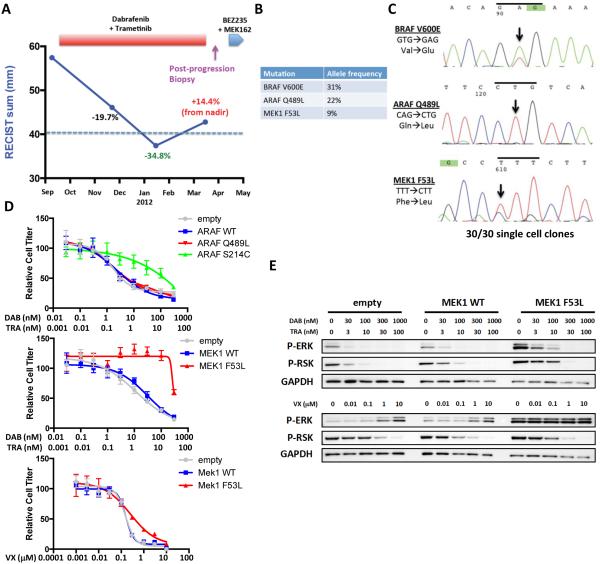
A third BRAF-mutant CRC patient was treated with combined RAF/MEK inhibition with dabrafenib and trametinib. The patient achieved a reduction in tumor burden that met criteria for a partial response by RECIST, but unfortunately developed disease progression after almost six months of therapy (Fig. 4A). The patient was then treated with the combination of a PI3K/mTOR inhibitor and a MEK inhibitor, but progressed rapidly through therapy. WES revealed that a tumor biopsy taken after progression on BRAF/MEK therapy retained the original BRAF V600E mutation, but also identified the presence of two candidate resistance mutations that were not present in the pre-treatment biopsy (Fig. 4B, Table S2). One mutation was a Q489L missense mutation in another RAF kinase, ARAF. ARAF mutations are rare in human cancer, but Q489 in ARAF corresponds to Q636 in BRAF, which is mutated in a small percentage of lung and colorectal cancers(22). The second mutation was a F53L missense mutation in MEK1 (MAP2K1) that occurs in helix A, a region of MEK1 and MEK2 previously found to be mutated in BRAF-mutant melanomas that have acquired resistance to RAF inhibitors or RAF/MEK inhibitor combinations(16, 21). The lower allele frequencies of the ARAF and MEK1 mutations suggested the possibility that these mutations were subclonal, perhaps representing two populations of cancer cells that developed resistance independently through distinct mechanisms, with each population harboring one mutation. While it is not possible to determine whether the ARAF and MEK1 mutations co-exist in the same tumor cells from the WES alone, we were able to utilize a patient-derived cell line generated from the post-progression biopsy. Sanger sequencing of the pooled cell line successfully identified the presence of the original BRAF V600E mutation, as well as the presence of both candidate resistance mutations (Fig. 4C). We generated 30 single cell clones to determine if individual tumor cells harbored different combinations of these three mutations. Surprisingly, we found that all 30 single cell clones harbored all three mutations, suggesting that the clone or clones that gave rise to this cell line harbored both candidate resistance mutations as well as the original BRAF V600E mutation. While this results does not preclude the possibility that a subpopulation of tumor cells may have developed one candidate resistance mutation first and that the second mutation may have arisen within a subclone of that population, it does make it highly unlikely that these two mutations existed independently in distinct tumor cells within the resistant lesion.
Figure 4. MEK1 F53L mutation drives clinical acquired resistance to combined RAF/MEK inhibition.
(A) Clinical time course of therapy for BRAF-mutant CRC patient #3 showing dates of therapy and timing of post-progression biopsy. Cumulative tumor diameter as measured by RECIST is shown throughout the treatment course. (B) List of key mutations identified in the post-progression biopsy with associated allele frequencies. (C) Sanger sequencing of a patient-derived cell line generated from the post-progression biopsy for BRAF V600E, ARAF Q489L, and MEK1 F53L. All mutations were found to be present in 30 of 30 single cell clones. (D) Cells expressing the indicated constructs were treated with dabrafenib plus trametinib or VX-11e as shown for 72h, and relative cell titer was determined. (E) Cells expressing wild-type MEK1 (MEK1 WT), MEK1 F53L, or empty vector control were treated with the indicated concentrations of drugs for 24h, and western blotting was performed with the indicated antibodies.
Consistent with these findings, we found that exogenous expression of ARAF Q489L did not markedly confer resistance to dabrafenib and trametinib relative to wild-type ARAF, whereas the previously identified activating S214C mutation in ARAF(23) effectively promoted resistance (Fig. 4D). Furthermore, ARAF S214C, but not ARAF Q489L led to MAPK pathway activation in 293T cells, relative to wild-type ARAF (Fig. S5A,B). While it is still possible that ARAF Q489L may have a subtle contribution to drug sensitivity, this mutation does not appear to be a primary driver of acquired RAF/MEK resistance in this setting.
Conversely, exogenous expression of MEK1 F53L led to robust resistance to dabrafenib and trametinib and to increased basal activation and phosphorylation of ERK, relative to wild-type MEK1 (Figs. 4D,E). MEK1 F53L expression also abrogated the ability of dabrafenib and trametinib to suppress MAPK signaling. Thus, MEK1 F53L is the likely driver of clinical acquired resistance in this patient’s tumor. Importantly, an ERK inhibitor retained the ability to suppress MAPK signaling and could mitigate resistance caused by MEK1 F53L expression. Similarly, an ERK inhibitor, alone or in combination with dabrafenib, was able to mitigate resistance in the tumor cell line derived from this patient (Fig. S5C). Since multiple mechanisms of acquired resistance to targeted therapy can arise within an individual patient(16), this case serves as an important example that careful validation of candidate resistance mechanisms will be a critical adjunct to properly interpret sequencing data as it pertains to the potential heterogeneity of acquired resistance.
DISCUSSION
In this study, we identified multiple molecular alterations within the MAPK pathway that lead to reactivation of MAPK signaling and acquired resistance to combined RAF/EGFR and RAF/MEK inhibition in BRAF-mutant CRC, both in vitro and in patient biopsies obtained upon development of clinical acquired resistance. The fact that all resistance mechanisms we identified promote reactivation of the MAPK pathway highlights the critical dependence of BRAF-mutant CRC on sustained MAPK signaling to maintain cancer cell proliferation and survival and underscores the MAPK pathway as a critical therapeutic target in this disease. Indeed, initial pharmacodynamic analysis of paired pre-treatment and on-treatment biopsies from BRAF-mutant CRC patients on clinical trials of RAF/EGFR or RAF/MEK inhibitor combinations have suggested that a major factor limiting response is that these inhibitor combinations still produce a lesser degree of MAPK pathway inhibition in BRAF-mutant CRC patients than single-agent RAF inhibitors in BRAF-mutant melanoma patients(11, 12). Conversely, the triple combination of RAF/EGFR/MEK inhibition produces a degree of MAPK inhibition closer to that observed with single-agent RAF inhibitor in BRAF-mutant melanoma, underscoring the importance of robust and sustained MAPK inhibition in BRAF-mutant cancers(12). These data are consistent with previous reports that the majority of acquired resistance mechanisms to RAF or RAF/MEK inhibition in BRAF-mutant melanoma—including NRAS alterations, BRAF amplification, MEK1 or MEK2 mutations, and BRAF splice isoforms—also lead to reactivation of MAPK signaling, as opposed to activation of alternative parallel signaling pathways(15, 16, 21). As some of these same resistance mechanisms were identified in our initial analysis of acquired resistance in BRAF-mutant CRC, including RAS alterations (though in KRAS, not NRAS), BRAF amplification, and MEK1 mutation, it is possible that other known resistance mechanisms in BRAF-mutant melanoma leading to MAPK reactivation may be found in other BRAF-mutant CRC patients. Indeed, the continued MAPK pathway dependence of BRAF-mutant CRC cells harboring molecular alterations promoting acquired resistance is supported by the fact that they remain sensitive to ERK inhibitors, which retain the ability to suppress MAPK signaling despite the presence of these alterations. While it is certainly possible that MAPK-independent mechanisms may lead to clinical acquired resistance in some cases of BRAF-mutant CRC, collectively these findings emphasize that effective targeting of the MAPK pathway is paramount in the therapeutic approach to this disease.
While our findings support MAPK pathway reactivation as a key event in the development of acquired resistance to RAF inhibitor combinations, it is striking that a different molecular mechanism of resistance was identified in each analysis, including KRAS mutation, KRAS amplification, BRAF amplification, and MEK1 mutation. The diversity of potential molecular alterations leading to acquired resistance suggests that therapeutic approaches targeting individual resistance alterations may be difficult to implement as a generalizable strategy to overcome resistance. However, the common thread among these diverse molecular alterations is that they converge on MAPK pathway reactivation as a basic mechanism for promoting resistance. Thus, it may be possible to find a key convergent signaling node that can effectively target resistance signals produced by many of these specific alterations. We found that an ERK inhibitor retained the ability to suppress MAPK signaling and could overcome each of the acquired resistance mechanisms identified. It has been previously demonstrated by our group and others that resistance to MEK inhibitors can be promoted by molecular alterations that activate the MAPK pathway upstream of MEK, including RAS mutation or amplification and BRAF amplification, likely by leading to increased pathway flux and MEK hyperactivation that can overcome the effects of MEK inhibitors(7, 24). However, in many of these cases of upstream MAPK pathway alterations, ERK inhibitors maintain their ability to suppress the MAPK pathway, as we have observed here(17, 18). Taken together, these findings suggest that ERK inhibitors, which are currently in early phase clinical trials, could be important components of future therapeutic strategies for BRAF-mutant CRC, either alone, or in combination with RAF and EGFR inhibitors. Further studies defining the common mechanisms of clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant CRC patients will be critical to improving therapies for this lethal subtype of CRC.
METHODS
Detailed methods are included in the Supplemental Materials.
Patient samples, cell lines, and reagents
Patient tumor specimens and normal blood were obtained from patients treated at the Massachusetts General Hospital under Institutional Review Board-approved studies. All patients provided written, informed consent, and studies were conducted in accordance with the Declaration of Helsinki.
VACO432 and COLO-320DM cells were obtained from the Massachusetts General Hospital Center for Molecular Therapeutics, which performs routine cell line authentication testing by single-nucleotide polymorphism and short tandem repeat analysis, and were passaged less than six months following receipt. Cells were grown in DMEM/F12 (GIBCO) with 10% FBS and assayed in DMEM/F12 with 5% FBS. Patient-derived cell line (patient 3) was generated and propagated from biopsy material in ACL-4 media (GIBCO) with 10% FBS and assayed in ACL-4 with 5% FBS. Cetuximab and Pantimumab were obtained from the Massachusetts General Hospital Pharmacy and diluted in PBS. Chemical inhibitors from the following sources were dissolved in DMSO: selumetinib, vemurafenib, dabrafenib, trametinib (Selleck Chemicals); Vx-11e (ChemieTek).
Computed tomography (CT) scans and RECIST measurements
Spiral CT scans were obtained using standard procedures in the Department of Radiology at the Massachusetts General Hospital as part of the routine clinical care of these patients. Response Evaluation Criteria in Solid Tumors (RECIST) measurements were performed by radiologists in the Tumor Imaging Metrics Core at the Dana-Farber/Harvard Cancer Center using standard methods. CT images corresponding to specific RECIST target lesions as defined by the Tumor Imaging Metrics Core were obtained for display.
Whole-exome and RNA sequencing
Whole-exome sequencing of matched pre-treatment and post-progression biopsies and normal blood was performed as previously described(16, 21, 25). All BAM files were deposited in dbGAP, accession number phs00803.v1.p1. Detailed methods for RNA sequencing from formalin-fixed paraffin-embedded tissue are included in the Supplemental Materials.
Western blot analysis and antibodies
Western blotting was performed using standard methods. After treatment with indicated drugs, cells were washed with cold PBS and lysed in the following lysis buffer: 20 mM Tris pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 10% glycerol, 1 mM EDTA, 1 mM EGTA, 5 mM sodium pyrophosphate, 50 mM NaF, 10 nM β–glycerophosphate, 1 mM sodium vanadate, 0.5 mM DTT, 4 μg/mL leupeptin, 4 μg/mL pepstatin, 4 μg/mL aprotinin, 1 mM phenylmethylsulfonyl fluoride. Lysates were centrifuged at 16,000 × g for 5 min at 4°C. Protein concentrations were determined by BCA assay (Thermo Scientific). Proteins were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Hybond-P, Amersham). Immunoblotting was performed per antibody manufacturer’s specifications. Antibodies for P-MEK, P-CRAF, ARAF, and BRAF (Cell Signaling) were used at 1:1000 dilution; P-ERK (Cell Signaling) was used at 1:2000 dilution; P-RSK (Millipore) was used at 1:10,000 dilution; KRAS (Santa Cruz) was used at 1:500 dilution; GAPDH (Millipore) was used at 1:1000 dilution. Protein detection on Western blots was performed using SuperSignal chemiluminescence (Thermo Scientific).
Fluorescence in situ hybridization (FISH)
FISH for BRAF was performed as described previously (7). FISH for KRAS was performed in the Massachusetts General Hospital Molecular Pathology Clinical Laboratory using a CLIA-approved clinical assay.
Generation of resistant cell lines
VACO432 cells were seeded at ~70% confluence in 10 cm plates in DMEM/F12 with 5% FBS and were treated continuously with either 3μM vemurafenib with 10μg/mL cetuximab (VACO-RE cells) or 3μM vemurafenib with 1μM selumetinib (VACO-RM cells) until cells capable of proliferating efficiently in the presence of drug were derived. Cells were maintained in fresh drug-containing media changed every 72-96 hours throughout.
Supplementary Material
SIGNIFICANCE.
RAF inhibitor combinations represent promising approaches in clinical development for BRAF-mutant CRC. Initial characterization of clinical acquired resistance mechanisms to these regimens identified several MAPK pathway alterations driving resistance by reactivating MAPK signaling, highlighting the critical dependence of BRAF-mutant CRCs on MAPK signaling and offering potential strategies to overcome resistance.
Acknowledgments
GRANT SUPPORT
This study is supported by grants from the NIH Gastrointestinal Cancer SPORE P50 CA127003 (to RBC, LAG, JAE, EMS); a Damon Runyon Clinical Investigator Award and 1K08CA166510 (to RBC); and NHGRI grant U54HG003067.
J.A.E. has received consulting fees from Roche, Genentech, and GSK, and has received consulting fees and research support from AstraZeneca and Novartis.
Footnotes
Disclosure of Potential Conflicts of Interest:
N.W. is a consultant/advisory board member, stockholder, and has ownership interest (including patents) in Foundation Medicine.
R.B.C. has received speaking honoraria from GSK.
No potential conflicts of interest were disclosed by the other authors.
REFERENCES
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:5931–7. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 3.Montagut C, Settleman J. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer letters. 2009;283:125–34. doi: 10.1016/j.canlet.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. The New England journal of medicine. 2014;371:1877–88. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 6.Kopetz S, Desai J, Chan E, Hecht JR, O'Dwyer PJ, Lee RJ, et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. ASCO Meeting Abstracts. 2010;28:3534. [Google Scholar]
- 7.Corcoran RB, Dias-Santagata D, Bergethon K, Iafrate AJ, Settleman J, Engelman JA. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Science signaling. 2010;3:ra84. doi: 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery. 2012;2:227–35. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montero-Conde C, Ruiz-Llorente S, Dominguez JM, Knauf JA, Viale A, Sherman EJ, et al. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer discovery. 2013;3:520–33. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 11.Corcoran RB, Atreya CE, Falchook GS, Infante JR, Hamid O, Messersmith WA, et al. Phase 1-2 trial of the BRAF inhibitor dabrafenib (D) plus MEK inhibitor trametinib (T) in BRAF V600 mutant colorectal cancer (CRC): Updated efficacy and biomarker analysis. ASCO Meeting Abstracts. 2014;32:3517. [Google Scholar]
- 12.Bendell JC, Atreya CE, Andre T, Tabernero J, Gordon MS, Bernards R, et al. Efficacy and tolerability in an open-label phase I/II study of MEK inhibitor trametinib (T), BRAF inhibitor dabrafenib (D), and anti-EGFR antibody panitumumab (P) in combination in patients (pts) with BRAF V600E mutated colorectal cancer (CRC) ASCO Meeting Abstracts. 2014;32:3515. [Google Scholar]
- 13.Tabernero J, Chan E, Baselga J, Blay J-Y, Chau I, Hyman DM, et al. VE-BASKET, a Simon 2-stage adaptive design, phase II, histology-independent study in nonmelanoma solid tumors harboring BRAF V600 mutations (V600m): Activity of vemurafenib (VEM) with or without cetuximab (CTX) in colorectal cancer (CRC) ASCO Meeting Abstracts. 2014;32:3518. [Google Scholar]
- 14.Geel RV, Elez E, Bendell JC, Faris JE, Lolkema MPJK, Eskens F, et al. Phase I study of the selective BRAFV600inhibitor encorafenib (LGX818) combined with cetuximab and with or without the {alpha}-specific PI3K inhibitor BYL719 in patients with advanced BRAF-mutant colorectal cancer. ASCO Meeting Abstracts. 2014;32:3514. [Google Scholar]
- 15.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer discovery. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatzivassiliou G, Liu B, O'Brien C, Spoerke JM, Hoeflich KP, Haverty PM, et al. ERK inhibition overcomes acquired resistance to MEK inhibitors. Molecular cancer therapeutics. 2012;11:1143–54. doi: 10.1158/1535-7163.MCT-11-1010. [DOI] [PubMed] [Google Scholar]
- 18.Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer discovery. 2013;3:742–50. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 19.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature communications. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer discovery. 2014;4:61–8. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meador CB, Micheel CM, Levy MA, Lovly CM, Horn L, Warner JL, et al. Beyond histology: translating tumor genotypes into clinically effective targeted therapies. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:2264–75. doi: 10.1158/1078-0432.CCR-13-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imielinski M, Greulich H, Kaplan B, Araujo L, Amann J, Horn L, et al. Oncogenic and sorafenib-sensitive ARAF mutations in lung adenocarcinoma. The Journal of clinical investigation. 2014;124:1582–6. doi: 10.1172/JCI72763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M, et al. Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells. Science signaling. 2011;4:ra17. doi: 10.1126/scisignal.2001752. [DOI] [PubMed] [Google Scholar]
- 25.Van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nature medicine. 2014;20:682–8. doi: 10.1038/nm.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.