Abstract
CD4 T cell immune responses such as interferon-γ and tumor necrosis factor-α secretion are necessary for Chlamydia immunity. We used an immunoproteomic approach in which Chlamydia trachomatis and Chlamydia muridarum-derived peptides presented by MHC class II molecules on the surface of infected dendritic cells (DCs) were identified by tandem mass spectrometry using bone marrow derived DCs (BMDCs) from mice of different MHC background. We first compared the C. muridarum immunoproteome in C3H mice to that previously identified in C57BL/6 mice. Fourteen MHC class II binding peptides from 11 Chlamydia proteins were identified from C3H infected BMDCs. Two C. muridarum proteins overlapped between C3H and C57B/6 mice and both were polymorphic membrane proteins (Pmps) which presented distinct class II binding peptides. Next we studied DCs from C57BL/6 mice infected with the human strain, C. trachomatis serovar D. Sixty MHC class II binding peptides derived from 27 C. trachomatis proteins were identified. Nine proteins were orthologous T cell antigens between C. trachomatis and C. muridarum and 2 of the nine were Pmps which generated MHC class II binding epitopes at distinct sequences within the proteins. As determined by antigen specific splenocyte responses outer membrane proteins PmpF, -G and -H and the major outer membrane protein (MOMP) were antigenic in mice previously infected with C. muridarum or C. trachomatis. Furthermore a recombinant protein vaccine consisting of the four Pmps (PmpEFGH) with MOMP formulated with a Th1 polarizing adjuvant significantly accelerated (p < 0.001) clearance in the C57BL/6 mice C. trachomatis transcervical infection model. We conclude that Chlamydia outer membrane proteins are important T cell antigens useful in the development of a C. trachomatis subunit vaccine.
Keywords: Chlamydia, Outer membrane proteins, Immunoproteomics, MHC, T cell, Epitope, Peptide, Antigen, Vaccine
1. Indrouction
CD4 T cell-mediated immunity is a major component of host defense against Chlamydia infection [1] and the identification of epitopes presented by MHC class II molecules should enable the development of a Chlamydia T cell vaccine [2]. Dendritic cells (DCs) are at the centre of initiation of T cell mediated immune responses [3]. DCs capture antigen in the periphery and migrate to regional lymph nodes where they present processed antigen on MHC molecules to naïve T cells to induce T cell mediated immune responses. Since T cells mainly recognize protein antigens, protective vaccine candidates are likely to be found within the proteome of an organism. An approach called immunoproteomics [4], in which peptides presented by immunoaffinity purified MHC molecules from infected DCs are identified by tandem mass spectrometry (MS/MS) allow genomic information to guide the delineation of the T cell immunoproteome of an organism.
We previously used immunoproteomics to identify epitopes presented by MHC class II molecules from C57BL/6 bone marrow derived DCs (BMDCs) infected with Chlamydia muridarum [2,5]. Chlamydia-specific CD4 T cells harvested from mice recovered from C. muridarum infection recognized these MHC class II-bound peptides in vitro [6] and the source proteins of these MHC class II-bound peptides accelerated clearance of C. muridarum genital tract infection when formulated as vaccine with a Th1 polarizing adjuvant consisting of cationic liposome and modified mycobacterial cord factor [7].
We are interested in identifying Chlamydia trachomatis proteins presented by MHC class II molecules. In this study we investigated the C. trachomatis immunoproteome using infected C57BL/6 murine DCs and compared the findings to the C. muridarum immunoproteome identified in two different inbred strains of mice (C57BL/6 and C3H). We found that outer membrane proteins were commonly identified as source proteins encoding MHC class II binding peptides in all three experimental conditions. When used as vaccine with a Th1 polarizing adjuvant recombinant outer membrane proteins accelerated clearance of C. trachomatis from transcervically infected C57BL/6 mice. We conclude that outer membrane proteins are important T cell antigens in both C. trachomatis and C. muridarum capable of presentation by multiple MHC class II molecules and which elicit protective immunity. They are therefore useful for vaccine development.
2. Methods
2.1. Chlamydia strains
C. muridarum strain Nigg and C. trachomatis serovar D were grown in HeLa 229 cells in Eagle's essential medium supplemented with 10% fetal calf serum (FCS). Elementary bodies (EBs) were purified from HeLa 229 cells on discontinuous density gradients of Renografin-76 (Squib Canada) as described previously [8].
2.2. Mice
Female C57BL/6 (H2b) and C3H/HeNCrl (C3H) (H2k) mice (8 to 10 weeks old) were purchased from Charles River Canada (Saint Constant, Canada). The mice were maintained and used in strict accordance with University of British Columbia guidelines for animal care.
2.3. Generation of BMDCs
Bone marrow derived dendritic cells (BMDCs) were generated as previously described [9]. Briefly, bone marrow cells flushed from the femurs of female C57BL/6 or C3H mice were cultured in Falcon petri dishes at 4 × 107 cells in 50ml DC medium. DC medium was IMDM supplemented with 10% FCS, 0.5 mM 2-ME, 4mM l-glutamine, 50μg/ml gentamicin, and 5% of culture supernatant of murine GM-CSF-transfected plasmacytoma X63-Ag8 and 5% of culture supernatant of murine IL-4 transfected plasmacytoma X63-Ag8 which contained 10ng/ml GM-CSF and 10ng/ml IL-4, respectively. On day 3, half of culture supernatants were removed and fresh DC medium was added. On day 5, nonadherent cells (purity of >50% CD11c+) were harvested and cultured in fresh DC medium for Chlamydia infection.
2.4. Purification of MHC class II-bound peptides
MHC class II-bound peptides were purified as described previously [2]. Briefly, 5 × 109 immature BMDCs were infected at a 1:1 multiplicity of infection with C. muridarum or C. trachomatis serovar D for 12 or 24 h. BMDCs were then solubilized in lysis buffer (1% 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate, 150mM NaCl, 20 mM Tris–HCl, pH 8, 0.04% sodium azide, protease inhibitors). MHC class II molecules were isolated using allele-specific anti-MHC monoclonal antibody affinity columns containing the monoclonal antibodies Y-3P (specific to I-A MHC class II allele of both C57BL/6 and C3H) and 14-4-4S (specific to I-E MHC class II allele of C3H). Purified MHC class II molecules were separated from the peptides using 0.2 N acetic acid and subjected to ultrafiltration through a 10-kDa cut-off membrane to remove high molecular weight material [10].
2.5. Identification of MHC class II-bound peptides
The MHC class II-bound peptides were further purified, concentrated, filtered and desalted using STop And Go Extraction tips [11]. Peptides were then analyzed by LC/MS/MS using an LTQ-Orbitrap Velos (Thermo Electron) on-line coupled to Agilent 1200 Series nanoflow HPLCs using nanospray ionization sources (Proxeon Biosystems, Odense, Denmark). Analytical columns were packed into 15 cm long, 50 μm inner diameter fused silica emitters using 3 μm diameter ReproSil Pur C18 AQ beads (Dr. Maisch, www.Dr-Maisch.com), joint with 2-cm-long, 100-μm-inner diameter fused silica trap column packed with 5 μm-diameter Aqua C-18 beads (Phenomenex, www.phenomenex.com) and a 20-μm-inner diameter fused silica gold coated spray tip with 6-μm-diameter opening. LC buffer A consisted of 0.5% acetic acid and buffer B consisted of 0.5% acetic acid and 80% acetonitrile. Gradients were run from 10% B to 32% B over 51 min, then from 32% B to 40% B in the next 5 min, then increased to 100% B over 2 min period, held at 100% B for 2.5 min, and then dropped to 0% B for another 20 min to recondition the column. The Velos was set to acquire a full range scan at 60,000 resolution in the Orbitrap, from which the ten most intense multiply-charged ions per cycle were isolated for fragmentation in the LTQ. Centroided fragment peak lists were processed with Proteome Discoverer v. 1.2 (ThermoFisher Scientific). The search was performed with Mascot algorithm v. 2.4 against a database comprised of the protein sequences from the mouse and Chlamydia proteome. The estimated false discovery rate was below 2%.
2.6. Molecular cloning, expression and purification of C. trachomatis recombinant proteins
The recombinant proteins CT143, CT144, CT375, CT424, RplF (L6), CT619, PmpG, PmpF, PmpE, PmpH, major outer membrane protein (MOMP) and GroEL1 (Hsp60) were cloned, expressed and purified as follows: CT143, CT144, CT375, CT424, rplF, CT619, pmpG, pmpF, pmpE, pmpH, MOMP and groEL1 DNA fragments were generated by PCR using genomic DNA isolated from C. trachomatis. PCR reactions were carried out using Herculase Enhanced DNA polymerase (Agilent Technologies). The PCR product was purified with the QIAquick PCR purification kit (Qiagen) and the purified DNA fragments were cloned into pET32a expression vector (Novagen) after restriction enzyme digestion with BamHI/NotI using standard molecular biology techniques. For pmpE, pmpF, pmpG and pmpH, only the first half of the gene (representing amino acids 18–520, 26–585, 25–512, and 24–520, respectively) were cloned into the vector for expression because of MHC binding peptides were all found within this domain. The accuracy of the sub cloned genes were confirmed by sequencing. Plasmids containing the CT143, CT144, CT375, CT424, rplF, CT619, pmpG, pmpF, pmpE, pmpH, MOMP and groEL1 genes were transformed into the E. coli strain BL21(DE3) (Strategene) where protein expression was carried out by inducing the lac promoter for expression of T7 RNA polymerase using isopropyl-β-d-thiogalactoside pyranoside. The expressed CT143, CT144, CT375, CT424, RplF, CT619, PmpG, PmpF, PmpE, PmpH, MOMP and GroEL1 proteins with N-terminal His-tag were purified by nickel column using the His bind purification system (Qiagen). LPS removal of these proteins was carried out by adding 0.1% Triton-114 in one of the wash buffers during purification.
2.7. Transcervical infection with C. trachomatis serovar D in mice and determination of Chlamydia titer
Mice were infected with C. trachomatis serovar D transcervically after two s.c. injections with 2.5 mg of medroxyprogesterone acetate (Depo-Provera; Pharmacia and Upjohn) at day 3 and day 10 prior to infection. Transcervical inoculation of C. trachomatis was performed using NSET (Non-Surgical Embryo Transfer Device for Mice, ParaTechs Product no. 60010) as described by Gondek et al. [12] Briefly, a speculum was inserted into the mouse vaginal tract and the NSET tip directly inserted into the upper genital tract. The C. trachomatis inoculum (2 × 107 IFU in 10 μl) was pipetted into the upper genital tract and the NSET device and speculum were removed. At day 6 post infection, vaginal swabs and whole genital tracts were collected and stored at −80°C for titration. Chlamydia titers in homogenates of genital tract tissue and in vaginal swabs were measured by inclusion counts on Hela cells as described previously [13].
2.8. ELISPOT assay
The IFN-γ ELISPOT assay was performed as described previously [14]. Briefly, 96-well MultiScreen-HA filtration plates (Millipore) were coated overnight at 4 °C with 2 μg/ml of murine IFN-γ specific monoclonal antibody (BD PharMingen, Clone R4-6A2). Mice were transcervically infected into the uterine cavity with 2 × 107 IFU live C. trachomatis serovar D. Fourteen or 21 days after infection, the splenocytes were harvested and stimulated in vitro with 1 μg/ml individual Chlamydia proteins or 5 × 105 IFU/ml heat killed-EB as a positive control. After 20 h incubation at 37×C and 5% CO2, the plates were washed and then incubated with biotinylated murine IFN-γ specific monoclonal antibodies (BD PharMingen, Clone XMG1.2) at 2 μg/ml. This was followed by incubation with streptavidin-alkaline phosphatase (BD PharMingen) at a 1:1000 dilution. The spots were visualized with a substrate consisting of 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium (Sigma-Aldrich).
2.9. Statistical analysis
The data were analyzed with the aid of GraphPad Prism software program. Data are presented as means ± standard errors of the means (SEM) (ELISPOTs assay). Wallis test was performed to analyze data on IFU (Chlamydia titer) from multiple groups and Mann-Whitney U test to compare median between pairs. p Values of <0.05 were considered significant.
3. Results
3.1. Identification of C. muridarum derived MHC class II-bound peptides from C3H mice
Proteins which contain epitopes that can be presented by multiple MHC class II alleles are ideal vaccine candidates. Such proteins may present sequence specific or “promiscuous” epitopes and appear to have properties favorable to entering the antigen processing and presentation system [15,16]. Using the immunoproteomic approach we previously identified 17 Chlamydia T cell antigens when C57BL/6 BMDCs were infected with the murine strain C. muridarum [2,5]. Thirteen of these antigens were tested in a murine genital tract model and eleven elicited protective immunity against C. muridarum genital tract infection [17]. We now use the immunoproteomic approach to identify C. muridarum derived MHC class II-bound peptides presented on BMDCs from C3H mice (H2 I-Ak and I-Ek MHC alleles). A total of 14 peptides derived from 11 C. muridarum source proteins were identified (Table 1). Two proteins, PmpG and PmpF, overlapped with the source protein antigens identified using BMDC derived from C57BL/6 mice. The peptide epitopes presented by the Pmps were different in sequence between C57BL/6 and C3H mice.
Table 1.
MHC class II (I-Ak or I-Ek)-bound C. muridarum-derived peptides and their source proteins identified when C3H bone marrow derived dendritic cells were infected with live C. muridarum.
| Peptide | Presenting MHC class II allele | Chlamydia muridarum locus# | Source protein | Protein abbreviation |
|---|---|---|---|---|
| DIVLSDMSISRQHAK | I-Ek | TC0035 | Hypothetical protein | TC0035 |
| LPVGNPAEPSLM | I-Ak | TC0052 | Major outer membrane protein | MOMP |
| LPVGNPAEPSLMIDG | ||||
| KKFISYAL | I-Ek | TC0243 | ABC transporter, permease protein | TC0243 |
| NISSDLQAHMIASKTHNQ NISSDLQAHMIASKTHNQIK ISSDLQAHMIASKTHNQI |
I-Ek | TC0262 | Polymorphic membrane protein F | PmpF |
| KDEGVVFFSKNIAAGKG | I-Ek | TC0263 | Polymorphic membrane protein G | PmpG |
| EDFYKQMRAFRR | I-Ek | TC0294 | Signal recognition particle protein | Ffh |
| IAAAGTGLL | I-Ak | TC0392 | Hypothetical protein | TC0392 |
| ITGIVPISPGVRA | I-Ak | TC0408 | Hypothetical protein | TC0408 |
| KLGKVLGPRNL | I-Ek | TC0592 | 50S Ribosomal protein L1 | RplA |
| VTNKVTLAMQGQKLEG | I-Ek | TC0741 | Translocated actin recruiting phosphoprotein33 | Tarp |
| GSFAPVGLN | I-Ak | TC0910 | Hypothetical protein | TC0910 |
3.2. Identification of C. trachomatis derived MHC class II-bound peptides from C57BL/6 mice
In order to formulate a vaccine against human C. trachomatis infection, we need to identify its T cell antigens and therefore extended the immunoproteomic approach to C. trachomatis. Sixty peptides derived from 27 different C. trachomatis proteins were identified when BMDCs derived from C57BL/6 mice were infected with C. trachomatis serovar D for 12 h (Table 2). Similar to the peptide repertoire identified in C. muridarum, the chromosomal location of genes encoding source proteins of the C. trachomatis derived epitopes were spread uniformly throughout the C. trachomatis genome. Interestingly, none of the source proteins for T cell antigens were encoded by genes located in the plasticity zone, the highly variable chromosomal region between C. muridarum and C. trachomatis genomes that encodes several of the species and biovar specific virulence factors [18].
Table 2.
MHC class II (I-Ab)-bound C. trachomatis serovar D-derived peptides and their source proteins identified when C57BL/6 bone marrow derived dendritic cells were infected with live C. trachomatis for 12 h.
| Peptide | Chlamydia trachomatis locus# | Source proteins | Protein abbrev. |
|---|---|---|---|
| YKLVYQNALSNFSGKK | CT045 | Leucyl aminopeptidase | PepA |
| GPKGRHVVIDKSFGSPQVTKDGVT | CT110 | Chaperonin GroEL1 41 | Hsp60 |
| EERVVGQPFAIAAVSDS | CT113 | Clp Protease ATPase | ClpB |
| DLKVTGPTIHTDLD | CT143 | Hypothetical protein 33 | CT143 |
| GKLIVTNPKSDISFGG | CT144 | Hypothetical protein 60 | CT144 |
| GSPGQTNYAAAKAGIIGFS | CT237 | 3-Ketoacyl-(acyl-carrier-protein) reductase | FabG |
| GTKTPIGTPIAVFSTEQ | CT247 | Dihydrolipoamide acetyltransferase 51 | PdhC |
| SPKEAAIAAARASLSPEEKR | CT289 | Hypothetical protein | CT289 |
| YDHIIVTPGANADILPE | CT375 | Predicted d-amino acid dehydrogenase | CT375 |
| FDGEKASVGAPTVGNAVVKG | CT420 | 50S ribosomal protein L21 33 | Rl21 |
| KLDGVSSPAVQESISESL | CT424 | Sigma regulatory factor | RsbV |
| TPSAVNPLPNPEIDS | CT472 | Hypothetical protein | CT472 |
| DSTHGSFAPQATFSDG | CT505 | Glyceraldehyde-3-phosphate dehydrogenase | GapA |
| VKGNEVFVTPAAHVVDRPG | CT514 | 50S ribosomal protein L6 | RplF |
| ETPGAAEGAEAQTASEQPSKENAEKQEENNED | CT559 | Yop proteins translocation lipoprotein | CdsJ |
| ADVLLLSPKASVSPGG | CT561 | Type III secretion translocase 46 | CdsL |
| IPFAKPDANLSAED | CT619 | Hypothetical protein | CT619 |
| KAPQFGYPAVQNSADS | CT622 | CHLPN 76 kDa homolog | CT622 |
| KEGEEDTAESAANEEPKAEASQEEE | CT664 | FHA domain; homology to adenylate cyclase | CT664 |
| IFDTTTLNPTIAGAGDVK | CT681 | Major outer membrane protein 45 | MOMP |
| TPVESTTPVAPEISVVNAK | CT759 | Muramidase (invasin repeat family) | NlpD |
| QVFQLITQVTGRSG | CT778 | Primosome assembly protein | PriA |
| ISYDYSSGNAEASSHN | CT837 | Hypothetical protein | CT837 |
| DAGVPIKAPVAGIAMG | CT842 | Polyribonucleotide Nucleotidyltransferase | Pnp |
| GSVVFSGATVNSADFH | CT869 | Polymorphic membrane protein E | PmpE |
| AMANEAPIAFIANVAG | CT871 | Polymorphic membrane protein G | PmpG |
| AEKGGGAIYAPTIDISTNGGS | CT872 | Polymorphic membrane protein H | PmpH |
There are 924 and 894 proteins encoded by the genomes of C. muridarum and C trachomatis, respectively, and in principle all of these could be processed and presented by antigen presenting cells. Strikingly only 3% (27 of 894) of the C. trachomatis proteome is actually detected via MHC class II molecules following infection of DCs by our method, a percentage similar to what we previously observed with C. muridarum [2]. The reasons for the limited presentation of Chlamydia peptides by antigen presenting cells are unknown and may be confounded by the sequestration of antigens within the Chlamydia inclusion of infected DCs [19].
Nine source proteins identified from C57BL/6 MHC class II–bound peptides overlap between the C. muridarum and C. trachomatis proteome: Comparison of source proteins among the MHC class II-bound Chlamydia peptides revealed 9 overlapping source proteins from the C. muridarum and C. trachomatis immunoproteome in C57BL/6 mice (Table 3 and Fig. 1). For T cell proteins (defined as containing MHC binding peptides) which overlapped between the C. trachomatis and C. muridarum proteome we expected that similar or identical epitopes would be derived from shared proteins which were highly conserved in sequence while different epitopes would be presented from proteins less conserved in sequence. Five of the six highly conserved proteins (>75% sequence identity) presented the same or very similar epitopes (identical: FabG, RsbV, CT375; similar: PdhC, RplF) although one source protein (GapA) from the highly sequence conserved group presented a completely different epitope despite its 95% amino acid sequence identity. Two of the three less conserved (<71% sequence identity) proteins (PmpE and PmpG) presented distinct epitopes located at different sequence positions within the surface exposed passenger domain of these outer membrane proteins (Fig. 2).
Table 3.
Nine overlapping source proteins between C. trachomatis and C. muridarum. Identical amino acid residues of the epitopes between the two strains are underlined and in bold. Percentage protein identity is for the whole length proteins of each comparison.
| C. trachomatis Locus#/protein abbreviation | C. trachomatis-derived peptides | C. muridarum locus# | C. muridarum-derived peptides | C. trachomatis/C. muridarum protein identity |
|---|---|---|---|---|
| CT143 | DLKVTGPTIHTDLD | TC0420 | DLNVTGPKIQTDVD | 75% |
| CT237 (FabG) | GSPGQTNYAAAKAGIIGFS | TC0508 | SPGQTNYAAAKAGIIGFS | 90% |
| CT247(PdhC) | GTKTPIGTPIAVFSTEQ | TC0518 | EGTKIPIGTPIAVFSTEQN | 87% |
| CT424 (RsbV) | KLDGVSSPAVQESISESL | TC0707 | KLDGVSSPAVQESISE | 96% |
| CT375 | YDHIIVTPGANADILPE | TC0654 | YDHIIVTPGANADIL | 85% |
| CT505 (GapA) | DSTHGSFAPQATFSDG | TC0792 | MTTVHAATATQSVVD | 95% |
| CT514 (RplF) | VKGNEVFVTPAAHVVDRPG | TC0801 | VKGNEVFVSPAAHIIDRPG | 96% |
| CT869 (PmpE) | GSVVFSGATVNSADFH | TC0261 | SRALYAQPMLAISEA | 69% |
| CT871 (PmpG) | AMANEAPIAFIANVAG | TC0263 | NAKTVFLSNVASPIYVDPA ASPIYVDPAAAGGQPPA |
71% |
Fig. 1.
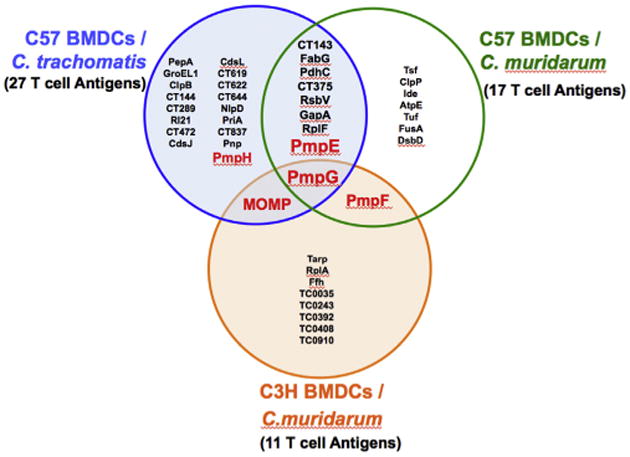
Venn diagram depicting the overlap of T cell antigens identified when BMDCs generated from mice of two different MHC background are infected with the human strain C. trachomatis and the mouse strain C. muridarum.
Fig. 2.
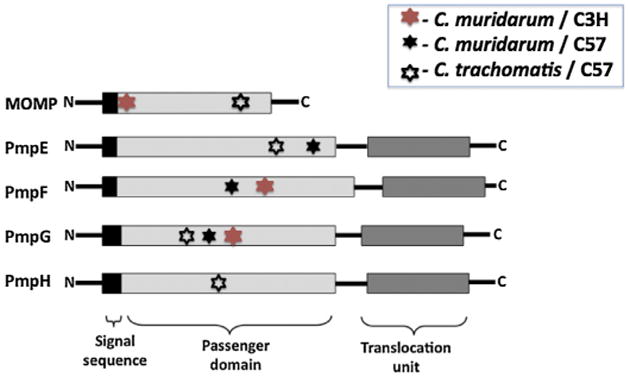
Location of MHC class II-bound peptides derived from C. muridarum (filled star) and C. trachomatis (open star) within the predicted passenger domains of the Pmp proteins identified using BMDCs from C57 (black star) or C3H (red star). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Determination of antigenicity of the Chlamydia proteins based on IFN-γ responses in C57BL/6 mice following C. trachomatis transcervical infection: Twelve proteins containing MHC class II-bound peptides identified by the immunoproteomic approach were evaluated for antigenicity in the C. trachomatis transcervical infection model. These included six overlapping source proteins between C. muridarum and C. trachomatis (CT143, CT375, CT424, RplF, PmpG, PmpE), two novel hypothetical source proteins (CT144, CT619), and PmpF, PmpH, MOMP and GroEL1 only identified in C. trachomatis (Table 2). To determine antigenicity in the context of in vivo infection, we performed IFN-γ ELISPOT assay using splenocytes from C57BL/6 mice following transcervical C. trachomatis infection since immunity in this model is due to IFN-γ secreting CD4 T cells [12]. Two and three weeks after transcervical inoculation of C trachomatis, mice were sacrificed and splenocytes were harvested and stimulated in vitro with the indicated recombinant protein. Media was used as a negative control and heat killed C. trachomatis serovar D elementary bodies (HK-EB) as a positive control. As shown in Fig. 3A, immune cells exposed to HK-EB developed the largest numbers of IFN-γ secreting cells where more than 1000 IFN-γ secreting cells were detected per 106 spleen cells. Cells stimulated with media as negative control showed nearly blank background levels indicating that IFN-γ secreting cells detected in the experimental system are Chlamydia antigen-specific. Relative to the negative control immune cells stimulated with individual Chlamydia protein exhibited positive IFN-γ responses (Fig. 3A). The IFN-γ responses in immune cells following stimulation with CT144, RplF (L6), CT169, PmpF, PmpH, MOMP and GroEL1 protein were strong (>50 spot forming cells [SFC] per 106 splenocytes); other antigens (PmpG, PmpE, CT143, CT375 and CT424) stimulated weak IFN-γ responses (<50 SFCs per 106 splenocytes). We conclude that outer membrane proteins such as MOMP, PmpF and PmpH are among the dominant C. trachomatis T cell antigens detected in this model system.
Fig. 3.
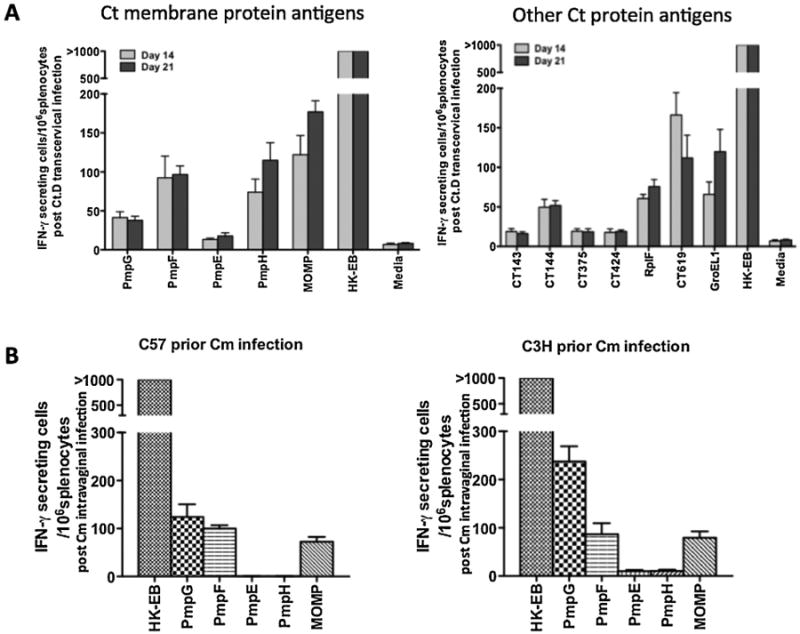
(A). T cell antigenicity of C. trachomatis (Ct) membrane protein antigens (left panel) and other identified T cell antigens (right panel) in immune C57BL/6 mice identified by IFN-γ ELISPOT assay. Mice were transcervically infected with 2 × 107 IFU live C. trachomatis serovar D. Fourteen or 21 days after infection, the splenocytes were harvested and stimulated in vitro for 20 h with 1 μg/ml individual Chlamydia proteins or 5 × 105 IFU/ml heat killed-EB as a positive control. The results represent the average of duplicate wells and are expressed as the means ± SEM of Chlamydia Ag-induced IFN-γ -secreting cells per 106 splenocytes for groups of five mice. (B). T cell antigenicity of C. muridarum (Cm) Pmps and MOMP in immune C57BL/6 (left panel) and C3H (right panel) mice identified by IFN-γ ELISPOT assay. Mice were intravaginally infected with 1500 IFU of C. muridarum. Three weeks later, splenocytes were harvested and stimulated in vitro for 20 h with 1 μg/ml individual Chlamydia proteins or heat killed-EB as a positive control.
Comparative antigenicity of the Chlamydia outer membrane proteins based on IFN-γ responses in C57BL/6 and C3H mice following C. muridarum intravaginal infection: After identifying source proteins from Chlamydia that generate MHC class II binding epitopes in two different genetic background, we determined antigenicity in the context of in vivo infection via IFN-γ ELISPOT assay using splenocytes from C57Bl/6 and C3H mice following intravaginal C. muridarum infection. As shown in Fig. 3B, immune cells exposed to HK-EB developed the largest number of IFN-γ secreting cells where more than 1000 IFN-γ secreting cells were detected per 106 spleen cells. The results demonstrate that IFN-γ responses in immune cells following stimulation with C. muridarum PmpG, PmpF and MOMP protein were immunodominant in both C57/BL6 and C3H.
3.3. Recombinant outer membrane protein vaccine accelerates clearance of C. trachomatis transcervical infection
We previously evaluated the protective efficacy of a C. muridarum outer membrane protein vaccine composed of Pmps and MOMP using the C. muridarum genital tract infection model in three different strains of mice and the multisubunit vaccine elicited protection as measured by accelerated clearance [20]. C. trachomatis does not infect the lower murine genital tract as efficiently as does C. muridarum and its clearance is mainly the result of innate immune response [21]. We therefore utilized the transcervical infection model established by Gondek et al. using C. trachomatis serovar D where CD4 T cells have been demonstrated to be essential to clearance [12]. We found that transcervical inoculation of C. trachomatis serovar D in C57BL/6 mice exhibited much higher Chlamydia loads detected by assaying homogenates of genital tract tissue in comparison to vaginal swabs (Fig. 4a). Transcervical infection by C. trachomatis serovar D was able to equally infect C57BL/6, Balb/c and C3H mice (Fig. 4b). Thus we used the transcervical infection mouse model to evaluate T cell vaccine efficiency against C. trachomatis serovar D challenge.
Fig. 4.
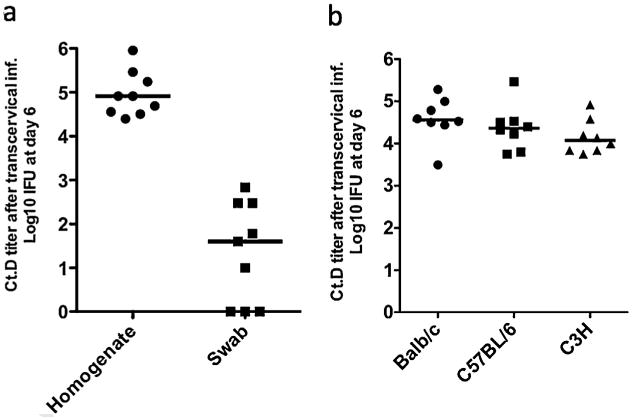
Transcervical infection of the upper genital tract with C. trachomatis serovar D. (a) Comparison of C. trachomatis serovar D (Ct.D) titer in homogenate of genital tract tissue with that of vaginal surface swab at day 6 post infection in C57BL/6 mice. Mice were infected with 2 × 107 IFU of C. trachomatis serovar D transcervically after two treatments with Depo-Provera at day 3 and day 10. Vaginal swabs and whole genital tracts were collected at day 6 post infection. Chlamydia titers in homogenates of genital tracts and in vaginal swab were measured by inclusion counts on Hela 229 cells. (b) Transcervical infection with C. trachomatis serovar D in Balb/c, C57BL/6 and C3H mice. Mice were infected with 2 × 107 IFU of C. trachomatis serovar D transcervically after two treatments with Depo-Provera at day 3 and day 10. Whole genital tracts were isolated and homogenized at day 6 post infection. Chlamydia titers in homogenates of genital tracts were measured by inclusion counts on Hela 229 cells.
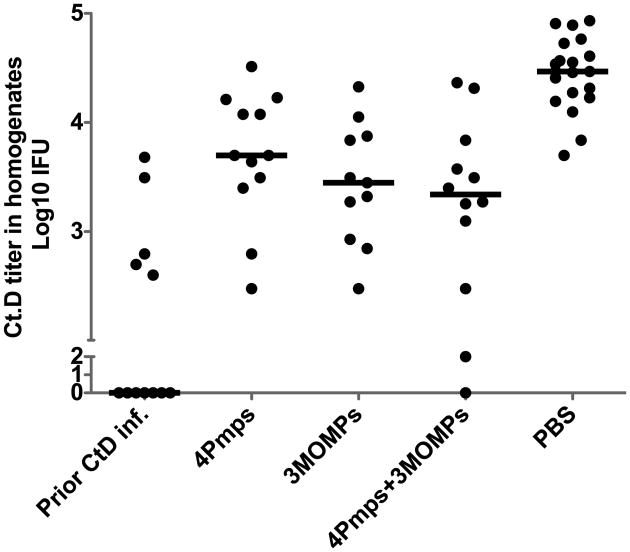
C. trachomatis serovars that cause human diseases can be clinically divided into three pathovars depending on their site of infection; trachoma (serovars A, B, Ba and C), sexually transmitted genital tract infection (serovars D through K) and lymphogranuloma venereum (serovars L1, L2 and L3). Pmps are an important component for chlamydial adhesion to host cells and sequences for six of the 9 Pmps correlate with the pathovar clustering [22]. MOMP is the serovar typing antigen, the major porin protein and the dominant antigen containing both B and T cell sites. Immunity directed to MOMP is determined by both serovar specific and serogroup specific antigenic determinants within the protein sequence [23]. We used four Pmps from C. trachomatis serovar D and MOMP from serovars D, J and F to represent serogroups B-complex, C-complex and GF-complex, respectively in a recombinant vaccine. We tested the efficacy of the multisubunit vaccine with the Th1 polarizing adjuvant DDA/MPL [24]. We tested both the Pmp/MOMP combination vaccine and the four Pmps and the three MOMPs as individual antigen components. Four weeks after the final immunization, C57BL/6 mice were challenged transcervically with C. trachomatis serovar D. Genital tracts were isolated and homogenized at day 6 post infection and C. trachomatis shedding was measured by inclusion counts on Hela 229 cells. Mice immunized with PBS were used as a negative control, and mice previously transcervically infected with C. trachomatis serovar D were used as a positive control. Strikingly mice previously infected with C. trachomatis serovar D were resistant to challenge infection, confirming that this model could be used to evaluate C. trachomatis vaccine candidates. All three vaccine groups demonstrated a significant reduction in C. trachomatis shedding compared to the PBS group (p< 0.001) (Fig. 5).
Fig. 5.
Vaccine elicited protection against C. trachomatis serovar D infection in C57BL/6 mice after immunization with different vaccine formulations with DDA/MPL adjuvant. Four weeks after the final immunization, mice were transcervically challenged with 2 × 107 IFU of C. trachomatis serovar D. Genital tracts were isolated and homogenized at day 6 post infection. Chlamydia titers in homogenates were measured by inclusion counts on Hela 229 cells. Mice immunized with PBS were used as a negative control, and mice previously infected with 2 × 107 IFU of C. trachomatis serovar D transcervically were used as a positive control. Horizontal bar represents median value for each group. All vaccine groups significantly reduced C. trachomatis shedding compared to the PBS group; p value <0.001.
4. Discussion
Identifying T cell antigens is of major importance to vaccinology for intracellular pathogens. The primary sequence of a protein is one determinant of T cell immunogenicity since the amino acid sequence influences antigen processing (protease susceptibility of the residues flanking the epitope) and determines anchoring of the peptide into MHC binding pockets. MHC class I and II differ in the importance of anchoring residues. In class I anchoring residues are critical to high affinity binding but for class II are less so [15]. Rather DM editing of the peptide complex is vital to high affinity class II binding [25]. Our data demonstrate that MHC allelic selection is clearly important for Chlamydia CD4 T cell antigens as seen when the C. muridarum immunoproteome is compared between C57BL/6 and C3H mice. Although 17 proteins yielded MHC class II binding peptides in C57BL/6 and 11 proteins generated binding peptides in C3H, only two C. muridarum protein (PmpG and PmpF) generated MHC class II binding epitopes of differing sequence in both strains of mice (Fig. 1). Furthermore 6 of the 9 antigenic orthologous proteins shared between the C. muridarum and C. trachomatis immunoproteome presented nearly identical peptide epitopes on the I-Ab allele of C57BL/6 mice (Table 3).
T cell immunogenicity also appears to be determined by properties intrinsic to the pathogen proteins such as cellular location, abundance, and kinetics of expression. Our results demonstrate that surface proteins are enriched among the Chlamydia T cell immunoproteome (Fig. 1). Of particular interest, MOMP and the polymorphic membrane family of proteins (PmpE, PmpF and PmpG) generated MHC class II binding epitopes at multiple sites within the sequence (Fig. 2) suggesting that these proteins are capable of presenting to the host immune system via multiple MHC binding epitopes. Pmps belong to a group of proteins known as type V autotransporters comprising an N-terminal signal sequence, a passenger domain and a translocation unit (Fig. 2) [26]. C. trachomatis and C. muridarum genomes encode nine different Pmps (PmpA to PmpI) [18,27]. A recent study reported by Becker and Hagemann showed that all nine C. trachomatis Pmps mediate adhesion to human epithelial cells [28].
Identification of four Pmps and MOMP as T cell antigens via an immunoproteomic analysis suggests that outer membrane proteins may have advantages over other groups of proteins in presenting to the immune system. This is supported by a recent quantitative proteomic analysis of C. trachomatis elementary bodies which revealed that the three Pmps we identified (PmpE, PmpG and PmpH) constituted 61% of the total Pmp protein abundance [29] and MOMP is already known to constitute over 60% of the total outer membrane protein abundance [8]. Thus high abundance proteins as well as outer membrane localization may play a role in favoring presentation.
PmpG is the most protective C. muridarum T cell antigen we identified via our immunoproteomic approach and two recent studies further validated the immunodominance of PmpG in the murine model [30,31]. These studies demonstrated that the PmpG epitope can be detected on splenic antigen presenting cells for at least 6 months after clearance of primary genital tract infection and a large fraction of the T cells activated during Chlamydia infection are PmpG-specific suggesting that PmpG in particular is intrinsically immunogenic.
In conclusion our results demonstrate both MHC class II allelic selection and intrinsic antigenic protein features contribute to T cell antigenicity of Chlamydia outer membrane proteins. MHC allele specificity appears to be more important than intrinsic features of antigenic proteins in Chlamydia perhaps because so few proteins enter the class II processing and presentation pathway from the Chlamydia vacuole. The shared T cell antigens we identified, in particular the Pmps and MOMP are likely relevant to Chlamydia immunobiology both in murine and human models and constitute potential C. trachomatis vaccine candidates.
Acknowledgments
This work was supported by a National Institutes of Health Grant R01AI076483 (to RCB) and a Canadian Institutes of Health Research operating grant MOP-77688 (to LJF). Mass spectrometry infrastructure used here was supported by the Canada Foundation for Innovation, the British Columbia Knowledge Development Fund and the BC Proteomics Network. LJF is the Canada Research Chair in Quantitative Proteomics.
References
- 1.Brunham RC, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol. 2005;5:149–61. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 2.Karunakaran KP, Rey-Ladino J, Stoynov N, Berg K, Shen C, Jiang X, et al. Immunoproteomic discovery of novel T cell antigens from the obligate intracellular pathogen Chlamydia. J Immunol. 2008;180:2459–65. doi: 10.4049/jimmunol.180.4.2459. [DOI] [PubMed] [Google Scholar]
- 3.Steinman RM, Pope M. Exploiting dendritic cells to improve vaccine efficacy. J Clin Invest. 2002;109:1519–26. doi: 10.1172/JCI15962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Jong A. Contribution of mass spectrometry to contemporary immunology. Mass Spectrom Rev. 1998;17:311–35. doi: 10.1002/(SICI)1098-2787(1998)17:5<311::AID-MAS1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 5.Yu H, Karunakaran KP, Kelly I, Shen C, Jiang X, Foster LJ, et al. Immunization with live and dead Chlamydia muridarum induces different levels of protective immunity in a murine genital tract model correlation with MHC class II peptide presentation and multifunctional Th1 cells. J Immunol. 186:3615–21. doi: 10.4049/jimmunol.1002952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu H, Jiang X, Shen C, Karunakaran KP, Brunham RC. Novel Chlamydia muridarum T cell antigens induce protective immunity against lung and genital tract infection in murine models. J Immunol. 2009;182:1602–8. doi: 10.4049/jimmunol.182.3.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H, Jiang X, Shen C, Karunakaran KP, Jiang J, Rosin NL, et al. Chlamydia muridarum T-cell antigens formulated with the adjuvant DDA/TDB induce immunity against infection that correlates with a high frequency of gamma interferon (IFN-gamma)/tumor necrosis factor alpha and IFN-gamma/interleukin-17 double-positive CD4+ T cells. Infect Immun. 78:2272–82. doi: 10.1128/IAI.01374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldwell HD, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–76. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su H, Messer R, Whitmire W, Fischer E, Portis JC, Caldwell HD. Vaccination against chlamydial genital tract infection after immunization with dendritic cells pulsed ex vivo with nonviable Chlamydiae. J Exp Med. 1998;188:809–18. doi: 10.1084/jem.188.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox A, Huczko EL, Engelhard VH, Shabanowitz J, et al. The application of mass spectrometry to the analysis of peptides bound to MHC molecules. In: Fernandez N, Butcher G, editors. MHC—a practical approach. 1997. pp. 142–60. [Google Scholar]
- 11.Ishihama Y, Rappsilber J, Mann M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J Proteome Res. 2006;5:988–94. doi: 10.1021/pr050385q. [DOI] [PubMed] [Google Scholar]
- 12.Gondek DC, Olive AJ, Stary G, Starnbach MN. CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. J Immunol. 2012;189:2441–9. doi: 10.4049/jimmunol.1103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bilenki L, Wang S, Yang J, Fan Y, Joyee AG, Yang X. NKT cell activation promotes Chlamydia trachomatis infection in vivo. J Immunol. 2005;175:3197–206. doi: 10.4049/jimmunol.175.5.3197. [DOI] [PubMed] [Google Scholar]
- 14.Ioannou XP, Griebel P, Hecker R, Babiuk LA, van Drunen Littel-van den Hurk S. The immunogenicity and protective efficacy of bovine herpesvirus 1 glycoprotein D plus Emulsigen are increased by formulation with CpG oligodeoxynucleotides. J Virol. 2002;76:9002–10. doi: 10.1128/JVI.76.18.9002-9010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chicz RM, Urban RG, Gorga JC, Vignali DA, Lane WS, Strominger JL. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barat S, Willer Y, Rizos K, Claudi B, Maze A, Schemmer AK, et al. Immunity to intracellular Salmonella depends on surface-associated antigens. PLoS Pathog. 2012;8:e1002966. doi: 10.1371/journal.ppat.1002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu H, Karunakaran KP, Jiang X, Shen C, Andersen P, Brunham RC. Chlamydia muridarum T cell antigens and adjuvants that induce protective immunity in Mice. Infect Immun. 80:1510–18. doi: 10.1128/IAI.06338-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28:1397–406. doi: 10.1093/nar/28.6.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rey-Ladino J, Jiang X, Gabel B, Shen C, Brunham RC. Survival of Chlamydia muridarum within dendritic cells (DCs) Infect Immun. 2007 doi: 10.1128/IAI.01618-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Karunakaran KP, Jiang X, Brunham RC. Evaluation of a multisubunit recombinant polymorphic membrane protein and major outer membrane protein T cell vaccine against Chlamydia muridarum genital infection in three strains of mice. Vaccine. 2014;32:4672–80. doi: 10.1016/j.vaccine.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sturdevant GL, Caldwell HD. Innate immunity is sufficient for the clearance of Chlamydia trachomatis from the female mouse genital tract. Pathog Dis. 2014;72:70–3. doi: 10.1111/2049-632X.12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borges V, Nunes A, Ferreira R, Borrego MJ, Gomes JP. Directional evolution of Chlamydia trachomatis towards niche-specific adaptation. J Bacteriol. 2012;194:6143–53. doi: 10.1128/JB.01291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang YX, Stewart S, Joseph T, Taylor HR, Caldwell HD. Protective monoclonal antibodies recognize epitopes located on the major outer membrane protein of Chlamydia trachomatis. J Immunol. 1987;138:575–81. [PubMed] [Google Scholar]
- 24.Yu H, Karunakaran KP, Jiang X, Shen C, Andersen P, Brunham RC. Chlamydia muridarum T cell antigens and adjuvants that induce protective immunity in mice. Infect Immun. doi: 10.1128/IAI.06338-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sant AJ, Chaves FA, Jenks SA, Richards KA, Menges P, Weaver JM, et al. The relationship between immunodominance, DM editing, and the kinetic stability of MHC class II:peptide complexes. Immunol Rev. 2005;207:261–78. doi: 10.1111/j.0105-2896.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 26.Bavoil PM, Wyrick PB. Chlamydia: genomics and pathogenesis. Horizon Biosci. 2006 [Google Scholar]
- 27.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, et al. Genome sequence of an obligate intracellular pathogen of humans:Chlamydia trachomatis. Science. 1998;282:754–9. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 28.Hagemann J, B E. All subtypes of the PMP adhesion family are implicated in chlamydial virulence and show species-specific function. 13th International symposium on human chlamydial infections. 2014:137. doi: 10.1002/mbo3.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saka HA, Thompson JW, Chen YS, Kumar Y, Dubois LG, Moseley MA, et al. Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol Microbiol. doi: 10.1111/j.1365-2958.2011.07877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li LX, McSorley SJ. B cells enhance antigen-specific CD4 T cell priming and prevent bacteria dissemination following Chlamydia muridarum genital tract infection. PLoS Pathog. 2013;9:e1003707. doi: 10.1371/journal.ppat.1003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson RM, Yu H, Kerr MS, Slaven JE, Karunakaran KP, Brunham RC. PmpG303-311, a protective vaccine epitope that elicits persistent cellular immune responses in Chlamydia muridarum-immune mice. Infect Immun. 2012;80:2204–11. doi: 10.1128/IAI.06339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]