Abstract
Macroautophagy (autophagy hereafter) may promote survival and growth of spontaneous tumors, including melanoma. We utilized a genetically engineered mouse model (GEMM) driven by oncogenic BRAFV600E and deficiency in the Pten tumor suppressor gene in melanocytes to test the functional consequences of loss of the essential autophagy gene autophagy-related-7, Atg7. Atg7 deficiency prevented melanoma development by BRAFV600E and allelic Pten loss, indicating that autophagy is essential for melanomagenesis. Moreover, BRAFV600E mutant, Pten-null, Atg7-deficient melanomas displayed accumulation of autophagy substrates and growth defects, which extended animal survival. Atg7-deleted tumors showed increased oxidative stress and senescence, a known barrier to melanomagenesis. Treatment with the BRAF inhibitor dabrafenib decreased tumor growth and induced senescence that was more pronounced in tumors with Atg7 deficiency. Thus Atg7 promotes melanoma by limiting oxidative stress and overcoming senescence, and autophagy inhibition may be of therapeutic value by augmenting the anti-tumor activity of BRAF inhibitors.
Keywords: Melanoma, autophagy, senescence, BRAFV600E, dabrafenib
Introduction
Malignant melanoma is the most dangerous cancer of the skin and extremely difficult to treat when disease recurs. In 2014, approximately 10,000 melanoma related deaths are expected to occur in the US with many thousands more in other nations (1). Recent advances have improved the treatment landscape of this disease (2–4), the use of which may allow five-year survival to approach 30–40% in the near future. Nonetheless, the majority of patients will die from their disease, making it imperative to build on these recent advances to develop more effective therapeutic strategies.
One of the most important events in the field has been the development of highly potent and specific inhibitors of the mitogen activated protein kinase (MAPK) pathway, as nearly half of melanomas harbor a mutation in BRAFV600E or BRAFV600K that leads to hyperactivation of the MAPK pathway which promotes tumor growth and survival (5). The selective BRAFV600E/K inhibitors vemurafenib and dabrafenib lead to significant improvements in clinical response rate (RR), progression-free survival (PFS), and overall survival (OS), compared to chemotherapy in BRAFV600E/K-mutant patients (3, 6). However, clinical resistance develops in nearly all patients, and durable disease remissions are uncommon. Thus, elucidation of critical tumor survival mechanisms that can be coordinately targeted along with BRAF inhibition remains an important area of investigation, with goals of improving the initial antitumor activity of BRAF inhibitors to prevent the emergence of resistance, or to effectively treat disease that is progressing.
Autophagy is one such mechanism that may be abrogated for therapeutic gain in patients with melanoma. Autophagy captures and degrades intracellular proteins and organelles to control their quality and to recycle their components to sustain metabolism in starvation (7). Levels of autophagy are normally low but are profoundly induced by starvation and other stressors. Although autophagy may be important for tumorigenesis in some settings, the mechanism remains to be defined. Autophagy suppresses p53 activation and p53-mediated tumor suppression, but the mechanism is not yet known (8). Autophagy also promotes tumorigenesis in the absence of p53. Autophagy is upregulated in and required for the survival of tumor cells in hypoxic tumor regions (9) and autophagy defects cause tumor cells to be greatly sensitized to metabolic stress in vitro independent of p53 status (10, 11). This suggests that the accumulated defective mitochondria and the inability to generate metabolic substrates from recycling create a metabolic liability that compromises tumorigenesis. Neither the role of defective mitochondria nor the critical substrates provided by autophagy are known. Nonetheless, this suggests that autophagy inhibition may be a valuable strategy for cancer therapy. This may be particularly relevant in melanoma, where indications of high basal levels of autophagy in malignant tumors have been reported (12–14), and have been found to predict inferior response to treatment and shorter survival (13). Indeed, concomitant inhibition of autophagy and either MAPK or PI3K/AKT/mTOR signaling has led to promising preclinical results, with increased cell death in both BRAFV600E-driven and BRAF wild type melanoma tumors (14–16). However, it is important to examine the loss of functional autophagy in spontaneously arising tumors in genetically engineered mouse models (GEMMs) that most closely resemble human disease. This is especially important given that melanomagenesis in these GEMMs occurs in the presence of an intact immune system and therapeutic immune checkpoint activation produces dramatic responses in a significant number of patients with recurrent melanoma (4).
Some cancers such as those driven by oncogenic KRAS and BRAF upregulate and require autophagy for their growth and survival. It is possible that these tumors, including melanoma, are more aggressive in nature and more dependent on alleviation of metabolic stress for their growth, and as such, may be most sensitive to disruption of autophagy. Deletion of the essential autophagy genes Atg7 or Atg5 in GEMMs for KRASG12D- and BRAFV600E-driven lung and pancreatic cancers causes accumulation of defective mitochondria and protein aggregates, tumor cell growth arrest and cell death, and progression to more benign disease (10, 11, 17–19). The anti-tumor activity of Atg7-deficiency is particularly profound in BRAFV600E-driven lung tumors where it produces a dramatic lifespan extension accompanying the reduction in tumor burden (11).
Whether autophagy is ubiquitously required for other BRAFV600E-driven cancers is not yet clear, although evidence suggests it is important to melanoma tumorigenesis. Deprivation of the amino acid leucine and blockade of adaptive autophagy activation in melanoma xenografts with activated RAS-MEK signaling, increases caspase-dependent apoptotic cell death and reduces tumor growth (20). Autophagy is also an important mechanism of the development of resistance to treatment in melanomas that harbor BRAFV600E mutations. Patients whose melanoma tumors showed higher levels of autophagosomes after BRAF or BRAF/MEK inhibition experienced lower response rates and shorter progression free survival, and this autophagy activation in preclinical models was dependent on MAPK-driven expansion of the ER stress response that can be abrogated with combined BRAF and autophagy inhibition (15).
We report here using GEMMs that deletion of Atg7 suppresses the growth and extends survival of mice bearing BRAFV600E and Pten null melanomas. This suggests that the anti-tumor activity of Atg7 deficiency extends to settings of Pten deficiency, a common scenario in human melanoma (21). The mechanism of tumor promotion by autophagy is suppression of senescence that correlated with reduction of oxidative stress, a known senescence trigger, which was also enhanced in the setting of targeted BRAF inhibition. This suggests that autophagy overcomes the senescence barrier to promote growth of melanomas and that inhibiting autophagy will have therapeutic value by promoting senescence in these tumors.
Results
Tumor-specific Atg7 deficiency prevents development of BRAFV600E-driven melanomas in the context of Pten heterozygosity
To test the hypothesis that autophagy is required for the growth of spontaneous, BRAFV600E-driven melanomas, we employed a GEMM where oncogenic BRAF is activated by the removal of Lox-P sites with Cre recombinase (22). Expression of the Cre transgene (Tyr-Cre/ERT2) is restricted to melanocytes by both the melanocyte-specific tyrosinase promoter and by the fusion of Cre to the tamoxifen-responsive element that is activated upon topical administration of 4-hydroxytamoxifen (4-HT). Since BRAFV600E activation alone is insufficient to cause melanoma in this model, a single floxed allele of the Pten tumor suppressor gene was introduced that is coordinately deleted in melanocytes with activation of BRAFV600E. Upon topical administration of 4-HT, Cre activates the BRAFV600E and deletes the Pten alleles specifically in melanocytes, initiating melanomagenesis that gradually progresses over 10 months to melanoma, presumably with spontaneous loss of heterozygosity of the remaining Pten allele (23) (Fig. 1). To address the role of autophagy in this setting, floxed alleles of the essential autophagy gene Atg7 (24) were introduced that can be coordinately deleted with BRAFV600E activation and heterozygous Pten deletion specifically in melanocytes. Adult mice with the genotypes of TgTyr-cre/ERT2/+; LSL-BRAFV600E/+; PtenFLOX/+; Atg7+/+ or Atg7FLOX/FLOX were administered 4-HT to the skin on the lower back to initiate melanomagenesis in the presence and absence of Atg7, and the mice were monitored for 40 weeks.
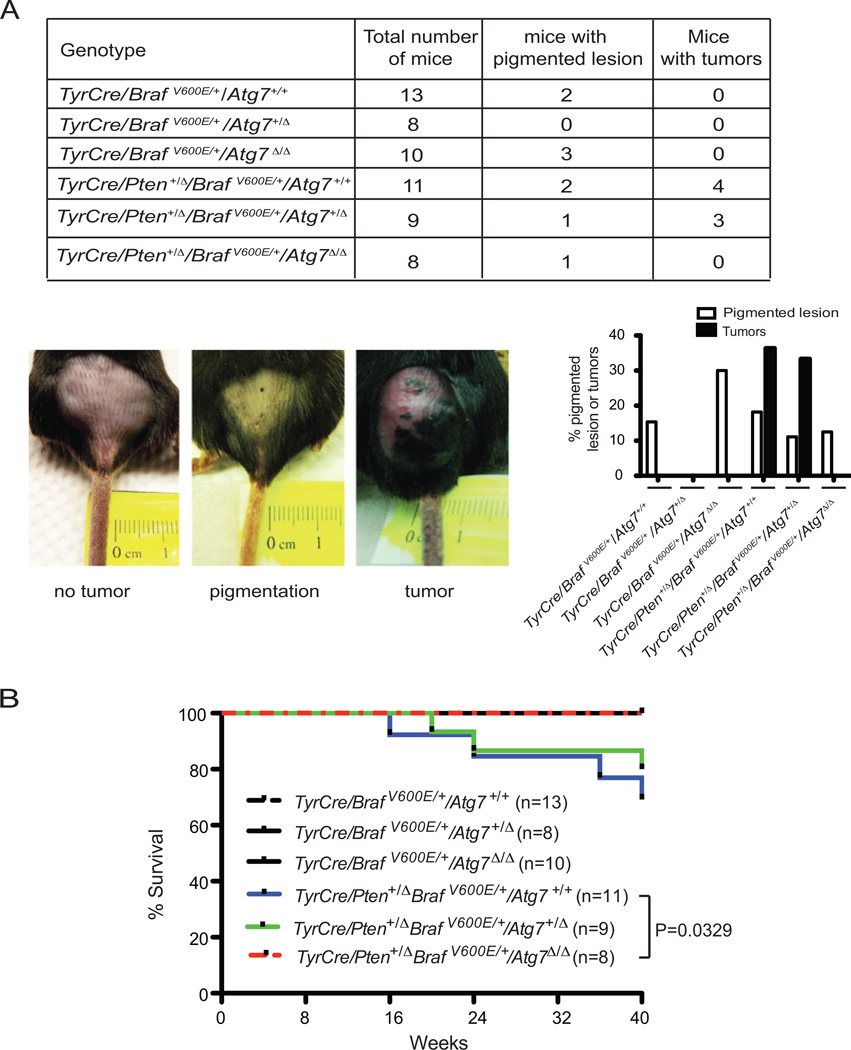
Figure 1. Tumor-specific Atg7 deletion in a BRAFV600E –driven and Pten heterozygous melanoma mouse model slows melanoma development.
A. The table shows numbers of 4-HT-induced mice and the breakdown of lesion type by genotype. The bar graph shows the percentage of mice with development of either pigmented lesions or melanoma by genotype during the 10-month post-induction period. Representative pictures of induced pigmented lesions and melanoma tumors on the lower back skin of the mice are shown. B. Kaplan-Meier survival curve of mice with the indicated tumor genotypes. Number of mice per group and P values (Log-rank Mantel-Cox test) are indicated.
As expected in the setting of functional autophagy (Atg7+/+), a subset of the mice gradually developed pigmented lesions, some of which progressed to melanoma tumors resulting in lethality in 4/11 mice (Fig. 1A and B). Heterozygosity for Atg7 (Atg7+/Δ) had no significant effect on tumorigenesis, consistent with previous results that retention of a single allele of Atg7 is sufficient for functional autophagy (11, 24). In contrast, there were fewer pigmented lesions with no tumors developing in the setting of Atg7 deficiency (Atg7Δ/Δ), and mouse survival was identical to wild type mice (Fig. 1A and B). Thus, Atg7 is required for development of melanomas driven by BRAFV600E and Pten loss-of-heterozygosity (Pten+/Δ). This is important as it suggests that autophagy inactivation blocks both initiation of, and gradual progression to, malignant melanoma in a physiological setting. The near complete absence of premalignant tissue with Atg7 deficiency made determining the mechanism of tumor suppression problematic. Moreover, the consequence of ATG7 ablation in the context of more aggressive disease was not known.
Atg7 facilitates growth and lethality of BRAFV600E-driven, Pten-deficient melanomas
To determine if Atg7 deficiency impedes aggressive melanoma growth, we tested the consequence of Atg7 ablation for the development and progression of BRAFV600E-driven, Pten-deficient melanomas. Adult mice with the genotypes of TgTyr-cre/ERT2/+; LSL-BRAFV600E/+; PtenFLOX/FLOX; Atg7+/+ or Atg7FLOX/FLOX then were administered 4-HT to the skin on the lower back to initiate melanomagenesis in the presence and absence of Atg7, and the mice were monitored for 6 months. As expected, melanomagenesis was greatly accelerated by complete deficiency in Pten (PtenΔ/Δ) in the context of activation of oncogenic BRAFV600E (Fig. 2A–D), which limited survival to less than 4 months (Fig. 2E) (23). The absence of a single allele of Atg7 caused a slight reduction in tumor volume (Fig. 2C), but had no significant effect on overall survival (Fig. 2E), consistent with partial deficiency in Atg7 retaining autophagy function. Loss of both Atg7 alleles, however, profoundly suppressed tumor growth (Fig. 2A–D). Indeed, Atg7 deficiency in melanomas extended mouse lifespan to a median survival of 14.5 weeks, compared with 10.5 weeks for mice bearing melanomas with Atg7 intact (Fig. 2E). Nonetheless, and in contrast to the setting above with tumors initiated with heterozygous deficiency in Pten (Fig. 1B), Atg7 deficiency did not completely suppress melanomagenesis and mice ultimately died from their tumors (Fig. 2E). In summary, these findings demonstrate for the first time that tumor-specific ablation of an essential autophagy gene suppresses growth of spontaneous melanomas driven by active Braf and Pten deficiency, significantly extending mouse survival.
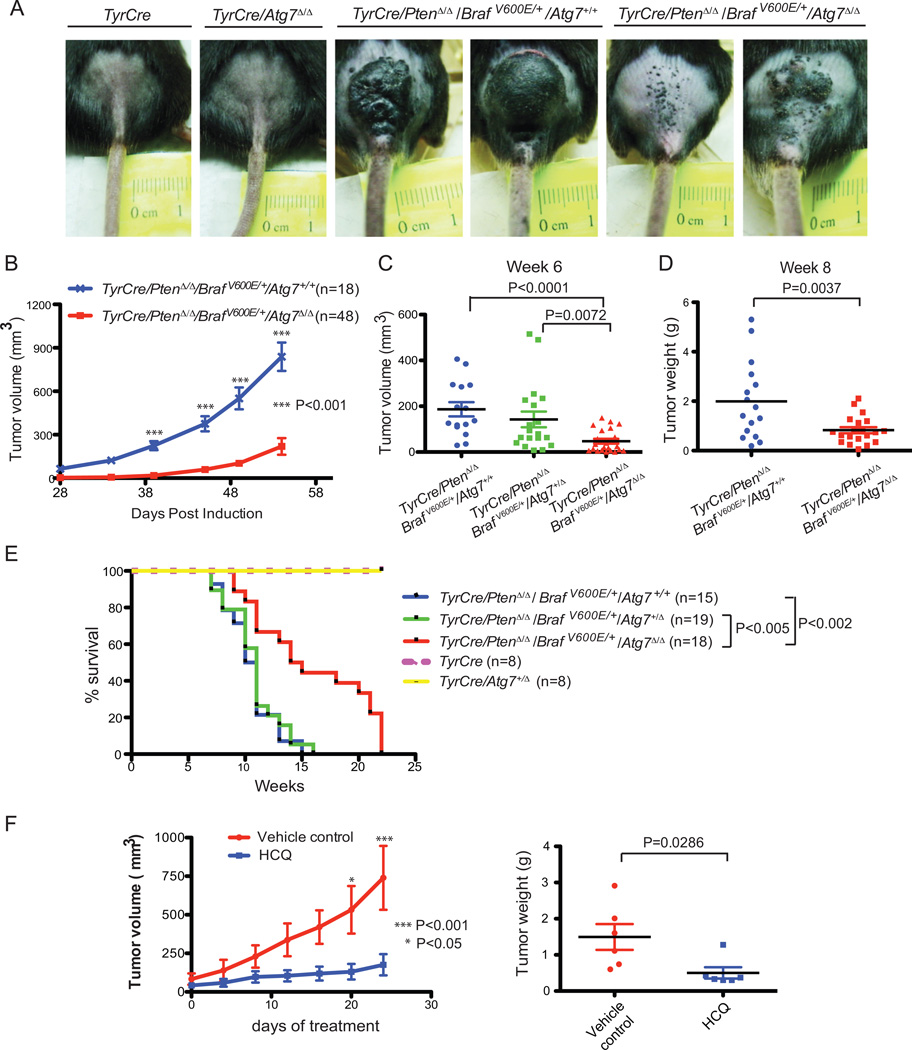
Figure 2. Tumor-specific Atg7 ablation in BRAFV600E–driven and Pten deficient melanomas impedes tumor growth and extends survival.
A. Representative pictures of the lower back skin of the mice 6 weeks post 4-HT induction. Mice with TyrCre/PtenΔ/Δ/BRAF/V600E/Atg7+/+ melanomas have greater tumor burden compared with those with Atg7 deletion. The control mice with the genotypes of TyrCre and TyrCre/Atg7Δ/Δ show no tumor burden. B. Tumor growth curves of mice with TyrCre/PtenΔ/Δ/BRAF/V600E/Atg7+/+ and TyrCre/PtenΔ/Δ/BRAFV600E/Atg7Δ/Δ melanomas. Tumors with Atg7 deletion demonstrate slower growth. The number of mice per group and P value (two way ANOVA, Bonferroni posttests) are indicated. C. Tumor size comparison. Tumor volumes were measured at week 6. D. Tumor weight comparison. In an independent experiment, 8 weeks after 4-HT induction, mice were euthanized and tumor samples were collected for weight measurement. E. Kaplan-Meier survival curve of mice with the indicated genotypes. The number of mice per group and P values (Log-rank Mantel-Cox test) are indicated. (F). Melanomas were induced with 4-HT to generate TyrCre/PtenΔ/Δ/BRAFV600E/Atg7+/+ tumors. When tumor volumes reached the size of approximately 100 mm3, HCQ (130mg/kg daily) or vehicle control were administered intraperitoneally. Tumor sizes were measured every 4 days. The tumor growth curve is indicated in the left panel with p values (two way ANOVA, Bonferroni posttests) and the weights of tumor samples collected the day after the last HCQ injection are indicated in the right panel with p value (t-test, unpaired two-tailed).
Recent evidence suggests that systemic rather than tumor-specific ablation Atg7 ablation in mice has more potent anti-tumor activity in Kras-driven lung cancers (17). To explore potential anti-melanoma activity of systemic autophagy deficiency, pharmacologic inhibition (as opposed to genetic ablation), of autophagy was tested by administration of the lysosomal autophagy inhibitor hydroxychloroquine (HCQ). Administration of HCQ (130mg/kg daily for 24 days) to the GEMM model with established melanomas driven by active Braf and Pten deficiency with Atg7 intact profoundly suppressed tumor growth (Fig. 2F). These data indicate that autophagy promotes the growth of aggressive melanomas by a tumor cell-autonomous mechanism and that systemic pharmacological autophagy inhibition with HCQ has anti-melanoma activity without apparent toxicity.
Atg7-deficient melanomas manifest autophagy defects
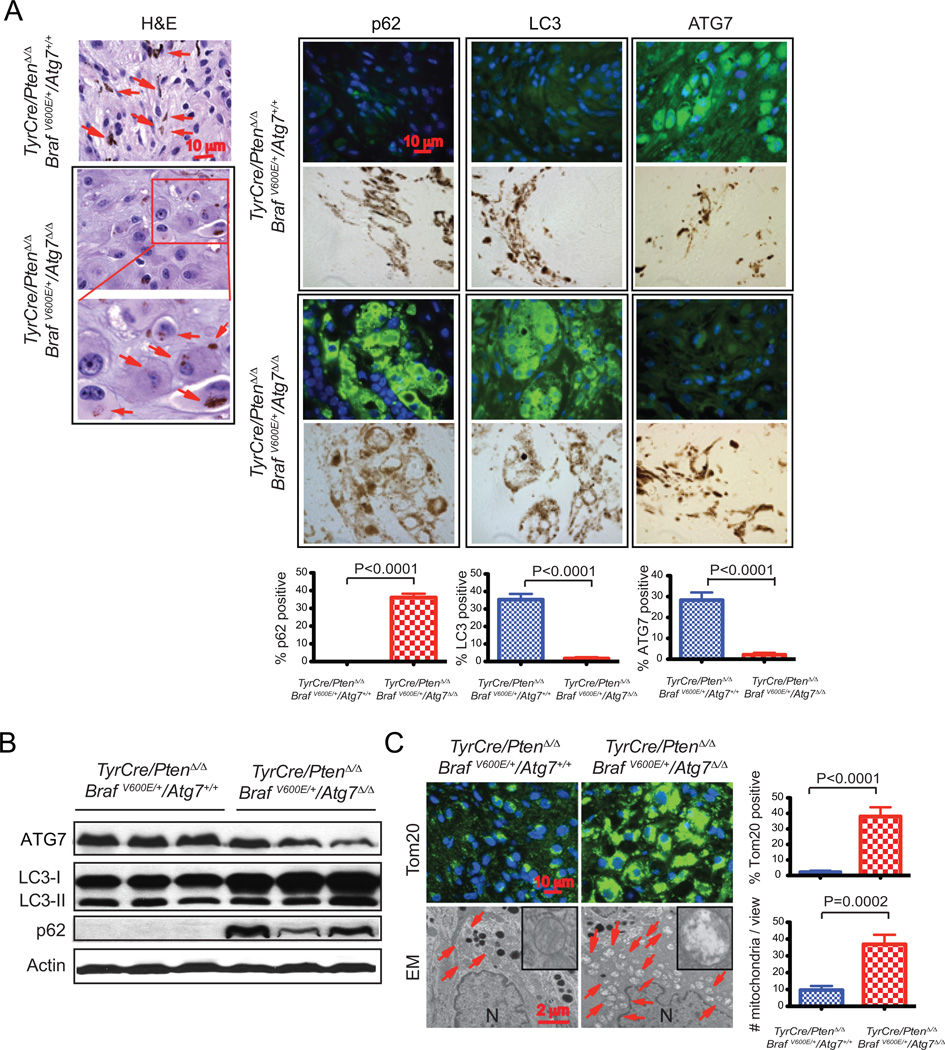
To address the functional consequences of Atg7 deficiency and loss of protein and organelle quality control to melanomas, tumors were examined histologically at 6, 9, and 12 weeks post melanoma induction. As the findings were similar throughout tumorigenesis, tumors 9 weeks post induction that are representative of all tumors examined are shown in Figure 3. In comparison to Atg7 wild type tumors, Atg7 null tumors contained melanocytes with a greatly expanded cytoplasm and the striking accumulation of the autophagy substrates p62 and LC3 (Fig. 3A). Cells with accumulated p62 and LC3 contained melanin in melanosomes indicating that they were tumor cells and not stromal in origin (Fig. 3A). Note that the accumulated LC3 in the Atg7-deficient tumors was diffusely localized consistent with a block of autophagy and not found in discrete puncta associated with LC3-I to LC3-II conversion and initiation of autophagosome formation (Fig. 3A). As expected, ATG7 protein expression was lost in null tumor cells (Fig 3A). Consistent with these findings, Western blot analysis of tumor tissue demonstrated reduction in ATG7, and accumulation of LC3-I and p62 in Atg7-deleted tumors (Fig. 3B). Note that tumors ablated for Atg7 contain a considerable stromal component that is not deleted for Atg7, explaining the presence of some ATG7 and LC3-II (Fig. 3B). In addition to accumulating autophagy protein substrates, electron microscopy revealed the dramatic accumulation of swollen, abnormal mitochondria that were also apparent in tumor sections stained for the mitochondrial outer membrane marker protein Tom20 (Fig. 3C). Thus, melanomas with Atg7 deletion display failure of both protein and organelle quality control similar to other GEMMs for cancer, where accumulation of defective mitochondria and reduced tumor burden have been observed in the setting of Atg7 deficiency (10, 11, 17).
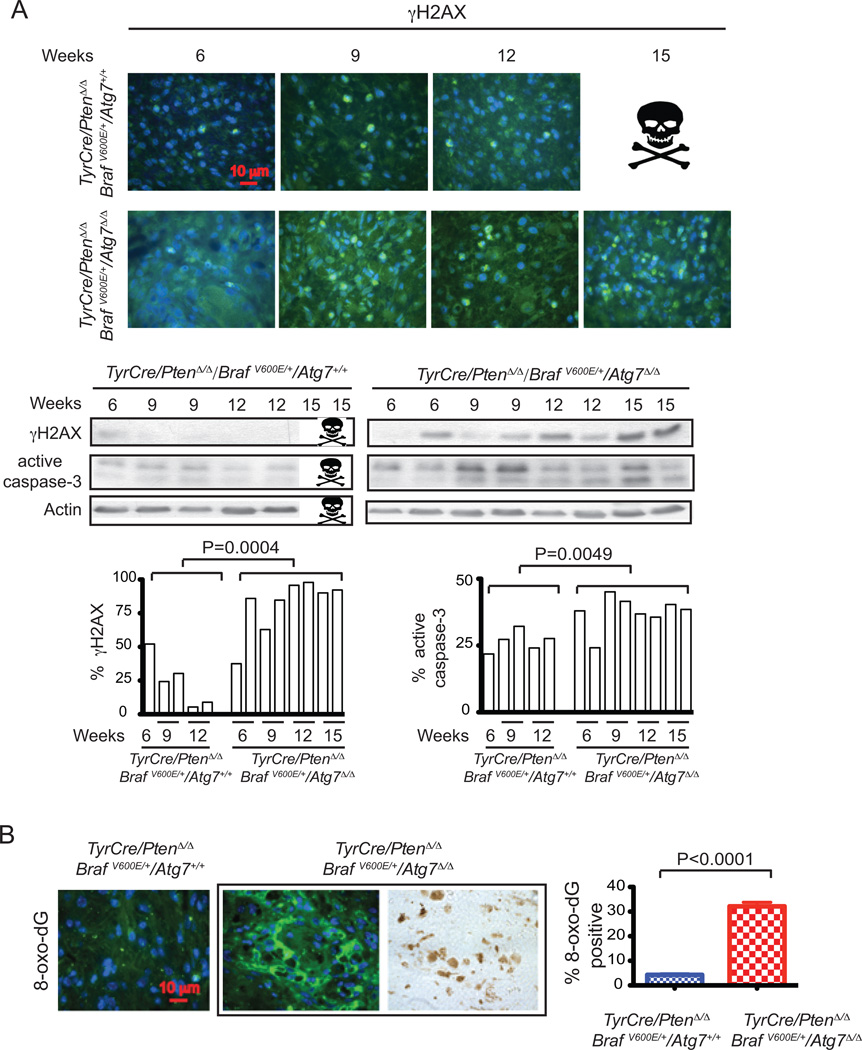
Figure 3. Atg7 deficiency is associated with accumulation of defective mitochondria in BRAFV600E–driven, Pten-deficient melanoma.
A. Representative images of TyrCre/PtenΔ/Δ/BRAFV600E/Atg7+/+ and TyrCre/PtenΔ/Δ/BRAFV600E/Atg7Δ/Δ tumor histology (hematoxylin and eosin [H&E] staining), and p62, LC3 and ATG7 immunofluorescence staining 9 weeks post tumor induction.. Arrows point to melanin. The positive stains for p62, LC3 and ATG7 co-localize with melanin (brown color), indicating these cells are melanoma cells. B. Western-blot for ATG7, LC3 and p62 of three representative samples. Actin serves as a protein loading control. C. Representative immunofluorescence staining of Tom20 and representative electron microscope images show abnormal mitochondria accumulation in BRAFV600E–driven, Pten-deficient melanoma, with quantification. (N) Nuclei. Arrows point to mitochondria.
Atg7 promotes melanoma proliferation and suppresses senescence and fibrosis
To address the mechanism by which ATG7 promoted melanoma growth, levels of proliferation and senescence were examined in matched wild type and deficient tumors. There were less Ki67 positive melanoma cells in the Atg7-deficient tumors compared to the wild type throughout the time course with the representative 9 weeks post melanoma induction data shown (Fig. 4A). None of the autophagy-deficient tumor cells were Ki67 positive as revealed by co-immunofluorescence staining with p62 (Fig. 4B). Thus loss of autophagy suppresses proliferation of melanoma cells that likely contributes to a reduction in tumor burden and increased mouse survival.
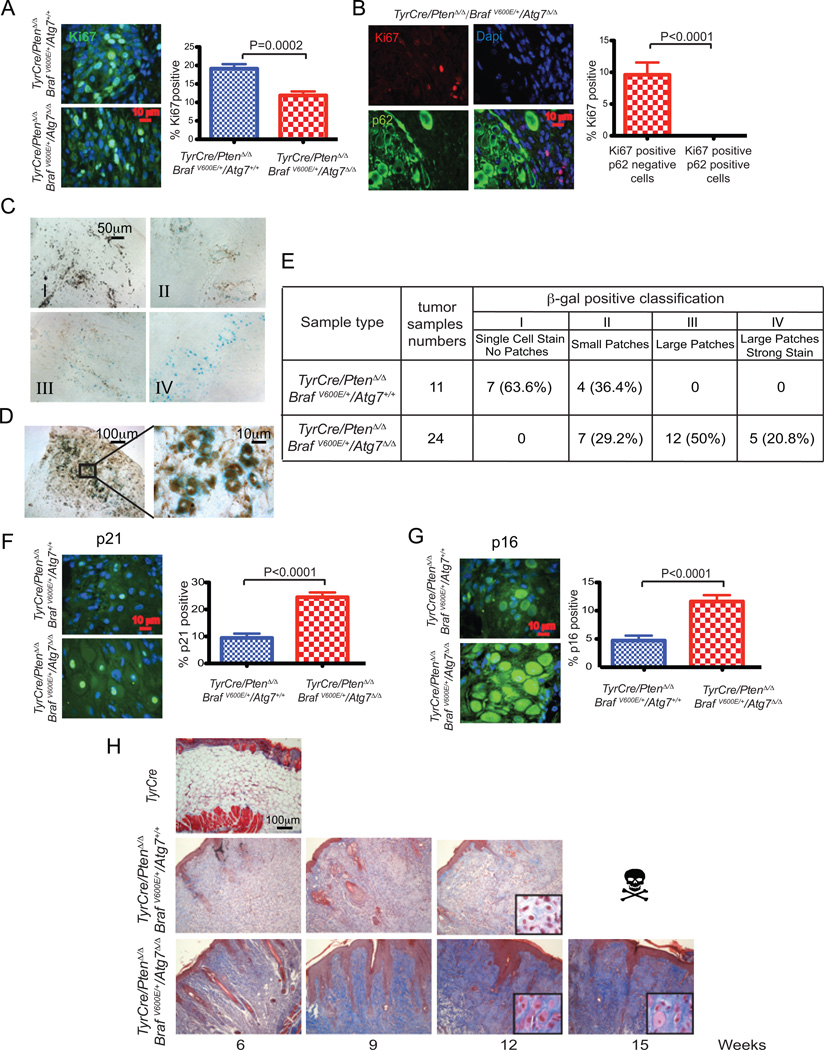
Figure 4. Atg7 promotes melanoma proliferation, and suppresses senescence and fibrosis.
All histology is from 9 weeks post melanoma induction. A. Representative images of Ki67 immunofluorescence staining and quantification. B. Representative images of co-immunofluorescence staining of Ki67 and p62 with quantification. p62 positive cells show no staining of Ki67. C. Representative images of different types of positive SA-β-gal staining. Type I: single cell positive stain. Type II: small groups (small patches) of cells show positive stain. Type III: large groups (large patches) of cells show positive stain. Type IV: large groups of cells show strong positive stain (large patches, strong stain). D. Image at high magnification shows positive SA-β-gal staining overlapping with melanin, indicating the cells are melanoma cells. E. Table shows the percentage of different types of positive SA-β-gal staining in all TyrCre/PtenΔ/Δ/BRAFV600E/Atg7+/+ and TyrCre/PtenΔ/Δ/BRAFV600E/Atg7Δ/Δ tumor tissue samples at 6, 9, and 12 weeks post melanoma induction. F. Representative images of p21 immunofluorescence staining and quantification shows augmented positive staining with Atg7-deficient melanoma samples. G. Representative images of p16 immunofluorescence staining and quantification show increased positive staining with Atg7-deficient melanoma samples. H. Representative images of Masson’s trichrome stain show more blue connective tissue and tumor cells with larger cytoplasm in Atg7-deficient melanoma samples than those that are wild type. The induced skin sample obtained from TyrCre mouse shows no tumor and serves as a negative control.
Reduced proliferation in melanoma is often associated with activation of senescence; therefore, the matched Atg7 wild type and deficient tumors were assessed for the activity of senescence-associated β-galactosidase (SA-β–gal) (25). Atg7 deficiency increased the level of SA-β–gal activity in tumor tissue resulting in more SA-β–gal-positive cells in larger patches with a greater level of activity compared to matched wild type tumors (Fig. 4C–E). Importantly, the pigmented melanosome-containing tumor cells were commonly positive for SA-β–gal in the tumors (Fig. 4D). Increased induction of the senescence-responsive CDK inhibitors p21 and p16 was also apparent in the Atg7-deficient compared to the wild type tumors (Fig. 4F,G), consistent with inhibition of proliferation and induction of senescence. Thus ATG7 expression is important for suppressing senescence in spontaneous BRAFV600E-driven and Pten-deficient melanomas, explaining why ATG7 deficiency compromises tumorigenesis.
An important consequence of increased senescence can be increased fibrosis (25). We noticed that Atg7-deficient tumors appeared to contain not only greatly enlarged tumor cells but also more extracellular matrix material and possibly non-tumor cells indicative of fibrosis. To test if part of the altered histology of Atg7-deficient tumors was due to increased fibrosis, Atg7-deficient and wild type tumors were stained with Masson’s trichrome stain that visualizes excessive collagen fibers or fibrin-like deposition associated with fibrosis (25). This revealed a dramatic increase in collagen fibers or fibrin like connective tissue (blue indicator) in the Atg7-deficent tumors compared to the wild type (Fig. 4H). Thus BRAFV600E-driven and Pten-deficient melanomas require ATG7 to suppress senescence that would otherwise limit tumor growth.
Atg7 suppresses oxidative stress
Senescence is triggered by many factors, and given that autophagy deficiency produces elevated oxidative stress and DNA damage in other settings (26, 27), which are known triggers of senescence, we assessed whether this was occurring at an increased level in Atg7-deficient compared to wild type melanomas. We first examined levels of the marker for DNA double strand breaks, γ-H2AX, in tumor tissue. Atg7-deficient melanomas have greatly increased numbers of γ-H2AX-positive tumor cells in comparison to wild type melanomas (Fig. 5A). Elevated γ-H2AX levels in Atg7-deficient tumors were also apparent when wild type and deficient tumors were examined by western blotting (Fig. 5A). Atg7-deficient tumors also had higher levels of active caspase-3 indicative of elevated apoptosis (Fig. 5A). Accumulation of damaged mitochondria in autophagy-deficient cells can result in elevated production of reactive oxygen species (ROS) that we hypothesized was the underlying basis for the increased senescence. While direct measurement of ROS in tumor tissue is problematic, the free radical-induced oxidative lesion marker, 8-Oxo-2'-deoxyguanosine (8-oxo-dG), can be readily measured. There was a striking increase in 8-oxo-dG in Atg7-deficient in comparison to wild type melanomas, indicative of elevated oxidative damage and consistent with elevated ROS production (Fig. 5B).
Figure 5. Functional autophagy suppresses oxidative stress and DNA damage.
A. Representative images of γ-H2AX immunofluorescence staining (upper panel), western blot of γ-H2AX and active caspase-3 of Atg7 wild type and deficient mouse melanoma samples 6, 9, 12, 15 weeks post induction Actin serves as a protein loading control (middle panel). The densitometric ratios from the western blots of γ-H2AX and active caspase-3 versus actin with the p value (t-test, unpaired two-tailed) at the indicated times are shown (lower panel). The images show more DNA damage response activation and apoptosis induction when Atg7 is deleted. B. Representative images of immunofluorescence staining for 8-oxo-dG (left panel) with quantification (right panel) and positivity for 8-oxo-dG co-localizes with melanin (brown color) indicating that these cells are melanoma cells.
Atg7 suppresses BRAF-inhibitor-induced senescence
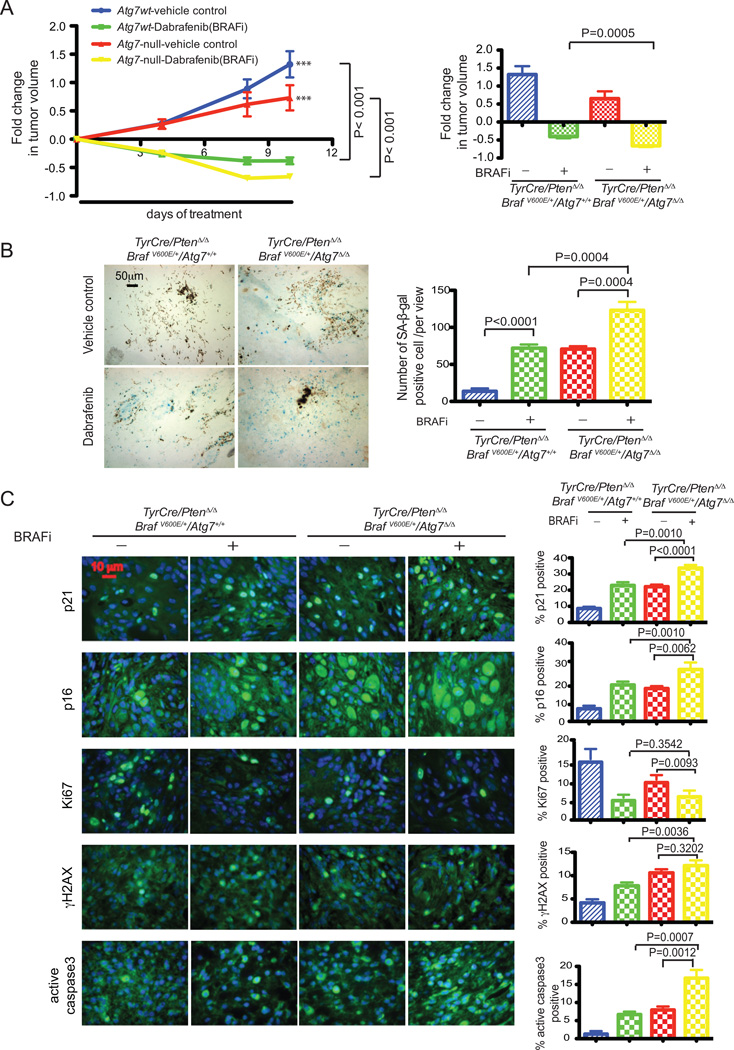
With the knowledge that ATG7 combats senescence and promotes growth of BRAFV600E-driven melanoma, we hypothesized that ATG7 also opposes senescence induced by pharmacologic BRAFV600E inhibition. To test this we examined matched wild type and Atg7 deficient tumors growth treated with the BRAFV600E/K inhibitor dabrafenib and its vehicle control. Melanoma tumors were induced with 4-HT. When tumor volumes reached 300–750 mm3, dabrafenib (20mg/kg daily) and its vehicle control were administered by oral gavage. As expected, BRAFV600E inhibition resulted in dramatic tumor volume reduction in both Atg7 null and wild type tumors. However, tumor reduction was more profound in Atg7 null, compared with wild type tumors (Fig. 6A). This more pronounced reduction in tumor growth was associated with a significantly greater induction of senescence as measured by SA-β-gal in Atg7 null tumors (Fig. 6B). Indeed, BRAF inhibitor–treated mice with Atg7 null tumors additionally demonstrated further evidence of increased senescence through induction of p21 and p16 (Fig. 6C). In addition, there was a significant increase in apoptosis as measured by active caspase-3. Although suppression of proliferation was similar to the wild type as measured by Ki67, and DNA damage response activation as measured by γ-H2AX was not significantly increased (Fig. 6C), this is possibly due to the timing of the analysis. Taken together, these findings demonstrate that senescence induced by treatment with BRAF inhibition in spontaneous melanomas is further increased by Atg7 deficiency, supporting the hypothesis that Atg7 overcomes senescence. It also must be considered that elimination of senescent cells by the immune system and promotion of apoptosis may also play a role in ATG7 null tumor shrinkage.
Figure 6. BRAF inhibition and autophagy deletion cooperate to impede melanoma tumor growth.
A. Melanomas were induced with 4-HT to generate TyrCre/PtenΔ/Δ/BRAFV600E/Atg7+/+ and TyrCre/PtenΔ/Δ/BRAFV600E/Atg7Δ/Δ tumors. When tumors reached the size of 300–750 mm3, dabrafenib (20mg/kg daily) and vehicle control were administered via oral gavage. Tumor sizes were measured and fold changes in tumor volumes were calculated. The tumor growth curve is indicated in the left panel with p values (two way ANOVA, Bonferroni posttests) and the fold change at last time point is indicated in the right panel with p value (t-test, unpaired two-tailed). Each data point represents the mean tumor volume changes from 10–13 mice. B. Representative images of SA-β-gal staining with tumor samples from mice treated with vehicle control or dabrafenib respectively for 4 days with samples collected at day 6 (left panel). Quantification of the images (right panel) revealed significantly more SA-β-gal staining in Atg7-deleted tumor samples treated with dabrafenib compared to vehicle control treated samples, or to the Atg7 wild type samples with dabrafenib or vehicle control treatment. P values were calculated by unpaired two-tailed t-test. Quantification was based on 5 samples of each group and at least of 3 views of each sample. C. Representative images of immunofluorescence staining for p21, p16, Ki67, γ-H2AX, active caspase-3 with quantification and their p values as indicated (right panel).
Discussion
It is known that autophagy plays a complex role in cancer and can both suppress and promote tumorigenesis (28). In general, it is thought that autophagy is used by cancers to promote tumor survival, with several lines of evidence supporting the role of deregulated autophagy in melanoma (29). Importantly, adult mice with systemic ablation of the Atg7 gene showed destruction of tumor tissues prior to normal tissues, underscoring the potential therapeutic utility of autophagy inhibition (17). We sought to determine the effects of the genetic ablation of Atg7 in a BRAFV600E-driven genetically engineered melanoma model that shares salient features of human disease to determine the role of ATG7 in promoting melanoma survival (23).
Our results demonstrate that Atg7 deficiency impeded the development of BRAFV600E-driven and Pten heterozygous melanomas, which extended mouse survival. We also found that Atg7-deficiency in more aggressive BRAFV600E-driven and Pten deficient melanomas produced slower growing and more senescent melanomas in comparison to those that were wild type for Atg7. Importantly, melanoma-specific deficiency in Atg7 dramatically increased the survival of these mice. These findings provide the proof of concept that ATG7 provides a melanoma-promoting function in the context of a spontaneously arising tumor with its normal microenvironment and in the presence of an intact immune system. It is also significant that this is the first GEMM where defects in autophagy in tumors with PTEN loss, as opposed to p53 loss, result in impaired tumor growth. This is an important observation due to the common frequency of PTEN loss and genetically normal Tp53 observed in human melanoma (21).
As in other GEMMs, evidence of dysfunctional mitochondria was observed in Atg7 deleted melanomas. Atg7 deficiency transformed the progression of BRAFV600E- and KRASG12D-driven lung tumors from adenomas to oncocytomas filled with defective mitochondria (10, 11), highlighting the important role of autophagy-dependent mitochondrial quality control in tumor progression. Further, p53 deletion in KRAS-driven lung tumors deficient for Atg7 reduced fatty acid oxidation and produced lipid accumulation, linking autophagy to lipid catabolism in cancer as well. While similar defects in mitochondrial quality control and elimination were observed here, Atg7-−deficient melanomas also displayed a unique phenotype of increased senescence and connective tissue deposition. This observed enhanced extracellular matrix formation and fibrosis in Atg7-deficient melanomas is a likely consequence of the role played by senescence in tissue remodeling (25), which was suppressed in melanoma tumors from animals with intact ATG7 function. As this occurred in an immunocompetent mouse model, it is possible that the fibrosis observed could be secondary to immune infiltration of the dying tumor. Despite the observed differences between these models, all Atg7-deficient tumors display accumulation of defective mitochondria and suppression of tumorigenesis.
Oncogene-induced senescence has been a well described mechanism of tumor suppression in many settings, and is thought to be caused by protein products of oncogenes resulting in activation of a tumor suppressor network (30). The senescence program is activated after an initial phase of cell proliferation and limits tumor growth. While others have found that Atg7 and Atg5 facilitate Ras-induced senescence in in vitro models (31) in melanoma, autophagy may be most critical for avoiding the senescence otherwise caused by BrafV600E activation and Pten deficiency, but also by BRAF inhibition and oncogene blockade (32). BRAFV600E-expressing melanocytes have been reported to display classic hallmarks of senescence (33), and abrogation of senescence through PTEN loss or PI3K pathway activation has been shown to contribute to melanomagenesis in BRAFV600E-driven melanoma models (34). Increased activation of this endogenous senescence program was noted in Atg7-deficient melanomas described here. This is consistent with the findings of others examining the deletion of Atg7 in melanocytes using the Cre-loxP system (35). Atg7 deletion suppressed lipidation of LC3 in melanocytes, which then entered into senescent premature growth arrest and accumulated ROS damage, ubiquitinated proteins and the polyubiquitin binding protein p62. The authors suggested that Atg7-dependent autophagy is dispensable for melanogenesis but necessary for achieving the full proliferative capacity of melanocytes, also consistent with our observation of delayed growth of Atg7-deficient melanomas.
Understanding the role of autophagy in the development of resistance to treatment, as opposed to simply in the process of tumorigenesis, is also critical in the development of novel therapeutics, which may target this process. Ma et al. reported that patients whose tumors developed resistance to BRAF inhibitors displayed higher levels of autophagosomes, and that treatment of multiple melanoma cell lines with BRAF inhibitors induced autophagy (15). Coordinate BRAF and autophagy inhibition with chloroquine-related compounds that interfere with lysosome function promotes tumor regression in BRAF-resistant xenografts, underscoring that autophagy is a likely mechanism of resistance to BRAF inhibition (15) Thus autophagy is important for melanoma survival that should be coordinately circumvented in targeted therapy to activated BRAF. The authors reported that this cytoprotective autophagy was activated by the binding of mutant BRAF to the ER stress gatekeeper GRP78, rapidly expanding the ER, followed by GRP78-PERK dissociation and a PERK-dependent ER stress response. These findings represent a different context compared with our examination of Atg7 in tumorigenesis, as they focus on a resistant model. However, these tumorigenic and resistance pathways are likely to be linked. Significant cross talk is known to exist between the ER response, generation of ROS and the unfolded protein response (UPR) that is critical for the pathogenesis of many diseases (36). Expansion of the ER stress response is a recognized consequence of autophagy inhibition (37); however activation of an uncontrolled ER stress response can also contribute to cell death (38) through increased generation of ROS. Our group and others have reported ROS-dependent cell death in the setting of autophagy inhibition in melanoma and kidney cancer models (14, 16, 39), and ROS is also a potent trigger of senescence. In both developing and resistant tumors, resulting activation of autophagy is potentially linked to a protein and organelle quality control response that is blunted when autophagy is inhibited or ablated, resulting in decreased tumor growth.
The clinical benefit of autophagy inhibition in human patients with cancer is an area of active investigation (40). Recent reports suggest that indeed, inhibition of autophagy combined with proteasome inhibitors or inhibitors of HDAC or mTOR produced clinical benefit in multiple tumor types (41–43). An intriguing case report documents an improved clinical outcome in a patient with a BRAFV600E mutant ganglioglioma (44). The patient initially responded to treatment with the BRAFV600 inhibitor vemurafenib but subsequently developed resistance; chemosensitivity was restored with the addition of the autophagy inhibitor HCQ. BRAF mutant CNS tumor cell lines were found to be autophagy dependent and coordinate BRAF and autophagy inhibition enhanced tumor cell death. These findings support our conclusions that autophagy, specifically ATG7, plays an important role in tumorigenesis and also those of others which suggest that autophagy inhibition may circumvent an important mechanism of resistance (15).
Whether upfront coordinate BRAF and autophagy inhibition could improve the initial activity of BRAFV600E inhibitors is an important clinical question. In this highly relevant melanoma model with an intact immune system, HCQ had potent anti-tumor activity as a single agent. This underscores that systemic autophagy inhibition with a currently available pharmacologic agent actively suppresses melanoma in the same setting where genetic ablation of autophagy compromises melanoma growth. It is noteworthy that this approach may have the highest clinical yield in patients whose tumors are most inherently reliant on autophagy (13, 15). This may be particularly applicable in patients whose tumors have PTEN aberrations, given the link between PTEN deficiency, resulting PI3K activation, and increased autophagy in human tumors (45, 46). Since PTEN and PI3K aberrations have been implicated in the early development of resistance to BRAF inhibitors (47–49), it is perhaps this population of patients that would best respond to concomitant BRAF and autophagy inhibition. This question will be explored in current clinical trials, and will be relevant to the design of future trials including newer, more potent autophagy inhibitors that have been reported (50).
In summary, Atg7 promotes the growth of BRAFV600E melanoma and inhibits activation of a senescence program, likely through the elimination of defective mitochondria and limiting oxidative stress. Tumors from GEMMs for melanoma with Atg7 deficiency demonstrated defects in autophagy and mitochondrial quality control, consistent with findings in other GEMMs for cancer, which indicate that this impaired mitochondrial function may be key to impairing tumor growth. Our observations provide genetic proof of concept evidence that autophagy is thus an important program in the survival and proliferation of BRAFV600E mutant, Pten-deficient melanoma. Abrogation of this pathway in autophagy-dependent tumors for therapeutic gain remains a promising clinical approach.
Materials and Methods
Mice
All animal care and experiments were carried out in compliance with Rutgers University Institutional Animal Care and Use Committee guidelines. Animals were obtained as follows: TyrCreER mice (The Jackson Laboratory, stock #: 013590), Atg7FLOX/FLOX mice (provided by Dr. M. Komatsu, Tokyo Metropolitan Institute of Medical Science), PtenFLOX/FLOX (The Jackson Laboratory, stock #: 004597) and BRAFca/ca (22). Mice were crossbred to generate the following mouse lines: (A). TyrCre/PtenFLOX/FLOX/BRAF+/+Atg7+/+. (B). PtenFLOX/FLOX/BRAFca/ca /Atg7+/+. (C). TyrCre/PtenFLOX/FLOX/BRAF+/+/Atg7FLOX/FLOX. (D). PtenFLOX/FLOX/BRAFca/ca / Atg7FLOX/FLOX. Crossbreeding (A) and (B) produces TyrCre/PtenFLOX/FLOX/BRAF ca/+Atg7+/+ mice, crossbreeding (A) and (D) produces TyrCre/PtenFLOX/FLOX/BRAFca/+AtgFLOX/+ mice, and crossbreeding (C) and (D) produces TyrCre/PtenFLOX/FLOX/BRAFca/+/Atg7FLOX/FLOX mice.
When mice reached 6–8 weeks of age, depilation was performed by shaving a small region of the back with an electric shaver to produce a 2 cm diameter region above the base of animal tails. A small, defined volume (3 µl) of 4-HT (10mM, 70% Z-isomer, Sigma) dissolved in ethanol (99%) was placed in the center spot of shaved skin for three consecutive days. Melanoma tumor volumes were measured and calculated with the following formula: volume=π/6 × L × W × H. For tumors that begin as multiple islands, individual volumes for those islands were added together for measurements prior to the eventual merging of these islands as a single tumor lesion.
Western Blotting
Collected tumor samples were ground in liquid nitrogen, lysed and probed with antibodies against ATG7 (Sigma-Aldrich; A2856), LC3 (Novus Biologicals; NB600-1384), p62, γ-H2AX (Cell Signaling Technology; 2577), active caspase-3 (Cell Signaling Technology; 9661) and β-actin (Sigma; A1978).
Immunofluorescence
Freshly collected tumor samples were fixed overnight in 10% formalin solution (Formalde-Fresh, Fisher Scientific) before transfer to 70% ethanol and sectioned. The tumor paraffin slides were deparaffinized, hydrated and incubated with primary antibody against LC3 (Nanotools, LC3-5F10), p62 (Enzo Life Sciences PW9860-0100), Atg7 (APG7, H-300, Santa Cruz Biotechnology, sc-33211), Tom20 (Santa Cruz Biotechnology; sc-11415), Ki67 (Abcam, ab-15580), p21 (BD Biosciences, 56431), p16 (Santa Cruz Biotechnology, M-156, sc-1207), γ-H2AX (Cell Signaling Technology; 2577), 8-oxo-dG (clone 2E2, Trevigen, 4354-mc-050). Quantification of the staining was calculated as the percentage of positive cells per representative field of view. Each data set was based on at least 5 samples of each group and at least of 5 representative field of views of each sample. P values are based on unpaired two-tailed t-test.
Electron microscopy
Small pieces of excised tumor tissue were immersed in fixation buffer (2.5% glutaraldehyde, 4% paraformaldehyde, 8 mM CaCl2, 0.1M cacodylate, pH 7.4) 24 hours at 4°C before handover for further processing, and analyzed by a JOEL 1200EX electron microscope.
Senescence-Associated β-Galactosidase Assessment
Tumor tissue samples were fixed with 10% formalin solution overnight before transfer to 15% sucrose for 4 hours and finally to 30% sucrose. 6-micron thick frozen tissue sections were prepared according to standard procedures. Senescence β-Galactosidase assay was performed following manufacturer's instructions (Senescence β-Galactosidase Staining Kit #9860).
Masson’s trichrome stain
Tumor paraffin slides were prepared as mentioned above and subjected to Masson’s trichrome stain according to manufacturer's instructions (Trichrome Stain Masson Kit, Sigma-Aldrich, HT15).
Drug treatment
Mice at the age of 6–8 weeks with genotypes of TyrCre/PtenFLOX/FLOX/BRAF ca/+/Atg7+/+ and TyrCre/PtenFLOX/FLOX/BRAFca/+/Atg7FLOX/FLOX were induced with 4-HT at the small region of the low back and randomized into four groups (two groups per genotype), and placed on respective treatment regimens. The dabrafenib (Chemie Tek) and its vehicle control were administered via oral gavage (20mg/kg daily). Tumor sizes were measured every three days. HCQ (Acros Organics) and its vehicle control were administered intraperitoneally (130mg/kg). Tumor sizes were measured every 4 days. The tumor volumes were calculated by the formula: volume=π/6 × L × W × H. Mice were weighed once per week.
Statement of Significance.
The essential autophagy gene Atg7 promotes development of BRAFV600E mutant, Pten-null melanomas by overcoming senescence, and deleting Atg7 facilitated senescence induction and anti-tumor activity of BRAF inhibition. This suggests that combinatorial BRAFV600E and autophagy inhibition may improve therapeutic outcomes in patients whose tumors have BRAFV600E/K mutations, an approach currently being explored in clinical trials.
Acknowledgements
The authors extend appreciation to The Rutgers Cancer Institute of New Jersey animal facility and Biospecimen Retrieval Service for assistance with sample slides preparation. We thank Dr. M. Komatsu for the Atg7FLOX/FLOX mice, Dr. M McMahon for the LSL-BRAFV600E mice, P. Estraghi for assistance with animal work, R. Patel for EM and Rutgers Cancer Institute of New Jersey Shared Resources and White laboratory members for helpful comments.
Grant support
This work was supported by the V Foundation for Cancer Research, NIH grants (R01 CA163591, R01 CA130893), AACR grant (59598), and the Val Skinner Foundation as well as a pilot grant from the Rutgers Cancer Institute of New Jersey (P30 CA072720).
Footnotes
Disclosure of potential conflict of interest: The authors have no potential conflicts of interest that pertain to this work.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ott PA, Hodi FS, Robert C. CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients. Clin Cancer Res. 2013;19:5300–5309. doi: 10.1158/1078-0432.CCR-13-0143. [DOI] [PubMed] [Google Scholar]
- 5.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 6.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 7.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155:1216–1219. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3:1272–1285. doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, Pawelek JM. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res. 2012;18:370–379. doi: 10.1158/1078-0432.CCR-11-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma XH, Piao S, Wang D, McAfee QW, Nathanson KL, Lum JJ, et al. Measurements of tumor cell autophagy predict invasiveness, resistance to chemotherapy, and survival in melanoma. Clin Cancer Res. 2011;17:3478–3489. doi: 10.1158/1078-0432.CCR-10-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie X, White EP, Mehnert JM. Coordinate autophagy and mTOR pathway inhibition enhances cell death in melanoma. PLoS One. 2013;8:e55096. doi: 10.1371/journal.pone.0055096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma XH, Piao SF, Dey S, McAfee Q, Karakousis G, Villanueva J, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest. 2014;124:1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rebecca VW, Massaro RR, Fedorenko IV, Sondak VK, Anderson AR, Kim E, et al. Inhibition of autophagy enhances the effects of the AKT inhibitor MK-2206 when combined with paclitaxel and carboplatin in BRAF wild-type melanoma. Pigment Cell Melanoma Res. 2014;27:465–478. doi: 10.1111/pcmr.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 19.Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, et al. A dual role for autophagy in a murine model of lung cancer. Nature communications. 2014;5:3056. doi: 10.1038/ncomms4056. [DOI] [PubMed] [Google Scholar]
- 20.Sheen JH, Zoncu R, Kim D, Sabatini DM. Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell. 2011;19:613–628. doi: 10.1016/j.ccr.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett DC. How to make a melanoma: what do we know of the primary clonal events? Pigment Cell Melanoma Res. 2008;21:27–38. doi: 10.1111/j.1755-148X.2007.00433.x. [DOI] [PubMed] [Google Scholar]
- 22.Dankort D, Filenova E, Collado M, Serrano M, Jones K, McMahon M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–384. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The Journal of cell biology. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munoz-Espin D, Serrano M. Cellular senescence: from physiology to pathology. Nat Rev Mol Cell Biol. 2014;15:482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- 26.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corazzari M, Fimia GM, Lovat P, Piacentini M. Why is autophagy important for melanoma? Molecular mechanisms and therapeutic implications. Semin Cancer Biol. 2013;23:337–343. doi: 10.1016/j.semcancer.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 30.Mooi WJ, Peeper DS. Oncogene-induced cell senescence--halting on the road to cancer. N Engl J Med. 2006;355:1037–1046. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- 31.Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haferkamp S, Borst A, Adam C, Becker TM, Motschenbacher S, Windhovel S, et al. Vemurafenib induces senescence features in melanoma cells. J Invest Dermatol. 2013;133:1601–1609. doi: 10.1038/jid.2013.6. [DOI] [PubMed] [Google Scholar]
- 33.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 34.Vredeveld LC, Possik PA, Smit MA, Meissl K, Michaloglou C, Horlings HM, et al. Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev. 2012;26:1055–1069. doi: 10.1101/gad.187252.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang CF, Gruber F, Ni C, Mildner M, Koenig U, Karner S, et al. Suppression of Autophagy Dysregulates the Antioxidant Response and Causes Premature Senescence of Melanocytes. J Invest Dermatol. 2014 doi: 10.1038/jid.2014.439. [DOI] [PubMed] [Google Scholar]
- 36.Zhang K. Integration of ER stress, oxidative stress and the inflammatory response in health and disease. Int J Clin Exp Med. 2010;3:33–40. [PMC free article] [PubMed] [Google Scholar]
- 37.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 39.Bray K, Mathew R, Lau A, Kamphorst JJ, Fan J, Chen J, et al. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PLoS One. 2012;7:e41831. doi: 10.1371/journal.pone.0041831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1391–1402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahalingam D, Mita M, Sarantopoulos J, Wood L, Amaravadi RK, Davis LE, et al. Combined autophagy and HDAC inhibition: a phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy. 2014;10:1403–1414. doi: 10.4161/auto.29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogl DT, Stadtmauer EA, Tan KS, Heitjan DF, Davis LE, Pontiggia L, et al. Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10:1380–1390. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levy JM, Thompson JC, Griesinger AM, Amani V, Donson AM, Birks DK, et al. Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov. 2014;4:773–780. doi: 10.1158/2159-8290.CD-14-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fan QW, Weiss WA. Autophagy and Akt promote survival in glioma. Autophagy. 2011;7:536–538. doi: 10.4161/auto.7.5.14779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Errafiy R, Aguado C, Ghislat G, Esteve JM, Gil A, Loutfi M, et al. PTEN increases autophagy and inhibits the ubiquitin-proteasome pathway in glioma cells independently of its lipid phosphatase activity. PLoS One. 2013;8:e83318. doi: 10.1371/journal.pone.0083318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Long GV, Fung C, Menzies AM, Pupo GM, Carlino MS, Hyman J, et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nature communications. 2014;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- 48.Lassen A, Atefi M, Robert L, Wong DJ, Cerniglia M, Comin-Anduix B, et al. Effects of AKT inhibitor therapy in response and resistance to BRAF inhibition in melanoma. Molecular cancer. 2014;13:83. doi: 10.1186/1476-4598-13-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deuker MM, Durban VM, Phillips WA, McMahon M. PI3'-Kinase Inhibition Forestalls the Onset of MEK1/2 Inhibitor Resistance in BRAF-Mutated Melanoma. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-14-0856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McAfee Q, Zhang Z, Samanta A, Levi SM, Ma XH, Piao S, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:8253–8258. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]