Abstract
Due to the active inhibition of the adipogenic programming, the default destiny of the developing lung mesenchyme is to acquire a myogenic phenotype. We have previously shown that perinatal nicotine exposure, by down-regulating PPARγ expression, accentuates this property, culminating in myogenic pulmonary phenotype, though the underlying mechanisms remained incompletely understood. We hypothesized that nicotine-induced PPARγ down-regulation is mediated by PPARγ promoter methylation, controlled by DNA methyltransferase 1 (DNMT1) and methyl CpG binding protein 2 (MeCP2), two known key regulators of DNA methylation. Using cultured alveolar interstitial fibroblasts and an in vivo perinatal nicotine exposure rat model, we found that PPARγ promoter methylation is strongly correlated with inhibition of PPARγ expression in the presence of nicotine. Methylation inhibitor 5-aza-2′-deoxycytidine restored the nicotine-induced down-regulation of PPARγ expression and the activation of its downstream myogenic marker fibronectin. With nicotine exposure, a specific region of PPARγ promoter was significantly enriched with antibodies against chromatin repressive markers H3K9me3 and H3K27me3, dose-dependently. Similar data were observed with antibodies against DNA methylation regulatory factors DNMT1 and MeCP2. The knock down of DNMT1 and MeCP2 abolished nicotine-mediated increases in DNMT1 and MeCP2 protein levels, and PPARγ promoter methylation, restoring nicotine-induced down regulation of PPARγ and upregulation of the myogenic protein, fibronectin. The nicotine-induced alterations in DNA methylation modulators DNMT1 and MeCP2, PPARγ promoter methylation, and its down-stream targets, were also validated in perinatally nicotine exposed rat lung tissue. These data provide novel mechanistic insights into nicotine-induced epigenetic silencing of PPARγ that could be exploited to design novel targeted molecular interventions against the smoke exposed lung injury in general and perinatal nicotine exposure induced lung damage in particular.
1. Background
During lung morphogenesis, under the paracrine endodermal influence, the mesenchymal default Wnt pathway is inhibited and the adipogenic pathway is up-regulated, resulting in the formation of lipid-laden alveolar interstitial adepithelial fibroblasts~lipofibroblasts (LIFs) [1-3]. Lipofibroblasts are vital for alveolar development, homeostasis, and injury repair [2] since they actively provide triglyceride substrate to alveolar epithelial type II (ATII) cells for surfactant synthesis [4], support ATII cell growth and differentiation [5] and act as an important defense against oxidant lung injury [6]. However, in the presence of altered mesenchymalepithelial signaling, e.g., following perinatal exposure to smoke/nicotine, pulmonary LIFs rapidly lose their lipogenic phenotype, transdifferentiating to a myogenic phenotype, i.e., myofibroblasts (MYFs) [7-9]. Transdifferentiated-LIFs (i.e., MYFs) are unable to maintain pulmonary epithelial cell growth and differentiation, resulting in failed alveolarization, seen in all chronic lung diseases, signifying the importance of LIFs in lung development and injury/repair [10, 11].
It is well-established that both pre- and postnatal exposure to maternal smoking results in detrimental long-term effects on lung growth and function [8, 12-15]. Although there are many agents in smoke that may be detrimental to the developing lung, there is ample evidence to support nicotine’s vital role in altering developing lung’s structure and function. Nicotine crosses the human placenta with minimal biotransformation [16]; it accumulates in fetal blood, maternal milk, amniotic fluid, and several fetal tissues, including the respiratory tract, and has been shown to have direct effects on pulmonary ATII cells and fibroblasts isolated from the developing lung [17-20]. Therefore, it is not surprising that perinatal nicotine exposure is an extensively-utilized model to study the effects of cigarette smoke on the developing lung. In fact, with the increasing use of electronic cigarettes, some of which contain nicotine in concentrations even higher than those found in traditional cigarettes [21][22], and with the nicotine patch during pregnancy as a common nicotine replacement therapy [23], interrogating the effects of prenatal nicotine exposure on the fetus is highly pertinent on its own. However, the mechanism (s) underlying nicotine’s effects on the developing fetus in general and the developing lung in particular remain incompletely understood.
Peroxisome proliferator-activated receptor γ (PPARγ), a ligand-dependent nuclear transcription factor, is implicated in a wide range of physiological processes [24] , such as metabolic homeostasis, adipogenesis, cellular and organ differentiation including the developing lung [25]. In particular, it is implicated in maintaining alveolar LIF differentiation. In in vitro and in vivo experimental models, nicotine down regulates PPARγ [7-9] . Since PPARγ is known to be epigenetically regulated [26-28], employing several in vitro and in vivo approaches, we tested the hypothesis that nicotine-mediated PPARγ down-regulation is epigenetically regulated. We demonstrate that nicotine alters PPARγ promoter methylation and the levels of its down stream myogenic targets, dose-dependently, and it recruits DNMT1 and MeCP2, two key epigenetic modulators, to form a repressive complex within a specific PPARγ promoter region. Furthermore, both specific and non-specific methylation blockers prevented the nicotine-mediated molecular alterations that characterize the alveolar interstitial fibroblast phenotype.
2. Methods
2.1 Perinatally nicotine exposed rat model and cultured primary fetal rat lung fibroblasts
Time-mated Sprague Dawley rat dams weighing 200–250 g received either placebo (diluent, n =6), or nicotine (1 mg/kg, subcutaneously; n =6) in 100-ml volumes daily from embryonic day 6 of gestation to postnatal day (PND) 21. After spontaneous delivery at term, the pups were allowed to breast feed ad libitum. Experimental animals were maintained on a 12-h light: 12-h dark cycle, pair-fed in accordance with the previous day’s food consumption by the nicotine-alone–treated group, and allowed free access to water. At PND 21, the pups were killed and lungs collected for further assays. All animal procedures were performed following National Institutes of Health guidelines, and were approved by the institutional Animal Care and Use Committees at both the LABioMed at Harbor-UCLA Medical Center.
2.2 Cell culture and experimental treatments
Embryonic human lung fibroblast cells WI-38 were grown in Dulbecco Minimum Essential Media with 10% fetal bovine serum; at 70% confluence, after overnight serum starvations, cells were cultured with or without nicotine (0, 10−9, or 10−6 M). For transfection, cells were grown in 6-well plates, and 24 h later, DNMT1 or MeCP2 siRNAs were transfected with oligofectamine (Life Technologies, Grand Island, NY). After 48 h, specific protein level was determined by Western blot analysis. For some experiments, 5-Aza-deoxycytidine (Aza-CdR) (Aza-CdR was dissolved in dimethyl sulfoxide (DMSO), Sigma, A2385, St. Louis, MO 63103) was added to the media at the indicated concentrations for 24 h. Rat e19 primary fetal rat lung fibroblasts were isolated and cultured as described previously [29].
2.3 Protein extraction and Western blotting
Cell and lung tissues were treated in RIPA buffer containing 1mM EDTA and PMSF (freshly added), supplemented with phophatase inhibitor (Sigma Aldrich, St. Louis, MO) and complete proteinase inhibitor cocktail (Roche Diagnostics., Indianapolis, IN). Fifty μg of total protein for each sample was subjected to 10% SDS gradient gels (Lonza, Rockland, ME). Western analysis was performed according to the previously described methods [29]. The primary antibodies, anti-PPARγ and DNMT1 (Santa Cruz Biotech, Dallas, Texas), MeCP2 (Millipore, Temecula, CA), as well as α-SMA and fibronectin (Santa Cruz Biotech, Dallas, Texas) were used diluted 1:500. The protein bands were quantified with Image J and normalized with internal control GAPDH.
2.4 Isolation of genomic DNA and bisulfite modification
Using gDNA Mini Kit (Boland Scientific, Paramount, CA), genomic DNA was isolated from cultured fibroblasts and lung tissue following manufacturer’s instructions. 1.0 μg of DNA was modified (converted C to T) using the EZ DNA methylation-Gold Kit (ZYMO Research, Orange, CA). Converted genomic-DNA was eluted in 20 μl of the elution buffer provided in the kit.
2.5 Methylation-specific PCR (MSP)
Methylation-specific PCR was used to detect methylation status of CpG islands in PPARγ gene promoter [30, 31]. 2.5 μl of the eluate of the bisulfite-modified DNA was used as template to amplify the specified fragment. PCR reactions were performed in 25-μl final volume with 2 units of Platinum Taq DNA polymerase (Invitrogen). The conditions included 30-40 cycles at 95°C for 30 sec, at 52°C for 30 sec, and at 72°C for 30 sec. For human M5 region (CpG-1), methylated PPARγ promoter (me) was amplified using the primer pair 5′ -TATTTTTGTTGAGGAGGAGGTTTC- 3′/5′ - GACTAAAAATCCTAACTACGCGCT - 3′; unmethylated PPARγ promoter (un) was amplified using the primer pair 5′-TATTTTTGTTGAGGAGGAGGTTTT- 3′/5′-TACAACTAAAAATCCTAACTACACACT - 3′. For human M6 region (CpG-2), methylated PPARγ promoter was amplified using the primer pair 5′- GAGTTTTATATTTCGGTTTTTTTAGATC- 3′/5′-AACTACCTAATATCGTTTACTCCTCG- 3′; unmethylated PPARγ promoter (un) was amplified using the primer pair 5′- GGAGTTTTATATTTTGGTTTTTTTAGATT - 3′/5′- AACTACCTAATATCATTTACTCCTCACC - 3′. For rat, methylated PPARγ promoter Island-3 was amplified using the primer pair 5′- GGTGATCGTTTAAGTAATTTGGTTC- 3′/5′-CCAATTAACCCTACCCTATACTCG- 3′; unmethylated PPARγ promoter Island-3 (un) was amplified using the primer pair 5′- GATTGTTTAAGTAATTTGGTTTGG - 3′/5′- CCAATTAACCCTACCCTATACTCAC - 3′. All MSP products were analyzed by electrophoresis on 2% agarose gels, quantified by Quantity One, and then plotted accordingly.
2.6 Chromatin Immunoprecipitation Assay
ChIP assay for DNMT1 and MeCP2 was carried out using 20 μg of cross-linked chromatin, prepared as described previously [32]. ChIP grade antibodies against tri-methylated H3K9me3 and H3k27me3, and H3k9Ac were bought from Millipore, Temecula, CA and 1.0 μg of antibody was used in each ChIP reaction with 50-100 μg of native chromatin, respectively. For human fibroblast cell line WI-38, the following primers covering the M5 (CpG-1) region were used for qChIP-PCR analysis: 5′ –AGTATGCTCCAGAAATAAGACTGG- 3′/5′ – GACAACATGGGTGGGGACGGAG - 3′. For rat in vivo nicotine exposure studies, island-3 region was covered: 5′ –CTCGTGAGGGGCACGGCCCGGC- 3′/5′ – TACCCGGTGGCTACTGGCGAGCC - 3′. Immunoglobin G (IgG) served as negative control. Average values of the eluates were normalized to average values of inputs and the ratio was plotted for analysis. Sonicated input DNA and IgG (or H2O) lanes served as positive and negative controls, respectively.
2.7 Immunofluorescence staining (single and double-staining)
Immunofluorescence staining on lung tissue sections was performed as previously described [33] and quantified using ImageJ software (National Institutes of Health, Bethesda, MD). Briefly, 5% of normal goat serum (NGS) in 1 x PBS was used as a blocking solution for 1 h at room temperature. The following primary antibodies were dissolved in 5% of NGS, DNMT1, 1:50 and MeCP2 1:100, and incubated for 1 h. After incubation, slides were washed with 1 x PBS for 5 min X 2 (no rocking) and incubated with secondary antibody (goat-anti Rabbit, red with Alexa Flour 594; green with Alexa Flour 488) in the dark for 30 min (anti rabbit, 1:100 in 5% of NGS). After washing 3 times with 1 x PBS, slides were mounted with mounting media with DAPI. For double-staining for PPARγ and DNMT1 or MeCP2, anti-PPARγ (1:100) was first used to stain overnight, then the slides were washed 3 times with 1 x PBS, then the they were blocked with Rabbit serum (5% in 1 x PBS) for 1 h, and finally incubated with another primary antibody (anti-DNMT1 or MeCP2 rabbit antibody) for one more hour. After incubation, slides were washed with 1 x PBS for 5 min X 3 (no rocking) and then incubated with above-mentioned secondary antibody in the dark for 30 min (anti rabbit, 1:100 in 5% of NGS). After washing 3 times with PBS, slides were mounted with mounting media with DAPI. The images were obtained using florescence microscope (Zeiss, Axioskop 40) for nuclei (DAPI, blue staining) and specific signal for DNMT1 and MeCP2 (red or green florescence).
2.8. Statistical analyses
All experiements were repeated at least 3 times. Differences between the groups were evaluated by one-way ANOVA followed by Newman-Keuls post-hoc test and unpaied Student’s t-test as needed. A P value of < 0.05 was considered to be statistically significant. Data are expressed as means ± SE.
3. Results
3.1. Nicotine exposure results in PPARγ promoter methylation
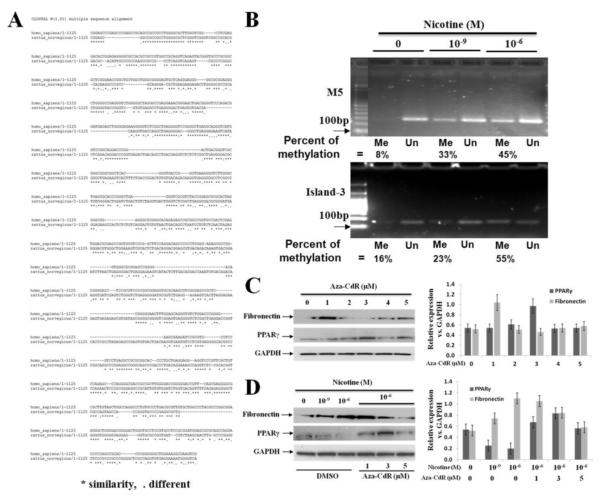
To determine the effect of nicotine on PPARγ promoter methylation in alveolar interstitial fibroblasts under in vitro conditions, human embryonic lung fibroblast cell line WI-38 and primary fetal rat lung fibroblasts were used. At 80-90% confluence, WI-38 cells were treated with or without nicotine (0, 10−9, and 10−6 M) for 24 h. Following which, genomic DNA was extracted and PPARγ promoter methylation was determined using MSP. We first used the following 4 previously published PPARγ promoter methylation-specific primers covering specific CpG-rich regions upstream of the transcription start site (TSS) M1 (−123 to +49), M2 (−235 to −151), M3 (−359 to −260) and M4 (−746 to −616) [34]. We found that M1, M2 and M3 regions were stably unmethylated regardless of exposure to nicotine (Fig. 1S-A). Interestingly, the M4 region was highly methylated whether or not it was exposed to nicotine (Fig. 1S-A). In order to further mechanistically explore nicotine-mediated down-regulation of PPARγ [7, 9, 35], using MethPrimer [36], we designed additional primers expanding coverage to two more CpG islands [37] further upstream, M5 region (CpG-1: −1268 to −1068, position in relation to TSS, chr3/ 123, 29428---123, 30333) and M6 region (CpG-2: −1686 to −1489; position in relation to TSS, chr3/ 123, 28995---123, 29227) in the PPARγ promoter. Nicotine exposure resulted in increased methylation of M5 region, dose-dependently (percent methylation: 8.2 ± 2.1%, 33.6 ± 2.7%, and 45.7 ± 4.9% following 24h exposure to 0, 10−9 M or 10−6 M nicotine, respectively; Fig.1B), while there was no effect on the methylation of M6 region (Fig. 1S-A), indicating the specificity and significance of M5 region for nicotine’s effect on PPARγ down-regulation and its likely role in alveolar interstitial fibroblast differentiation.
Fig. 1.
Site-specific epigenetic regulation of PPARγ promoter in the human and rat lung alveolar interstitial fibroblasts: (A) The human PPARγ promoter M5 (CpG-1, human Chr4: 123,29428 to 123,30333) and rat Island-3 regions (rat chr4: 210,563,793 to 210,564026) were aligned at http://uswest.ensembl.org/Homo_sapiens/Location/Compara_Alignments?align=612&db=core&t=ENST00000287820&g=ENSG00000132170&r=3%3A12329428-12330333. Sequence homology was calculated with the FASTA function of Genetics Computer Group. The sequences show more than 70% homology between the two regions; (B) Methylation-specific-PCR showed nicotine-mediated increase in human M5 region (upper panel) and in rat island-3 region (lower panel) PPARγ promoter methylation dose-dependently (DNA was extracted from human WI-38 cells and primary embryonic day 19 rat lung fibroblasts, respectively); the bands were quantified with NIH Image J software and percent methylation was calculated as = Me / (Me + Un); (C) The optimal concentration of Aza-CdR for methylation inhibition and its downstream effects on fibronectin and PPARγ protein levels was explored, which was determined to be 3 μM; this concentration was used for further experiments; the quantified data normalized to GAPDH protein levels are shown; (D) Aza-CdR restores the nicotine-induced down-regulation of PPARγ and up-regulation of fibronectin, dose-dependently; the quantified data normalized to GAPDH protein levels are shown. The representative Western bands from at least three independent experiments are shown; *p < 0.05 vs. control.
We next determined the effect of nicotine on PPARγ promoter methylation using cultured primary fetal rat lung fibroblasts. Methylation specific primers were designed to cover three CpG islands of rat PPARγ promoter Island-1 (chr4/ 210, 562586 to 210, 563185); Island-2 (chr4/ 137, 317002 to 137, 317450); and Island-3 (chr4/ 210, 563937 to 210, 564106). Nicotine exposure resulted in increased PPARγ promoter methylation in island-3 region, dose-dependently (percent methylation: 12.3 ± 2.4%, 23.5 ± 3.8%, and 55.2 ± 6.0% following 24h exposure to 0, 10−9 M or 10−6 M nicotine, respectively, Fig.1B), while the other two CpG islands remained unmethylated (Fig. 1S-B). Interestingly, we found more than 70 % homology between the human M5 region and the rat island-3 region of PPARγ promoter methylation sites affected by nicotine (Fig.1A), suggesting evolutionary conservance of this region in determining nicotine’s effect on alveolar fibroblasts.
Having demonstrated nicotine-induced site-specific PPARγ promoter methylation, we next determined if blocking PPARγ promoter methylation inhibits nicotine’s effect on alveolar fibroblast differentiation. WI-38 cells were treated with DNMT1 inhibitor Aza-CdR, with or without nicotine for 24 h. Under basal conditions, i.e., without nicotine, Aza-CdR increased PPARγ protein levels dose-dependently, with an optimal effect at 3 μM concentration (Fig. 1C). At this concentration of Aza-CdR, nicotine-mediated decrease in PPARγ and an increase in fibronectin protein levels were restored (Fig. 1D). In addition, highly methylated human PPARγ promoter M4 region was also regulated by Aza-CdR (Fig.1S-C). Since Aza-CdR selectively degrades DNMT1 by a proteosomal pathway [38], higher PPARγ protein levels in our studies on Aza-CdR treatment are possibly due to decreased promoter methylation resulting from DNMT1 degradation caused by Aza-CdR. Taken together, these data highlight the importance of promoter regions M4 and M5 in human PPARγ gene for its expression and activation.
3.2. Knock down of methylation mediators MeCP2 and DNMT1 decreases PPARγ methylation
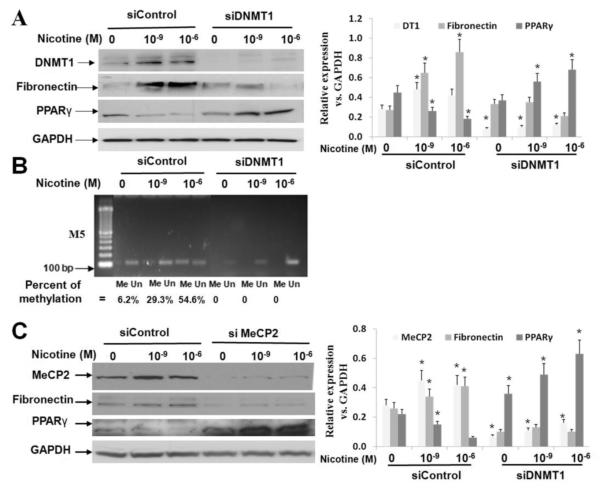
To determine whether MeCP2 and DNMT1, two known methylation modulators are determinants of PPARγ promoter methylation [39], these genes were silenced in WI-38 cells using specific siRNAs. DNMT1 knockdown in the presence or absence of nicotine (Fig. 2A) showed that nicotine increased DNMT1 protein levels, but decreased PPARγ and increased fibronectin protein levels. All changes were blocked by DNMT1 knockdown (percent methylation: 6.2 ± 1.9 % , 29.3 ± 3.0%, 54.6 ± 7.0 % following 24 h exposure to 0, 10−9 M or 10−6 M nicotine, respectively, Fig. 2B). Similarly, knock-down of MeCP2 with or without nicotine demonstrated that nicotine exposure increased MeCP2 and fibronectin protein levels, but decreased PPARγ protein levels, whereas, knocking down of MeCP2 restored PPARγ and its downstream target fibronectin protein levels (Fig. 2C).
Fig. 2.
DNMT1 and MeCP2 are activated by nicotine, and their knock down (siRNA) abolishes PPARγ promoter methylation, restoring nicotine-induced changes in PPARγ and fibronectin protein levels: (A) On nicotine exposure of WI-38 cells, DNMT1 and fibronectin protein levels increase, PPARγ levels decrease; DNMT1 knock-down using siRNA restores the nicotine-induced down-regulation of PPARγ and up-regulation of fibronectin; the quantified data normalized to GAPDH protein levels are shown; (B) Methylation specific-PCR shows that DNMT1 knock-down decreases PPARγ promoter methylation even under basal conditions, i.e., without nicotine exposure; the bands were quantified with NIH Image J software and percent methylation was calculated as = Me / (Me + Un); (C) MeCP2 and fibronectin protein levels increase, but PPARγ protein levels decrease on nicotine exposure; MeCP2 knock down restores nicotine-induced changes in PPARγ and fibronectin protein levels; the quantified data normalized to GAPDH protein levels are shown. The representative data from at least three independent experiments are shown, *p < 0.05 vs. control.
3.3. Nicotine recruits DNA methylation mediators DNMT1 and MeCP2 to a specific region in PPARγ promoter
MeCP2 and DNMT1 are known to regulate PPARγ transcription [40-42] by physical interactions with the PPARγ promoter [39] , via a repressive complex that includes DNMT1 and MeCP2 binding to specific CpG sites in the PPARγ promoter. Therefore, using ChIP-PCR, we next examined the physical interaction between the epigenetic modulators DNMT1 and MeCP2, and PPARγ promoter.
3.3.1. ChIP-PCR demonstrates nicotine-mediated enrichment of a specific PPARγ promoter region with DNMT1 and MeCP2
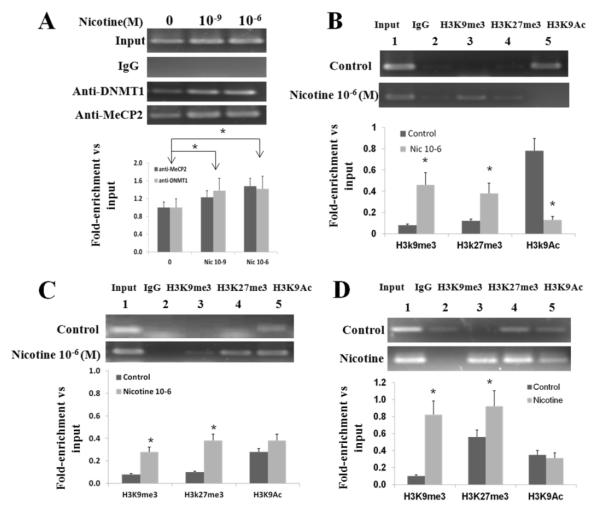
To test whether MeCP2 and/or DNMT1 bind to the PPARγ promoter in WI-38 cells, antibodies anti-MeCP2 and anti–DNMT1 were used in a ChIP assay with chromatin extracts from the control and nicotine treated WI-38 cells. As shown in Fig. 3A, a DNA fragment located within M5 region was amplified by PCR using immunoprecipitated DNA as a template together with primers spanning the PPARγ promoter region. Nicotine exposure increased the signal intensity, dose-dependently, with more than 20 % increase on 10−6 M nicotine exposure for 24h, indicating increased binding of DNMT1 and MeCP2 within PPARγ promoter (M5 region). These data support nicotine-mediated physical interaction between the PPARγ promoter and the methylating intermediates DNMT1 and MeCP2.
Fig. 3.
ChIP-PCR analysis shows enrichment of a specific PPARγ promoter region (human M5 or rat island-3). Chromatin was extracted from human WI38 cells and from rat fibroblasts following exposure to nicotine; (A) ChIP was performed with anti-MeCP2 and anti-DNMT1 antibodies following nicotine treatment of WI-38 cells; dose-dependent enrichment is shown; (B) ChIP-PCR was performed with anti-H3K9me3, H3K27me3 and H3K9Ac antibodies to examine the specific PPARγ promoter fragment enrichment in the presence of nicotine. The data show significant enrichment of H3K9me3 and H3K27me3, but a significant decrease in enrichment for H3K9Ac at the PPARγ promoter M5 region (human); (C) ChIP-PCR was also performed with anti-H3K9me3, H3K27me3, and H3K9Ac antibodies to examine the specific PPARγ promoter fragment enrichment in cultured primary fetal rat lung fibroblasts. The data show significant enrichment of H3K9me3 and H3K27me3 (trimethylation) of PPARγ promoter island-3, but no change was observed for H3K9Ac; (D) Using anti-H3K9me3, anti-H3K27me3 and anti-H3K9Ac, ChIP-PCR was performed on chromatin extracted from the in vivo perinatally nicotine exposed rat lung tissue to examine their enrichment in PPARγ promoter (rat island-3 region) under in vivo conditions. Similar to the human data (B and C), there was significant enrichment of H3K9me3 and H3K27me3, although there was no change in H3K9Ac enrichment. The representative gels are shown; experiments were performed in triplicates; data are mean ± SE. The ratio of the intensity of each band to input was calculated. *p < 0.05 vs. control; N=3
3.3.2. Specific repressive chromatin marks (H3K9me3, H3K27me3 and H3K9Ac) are associated with PPARγ inhibition with nicotine treatment
Using a quantitative ChIP assay, H3K9me3 and H3k27me3, two known gene repressors, and H3K9Ac, a known gene activator, were used to gain further insights to the mechanism(s) by which DNA modifications might affect PPARγ expression. With the promoter segment extending to M5 region, the chromatin was prepared from nicotine treated WI-38 cells, and then ChIP-PCR was performed with anti-H3K9me3, H3K27me3 and anti-H3K9Ac, using immunoprecipitated DNA as a template. Fig. 3B shows significant enrichment of H3K9me3 and H3K27me3 on nicotine exposure, but a decrease in enrichment for H3K9Ac. These changes suggest that nicotine promotes increased H3K9Me3 and H3K27me3 methylation by recruiting transcription repressors DNMT1 and MeCP2, indicating a key roles for H3K9 and H3K27 trimethylations in mediating nicotine’s effect on PPARγ expression and the resultant fibroblast differentiation. We also interrogated these data in cultured primary fetal rat lung fibroblasts. As above, ChIP-PCR was performed with anti-H3K9me3, H3K27me3 and anti-H3K9Ac. DNA fragment within the rat island-3 region was amplified, showing significantly increased enrichment H3K9Me3 and H3K27me3, but no significant change for H3K9Ac following 10−6 M nicotine exposure for 24 h (Fig. 3C).
Finally, using ChIP-PCR, we examined the PPARγ methylation status in perinatally nicotine exposed rat lung tissue. Again, as above, chromatin was prepared and ChIP-PCR was performed with anti-H3K9me3, H3K27me3 and anti-H3K9Ac antibodies; the rat PPARγ CpG island-3 fragment clearly enriched with anti H3K9me3 and H3K27me3 antibodies, while no significant enrichment with anti-H3K9Ac antibodies (Fig. 3D). in nicotine treated lungs was observed.
3.4. PPARγ, DNMT1 and MeCP2 interactions coordinate the in vivo nicotine-induced changes in lung PPARγ expression
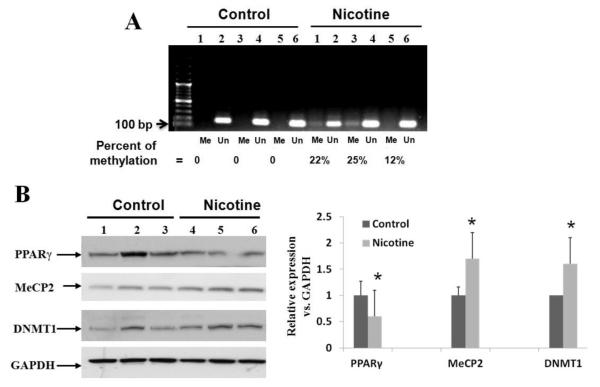
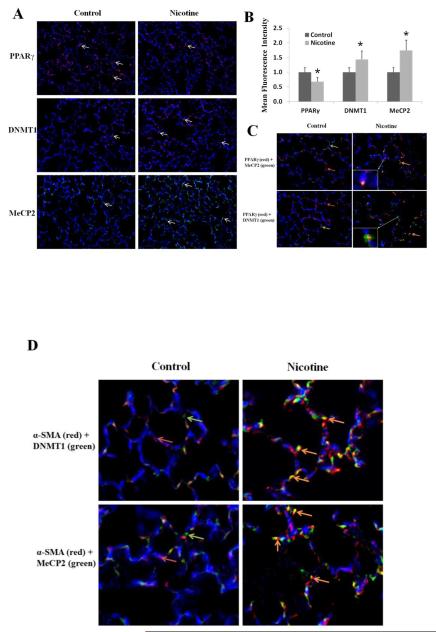
To further validate the in vitro data outlined above to the in vivo setting, PND21 lung tissue, obtained from rat pups perinatally exposed to nicotine, was analyzed. First, we assessed PPARγ methylation using MSP assay and found clearly increased PPARγ promoter methylation in the perinatally nicotine exposed lung tissue (percent methylation increased by an average of 19.6 ± 6.8% vs. the control group, Fig. 4A). Secondly, further corroborating our in vitro data, by Western blot analysis, PPARγ protein level decreased, but DNMT1 and MeCP2 protein levels increased in the perinatally nicotine exposed lungs (Fig. 4B). These data were also further corroborated by immunohistochemistry. The staining of DNMT1 and MeCP2 increased, while the staining for PPARγ decreased in lung tissue sections of the perinatally nicotine exposed lungs (Fig. 5A and 5B). Co-staining for DNMT-1 and MeCP2 with PPARγ shows co-localization of PPARγ and MeCP2, and PPARγ and DNMT1, while no or very little co-staining was seen under control conditions (Fig. 5C). Furthermore, co-staining with a mesenchymal specific marker α-SMA showed co-localization and increased signal intensity of DNMT1 and MeCP2 staining with α-SMA staining (Fig.5D). Overall, consistent with the Western data outlined above, on perinatal nicotine exposure, epigenetic modulators DNMT1 and MeCP2 showed up-regulation and co-localization with PPARγ and α-SMA, confirming the mesenchymal specificity of nicotine’s these epigenetic effects.
Fig. 4.
In vivo perinatal nicotine exposure results in increased PPARγ promoter methylation in nicotine exposed rat lung: (A) Using methylation specific-PCR, nicotine-mediated increase in methylation of the rat island-3 region is shown; the bands were quantified with NIH Image J software and percent methylation was calculated as = Me / (Me + Un); (B) Similar to our findings under in vitro conditions (Fig. 2), perinatal nicotine exposure resulted in increased DNMT1 and MeCP2, but decreased PPARγ protein levels on Western analysis of the whole lung lysates obtained from the lungs of postnatal day 21 rats. The data for each condition were generated from at least 6 animals, each from a separate litter. The protein bands, quantified and normalized to GAPDH band intensities, are shown. *p < 0.05 vs. control.
Fig. 5.
Effect of perinatal nicotine exposure on PPARγ epigenetic markers in paraformaldehyde-fixed paraffin-embedded lung tissue sections obtained from postnatal day 21 rats: (A) Nicotine administration significantly reduced staining for PPARγ (red), but increased staining for DNMT1 (red) and MeCP2 (green); Magnification 20X; (B) mean fluorescence intensity of the specific signal was quantified using NIH Image J software and plotted; (C) co-staining for either PPARγ and DNMT1 (green arrow, lower panel) or PPARγ (red arrows) and MeCP2 (green arrow, upper panel) shows co-localization (orange arrows) of PPARγ with both DNMT1 and MeCP2, stained independently. Perinatal nicotine exposure significantly reduced staining for PPARγ (red), but increased staining for DNMT1 (green, lower panels) and MeCP2 (green, upper panels), magnification 40X; (D) co-staining with a mesenchymal specific marker α-SMA (red) showed co-localization (orange arrows) and increased signal intensity of DNMT1 (green, upper panel) and MeCP2 (green, lower panel) staining, magnification 40X. Representative images of at least 3 independent experiments for each condition are shown, *p < 0.05 vs. control.
4. Conclusions
In this study, using well-documented in vitro and in vivo models, we have determined key epigenetic changes in PPARγ promoter methylation in association with nicotine-mediated alveolar interstitial fibroblast transdifferentiation. Our data clearly supports that nicotine-induced modulation of alveolar interstitial fibroblast differentiation, i.e., decreased PPARγ and increased fibronectin protein levels, indicating myofibroblast differentiation, is mediated by alterations in PPARγ promoter methylation via its interactions with MeCP2 and DNMT1, which in turn also modulate H3K9me3 and H3K27me3 methylation.
Using cultured embryonic alveolar interstitial fibroblast cell line WI-38 and primary rat lung fibroblasts, and lung tissue from the in vivo perinatally nicotine exposed rat pups, we found a clear evidence for increased PPARγ promoter methylation and its physical interaction with key methylation modulating intermediates MeCP2 and DNMT1. This was accompanied by nicotine-induced down-regulation of PPARγ and up-regulation of fibronectin. Examining, PPARγ promoter methylation by MSP, spanning the entire promoter region just upstream of the TSS, we found that CpG region 4 (M4) was highly methylated with or without nicotine treatment, whereas the three other previously studied regions remained unmethylated (M1, 2, 3) with or without nicotine exposure. This forced us to examine even further upstream of the PPARγ promoter. Fortuitously, with nicotine exposure, we found dose-dependent increase in PPARγ promoter methylation in the M5 region, indicating the likely significance of this region in mediating nicotine’s effects on alveolar fibroblast differentiation. Similarly, the methylation of rat island-3 region also showed dose-dependent increase; alignment of the rat and human genomic sequences indicated more than 70% homology between the human M5 and the rat island-3 regions, suggesting an evolutionary conservance of this region in determining nicotine’s effects on the developing lung. Methylation inhibitor Aza-CdR restored the nicotine-induced down regulation of PPARγ and the up-regulation of fibronectin protein levels, supporting PPARγ promoter’s methylation playing a key role in determining alveolar interstitial fibroblast phenotype. Further exploration of the intermediates involved in nicotine-induced PPARγ methylation, ChIP-PCR revealed physical interactions between DNMT1 and MeCP2, two known methylation mediators [43], with specific regions (H3K9me3 and H3K27me3) of the PPARγ promoter. Further, confirming that MeCP2 and DNMT1 levels regulate PPARγ methylation, their down-regulation using siRNAs resulted in the up-regulation of PPARγ and down-regulation of fibronectin, strongly suggesting that nicotine’s effects on PPARγ and its downstream targets are mediated via the recruitment of DNMT1 and MeCP2. MeCP2 is known to bind preferentially to methylated DNA regions, depending on the context [43], serving as either a transcriptional repressor or activator, our study indicates that on exposure to nicotine, MeCP2 plays a repressive role in PPARγ expression.
To gain further mechanistic insights to epigenetic modifications affecting PPARγ levels, quantitative ChIP assay was performed to determine the PPARγ promoter methylation with the promoter segment extending into the M5 and M6 regions in human WI-38 cells. The repressive chromatin marks H3K9me3 and H3K27me3 were significantly enriched on exposure to nicotine, consistent with the previous observation that an H3K9me3-containing co-repressor complex inactivates PPAR function through histone H3K9 methylation [44]. Lastly, validating our in vitro data in an in vivo setting, perinatal nicotine exposure resulted in increased DNMT1 and MeCP2 levels, in association with decreased PPARγ protein levels.
Our data provide novel mechanistic insights to the pulmonary phenotype seen in infants exposed to nicotine as such or as a constituent of cigarette smoke. These are highly relevant findings since despite all efforts a large number of woman still smoke while pregnant, resulting in the births of at least 400,000 smoke-exposed infants per year in the US alone [45][46]. Furthermore, pregnant women are also commonly exposed to nicotine as part of the nicotine replacement therapy [47] and/or via the use of electronic cigarettes since the popularity of electronic cigarettes has risen dramatically in recent years [48]. This is particularly concerning since our recent data suggests that the risk of the transmission of perinatal smoke exposure-induced lung phenotype is not restricted only to the exposed offspring, but is also transmitted to non-exposed second and third generation progeny [49, 50]. The data included herein set the stage to examine the efficacy of specific DNA methylation modulators to prevent the everlasting consequences of nicotine/smoke exposure on the developing lung.
In conclusion, we demonstrate that epigenetic events play a major role in mediating nicotine’s effect on PPARγ expression, a key nuclear transcripton factor that determines the lung fibroblast phenotype. Though, translationally, still challenging at this stage, modulation of specific PPARγ promoter methylation sites, as uncovered by our studies, can be a novel targeted strategy to block smoke exposed lung injury in general and perinatal nicotine exposure induced lung damage in particular.
Supplementary Material
Highlights.
Perinatal smoke/nicotine exposure is a well-established factor for affecting lung growth and differentiation by altering specific developmental signaling pathways necessary for fetal lung development, resulting in childhood asthma. Though we have previously shown that perinatal nicotine exposure, by down-regulating PPARγ expression accentuates the myogenic pulmonary phenotype, the underlying molecular mechanisms remain incompletely understood. Using, previously validated, both in vitro and in vivo models, here we provide novel mechanistic insights into nicotine-induced epigenetic silencing of PPARγ that could be exploited to design novel targeted molecular interventions against the smoke/nicotine exposed lung injury in general and perinatal nicotine exposure induced lung damage in particular.
Acknowledgments
Grant Support: NIH (HL075405, HL55268, HD51857, HD058948, HL107118, and HD071731) and TRDRP (23RT-0018)
Abbreviations
- α-SMA
Alpha Smooth Muscle Actin
- ATII
Alveolar epithelial type II
- Aza-CdR
5-aza-2′-deoxycytidine
- ChIP
Chromatin Immunoprecipitation
- DMSO
Dimethyl sulfoxide
- DNMT1
DNA methyltransferase 1
- LIFs
Lipofibroblasts
- MSP
Methylation-specific-PCR
- MeCP2
Methyl CpG binding protein 2
- MYFs
Myofibroblasts
- PPARγ
Peroxisome proliferator-activated receptor gamma
- PND
Postnatal day
- TSS
Transcription start site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Torday JS, Rehan VK. Up-regulation of fetal rat lung parathyroid hormone-related protein gene regulatory network down-regulates the Sonic Hedgehog/Wnt/betacatenin gene regulatory network. Pediatr. Res. 2006;60:382–388. doi: 10.1203/01.pdr.0000238326.42590.03. [DOI] [PubMed] [Google Scholar]
- [2].Torday JS, Rehan VK. Developmental cell/molecular biologic approach to the etiology and treatment of bronchopulmonary dysplasia. Pediatr. Res. 2007;62:2–7. doi: 10.1203/PDR.0b013e31806772a1. [DOI] [PubMed] [Google Scholar]
- [3].Rehan VK, Torday JS. The Lung Alveolar Lipofibroblast: An Evolutionary Strategy Against Neonatal Hyperoxic Lung Injury. Antioxid. Redox. Signal. 2014 doi: 10.1089/ars.2013.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Torday J, Hua J, Slavin R. Metabolism and fate of neutral lipids of fetal lung fibroblast origin. Biochim. Biophys. Acta. 1995;1254:198–206. doi: 10.1016/0005-2760(94)00184-z. [DOI] [PubMed] [Google Scholar]
- [5].Torday JS, Torres E, Rehan VK. The role of fibroblast transdifferentiation in lung epithelial cell proliferation, differentiation, and repair in vitro. Pediatr. Pathol. Mol. Med. 2003;22:189–207. doi: 10.1080/pdp.22.3.189.207. [DOI] [PubMed] [Google Scholar]
- [6].Torday JS, Torday DP, Gutnick J, Qin J, Rehan V. Biologic role of fetal lung fibroblast triglycerides as antioxidants. Pediatr. Res. 2001;49:843–849. doi: 10.1203/00006450-200106000-00021. [DOI] [PubMed] [Google Scholar]
- [7].Rehan VK, Wang Y, Sugano S, Romero S, Chen X, Santos J, Khazanchi A, Torday JS. Mechanism of nicotine-induced pulmonary fibroblast transdifferentiation. Am. J. Physiol Lung Cell Mol. Physiol. 2005;289:L667–L676. doi: 10.1152/ajplung.00358.2004. [DOI] [PubMed] [Google Scholar]
- [8].Rehan VK, Asotra K, Torday JS. The effects of smoking on the developing lung: insights from a biologic model for lung development, homeostasis, and repair. Lung. 2009;187:281–289. doi: 10.1007/s00408-009-9158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Krebs M, Sakurai R, Torday JS, Rehan VK. Evidence for in vivo nicotine-induced alveolar interstitial fibroblast-to-myofibroblast transdifferentiation. Exp. Lung Res. 2010;36:390–398. doi: 10.3109/01902141003714023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rehan VK, Torday JS. PPARgamma Signaling Mediates the Evolution, Development, Homeostasis, and Repair of the Lung. PPAR. Res. 2012;2012:289867. doi: 10.1155/2012/289867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Torday JS, Rehan VK. Developmental cell/molecular biologic approach to the etiology and treatment of bronchopulmonary dysplasia. Pediatr. Res. 2007;62:2–7. doi: 10.1203/PDR.0b013e31806772a1. [DOI] [PubMed] [Google Scholar]
- [12].Wongtrakool C, Wang N, Hyde DM, Roman J, Spindel ER. Prenatal nicotine exposure alters lung function and airway geometry through alpha7 nicotinic receptors. Am. J. Respir. Cell Mol. Biol. 2012;46:695–702. doi: 10.1165/rcmb.2011-0028OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Maritz GS, Harding R. Life-long programming implications of exposure to tobacco smoking and nicotine before and soon after birth: evidence for altered lung development. Int. J. Environ. Res. Public Health. 2011;8:875–898. doi: 10.3390/ijerph8030875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sekhon HS, Keller JA, Benowitz NL, Spindel ER. Prenatal nicotine exposure alters pulmonary function in newborn rhesus monkeys. Am. J. Respir. Crit Care Med. 2001;164:989–994. doi: 10.1164/ajrccm.164.6.2011097. [DOI] [PubMed] [Google Scholar]
- [15].Hylkema MN, Blacquiere MJ. Intrauterine effects of maternal smoking on sensitization, asthma, and chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 2009;6:660–662. doi: 10.1513/pats.200907-065DP. [DOI] [PubMed] [Google Scholar]
- [16].Luck W, Nau H. Nicotine and cotinine concentrations in serum and milk of nursing smokers. Br. J Clin. Pharmacol. 1984;18:9–15. doi: 10.1111/j.1365-2125.1984.tb05014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Maritz GS, Thomas RA. Maternal nicotine exposure: response of type II pneumocytes of neonatal rat pups. Cell Biol. Int. 1995;19:323–331. doi: 10.1006/cbir.1995.1075. [DOI] [PubMed] [Google Scholar]
- [18].Roman J, Ritzenthaler JD, Gil-Acosta A, Rivera HN, Roser-Page S. Nicotine and fibronectin expression in lung fibroblasts: implications for tobacco-related lung tissue remodeling. FASEB J. 2004;18:1436–1438. doi: 10.1096/fj.03-0826fje. [DOI] [PubMed] [Google Scholar]
- [19].Rehan VK, Wang Y, Sugano S, Romero S, Chen X, Santos J, Khazanchi A, Torday JS. Mechanism of nicotine-induced pulmonary fibroblast transdifferentiation. Am. J. Physiol Lung Cell Mol. Physiol. 2005;289:L667–L676. doi: 10.1152/ajplung.00358.2004. [DOI] [PubMed] [Google Scholar]
- [20].Rehan VK, Wang Y, Sugano S, Santos J, Patel S, Sakurai R, Boros LG, Lee WP, Torday JS. In utero nicotine exposure alters fetal rat lung alveolar type II cell proliferation, differentiation, and metabolism. Am. J. Physiol Lung Cell Mol. Physiol. 2007;292:L323–L333. doi: 10.1152/ajplung.00071.2006. [DOI] [PubMed] [Google Scholar]
- [21]. http://www.iom.edu/~/media/Files/Activity%20Files/PublicHealth/Modified-Risk-Tobacco/MRTP_rb.pdf.
- [22].Rees VW, Connolly GN. Potentially Reduced Exposure Tobacco Products: A Public Health Information Guide. Harvard School of Public Health; 2008. [Google Scholar]
- [23].Clark SM, Nakad R. Pharmacotherapeutic management of nicotine dependence in pregnancy. Obstet. Gynecol. Clin. North Am. 2011;38:297–311. x. doi: 10.1016/j.ogc.2011.02.017. [DOI] [PubMed] [Google Scholar]
- [24].Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Prog. Lipid Res. 2006;45:120–159. doi: 10.1016/j.plipres.2005.12.002. [DOI] [PubMed] [Google Scholar]
- [25].Knouff C, Auwerx J. Peroxisome proliferator-activated receptor-gamma calls for activation in moderation: lessons from genetics and pharmacology. Endocr. Rev. 2004;25:899–918. doi: 10.1210/er.2003-0036. [DOI] [PubMed] [Google Scholar]
- [26].Fujiki K, Kano F, Shiota K, Murata M. Expression of the peroxisome proliferator activated receptor gamma gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC. Biol. 2009;7:38. doi: 10.1186/1741-7007-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sookoian S, Rosselli MS, Gemma C, Burgueno AL, Fernandez GT, Castano GO, Pirola CJ. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor gamma coactivator 1alpha promoter. Hepatology. 2010;52:1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- [28].Yideng J, Zhihong L, Jiantuan X, Jun C, Guizhong L, Shuren W. Homocysteine-mediated PPARalpha,gamma DNA methylation and its potential pathogenic mechanism in monocytes. DNA Cell Biol. 2008;27:143–150. doi: 10.1089/dna.2007.0658. [DOI] [PubMed] [Google Scholar]
- [29].Sakurai R, Cerny LM, Torday JS, Rehan VK. Mechanism for nicotine-induced up-regulation of Wnt signaling in human alveolar interstitial fibroblasts. Exp. Lung Res. 2011;37:144–154. doi: 10.3109/01902148.2010.490288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhao Q, Fan YC, Zhao J, Gao S, Zhao ZH, Wang K. DNA methylation patterns of peroxisome proliferator-activated receptor gamma gene associated with liver fibrosis and inflammation in chronic hepatitis B. J. Viral Hepat. 2013;20:430–437. doi: 10.1111/jvh.12048. [DOI] [PubMed] [Google Scholar]
- [31].Jeronimo C, Usadel H, Henrique R, Silva C, Oliveira J, Lopes C, Sidransky D. Quantitative GSTP1 hypermethylation in bodily fluids of patients with prostate cancer. Urology. 2002;60:1131–1135. doi: 10.1016/s0090-4295(02)01949-0. [DOI] [PubMed] [Google Scholar]
- [32].Zhang L, Chen H, Gong M, Gong F. The chromatin remodeling protein BRG1 modulates BRCA1 response to UV irradiation by regulating ATR/ATM activation. Front Oncol. 2013;3:7. doi: 10.3389/fonc.2013.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Liu J, Sakurai R, O’Roark EM, Kenyon NJ, Torday JS, Rehan VK. PPARgamma agonist rosiglitazone prevents perinatal nicotine exposure-induced asthma in rat offspring. Am. J. Physiol Lung Cell Mol. Physiol. 2011;300:L710–L717. doi: 10.1152/ajplung.00337.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pancione M, Sabatino L, Fucci A, Carafa V, Nebbioso A, Forte N, Febbraro A, Parente D, Ambrosino C, Normanno N, Altucci L, Colantuoni V. Epigenetic silencing of peroxisome proliferator-activated receptor gamma is a biomarker for colorectal cancer progression and adverse patients’ outcome. PLoS. One. 2010;5:e14229. doi: 10.1371/journal.pone.0014229. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [35].Rehan VK, Sakurai R, Wang Y, Santos J, Huynh K, Torday JS. Reversal of nicotine-induced alveolar lipofibroblast-to-myofibroblast transdifferentiation by stimulants of parathyroid hormone-related protein signaling. Lung. 2007;185:151–159. doi: 10.1007/s00408-007-9007-0. [DOI] [PubMed] [Google Scholar]
- [36].Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- [37].Pancione M, Sabatino L, Fucci A, Carafa V, Nebbioso A, Forte N, Febbraro A, Parente D, Ambrosino C, Normanno N, Altucci L, Colantuoni V. Epigenetic silencing of peroxisome proliferator-activated receptor gamma is a biomarker for colorectal cancer progression and adverse patients’ outcome. PLoS. One. 2010;5:e14229. doi: 10.1371/journal.pone.0014229. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [38].Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [39].Hite KC, Adams VH, Hansen JC. Recent advances in MeCP2 structure and function. Biochem. Cell Biol. 2009;87:219–227. doi: 10.1139/o08-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Joss-Moore LA, Wang Y, Ogata EM, Sainz AJ, Yu X, Callaway CW, McKnight RA, Albertine KH, Lane RH. IUGR differentially alters MeCP2 expression and H3K9Me3 of the PPARgamma gene in male and female rat lungs during alveolarization. Birth Defects Res. A Clin. Mol. Teratol. 2011;91:672–681. doi: 10.1002/bdra.20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mann J, Chu DC, Maxwell A, Oakley F, Zhu NL, Tsukamoto H, Mann DA. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology. 2010;138:705–14. 714. doi: 10.1053/j.gastro.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhu NL, Wang J, Tsukamoto H. The Necdin-Wnt pathway causes epigenetic peroxisome proliferator-activated receptor gamma repression in hepatic stellate cells. J. Biol. Chem. 2010;285:30463–30471. doi: 10.1074/jbc.M110.156703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sugii S, Evans RM. Epigenetic codes of PPARgamma in metabolic disease. FEBS Lett. 2011;585:2121–2128. doi: 10.1016/j.febslet.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. http://www.cdc.gov/tobacco/data_statistics/fact_sheets/adult_data/cig_smoking/index.htm.
- [46]. http://www.childtrends.org/?indicators=mothers-who-smoke-while-pregnant.
- [47].Clark SM, Nakad R. Pharmacotherapeutic management of nicotine dependence in pregnancy. Obstet. Gynecol. Clin. North Am. 2011;38:297–311. x. doi: 10.1016/j.ogc.2011.02.017. [DOI] [PubMed] [Google Scholar]
- [48].Arrazola RA, Kuiper NM, Dube SR. Patterns of current use of tobacco products among U.S. high school students for 2000-2012--findings from the National Youth Tobacco Survey. J. Adolesc. Health. 2014;54:54–60. doi: 10.1016/j.jadohealth.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rehan VK, Liu J, Naeem E, Tian J, Sakurai R, Kwong K, Akbari O, Torday JS. Perinatal nicotine exposure induces asthma in second generation offspring. BMC. Med. 2012;10:129. doi: 10.1186/1741-7015-10-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rehan VK, Liu J, Sakurai R, Torday JS. Perinatal nicotine-induced transgenerational asthma. Am. J. Physiol Lung Cell Mol. Physiol. 2013;305:L501–L507. doi: 10.1152/ajplung.00078.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.