Abstract
MLL -rearrangements generate MLL-fusion proteins that bind DNA and drive leukemogenic gene expression. This gene expression program is dependent on the histone 3 lysine 79 (H3K79) methyltransferase DOT1L, and small molecule DOT1L inhibitors show promise as therapeutics for these leukemias. However, the mechanisms underlying this dependency are unclear. We conducted a genome-scale RNAi screen and found that the histone deacetylase SIRT1 is required for the establishment of a heterochromatin-like state around MLL-fusion target genes after DOT1L inhibition. DOT1L inhibits chromatin localization of a repressive complex composed of SIRT1 and SUV39H1, thereby maintaining an open chromatin state with elevated H3K9 acetylation and minimal H3K9 methylation at MLL-fusion target genes. Furthermore, the combination of SIRT1 activators and DOT1L inhibitors shows enhanced activity against MLL-rearranged leukemia cells. These results indicate that the dynamic interplay between chromatin regulators controlling activation and repression of gene expression could provide novel opportunities for combination therapy.
INTRODUCTION
Eukaryotic genomes are intricately organized into chromatin, which is composed of genomic DNA wrapped around nucleosomes and packaged in a highly orchestrated fashion. One mode of regulation that controls chromatin organization is the covalent modification (such as acetylation and methylation) of specific amino acid residues in histones found within the nucleosomes1. These posttranslational histone modifications are known to modulate the local structure of chromatin and influence gene expression by regulating the accessibility of gene loci to transcriptional machinery2. For example, acetylation of histone H3 lysine 9 (H3K9) is enriched in more accessible chromatin regions designated for active gene transcription. On the other hand, methylation of H3K9 is associated with tightly packaged and repressed genomic regions known as heterochromatin3. The dynamic remodeling of histone modifications by nuclear proteins including histone acetyltransferases, deacetylases, methyltransferases as well as demethylases, ensures appropriate gene expression during cellular division and organismal development.
While most of the histone modifications are found on the exposed histone N-terminal tails, methylation of H3K79 is present on the surface of the highly structured nucleosome core4. Thus far, the only known enzyme that catalyzes methylation of H3K79 is DOT1L (Disruptor Of Telomeric-silencing 1 Like), which was initially identified in S. cerevisiae5,6. Recent genome-wide analysis of the mammalian epigenome have shown that H3K79 methylation is broadly associated with actively transcribed genes and is not frequently found in intergenic regions7–9. The interaction between DOT1L and a number of nuclear protein complexes involved in the control of gene expression further supports the relationship between H3K79 methylation and gene expression10–17. However, recent studies have shown that genetic inactivation of Dot1L and loss of all H3K79 methylation in mouse models does not lead to widespread collapse of transcription8. Thus, the exact biological function of DOT1L and H3K79 methylation in the control of mammalian gene expression remains unclear.
An essential role for DOT1L and H3K79 methylation has been documented in leukemias with rearrangement of the Mixed Lineage Leukemia gene (MLL-r), which accounts for approximately 5–10% of human acute leukemia cases and is generally associated with poor prognosis18. Studies using mouse genetic models have shown that leukemias driven by MLL-r are highly dependent on Dot1L for leukemia initiation and maintenance, whereas many other types of transformed hematopoietic cells are insensitive to complete loss of Dot1L and H3K79 methylation8,19–23. Epigenomic studies revealed that MLL-fusion targets (genes directly bound by MLL-fusion proteins) are associated with aberrantly high levels of H3K79 dimethylation (H3K79me2) in MLL-r leukemias24–26. These MLL-fusion targets include developmental genes such as MEIS1 and HOXA cluster genes, which are known to induce leukemia if ectopically expressed27. Since DOT1L interacts with multiple MLL-translocation partners such as AF9, ENL, and AF10, it has been suggested that these MLL-fusion proteins may drive ectopic gene expression through recruiting excessive DOT1L activity to their target loci10–16,28. These previous discoveries have established a foundation for disease-specific epigenetic therapies against the highly malignant MLL-r leukemias. Indeed, small molecular inhibitors for DOT1L (e.g. EPZ4777, EPZ5676 and others) have been developed, one of which is currently undergoing Phase I clinical trials29–34. Despite the promising progress toward DOT1L inhibitor therapy for individuals with MLL-r leukemia, the exact mechanisms by which DOT1L and H3K79 methylation maintain leukemic gene expression remain elusive. Understanding the unique subordination of the MLL-r leukemic program to DOT1L could not only uncover additional therapeutic opportunities, but also provide information regarding the fundamental function of DOT1L and H3K79 methylation in mammalian gene regulation.
Here, we conducted a genome-scale RNAi screen in murine MLL-AF9 leukemia cells engineered to conditionally excise Dot1L so we could identify genes that, when suppressed, would rescue Dot1L dependence. This unbiased approach discovered Sirtuin 1 (Sirt1), a mammalian orthologue of the yeast Sir2 histone deacetylase35,36, to be a critical “antagonist of Dot1L” in MLL-r leukemia. Mechanistically, we found that inhibition of DOT1L in MLL-fusion leukemias leads to a Sirt1-dependent decrease of H3K9 acetylation, gain of H3K9 methylation and loss of chromatin accessibility at MLL-fusion target genes. Thus Sirt1 is necessary for acquisition of a repressed chromatin state upon Dot1L inhibition. This led us to demonstrate the potential for combination of SIRT1 activators and DOT1L inhibitors as a novel therapeutic strategy against difficult-to-treat MLL-r leukemias.
RESULTS
Genome-scale RNAi screen identifies Sirt1 as an “antagonist of Dot1L”
We hypothesized that there are other effectors required for the decrease in MLL-fusion driven gene expression that occurs upon Dot1L inhibition, and that genetic suppression of these effectors would reverse the antiproliferative effect of DOT1L inactivation in MLL-fusion leukemias. To identify these negative regulators of the DOT1L pathway, or “antagonists of Dot1L,” we conducted a pooled screen by introducing a mouse genome-scale shRNA library (containing 92,425 hairpins targeting 16,924 mouse genes)37,38 into Dot1Lfl/fl-MLL-AF9 leukemic cells8 harboring tamoxifen-inducible Cre recombinase (CreER) (Fig. 1a). We confirmed the bi-allelic excision of Dot1L and loss of H3K79me2 in these cells following induction of Cre recombinase activity by tamoxifen treatment (Fig. 1b). We then assessed the relative frequencies of each integrated shRNA sequence before and after Dot1L gene excision by massively parallel sequencing (Hi-seq). Since inactivation of Dot1L induced myeloid differentiation and severely inhibited proliferation of MLL-AF9 leukemic cells (Fig. 1c,d), shRNA constructs that rendered a growth or survival advantage to these cells were expected to be enriched in the screen after tamoxifen-induced Dot1L deletion. Analyses that compared hairpin frequency on day 9 and day 0 identified 934 significantly enriched shRNA constructs (more than 4-fold increase; p ≤ 0.05) after Dot1L deletion (Fig. 1e and Supplementary Table 3). Remarkably, we found three shRNAs targeting Sirtuin 1 (Sirt1) in this top 1% of enriched hairpins, making Sirt1 our leading candidate “antagonist of Dot1L” in MLL-AF9 leukemia (additional candidates are shown in Supplementary Fig. 1).
Figure 1.
Genome-scale RNAi screen for “antagonists of Dot1L” in MLL-AF9 leukemia. (a) Schematic outline of a genome-scale shRNA library screen coupled with high-throughput sequencing (HiSeq) in mouse MLL-AF9 leukemia cells harboring Dot1lfl/fl alleles and tamoxifen-inducible Cre recombinase (CreER). (b) Genotyping PCR for Dot1L engineered allele and immunoblot for histone H3 modifications in MLL-AF9-Dot1lfl/fl-CreER leukemia cells after tamoxifen-induced Dot1L-excision. (c) Wright-Giemsa stain of MLL-AF9-Dot1lfl/fl-CreER leukemia cells before and after tamoxifen treatment. Scale bar, 20 μm. (d) Cell number expansion of Dot1lfl/fl-CreER (red) and Dot1lwt/wt-CreER (blue) MLL-AF9 leukemia cells cultured in tamoxifen. (e) Volcano plot depicts the changes in representation (x-axis) and significance (y-axis) of each shRNA construct in the screen before versus after tamoxifen-induced Dot1L deletion. Total library (gray; 92,425 shRNA), enriched shRNA (red; more than 4-fold increase and p value ≤ 0.05 in the six replicates; 934 shRNA) and sh-Sirt1 (blue; five shRNA) are highlighted. (f) Relative blast colony number from sh-LUC or sh-Sirt1 transduced MLL-AF9-Dot1lfl/fl-CreER leukemic cells cultured in ethanol (green) or tamoxifen (red). The numbers of blast colonies were normalized to the total colony count in control cultures transduced with the same shRNA construct. Data represent the observed values and mean ± s.d. of (d) three independent experiments and (f) four replicates. *P < 0.01 using Student’s t-test.
Sirt1 mediates silencing of the MLL-AF9 leukemic program upon Dot1L inactivation
To validate our genome-scale shRNA library screen results, we assessed whether the Sirt1 shRNAs that were selected for in the screen also suppressed Sirt1 expression. We also performed colony-forming assays. We found that the three shRNAs selected for in the screen suppressed Sirt1 expression and depletion of Sirt1 by these individual shRNAs (sh-Sirt1) maintained more MLL-AF9 driven blast-like colonies after Dot1L deletion, as compared to the control cultures transduced with sh-LUC (Fig. 1f and Supplementary Fig. 1c, d). Of note, depletion of Sirt1 alone did not influence the proliferation and blast-like colony potential of these leukemic cells. Additionally, we subjected the MLL-AF9 leukemia cells to EPZ4777, a selective small molecular DOT1L inhibitor29, and found that suppression of Sirt1 in MLL-AF9 leukemic cells reduced their sensitivity to DOT1L inhibition (Fig. 2a,b and Supplementary Fig. 2). Similarly, small molecule inhibitors of SIRT1 including Ex527 and suramin39 desensitized MLL-AF9 leukemic cells to Dot1L inhibition, suggesting that Sirt1’s enzymatic activity is important for the suppression of MLL-AF9 leukemic cells caused by DOT1L inhibition (Fig. 2c). On the other hand, forced expression of Sirt1 by retroviral transduction re-sensitized the Sirt1 knockdown cells to EPZ4777 treatment (Fig. 2d,e).
Figure 2.
Sirt1 mediates the response of MLL-AF9 leukemia cells to DOT1L inhibitor EPZ4777. (a,c,h,i) Effect of EPZ4777 on the proliferation of mouse MLL-AF9 leukemia cells transduced with (a) sh-Sirt1 (red) or sh-LUC (green), (h) MSCV-puro-Meis1 (red), Hoxa7 (blue), or empty vector (green), and (i) MSCV-puro-Meis1 plus MSCV-ires-Tomato-Hoxa7 (red) or dual empty vectors (green), as well as (c) co-treated with Ex527 (red), suramin (blue) or DMSO (green). (b,d) Immunoblot of (b) Sirt1, H3K79me2, histone H3 and tubulin in MLL-AF9 leukemic cells transduced with sh-LUC or sh-Sirt1 and cultured in EPZ4777, and (d) Sirt1 and tubulin in sh-Sirt1 transduced MLL-AF9 cells further infected with MSCV-ires-Tomato empty vector (MIT-Vec) or Sirt1 (MIT-Sirt1) virus. (e) Relative number of the cells described in (d) cultured in EPZ4777 (red) or DMSO (green). (f) Microarray and GSEA analyses showing changes in expression of “EPZ4777_down gene set” (978 genes; Supplementary Table 3) in sh-LUC transduced MLL-AF9 cells cultured in EPZ4777 versus DMSO (left panel), as well as sh-Sirt1 versus sh-LUC transduced MLL-AF9 cells cultured in EPZ4777 (right panel). Heatmaps showing genes comprising the early leading edge (top 50 genes) of the GSEA plots. (g) RT-qPCR of Hoxa7 and Meis1 in sh-LUC or sh-Sirt1 transduced MLL-AF9 leukemic cells cultured in EPZ4777 (red) or DMSO (green). Cells were cultured in the presence of EPZ4777 or DMSO for (f,g) 6 days and (a,c,e,h,i) 9 days, respectively. Data represent the observed values and mean ± s.d. of (a,c,e,h,i) three replicates, and (g) three independent experiments. *P < 0.05; **P < 0.01 to control group using Student’s t-test.
Genes directly regulated by the MLL-AF9 fusion proteins are highly dependent on Dot1L for continued expression8. Therefore, we assessed whether depletion of Sirt1 protects MLL-AF9 cells through maintaining the expression of MLL-AF9 driven genes after Dot1L inhibition. We used microarrays and Gene Set Enrichment Analysis (GSEA) and found that the expression of the EPZ4777_down gene set (978 genes; Supplementary Table 3)29 was significantly (P > 0.01) maintained in EPZ4777 treated cells transduced with sh-Sirt1 (Fig. 2f). Of interest, the top two rescued MLL-AF9 target genes (129 genes previously found to be bound by MLL-AF9 fusion protein in mouse MLL-AF9 leukemia; Supplementary Table 3)8 in the GSEA plot were Hoxa7 and Meis1. Reverse transcription and quantitative PCR (RT-qPCR) confirmed that both Hoxa7 and Meis1 mRNA levels were drastically reduced upon Dot1L inhibition, whereas the expression of these two genes was significantly maintained by knockdown of Sirt1 in combination with DOTL1 inhibition (Fig. 2g). Furthermore, we validated that ectopic expression of either Hoxa7 or Meis1 (or the combination) mimicked the effect of Sirt1-knockdown on reducing the sensitivity of MLL-AF9 leukemic cells to DOT1L inhibitor (Fig. 2h and Supplementary Fig. 3). These data validate Sirt1 as a critical component of the cellular machinery that is essential for suppression of the MLL-AF9 leukemic program upon DOTL1 inhibition.
Sirt1 is required for H3K9 deacetylation in response to Dot1L inhibition
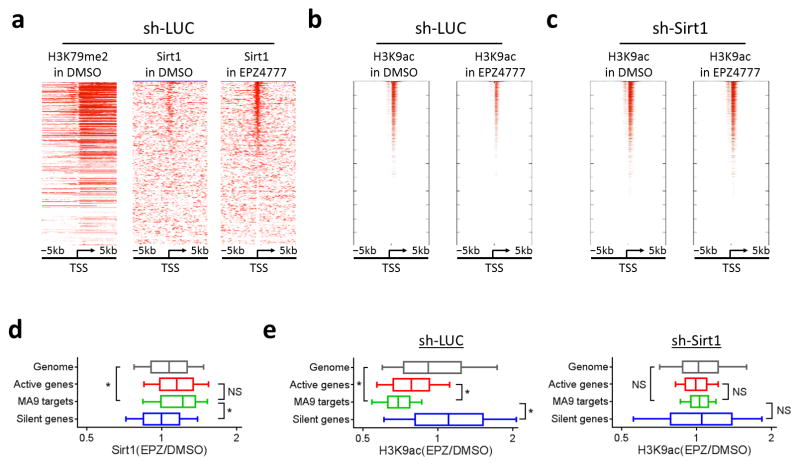
To investigate the mechanisms by which Sirt1 participates in silencing gene expression after Dot1L inhibition, we performed chromatin immunoprecipitation coupled with high throughput sequencing (ChIP-seq) for Sirt1 and H3K79me2 in mouse MLL-AF9 leukemia cells. We observed that EPZ4777 treatment increased the Sirt1 protein occupancy around the transcriptional start site, or TSS, of about one third of annotated genes in the mouse genome (Fig 3a and Supplementary Figs. 4–6). Interestingly, genes that were occupied by the Sirt1 protein after Dot1L inhibition highly correlated with the initial levels of H3K79me2 (positively correlated with actively expressed genes) around their promoter proximal regions. Because Sirt1 is able to deacetylate multiple lysine positions in histones, we further performed ChIP-seq for H3K9ac, a chromatin substrate of Sirt1 that is highly associated with the TSS of actively transcribed genes, in mouse MLL-AF9 leukemia cells. We found that the distribution of H3K9ac at the active TSS overlapped with the loci where the recruitment of Sirt1 was observed after suppression of Dot1L (Fig. 3b). Concomitant with the increased Sirt1 occupancy on the active TSS, we observed reduced H3K9ac on these loci after DOT1L inhibitor treatment (Fig. 3b). Importantly, while depletion of Sirt1 minimally affected the level and distribution of H3K9ac under normal conditions, it completely blocked the loss of H3K9ac from the active TSS induced by DOT1L inhibitor treatment (Fig. 3c). These results suggest that a function of Dot1L activity is to antagonize Sirt1-dependent deacetylation of H3K9 on the actively expressed genes.
Figure 3.
Sirt1 localizes to active genes and mediates deacetylation of H3K9 in response to Dot1L inhibition. (a–c) Heatmaps showing ChIP-seq signal of (a) H3K79me2 and Sirt1, and (b,c) H3K9ac at TSS ± 5 kb regions for all genes in MLL-AF9 leukemic cells transduced with (a,b) sh-LUC or (c) sh-Sirt1. Genes are ranked according to ChIP-seq signal of Sirt1 in EPZ4777 from high (top) to low (bottom). (d,e) Boxplots showing changes in ChIP-seq signal of (d) Sirt1 and (e) H3K9ac at TSS ± 2 kb regions of genome (gray; 18,420 genes), active genes (red; 4,560 genes), MLL-AF9 targets (green; 129 genes) and silent genes (blue; 4,560 genes) in mouse MLL-AF9 leukemic cells. Cells were cultured in DMSO or EPZ4777 for 6 days. Data represent mean ± s.d. NS, not significant; *P < 0.001 to MLL-AF9 targets using Welch’s t-test.
To investigate if Sirt1 is part of the mechanism for selective suppression of MLL-fusion driven gene expression after Dot1L inhibition, we compared Sirt1 occupancy after DOT1L inhibition at the MLL-AF9 targets (129 genes; Supplementary Table 3)8, to Sirt1 occupancy after DOT1L inhibition at all actively expressed or silent genes (4,560 genes each; determined based on the Affymetrix array) in mouse MLL-AF9 leukemic cells (Fig. 3d). We observed a similar level of Sirt1 recruitment to MLL-AF9 targets as compared to the active genes upon EPZ4777 treatment thus there does not appear to be enhanced recruitment of Sirt1 to MLL-AF9 targets as compared to the larger group of active genes. However, when we compared the deacetylation of H3K9 upon Dot1L-inhibition, we identified a stronger loss of H3K9ac at the MLL-AF9 target genes compared to the active gene set (Fig. 3e). Moreover, the removal of H3K9ac at MLL-AF9 targets was dependent on the presence of Sirt1, as knockdown of Sirt1 completely blocked the loss of H3K9ac at these TSS normally induced by DOT1L inhibitor treatment (Fig. 3e). These data show that the MLL-AF9 target genes are more sensitive to Sirt1-mediated histone deacetylation than other actively expressed genes.
Unique H3K9 epigenomic signature at MLL-AF9 target genes
Our data identified a significantly stronger loss of H3K9ac at the MLL-AF9 target genes compared to other active gene loci after Dot1L inhibition. Interestingly, the majority of MLL-AF9 target genes in these leukemic cells possessed not only the highest H3K79me2 levels in the genome as previously reported8, but they also showed elevated H3K9ac levels relative to other expressed loci (Fig. 4a,c,d and Supplementary Figs. 7). We also note that H4K16ac (another known Sirt1 substrate) showed the same elevated pattern as H3K9ac (Supplementary Fig. 8). On the contrary, the MLL-AF9 target gene set exhibited a scattered distribution of these two histone marks among expressed genes in normal Lin−Sca1+cKit+ (LSK) hematopoietic stem/progenitor cells (Fig. 4b). Because the leukemic cells used in this study were established through transformation of mouse LSK cells by MLL-AF9, the current analyses suggest that the unique H3K9 epigenomic signature observed at the MLL-AF9 targets in these leukemias is attributable to the presence of MLL-fusion oncoprotein at those loci. Indeed, we found that the genomic regions showing exaggerated H3K9achi status overlapped remarkably with the previously defined MLL-AF9 occupied peaks (Fig. 4c)8. Similarly, the increased H3K9ac level at MLL-AF9 target loci was confirmed in human MLL-AF9 cell line models (Fig. 4e–g). In fact, the two most well-studied MLL-fusion target loci, HOXA cluster and MEIS1, represent two of the most enlarged H3K9achi domains in these MLL-r leukemias (Fig. 4h).
Figure 4.
Unique H3K9 epigenomic signature at MLL-AF9 bound gene loci in MLL-fusion leukemia. (a,b) Scatterplots showing ChIP-seq signals for H3K79me2 (x-axis) and H3K9ac (y-axis) in mouse (a) MLL-AF9 leukemic cells, and (b) LSK cells sorted from normal mouse bone marrow. Hoxa cluster genes and Meis1 are highlighted in black circles. (c,h) Screen shots showing ChIP-seq profiles of MLL-AF9 fusion protein (black), H3K79me2 (blue) and H3K9ac (red) at select MLL-AF9 bound target (HOXA cluster and MEIS1), active gene (GAPDH) and silent gene (HBB/OLFR) loci in (c) mouse and (h) human MLL-AF9 leukemic cell lines. The core occupied regions for MLL-AF9 fusion protein in mouse MLL-AF9 leukemia are highlighted (c; green-dashed box). (d–g) Boxplots showing ChIP-seq signal of H3K9ac in (d) mouse and (e–g) human MLL-AF9 leukemic cells including (e) Molm13, (f) Nomo1 and (g) MonoMac6 cells. (a,b,d–g) Data showing ChIP-seq signals of H3K79me2 or H3K9ac at TSS ± 2 kb regions of genome (gray; 18,240 genes), active genes (red; 4,560 genes), MLL-AF9 targets (green; 129 genes) and silent genes (blue; 4,560 genes). (d–g) Data represent mean ± s.d. *P < 0.001 to MLL-AF9 targets using Welch’s t-test.
Involvement of H3K9 methylation by Suv39h1 in Sirt1-mediated silencing of leukemic genes
It has been reported that mammalian SIRT1 physically interacts with other proteins that modify histones and is found in complexes that mediate chromatin silencing. We therefore investigated dimethylation of histone 3 lysine 9 (H3K9me2) and trimethylation of histone 3 lysine 27 (H3K27me3), both of which have been connected to Sirt1-associated gene silencing40–42. Concomitant with the reduction of H3K9ac, inhibition of Dot1L increased the amount of both H3K9me2 and H3K27me3 around the TSS of MLL-AF9 bound genes, suggesting potential involvement of multiple epigenetic repressive mechanisms in silencing the MLL-fusion target loci (Fig. 5a). Interestingly, depletion of Sirt1 completely blocked the removal of H3K9ac and selectively inhibited the accumulation of H3K9me2 at MLL-AF9 targets after Dot1L inhibition, whereas the gain of H3K27me3 at those loci remained largely unaffected (Fig. 5a). Thus Sirt1 function is more critical for H3K9 methylation than for H3K27 methylation subsequent to Dot1L inhibition at the MLL-AF9 targets.
Figure 5.
Methylation of H3K9 by Suv39h1 is involved in Sirt1-mediated silencing of the MLL-AF9 leukemic program upon suppression of Dot1L. (a) Scatterplots and boxplots showing changes in ChIP-seq signals for H3K9ac (x-axis; both panels), H3K9me2 (y-axis; left panel) and H3K27me3 (y-axis; right panel) at TSS ± 2 kb regions of genome (gray; 18,240 genes) and MLL-AF9 targets (129 genes) in sh-LUC (red) or sh-Sirt1 (blue) transduced mouse MLL-AF9 leukemic cells cultured in EPZ4777 versus DMSO. (b) Venn diagram showing the overlap genes in “SIRT1-interacting proteins” and “candidate antagonists of Dot1L”. (c) Effect of EPZ4777 on the proliferation of MLL-AF9 leukemic cells transduced with sh-LUC (green), sh-Sirt1 (red) or sh-Suv39h1 (blue). (d) Immunoblot of Suv39h1, Sirt1, H3K9me2 and histone H3 in MLL-AF9 leukemic cells transduced with sh-LUC or sh-Suv39h1. (e,g,h) ChIP-qPCR of (e) H3K9me2, (g) Sirt1, and (h) Suv39h1 for Hoxa7 and Meis1 gene TSS regions in MLL-AF9 leukemic cells transduced with sh-LUC, sh-Sirt1, or sh-Suv39h1. (f) RT-qPCR of Hoxa7 and Meis1 in sh-LUC or sh-Suv39h1 transduced MLL-AF9 leukemic cells. (i) Bar-graph and boxplot showing changes in ATAC-seq signals at TSS ± 2 kb regions of MLL-AF9 targets (129 genes) in sh-LUC (green), sh-Sirt1 (red) and sh-Suv39h1 (blue) transduced MLL-AF9 leukemic cells cultured in EPZ4777 versus DMSO. Individual MLL-AF9 target genes in bar-graph are ranked according to ATAC-seq (EPZ/DMSO) ratio in sh-LUC cells from high (left) to low (right). Cells were cultured in the presence of EPZ4777 or DMSO for (a,e–i) 6 days and (c) 9 days, respectively. Data represent the observed values and mean ± s.d. of (c,e) two and (g,h) four replicates, and (f) three independent experiments. NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001 using (a,i) Welch’s t-test and (c, e–h) Student’s t-test.
To identify effectors responsible for Sirt1-dependent H3K9 methylation and gene silencing, we hypothesized that knockdown of such effectors would protect MLL-AF9 target gene expression after inactivation of Dot1L. We overlapped a list of SIRT1 interacting proteins (Supplementary Table 3) with the “candidate antagonists of Dot1L,” which we defined as genes having two or more shRNAs enriched more than 2-fold in the top 10 % of the shRNA library screen described in Fig. 1. Remarkably, out of the 17 overlapping candidates, one of the genes, Suv39h1, encodes a H3K9 methyltransferase (Fig. 5b). Suppression of Suv39h1 desensitized MLL-AF9 leukemic cells to Dot1L inhibition, thus phenocopying Sirt1 depletion (Fig. 5c). Suppression of Suv39h1 resulted in a slight reduction of global H3K9me2 in MLL-AF9 leukemic cells and decreased the basal level of H3K9me2 at the TSS of Hoxa7 and Meis1 (Fig. 5d,e). Importantly, depletion of Suv39h1 completely blocked the accumulation of H3K9me2 (Fig. 5e) around the TSS of Hoxa7 and Meis1 and significantly maintained the expression (Fig. 5f) of these two MLL-AF9 targets after Dot1L inhibition. Thus, Suv39h1 is important for H3K9 methylation and silencing of the MLL-AF9 driven leukemic genes upon suppression of Dot1L. Furthermore, we found concomitant increases of both Sirt1 and Suv39h1 occupancy at Hoxa7 and Meis1 loci after Dot1L suppression (Fig. 5g,h and Supplementary Fig. 4). Remarkably, the localization of Suv39h1 to these two MLL-AF9 target loci was dependent on the presence of Sirt1 as suppression of Sirt1 diminished Suv39h1 binding to chromatin after Dot1L inhibition (Fig. 5h). Because H3K9 methylation is frequently associated with chromatin compaction and heterochromatin, we next investigated whether Sirt1 and Suv39h1 affect the accessibility of chromatin by ATAC-seq43. While Dot1L inhibition significantly reduced transposase accessibility for the majority of MLL-AF9 targets (Fig. 5i and Supplementary Figs. 9 and 10), knockdown of either Sirt1 or Suv39h1 sustained the accessibility at these loci (Fig. 5i). Collectively our data demonstrate that upon inhibition of Dot1L, Sirt1 is required for Suv39h1 chromatin localization and subsequent H3K9 dimethylation which instructs a less accessible chromatin state in favor of gene silencing at genes directly regulated by MLL-fusions (Supplementary Fig. 13).
Pharmacological activation of Sirt1 sensitizes MLL-r leukemia to DOT1L inhibition
Based on the enlarged H3K9achi domains observed at HOXA and MEIS1 loci in MLL-AF9 leukemias (Fig. 4c,d), and since Sirt1 is required for deacetylation of H3K9, accumulation of H3K9 methylation and silencing of MLL-AF9 target genes, we explored whether pharmacological activation of Sirt1 enhances efficacy of DOT1L inhibitors against MLL-r leukemias. While EPZ4777 treatment alone suppressed the expression of Hoxa7 and Meis1, the combination of SRT1720 (a potent SIRT1 agonist)44,45 with EPZ4777 further diminished the expression of these two genes (Fig. 6a). Concomitantly, we observed a further reduction of the H3K9ac levels at the TSS of Hoxa7 and Meis1 loci in cells receiving the SRT1720 and EPZ4777 combination treatment (Fig. 6b).
Figure 6.
SIRT1 activator SRT1720 sensitizes MLL-r leukemia to DOT1L inhibitor EPZ4777. (a–e) Mouse MLL-AF9 leukemic cells were treated with DMSO (blue), SRT1720 alone (1 μM; red), EPZ4777 alone (10 μM; orange) or the combination of SRT1720 plus EPZ4777 (green) for 6 days in tissue culture. (a) RT-qPCR and (b) H3K9ac-ChIP-qPCR for Hoxa7 and Meis1 genes. (c) Kaplan-Meier survival curves of mice transplanted with pretreated cells. (d) Percentage of GFP-positive MLL-AF9 leukemic blasts in the peripheral blood of the mice described in (c) on day 15 post-transplantation. (e) Relative blast colony count of pretreated cells further cultured in methylcellulose without the small molecules. (f,g) Effect of EPZ4777 on the proliferation of (f) mouse MLL-AF9 leukemic cells, and (g) human Molm13 (MLL-AF9), MV4-11 (MLL-AF4), SEMK2 (MLL-AF4), Kasumi-1 (AML1-ETO) and HL-60 leukemic cell lines. Cells were co-treated with the indicated concentration of EPZ4777 and either DMSO (red) or SRT1720 (1 μM; green) for (f) 3, 6 and 10 days, and (g) 17 days, respectively. Data represent the observed values and mean ± s.d. of (a,b,f,g) two and (e) six replicates, and (d) ten mice per group. *P < 0.05 and #P <0.01 compared to DMSO group; **P < 0.05 and ##P < 0.01 compared to both DMSO and EPZ4777 alone groups using (a,b,d,e) Student’s t-test and (c) Mantel-Cox test.
To examine the impact of ex vivo SRT1720 and EPZ4777 treatment on the ability of the treated cells to establish leukemia in mice, we transplanted equal numbers of viable cells from each treatment group into sub-lethally irradiated recipient mice (Fig. 6c). Survival analysis revealed that SRT1720 pre-treatment had no effect on leukemia latency, whereas EPZ4777 pre-treatment significantly prolonged survival of injected mice. Impressively, the onset of leukemia was further delayed in the case of SRT1720 and EPZ4777 combination treatment (median survival 49 days). Additionally, the percentage of MLL-AF9 leukemia cells in the peripheral blood was significantly lower in the mice that received cells treated with both SRT1720 and EPZ4777 as compared to mice that received cells that were treated with either small molecule individually (Fig. 6d). Consistent with these results, the colony-forming potential (an in vitro assessment of leukemia initiating capacity) of the combinatorially treated cells was drastically impaired as compared to cells treated with either the DOT1L inhibitor or SIRT1 agonist alone (Fig. 6e).
Interestingly, SRT1720 treatment substantially reduced both the “exposure time” and “dose” of EPZ4777 required to induce an antiproliferative effect on MLL-AF9 leukemic cells (Fig. 6f). Finally, we validated that addition of SRT1720 enhances the potency of EPZ4777 against human leukemia cell lines containing MLL-AF9 (Molm13) and MLL-AF4 (MV4-11 and SEMK2) gene translocations (Fig. 6g and Supplementary Fig. 11). Importantly, we did not observe a synthetic toxicity induced by EPZ4777 plus SRT1720 in non-MLL rearranged leukemia cells (Kasumi-1 and HL-60). Together, our findings suggest that individuals with leukemia bearing MLL translocations may benefit from synergistic therapies that simultaneously target both DOT1L and other epigenetic regulators like SIRT1.
DISCUSSION
The dependency of the MLL-fusion driven gene expression program on DOT1L provides a potential therapeutic opportunity for MLL-r leukemias. A better understanding of DOT1L dependence might provide new therapeutic opportunities and also shed light on epigenetic mechanisms of gene regulation. Our genome-scale “suppressor” screen identified Sirt1 as important for Dot1L dependence in MLL-AF9 driven acute myeloid leukemia. We further demonstrate that Dot1L contributes to the maintenance of MLL-fusion target gene expression by antagonizing an H3K9-mediated silencing mechanism instructed by Sirt1, which can be exploited to enhance the efficacy of DOT1L inhibitory therapy.
It has been proposed in S. cerevisiae that the presence of H3K79 methylation across the majority of the yeast genome marks regions of euchromatin thereby relegating Sir2 binding to genomic regions, like the mating type loci, that lack H3K79 methylation46–49. Yeast Sir2 represses gene expression via deacetylation of local histones and promotion of histone modifications that facilitate heterochromatin formation50,51. We found that inactivation of Dot1L in MLL-AF9 leukemia enhances Sirt1 occupancy at most genes that were previously decorated with H3K79 methylation, thus demonstrating an antagonistic relationship between Dot1L and Sirt1 in the mammalian epigenome much like in yeast. However, significant transcriptional silencing was observed only for limited loci that were highly enriched for genes bound by MLL-AF9, suggesting another layer of regulation. Along these lines, we found an enlarged H3K9achi signature at MLL-AF9 bound TSS in both human and mouse MLL-r leukemias (Fig. 4). Of note, a recent study discovered that the super elongation complex member AF9 (which also interacts with DOT1L) directly binds H3K9ac with high affinity52, thus providing a direct link between H3K9ac, H3K79me2, and active gene transcription. Importantly, our data indicate that MLL-fusion proteins prevent SIRT1-dependent deacetylation of H3K9 at least in part by constitutively recruiting DOT1L activity, thereby establishing an H3K79me2hi and H3K9achi histone modification state. It is also important to note that H4K16ac (another Sirt1 chromatin substrate associated with gene transcription) follows the same pattern as H3K9ac. This unique ability of MLL-fusion complexes may uncouple their target genes from normal transcriptional regulation, therefore enabling continuous expression of the leukemic program.
Concomitant with Sirt1-dependent histone deacetylation, we found Suv39h1-mediated H3K9 dimethylation at MLL-AF9 target loci after Dot1L inhibition. Given that H3K9 methylation is associated with regions of heterochromatin53, we postulate that the ectopic expression of MLL-fusion target genes in leukemia is dependent on open and accessible chromatin that is maintained by the ability of Dot1L and H3K79 methylation to repel Sirt1. This hypothesis is supported by the fact that, in addition to the changes in H3K9 modifications, we also found a Sirt1-dependent decrease in chromatin accessibility at MLL-AF9 target genes after Dot1L inhibition. Our findings are in agreement with previous reports showing that direct recruitment of Sirt1 to a promoter in mammalian cells suppresses gene expression through local facultative heterochromatin-mediated gene silencing36,51. Therefore it appears that the MLL-AF9 fusion protein subverts an evolutionarily conserved mechanism of gene regulation and that a major role of Dot1L in the maintenance of MLL-r leukemias is to prevent Sirt1-dependent repressive mechanisms. While we have focused extensively on H3K9 modifications, it is important to note that H3K27 trimethylation also increases at MLL-AF9 target genes after Dot1L inhibition (Fig. 5a) and therefore Dot1L may be inhibiting multiple forms of gene repression.
Our study identifies a unique requirement for Sirt1 in silencing MLL-AF9 target genes (including the HOXA locus) specifically after Dot1L inhibition. This finding is in agreement with recent studies showing a role of SIRT1 in repressing MLL1-regulated genes (including HOXA genes) in normal hematopoietic cells54,55. Other studies have reported that inhibitors of SIRT1 can suppress acute myeloid leukemias that harbor a FLT3-ITD through activation of the p53 pathway56,57. Although we confirmed that modulation of p53 activity by Sirt1 does occur in the MLL-AF9 leukemia without FLT3-ITD, this mechanism does not play a dominant role in the proliferation and survival of these leukemic cells (data not shown). Instead, our data demonstrate that Sirt1 is upstream of a series of changes in chromatin elicited by Dot1L inhibition in MLL-AF9 leukemia. Interestingly, SIRT1 not only directly recruits SUV39H1, but also activates SUV39H1’s methyltransferase activity by deacetylating the SET domain40. This fact combined with our findings suggests that activation of SIRT1 might facilitate silencing of leukemogenic gene expression upon inhibition of DOT1L. Indeed small molecule activators of SIRT1 enhanced the efficacy of DOT1L inhibitor treatment against MLL-AF9 leukemic cells via a more rapid and complete silencing of MLL-AF9 target genes. These findings have immediate clinical relevance since DOT1L inhibitors are now in clinical trials for individuals with MLL-rearranged leukemia. Important clinical responses have been noted with DOT1L inhibitor treatment alone58, but there is no doubt that combination approaches will be necessary. Our data suggest that targeting multiple epigenetic mechanisms controlling the expression of the MLL-fusion driven leukemogenic program should lead to greater therapeutic responses. Furthermore, this study also raises important concerns that incorrect combinations of anti-cancer therapeutics targeting epigenetic modulators, in this case DOT1L inhibitors with SIRT1 inhibitors (Fig. 2c), may diminish clinical benefit due to antagonism of their downstream effects.
In summary, our findings demonstrate that DOT1L and H3K79me2 antagonize gene-repressive mechanisms to control mammalian gene expression. MLL-AF9 fusion proteins take advantage of this unique form of gene regulation to maintain a developmental program in cells where the program should be repressed59. These studies prompt further experiments to define the developmental role of the DOT1L-SIRT1 axis in normal hematopoiesis, and how specific combinations of histone modifications coordinate gene expression in a broader spectrum of biological processes such as body patterning, metabolism, aging and other cancers.
ONLINE METHODS
Reagents
For shRNA and primer sequences please refer to Supplementary Table 1 and 2. The pLKO shRNA library and individual constructs were obtained from the RNAi Consortium at the Broad Institute. The sh-LUC (sequence D11), sh-Sirt1 (sequence A1) and sh-Suv39h1 (sequence C2) were used in all the shRNA experiments unless indicated otherwise. Antibodies for Sirt1 (Abcam; ab12193), Suv39h1 (Cell Signaling; #8729), H3K9me2 (Abcam; ab1220)60, H3K9ac (Abcam; ab4441)60, H3K27me3 (Upstate; 07-449)60, H3K36me3 (Abcam; ab9050)60, H3K79me2 (Abcam; ab3594)60 were used for immunoblot and immunoprecipitation experiments. EPZ4777 was obtained from Epizyme and J. Bradner. Ex527 and suramin were from Santa Cruz. SRT1720 was from Selleck Chemicals. EpiQ Chromatin Analysis Kit was from Bio-Rad.
Cell culture
Mouse MLL-AF9 leukemic cells with Dot1lfl/fl or wild-type genotypes were generated by transformation of mouse bone marrow Lin−Sca1+cKit+ (LSK) cells with retrovirus expressing MLL-AF9 fusion protein and transplanted into sublethally irradiated recipient mice as described previously8. The leukemic blasts harvested from the diseased mice were cultured in vitro in IMDM plus 15% FBS supplemented with 20 ng/ml murine SCF (PeproTech), 10 ng/ml murine IL-3 (PeproTech) and 10 ng/ml murine IL-6 (PeproTech). Human leukemic cell lines Molm-13, MV4-11 and HL-60 were maintained in RPMI plus 10% FBS. All cell culture medium contained L-Glutamine (2mM; Gibco), penicillin (100 units/ml; Gibco), streptomycin (100 ug/ml; Gibco) and plasmocin (5 ug/ml; InvivoGen). Human cell lines including HL-60, MV4-11 and Molm-13 were tested in March - May, 2013 for authentication by short tandem repeat (STR) profiling performed by ATCC. Live cell counts were obtained by high-throughput flow cytometry with SYTOX-blue cell death dye exclusion (Life Technologies) using an LSRFortessa-HTS Analyzer (BD biosciences).
Genome-scale shRNA screen
A monoclonal mouse leukemic MLL-AF9-Dot1lfl/fl cells8 expressing CreER were transduced with a pool of pLKO lentiviral library (92,425 shRNA constructs targeting 16,924 mouse genes) obtained from the RNAi Consortium at the Broad Institute. The amount of virus and cell number was pre-titrated to obtain 7 × 106 infected cells at a 14 ± 0.2 % transduction efficiency for each replicate. Cells were then cultured in methylcellulose (M3234; Stem Cell Technology) supplied with 10 ng/ml murine IL-3 (PeproTech) and 100 nM tamoxifen (Sigma) for 9 days. The integrated shRNA sequences before (T0) and after 9 days Dot1L gene excision (T9) were assessed by high throughput sequencing conducted by the RNAi Consortium at the Broad Institute. The screen was performed with six individually infected and processed replicates.
Immunoblot
Cells were harvested and lysed in SDS-PAGE sample buffer at five million cells per milliliter. Twenty microliter of cell lysate was loaded in each lane. Proteins were resolved by 10% NuPAGE (Invitrogen), transferred to nitrocellulose membrane (Novex), and probed with the indicated antibodies.
Chromatin immunoprecipitation and high-throughput sequencing (ChIP-seq)
ChIP-seq was performed as previously described8,24. Briefly, cell samples were crosslinked by 1% formaldehyde for 10 minutes, and the reaction was stopped by addition of glycine to 125 mM final concentration. The fixed cells were lysed in SDS buffer, and the chromatin was fragmented by sonication. The sheared chromatin was incubated with the indicated antibodies, and recovered by binding to protein A/G agarose (Millipore). Eluted DNA fragments were used directly for qPCR or subjected to high-throughput sequencing using HiSeq 2000 platform (Illumina) for the mouse MLL-AF9 leukemia. The ChIP-seq of H3K9ac in human MLL-r cells was sequenced using Heliscope platform (Helicos Biosciences).
Assay for transposase-accessible chromatin using sequencing (ATAC-seq)
ATAC-seq was performed as previously described43. To examine the initial changes in chromatin, we harvested nuclei from mouse MLL-AF9 cells treated with 1μM EPZ4777 for 6 days, a time point prior to when any remarkable growth suppression or cell death was observed. For each sample, cell nuclei were prepared from 50,000 cells, and incubated with 2.5 μL transposase (Illumina) in a 50 uL reaction for 30 min at 37°C. Following purification of transposase fragmented DNA, the library was amplified by PCR and subjected to high-throughput sequencing using HiSeq 2000 platform (Illumina).
Animal
Mouse MLL-AF9 secondary leukemic cells were treated with DMSO, SRT1720 (1 μM), EPZ4777 (10 μM) or the combination of SRT1720 and EPZ4777 for 6 days in tissue culture. For each drug group, 900,000 pretreated cells were transplanted into sub-lethally irradiated (450 cGy) 6-week-old age-matched female C57BL/6 mice (Taconic) via tail vein injection. The animals were randomly assigned to the experimental groups. Ten mice per group were chosen to have an estimated 80% power in detecting a greater than 1.50 standard deviation difference in means at a significance level of alpha = 0.05 using a two-sided test. Kaplan-Meier survival curves of mice and the t-test were performed using Prism 6 program (GraphPad). No animal was excluded from any of the analyses. All the mouse experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at Memorial Sloan-Kettering Cancer Center.
Data Analysis and Statistical Methods
High throughput reads were aligned to mouse genome assembly mm9 using as previously described8,61. Reads that aligned to multiple loci in the genome were discarded. The ChIP-Seq signal for each gene was quantified as total number of reads per million in the region 1kb upstream of TSS to 4kb downstream of TSS for H3K79me2 and 2kb upstream of TSS to 2 kb downstream of TSS for H3K9ac, H3K9me2, H3K27me3 and Sirt1. GeneChip Mouse Genome 430 2.0 Arrays (Affymetrix) were used for genome-wide expression profiling and probe sets were mapped to genes using library files downloaded from the Affymetrix website (http://www.affymetrix.com). GSEA62 was performed using www.broadinstitute.org/gsea. The integrative analysis of histone modification levels and gene expression was performed using iCanPlot (http://www.icanplot.org)63. A total of 18,240 annotated genes in the mouse genome were used in this study. Gene expression was determined based on the Affymetrix array in MLL-AF9 leukemia. Active (top quartile; 4,560 genes) and silent (bottom quartile; 4,560 genes) genes were selected for analysis. MLL-AF9 targets (129 genes) were defined by MLL-AF9-ChIP-seq as previously reported8. For statistics performed using Student’s t-test, a two-sample equal variance with normal distribution was assumed. The investigators were not blinded to the sample groups for all experiments. The whiskers of boxplots represent 10 – 90 percentile of the data.
Supplementary Material
Acknowledgments
We would like to thank J. Brady and Z. Feng for administrative assistance. This work was supported by the Leukemia and Lymphoma Society, Gabrielle’s Angel Research Foundation and NIH Grants CA66996, CA140575, and CA176745 to S.A.A..
Footnotes
Accession codes. All microarray and ChIP-seq data used in this study have been deposited into the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) with accession code GSE61022.
AUTHOR CONTRIBUTIONS
C.W.C. and S.A.A. conceived the study and wrote the paper; R.P.K and A.U.S. conducted genome-wide data analyses; A.J.D. and N.Z., performed ChIP-seq experiments; E.R., S.H.C., H.X., X.W. and C.D. performed molecular biology, cell culture and animal experiments; J.G.D, D.E.R. and W.C.H. processed shRNA library screen; K.M.B. generated the Dot1L mouse model and conceived experiments; J.Q. and J.E.B. synthesized and supplied EPZ4777 and provided conceptual input.
COMPETING FINANCIAL INTERESTS
S.A.A. is a consultant for Epizyme Inc.. The remaining authors report no competing financial interests.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nature reviews Molecular cell biology. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 3.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 4.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 5.Singer MS, et al. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Q, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Current biology : CB. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 7.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Bernt KM, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steger DJ, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Molecular and cellular biology. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Human molecular genetics. 2007;16:92–106. doi: 10.1093/hmg/ddl444. [DOI] [PubMed] [Google Scholar]
- 11.Mohan M, et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes & development. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller D, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110:4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121:167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Xia X, Reisenauer MR, Hemenway CS, Kone BC. Dot1a-AF9 complex mediates histone H3 Lys-79 hypermethylation and repression of ENaCalpha in an aldosterone-sensitive manner. The Journal of biological chemistry. 2006;281:18059–18068. doi: 10.1074/jbc.M601903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park G, Gong Z, Chen J, Kim JE. Characterization of the DOT1L network: implications of diverse roles for DOT1L. The protein journal. 2010;29:213–223. doi: 10.1007/s10930-010-9242-8. [DOI] [PubMed] [Google Scholar]
- 16.Shen C, Jo SY, Liao C, Hess JL, Nikolovska-Coleska Z. Targeting recruitment of disruptor of telomeric silencing 1-like (DOT1L): characterizing the interactions between DOT1L and mixed lineage leukemia (MLL) fusion proteins. The Journal of biological chemistry. 2013;288:30585–30596. doi: 10.1074/jbc.M113.457135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SK, et al. Human histone H3K79 methyltransferase DOT1L protein [corrected] binds actively transcribing RNA polymerase II to regulate gene expression. The Journal of biological chemistry. 2012;287:39698–39709. doi: 10.1074/jbc.M112.384057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annual review of pathology. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117:4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117:6912–6922. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, et al. Abrogation of MLL-AF10 and CALM-AF10-mediated transformation through genetic inactivation or pharmacological inhibition of the H3K79 methyltransferase Dot1l. Leukemia. 2013;27:813–822. doi: 10.1038/leu.2012.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deshpande AJ, et al. Leukemic transformation by the MLL-AF6 fusion oncogene requires the H3K79 methyltransferase Dot1l. Blood. 2013;121:2533–2541. doi: 10.1182/blood-2012-11-465120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang MJ, et al. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer research. 2010;70:10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krivtsov AV, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernt KM, Armstrong SA. A role for DOT1L in MLL-rearranged leukemias. Epigenomics. 2011;3:667–670. doi: 10.2217/epi.11.98. [DOI] [PubMed] [Google Scholar]
- 26.Guenther MG, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes & development. 2008;22:3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buske C, Humphries RK. Homeobox genes in leukemogenesis. International journal of hematology. 2000;71:301–308. [PubMed] [Google Scholar]
- 28.Deshpande AJ, et al. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer cell. 2014;26:896–908. doi: 10.1016/j.ccell.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daigle SR, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daigle SR, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122:1017–1025. doi: 10.1182/blood-2013-04-497644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anglin JL, et al. Synthesis and structure-activity relationship investigation of adenosine-containing inhibitors of histone methyltransferase DOT1L. Journal of medicinal chemistry. 2012;55:8066–8074. doi: 10.1021/jm300917h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu W, et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nature communications. 2012;3:1288. doi: 10.1038/ncomms2304. [DOI] [PubMed] [Google Scholar]
- 33.Basavapathruni A, et al. Nonclinical pharmacokinetics and metabolism of EPZ-5676, a novel DOT1L histone methyltransferase inhibitor. Biopharmaceutics & drug disposition. 2014;35:237–252. doi: 10.1002/bdd.1889. [DOI] [PubMed] [Google Scholar]
- 34.Klaus CR, et al. DOT1L inhibitor EPZ-5676 displays synergistic antiproliferative activity in combination with standard of care drugs and hypomethylating agents in MLL-rearranged leukemia cells. The Journal of pharmacology and experimental therapeutics. 2014;350:646–656. doi: 10.1124/jpet.114.214577. [DOI] [PubMed] [Google Scholar]
- 35.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochemical and biophysical research communications. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 36.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Molecular cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 37.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 38.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nature methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 39.Cen Y. Sirtuins inhibitors: the approach to affinity and selectivity. Biochimica et biophysica acta. 2010;1804:1635–1644. doi: 10.1016/j.bbapap.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 40.Vaquero A, et al. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 41.Murayama A, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 42.Kuzmichev A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milne JC, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitchell SJ, et al. The SIRT1 activator SRT1720 extends lifespan and improves health of mice fed a standard diet. Cell reports. 2014;6:836–843. doi: 10.1016/j.celrep.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martino F, et al. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Molecular cell. 2009;33:323–334. doi: 10.1016/j.molcel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi YH, et al. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Molecular cell. 2011;42:118–126. doi: 10.1016/j.molcel.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitada T, et al. Mechanism for epigenetic variegation of gene expression at yeast telomeric heterochromatin. Genes & development. 2012;26:2443–2455. doi: 10.1101/gad.201095.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 51.Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Molecular cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, et al. AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell. 2014;159:558–571. doi: 10.1016/j.cell.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Molecular cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh SK, et al. Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance. The Journal of experimental medicine. 2013;210:987–1001. doi: 10.1084/jem.20121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mishra BP, et al. The histone methyltransferase activity of MLL1 is dispensable for hematopoiesis and leukemogenesis. Cell reports. 2014;7:1239–1247. doi: 10.1016/j.celrep.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sasca D, et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood. 2014;124:121–133. doi: 10.1182/blood-2013-11-538819. [DOI] [PubMed] [Google Scholar]
- 57.Li L, et al. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute Myeloid Leukemia stem cells. Cell stem cell. 2014;15:431–446. doi: 10.1016/j.stem.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stein EM, et al. The DOT1L Inhibitor EPZ-5676: Safety and Activity in Relapsed/Refractory Patients with MLL-Rearranged Leukemia. 56th American Society of Hematology Annual Meeting & Exposition; San Francisco, CA. 2014. [Google Scholar]
- 59.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 60.Egelhofer TA, et al. An assessment of histone-modification antibody quality. Nature structural & molecular biology. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinha AU, Armstrong SA. iCanPlot: visual exploration of high-throughput omics data using interactive Canvas plotting. PloS one. 2012;7:e31690. doi: 10.1371/journal.pone.0031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.